Abstract
Translation of mRNA into protein is a fundamental step in eukaryotic gene expression requiring the large (60S) and small (40S) ribosome subunits and associated proteins. By modern proteomic approaches, we previously identified a novel 40S-associated protein named Asc1p in budding yeast and RACK1 in mammals. The goals of this study were to establish Asc1p or RACK1 as a core conserved eukaryotic ribosomal protein and to determine the role of Asc1p or RACK1 in translational control. We provide biochemical, evolutionary, genetic, and functional evidence showing that Asc1p or RACK1 is indeed a conserved core component of the eukaryotic ribosome. We also show that purified Asc1p-deficient ribosomes have increased translational activity compared to that of wild-type yeast ribosomes. Further, we demonstrate that asc1Δ null strains have increased levels of specific proteins in vivo and that this molecular phenotype is complemented by either Asc1p or RACK1. Our data suggest that one of Asc1p's or RACK1's functions is to repress gene expression.
The eukaryotic 80S ribosome, consisting of small (40S) and large (60S) subunits, is the catalytic and regulatory macromolecular complex responsible for the decoding of mRNA into polypeptides. Together, the small and large ribosomal subunits contain the 18S, 28S (25S in yeast), 5.8S, and 5S rRNAs along with a large number of proteins. The structure and function of the ribosome have been extensively studied for decades, and a 15 Å cryoelectron microscopic map of the yeast ribosome is available (51). While the catalytic activity of the ribosome's 28S rRNA in peptide bond formation is well established, the function and regulatory activity of the ribosomal proteins are still largely unknown (12).
Advanced mass spectrometry (MS) analysis has facilitated the identification of novel ribosomal proteins. In a proteomic screen of the Saccharomyces cerevisiae 40S, 60S, and 80S components, we identified a novel component of the 40S and 80S subunits, Asc1p (ASC1, YMR116C, CPC2, BEL1), which remains associated with the small subunit in the presence of 1 M KCl (34). Under these stringent conditions, transient translation factors and ribosome biogenesis factors present in lower salt concentrations are shed from the 40S component. In these experiments, Asc1p was present at a concentration equimolar to that of the other ribosomal proteins (34). By established criteria for defining ribosomal proteins, Asc1p can be classified as a novel core 40S ribosomal component (31). We also demonstrated that RACK1 (receptor for activated C kinase 1), a protein with 52% sequence identity to Asc1p, is localized to the 40S and 80S components and polysomes in human cells (34). These observations have been confirmed by several other studies (2, 4, 6, 26, 50).
RACK1 was originally identified as a protein with sequence similarity to the guanine nucleotide-binding protein β subunit and other proteins containing Trp-Asp (WD) repeat domains (20). RACK1 was later shown to associate both in vitro and in vivo with activated protein kinase CβII (PKCβII) (46) and is hypothesized to function as an anchoring protein that localizes activated PKC to the insoluble cell fraction. A plethora of independent studies have attempted to define the molecular function of RACK1. Yeast two-hybrid and coimmunoprecipitation methods show RACK1 interacting with a large number of cellular proteins with roles in signal transduction (5, 9, 10, 15, 17, 21, 22, 29, 30, 32, 33, 36, 38, 41-43, 56, 62). Because of its apparent ability to interact with a number of signaling molecules, RACK1 is perceived to play a crucial role in a multitude of biological processes.
Consistent with our previous results showing association of Asc1p or RACK1 with the ribosome, more recent studies have implicated ASC1 and RACK1 in translational control. General translational inhibition occurs upon amino acid starvation when the eIF2α kinase Gcn2p is activated (11). Yeast strains deficient in Gcn2p fail to initiate the amino acid starvation response. An asc1 mutation has been shown to restore the amino acid starvation response in a gcn2Δ mutant strain (24). Further, Schizosaccharomyces pombe cpc2Δ (asc1Δ) null mutants appear to have a subset of genes that are transcriptionally and translationally repressed relative to those of the wild type (50). The cpc2Δ (asc1Δ) null strains were also sensitive to puromycin, a drug that causes premature termination of translation, suggesting that translational fidelity has been reduced (50). Moreover, in mammalian COS cells, transient overexpression of RACK1 stimulated translation in vivo (4).
In this report, we provide biochemical, evolutionary, genetic, and functional data showing that Asc1p or RACK1 is a core 40S ribosomal protein in eukaryotes. Further, our results suggest that Asc1p and RACK1 function to repress gene expression. This novel expression phenotype provides insight into a potential regulatory mechanism in eukaryotic gene expression.
MATERIALS AND METHODS
Plasmids and yeast strains.
Strain construction, genetic manipulations, and yeast medium preparation were carried out by standard methods (49). To construct plasmid pASC1, a 2.13-kb BamHI-XbaI yeast genomic fragment from lambda clone λPM5992 (ATCC 70652) containing the entire ASC1 locus was cloned into the BamHI and XbaI sites of pRS416 (8, 40, 44). Plasmid pME1867 (renamed pRACK1 in these studies), containing the rat cDNA for RACK1 expressed under the control of the ASC1 promoter and terminator sequences in a pRS316 vector backbone, was a gift from Gerhard Braus (24). To construct plasmid pET100-ASC1, PCR primers A-ASC1 (CACCATGGCATCTAACGAAGTTTTAG), B-ASC1 (TTAAGTTCCAAGCCTTAACCATTTTGTCGTTACCGGC), C-ASC1 (ACAAAATGGTTAAGGCTTGGAACTTAAACCAATTCC), and D-ASC1 (TTAGTTAGCAGTCATAACTTGCC) were used in a crossover PCR to amplify an intronless ASC1 DNA fragment encoding the complete Asc1p protein (6). The intronless ASC1 fragment was cloned into the pET100/D-TOPO bacterial expression vector (Invitrogen) to create plasmid pET100-ASC1 encoding an N-terminally tagged His6::ASC1 fusion protein expressed from a T7 promoter. To construct plasmid p426-GPD::His6-ASC1, a Klenow-treated 1.1-kb NdeI-SacI fragment from plasmid pET100-ASC1 containing the His6-ASC1 fusion was cloned into the SmaI site of p426GPD (39). All constructs were confirmed by DNA sequencing.
The yeast strains used were BY4743 (MATa/MATα his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 met15Δ0/MET15 LYS2/lys2Δ0 ura3Δ0/ura3Δ0) (60), YDM36556 (MATa/MATα his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 met15Δ0/MET15 LYS2/lys2Δ0 ura3Δ0/ura3Δ0 Δasc1::Kanr/Δasc1::Kanr) (60), AL150 (YDM36556 with pRS416), AL156 (YDM36556 with pASC1), AL141 (YDM36556 with pRACK1), AL190 (BY4743 with pRS316), AL191 (BY4743 with pASC1), AL140 (BY4743 with pRACK1), AL143 (YDM36556 with p426-GPD::His6-ASC1), AL146 (YDM36556 with pYES-DEST52), AL103 (MATa his7 ura3-52), AL030 (MATa his7 ura3-52 asc1Δ) (34), AL185 (YDM36556 plus p180 [GCN4 5′ untranslated region {UTR}-lacZ reporter plasmid]) (23), and AL183 (BY4743 with p180). Strains containing chromosomal deletions of asc1 were confirmed by PCR of yeast genomic DNA, PCR of yeast cDNA, and absence of Asc1p by two-dimensional (2D) gel electrophoresis.
Polysome analysis.
Yeast cell extracts were prepared essentially as previously described (37). Briefly, yeast strains were grown in synthetic complete medium without uracil (SC−URA) to an optical density at 600 nm (OD600) of 0.6, and 5 ml of cells was lysed with 0.5-mm glass beads in 250 μl of lysis buffer (10 mM Tris-HCl [pH 8.0] 140 mM NaCl, 1.5 mM MgCl2, 0.5% NP-40, 200 U of RNasin [Promega] per ml). Extracts were centrifuged in a microcentrifuge for 1 min at 20,000 × g. Supernatants were supplemented with 250 μl of 2× translation stop buffer (20 mM dithiothreitol [DTT], 665 μg of heparin per ml, 150 μg of cycloheximide per ml) and 1 tablet of mini-Complete protease inhibitor (Roche) per 5 ml of buffer. The supplemented extracts were centrifuged for 5 min at 20,000 × g. Supernatants (200 μl) were gently layered on top of a 15 to 40% sucrose gradient cast in 10 mM Tris-HCl (pH 7.5), 140 mM NaCl, 1.5 mM MgCl2, 10 mM DTT, 100 μg of cycloheximide per ml, and 0.5 mg of heparin per ml. Gradients were centrifuged in a Beckman tabletop ultracentrifuge (TlS 55) for 75 min at 50,000 rpm. Fractions were collected from the top of the gradients and partitioned for RNA isolation, Western blotting, or MS analysis (34, 37). RNA was isolated from sucrose gradient fractions and yeast strains, with TRI-reagent LS (Molecular Research Center Inc. [MRC]) in accordance with the manufacturer's protocol. RNA from gradient fractions was loaded and run on nondenaturing 1% agarose gels cast in 1× Tris-acetate-EDTA and stained with ethidium bromide.
Western analysis and direct analysis of large protein complexes (DALPC).
Western analysis was performed on polysome profile fractions from S. cerevisiae cells, human HEK293 cells, mouse NT2 cells, and in vitro translation extracts from S. cerevisiae strains. Sucrose gradient fractions or in vitro translation extracts were mixed with Laemmli buffer, heated for 5 min at 100°C, loaded onto NuPAGE 10% Bis-Tris gels, and separated with 1× morpholinepropanesulfonic acid-sodium dodecyl sulfate running buffer (Invitrogen). For Western analysis, NuPAGE gels were transferred to nitrocellulose membranes and blocked overnight in Tris-buffered saline containing 0.1% Tween and 10% nonfat dry milk. Western blots were probed with either affinity-purified rabbit polyclonal antibodies to Asc1p generated against full-length recombinant His6-tagged Asc1p (Bethyl Laboratories, Montgomery, Tex.), mouse RACK1 monoclonal antibodies (BD Biosciences), Aip1p polyclonal antibodies (45), or Rpl3p monoclonal antibodies (59). Western blots were washed three times in Tris-buffered saline containing 0.1% Tween and then incubated with the appropriate horseradish peroxidase-tagged secondary antibody (Promega). Blots were developed with ECL Plus reagent (Amersham-Pharmacia). Asc1p antibody specificity was confirmed by Western blotting of Asc1p positive control antigen and whole-cell lysates from wild-type and asc1Δ null yeast strains (data not shown). The MS approach termed DALPC was performed essentially as described with Drosophila melanogaster embryos and C. elegans strain N2 pooled ribosomal and nonribosomal fractions (34, 47).
Genetic complementation of yeast asc1Δ mutant strains.
Yeast strains were grown in either SC−URA or SC+URA to an OD600 of 0.6. Cells were counted with a hemocytometer, adjusted to the same concentration, and serially diluted (10-fold). Five microliters of each 10-fold serial dilution (108 to 105) was spotted onto SC−URA plates. The plates were incubated at 30 or 37°C for 72 h and photographed.
In vitro translation assays.
To prepare translation extracts, 2 liters of yeast was grown in yeast extract-peptone-dextrose to an OD600 of 2. Cells were washed five times in ribosome buffer lacking protease inhibitors (30 mM HEPES [pH 7.4], 100 mM potassium acetate, 2 mM magnesium acetate, 2 mM fresh DTT, 8.5% mannitol). After washing, 8 g of cells (wet weight) was lysed in 15 ml of ribosome buffer containing mini-Complete protease inhibitor (Roche) with 48 g of 0.5-mm glass beads. Cells were lysed in 50-ml sterile Falcon tubes by rigorous rocking in a 6-in. arc for 1-min intervals (five times) with 1-min intervals on ice between periods of rocking. Extracts were cleared by centrifugation twice at 20,000 × g for 10 min. Five milliliters of extract was loaded onto a 75-ml bed volume Sepharose G-25 column. The sample was fractioned with an isocratic buffer (ribosome buffer plus protease inhibitors) flowing at 0.5 ml/min. The flowthrough fractions (0.5 ml) with an OD260 of >90 were pooled and used for the in vitro translation assays (3).
Plasmid T3 lucpA, originally created by Peter Sarnow's laboratory (25), was kindly provided by Alan Sachs. T3 lucpA was purified with a QIAGEN miniprep and linearized with BamHI. The linearized plasmid was purified with a QIAquick PCR cleanup kit (QIAGEN). Capped luciferase mRNAs were synthesized with the Amplicap T3 high-yield message maker kit (Epicentre) with purified, linearized, T3 lucpA DNA as the template. The capped luciferase mRNAs were purified prior to in vitro translation with RNeasy spin columns (QIAGEN). Uncapped luciferase mRNA was purchased from Promega. Total RNA from wild-type yeast strain BY4743 grown to an OD600 of 1.0 was isolated with TRI-reagent (MRC). Following isolation of total RNA, poly(A)+ mRNAs were isolated with an Oligotex mRNA isolation kit (QIAGEN). In vitro translation assays were conducted as described previously (54).
Assay for β-galactosidase activity.
The p180 plasmid containing the 5′ UTR of GCN4 cloned in front of the lacZ gene was transformed into yeast strains BY4743 and YDM36556 (23). Strains were grown in SC−URA to an OD600 of 0.6. Cells were then pelleted by centrifugation at 9,000 × g for 5 min. Cells were lysed by bead beating in the 1× lysis buffer provided by the manufacturer (Promega). After lysis, extracts were centrifuged at 20,000 × g for 2 min. Following centrifugation, supernatants were assayed for β-galactosidase activity by the manufacturer's (Promega) protocol and measured for OD420 and OD280. Relative β-galactosidase activity was standardized by dividing the β-galactosidase activity (OD420) by the respective OD280 of each individual sample.
Purification of recombinant Asc1p.
One liter of Escherichia coli strain BL21 containing plasmid pET100-ASC1 was grown to an OD600 of 0.6 in Luria-Bertani medium. Protein expression was induced with isopropyl-β-d-thiogalactopyranoside at 37°C for 1 h. Cells were harvested and lysed by sonication, and the recombinant protein was purified in accordance with the PROBOND kit (Invitrogen) native purification instructions. To prepare Asc1p protein for in vitro translation reactions, recombinant Asc1p protein was dialyzed exhaustively against ribosome buffer containing protease inhibitors.
2D difference gel electrophoresis (2D DIGE) analysis.
One liter of each strain of yeast was grown in SC−URA to an OD600 of 0.6. Cells were harvested and lysed by bead beating in lysis buffer (10 mM Tris-HCl [pH 8.0], 140 mM NaCl, 1.5 mM MgCl2, 0.5% NP-40, 200 U of RNasin [Promega] per ml). After lysis, extracts were centrifuged at 20,000 × g for 10 min. Extracts were then partitioned for protein and mRNA analysis. Samples designated for 2D analysis were digested with RNase A and DNase and centrifuged again at 20,000 × g for 10 min prior to protein extraction. Three independent samples for each strain were prepared and analyzed.
2D DIGE analysis with a mixed-sample internal standard was carried out essentially as described previously (1, 13). Triplicate protein samples for each strain were individually labeled with either Cy3 or Cy5. To control for bias in the fluorescent dyes, we reversed the Cy3 and Cy5 dyes used to label the whole-cell extracts in one of each of the experimental replicates of the four strains being compared. The Cy2-labeled mixed-sample internal standard was composed of an equal portion of all 12 of the extracts used in the experiment. The Cy2 standard was used to normalize protein abundances across different gels and to control for gel-to-gel variation (1, 13). Equal amounts of Cy3-labeled sample, Cy5-labeled sample, and Cy2-internal control were mixed together and run on individual 2D gels. Triplicate Cy2-, Cy-3-, and Cy-5-labeled samples were then loaded onto a total of six 24-cm pH 4 to 7 immobilized pH gradient strips (Amersham Biosciences) and subjected to 2D gel electrophoresis with the IPGphor and DALT-twelve systems in accordance with the manufacturer's (Amersham Biosciences) protocols.
2D DIGE gels were scanned with a Typhoon 9410 variable-mode imager with the recommended mutually exclusive emission and excitation wavelengths for each Cy dye (Amersham Biosciences). 2D DIGE analysis was performed with DeCyder version 5.0 software (Amersham Biosciences), which uses a triple-codetection algorithm to generate the same protein spot-feature boundary for individual Cy2, Cy3, and Cy5 signals. The Cy3/Cy2 and Cy5/Cy2 ratios were then calculated and compared among the six DIGE gels, allowing for the application of Student t test statistical analyses to triplicate samples from all of the strains despite separation on different DIGE gels (1, 13).
In our analysis of in vivo protein levels, we considered 2D gel features to be a protein only if the spot was confirmed with both Cy labels and was present on every gel. Only proteins with ≥1.5-fold differences in abundance were considered significant. Differences in protein levels between wild-type and asc1Δ null samples were considered statistically significant only if the difference fell within the 95% confidence interval as determined by the Student t test.
Identification of 2D DIGE proteins.
A SyproRuby (Molecular Probes) poststain image (similarly acquired with a Typhoon 9410) was used to ensure accurate robotic protein excision for subsequent trypsin in-gel digestion with the ProSpot spot-handling workstation (Amersham Biosciences). Protein identifications were done by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) MS and TOF-TOF tandem MS with a Voyager 4700 MALDI-TOF-TOF mass spectrometer (Applied Biosystems). Protein identifications were based on the acquired mass spectral data combined with database interrogation by the MASCOT algorithm (Matrix Science).
Multiplex RT-PCR and real-time quantitative RT-PCR.
Lysate-matched RNA extracts from the 2D DIGE protocol were analyzed for levels of mRNA transcripts encoding identified proteins. The extracts were treated with molecular biology grade DNase (Invitrogen) prior to reverse transcription (RT). RNA was reverse transcribed with oligo(dT) priming (Perkin-Elmer) and Superscript II (Invitrogen). Specific PCR primers were designed to the cDNA transcripts of interest with dsGENE software (Accelrys). Triplex PCR was performed by previously described protocols (18). The primer pairs used for RT-PCR were TDH3 (glyceraldehyde-3-phosphate dehydrogenase [GAPDH]) (5′ TCTTCCATCTTCGATGCTGCCG and 5′ AGCCTTGGCAACGTGTTCAACC), APE2 (5′ ACCAAAGGAAACCCAGGATGCG and 5′ AGCAGCTTTTTCAACGTCTGCG), AIP1 (5′ CGTCCTTGTGAGCATTCAACGC and 5′ TCTTCGCGCAAAACCCTCGTAC), DKA1 (5′ AAGCACGGCATTCTGGAGGATG and 5′ TCTTGGGGAACGTACGCATTCG), ENO2 (5′ TAGAGCCGCTGCTGCTGAAAAG and 5′ TTGGAGCAACACCACCTTCGTC) TPS1 (5′ TACAGGTTGCAGTGCCAAGTCG and 5′ ATTGTGCGGCACCTGTGAACTC), ALD3 (5′ AAAGCTGCCAGGGCTGCTTTTG and 5′ TATTGAACTTGTCGACCGCCCC), and CTT1 (5′ ACACCAGACACTGCAAGAGACC and 5′ TACGCGTTCATACTAGCCCACG). PCR samples were taken at cycles 17, 20, 23, 26, and 30. PCR products were run on 6% polyacrylamide gels cast in 0.5× Tris-borate-EDTA and stained with ethidium bromide.
For real-time RT-PCR, cDNA samples were prepared as described above. cDNAs were amplified in an IQ SYBR green supermix by the manufacturer's recommended protocol (Bio-Rad). Samples were analyzed in a Bio-Rad iCycler with the following temperature cycle: 95°C for 10 s, 64°C for 1 min. PCR samples were cycled 32 times. To quantitate relative mRNA levels between yeast strains, mRNA levels were divided by the calculated TDH3 (GAPDH) mRNA levels for each strain as a standard. For all of the cDNAs amplified, a stepwise melting curve protocol of 0.5°C was performed after PCR to confirm the presence of a single PCR product.
RESULTS
Mammalian RACK1 and S. cerevisiae Asc1p are orthologous ribosomal proteins.
In a past study we showed that Asc1p localizes to the 40S and 80S subunits in S. cerevisiae and RACK1 localizes to the 40S subunit and polysomes in human HeLa cells (34). Sequence similarity and ribosome localization suggested that the two proteins are orthologous (http://linklab.mc.vanderbilt.edu). To further examine if RACK1 and Asc1p are biochemically orthologous ribosomal proteins, we performed a series of competitive and noncompetitive polysome profiling experiments with S. cerevisiae.
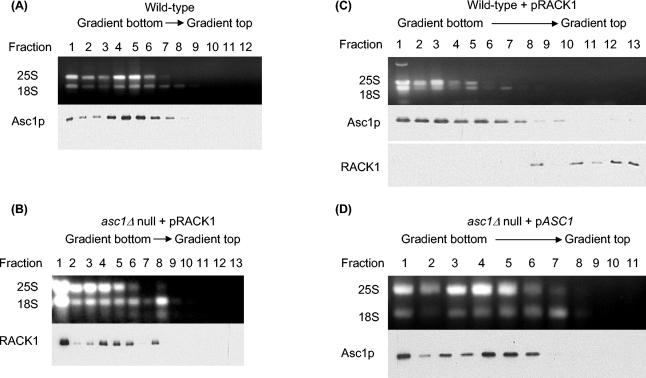
Polysome profiling results showed that in wild-type S. cerevisiae, Asc1p localized to ribosomal fractions but was absent from nonribosomal fractions (Fig. 1A). We reasoned that if RACK1 and Asc1p are orthologs, then in the absence of Asc1p, RACK1 should localize to ribosomes in yeast. When we expressed rat RACK1 (99% identical to human RACK1) in an asc1Δ null yeast strain, we found that RACK1 localized to ribosomal fractions in a polysome profile but was absent from nonribosomal fractions (Fig. 1B). This result indicated that RACK1 associated with the yeast ribosome similarly to Asc1p and therefore might perform similar functions within the ribosome. Interestingly, when we expressed RACK1 in a wild-type yeast background we found that Asc1p was still included in ribosomal fractions but RACK1 was excluded (Fig. 1C). Because these results suggest that Asc1p and RACK1 compete for the same ribosomal position, we refer to this phenomenon as species-competitive-protein exclusion (compare Fig. 1B and C).
FIG. 1.
Polysome profiles showing that Asc1p and RACK1 are biochemically orthologous ribosomal proteins. (A) Polysome profile showing that yeast Asc1p localizes to the polysome fractions and is absent in the nonribosomal fractions. Cell lysate from yeast wild-type strain AL190 was fractionated by sucrose gradient centrifugation, and fractions were collected. An aliquot of each fraction was analyzed by agarose gel electrophoresis to identify fractions with 25S and 18S rRNAs. Western analysis of the fractions with anti-Asc1p antibodies shows that Asc1p is in the ribosome fractions (lanes 1 to 8) and is absent from the nonribosomal fractions (lanes 9 to 12). (B) Polysome profile showing that RACK1 expressed in a yeast asc1Δ null strain localizes to the ribosomal fractions and is absent from the nonribosomal fractions. Cell lysate from yeast strain AL141 was fractionated by sucrose gradient centrifugation and analyzed as described for panel A except that anti-RACK1 antibody was used for Western analysis. (C) Polysome profile showing exclusion of RACK1 from ribosomal fractions in a wild-type yeast strain. Cell lysate from yeast strain AL140 was fractionated by sucrose gradient centrifugation and analyzed as described for panel A except that anti-RACK1 and anti-Asc1p antibodies were used in separate Western blot assays. (D) Polysome profile of an asc1Δ null strain complemented by expression of ASC1. Cell lysate from yeast strain AL156 was fractionated by sucrose gradient centrifugation and analyzed as described for panel A.
Localization of RACK1 to ribosomes is evolutionarily conserved in eukaryotes.
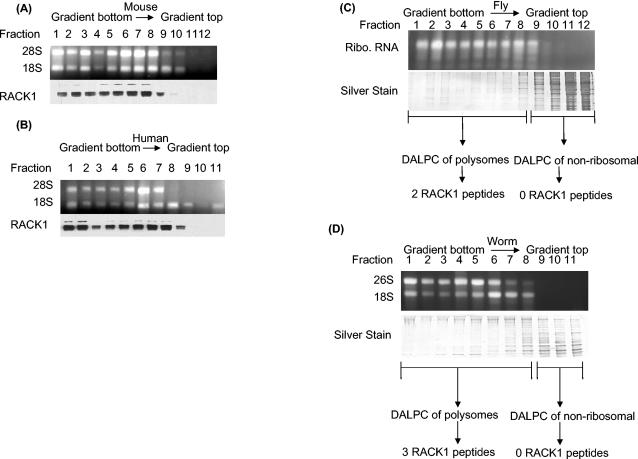
Since the polysome profiling experiments suggested that Asc1p and RACK1 are biochemically orthologous ribosomal proteins, we wanted to investigate whether RACK1 is a conserved eukaryotic ribosomal protein. Alignment of yeast Asc1p with four putative orthologs from different species, Caenorhabditis elegans (NP_501859), Drosophila melanogaster (NP_477269), Mus musculus (BAA06185), and Homo sapiens (NP_006089), revealed a high level of sequence similarity among the different proteins (http://linklab.mc.vanderbilt.edu). To test if localization of these Asc1p-RACK1 orthologs is conserved among eukaryotic species, we performed polysome profiling of mouse, human, fly, and nematode extracts (Fig. 2A to D, respectively). By Western analysis, a RACK1 monoclonal antibody recognized a major and a minor band in the ribosomal fractions of M. musculus and H. sapiens. In both organisms, the RACK1 antibody failed to detect a protein in the nonribosomal fractions. Because the RACK1 antibody poorly recognized a cognate fly and worm protein, we analyzed D. melanogaster and C. elegans polysomes and nonribosomal fractions by the MS approach termed DALPC (34, 47, 63) (Fig. 2C and D). DALPC identified peptides derived from the putative othologous RACK1 proteins in the polysomal but not the nonribosomal fractions for both D. melanogaster and C. elegans. Together, our data show that in the four eukaryotes tested, ASC1 and RACK1 were found in the ribosomal fractions and were absent from the nonribosomal fractions. Collectively, these results suggest that both the sequences and localization of ASC1 and RACK1 to ribosomal fractions are conserved in eukaryotes.
FIG. 2.
Polysome profiling of RACK1 protein showing its polysomal localization in four eukaryotic species. (A) Mouse polysome profile showing that RACK1 localizes to the polysome fractions and is absent from the nonribosomal fractions. A cell lysate from mouse NT2 cells was fractionated by sucrose gradient centrifugation, and fractions were collected. An aliquot of each fraction was analyzed by agarose gel electrophoresis to show fractions with 28S and 18S rRNAs. Western analysis of the fractions with anti-RACK1 antibodies shows the distribution of RACK1. (B) Human polysome profile showing that RACK1 localizes to the polysome fractions and is absent from the nonribosomal fractions. A cell lysate from human HEK293 cells was fractionated by sucrose gradient centrifugation and analyzed as described for panel A. (C) DALPC analysis of the Drosophila polysome profile shows that the putative RACK1 ortholog (NP_477269) localizes to the polysome fractions and is absent from the nonribosomal fractions. Each fraction was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis to visualize the amount of protein in each fraction. Polysome and nonribosomal fractions were separately pooled and digested with trypsin, and proteins were identified by the DALPC-MS approach (34, 47, 63). The Drosophila 28S and 18S rRNAs typically comigrate as a doublet (27). Ribo., ribosomal. (D) DALPC analysis of a nematode polysome profile shows that the putative worm RACK1 ortholog (NP_501859) localizes to the polysome fractions and is absent from the nonribosomal fractions. A lysate from wild-type worms was fractionated by sucrose gradient centrifugation, and fractions were collected. An aliquot of each fraction was analyzed by agarose gel electrophoresis to identify fractions with 28S and 18S rRNAs. Protein samples were analyzed as described for panel C.
S. cerevisiae ASC1 and mammalian RACK1 are genetically orthologous.
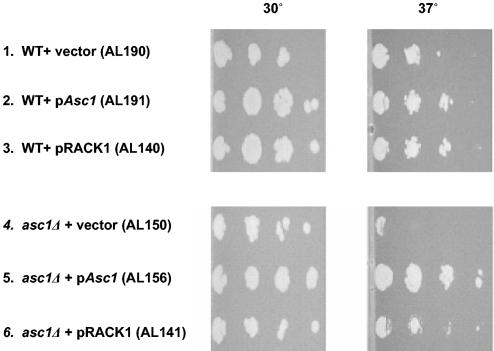
Because polysome profiling experiments with yeast and higher eukaryotes indicated that Asc1p and RACK1 are biochemically conserved orthologous ribosomal proteins, we wanted to test to see if ASC1 and RACK1 are genetically orthologous genes. We found that an asc1Δ null yeast strain had a temperature-sensitive growth defect (Fig. 3, line 4). With a plate dilution assay, we tested whether CEN plasmids expressing ASC1 (pASC1) or RACK1 (pRACK1) at the endogenous level of ASC1 could complement the temperature-sensitive phenotype in an asc1Δ null strain. Complementation in the asc1Δ null strain was complete for ASC1 and partial for RACK1 (Fig. 3, lines 5 and 6). These results are in excellent agreement with those of a previous study (24). Polysome profile analysis of the yeast asc1Δ null plus pASC1 complementing strain demonstrated that ASC1 expression is restored and the complementing Asc1p protein is localized to the ribosomes (Fig. 1D). These data indicate that yeast ASC1 and RACK1 are genetically orthologous.
FIG. 3.
Genetic complementation of the temperature-sensitive growth defect in a yeast asc1Δ null strain by ASC1 and RACK1. The indicated yeast strains were grown to logarithmic phase in liquid medium, spotted in a dilution series onto SC−URA plates, grown at two different temperatures for 72 h, and then photographed. Strains 1 to 3 are wild-type (WT) controls. Experiments with strains 4 to 6 were performed in the asc1Δ null background. The genotypes of the strains are described in Materials and Methods.
Asc1p-deficient ribosomes have increased translational activity.
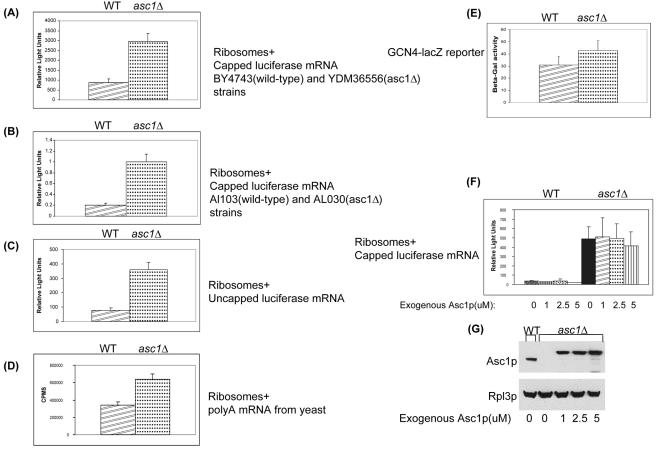
Because our data indicated that RACK1 and Asc1p are conserved eukaryotic ribosomal proteins and earlier studies had implicated Asc1p in the amino acid starvation response, we wanted to determine the role of Asc1p or RACK1 in translation. To evaluate the activity of Asc1p in translation, we performed in vitro translation assays with extracts from wild-type and asc1Δ null yeast strains (54). In three independent experiments translating a capped and polyadenylated luciferase reporter mRNA, the asc1Δ null strain ribosomes had translational activity 3- to 10-fold higher than that of wild-type ribosomes (Fig. 4A and F). To show that the increased translational activity was not strain dependent, in vitro translation assays were performed with extracts from genetically independent wild-type and asc1Δ null strains (34). Similar to the earlier strains, the second asc1Δ null strain had higher in vitro translational activity than did the isogenic wild-type strain (Fig. 4B).
FIG. 4.
Up-regulation of translational activity in asc1Δ extracts. (A) In vitro translation of capped and polyadenylated luciferase reporter mRNA with extracts prepared from wild-type (WT) BY4743 and isogenic asc1Δ null strain YDM36556. Translational activity was determined by measuring luminescence (relative light units) after 30 min of incubation at 26°C. Error bars indicate standard deviations. (B) In vitro translation of capped and polyadenylated luciferase reporter mRNA with extracts prepared from wild-type AL103 and isogenic asc1Δ null strain AL030. (C) In vitro translation of uncapped and polyadenylated luciferase reporter mRNA with extracts prepared from wild-type BY4743 and isogenic asc1Δ null strain YDM36556. (D) In vitro translation of poly(A)-enriched mRNAs with extracts prepared from wild-type BY4743 and isogenic asc1Δ null strain YDM36556. The extracts were incubated with whole wild-type yeast mRNA and [35S]methionine. Translational activity was determined by measuring the counts per min after 30 min of incubation at 26°C. (E) In vivo analysis of GCN4 translation. Wild-type BY4743 and asc1Δ null YDM36556 strains were transformed with a GCN4 5′ UTR-lacZ reporter plasmid (p180) (23). Cells were grown in SC−URA to an OD600 of 0.6 and assayed for β-galactosidase activity as described in Materials and Methods. (F) In vitro translation of capped and polyadenylated luciferase reporter mRNA in the presence of an increasing concentration of recombinant Asc1p protein. Protein extracts were prepared from wild-type BY4743 and isogenic asc1Δ null strain YDM36556. The extracts were incubated with luciferase mRNA and the indicated amounts of recombinant Asc1p protein. Translational activity was determined by measuring luminescence (relative light units) after 30 min of incubation at 26°C. (G) Western analysis of in vitro extracts with anti-Asc1p and anti-Rpl3p antibodies. As a loading control for the in vitro translation assay, Asc1p and Rpl3p levels were measured. Poly-His-tagged recombinant Asc1p migrates more slowly than endogenous Asc1p. Rpl3p levels suggest that 60S subunit levels were similar between wild-type and asc1Δ translation extracts.
To verify that the asc1Δ null increased translational activity phenotype was independent of the reporter molecule, we translated uncapped luciferase, capped luciferase, wild-type poly(A)+ mRNAs, and a GCN4 5′ UTR-lacZ reporter. We found that translational activity is elevated in the asc1Δ null strain compared to that in the wild-type, control strains (Fig. 4A through F).
In an attempt to modulate the higher translational activity of the ribosomes lacking Asc1p, we added recombinant Asc1p protein to our in vitro extracts (http://linklab.mc.vanderbilt.edu). Expression of the recombinant protein in asc1Δ null strains complemented the temperature-sensitive growth defect (http://linklab.mc.vanderbilt.edu). However, addition of exogenous Asc1p protein to the in vitro translation extracts failed to repress translational activity (Fig. 4F).
Since the recombinant protein failed to restore translational activity to wild-type levels, we sought to determine if our recombinant protein was added at wild-type stoichiometric levels. Western analysis of in vitro translation extracts showed the levels of exogenous Asc1p equal to or greater than that found in wild-type ribosome preparations (Fig. 4G). When relative levels of Rpl3p protein were compared between extracts, the amounts of ribosomes appeared to be similar (Fig. 4G). These data suggest that equal amounts of ribosomes were present in each extract for translation. Collectively, these data show that ribosomes lacking Asc1p have increased (derepressed) translation activity, suggesting that Asc1p acts as a translational repressor.
Asc1p or RACK1 functionally complements increased protein levels in asc1Δ null strains.
Because the in vitro data above suggested that Asc1p functions as a translational repressor, we sought to analyze in vivo changes in protein levels among wild-type, asc1Δ null, asc1Δ null plus pASC1, and asc1Δ null plus pRACK1 complemented strains. We used 2D DIGE to quantify any changes in the in vivo protein levels for a large population of proteins (1, 13). In the 2D DIGE experiments, whole-cell lysates were prepared from three independent cultures for each of the four strains described above (i.e., 12 independent samples). The lysates were labeled prior to electrophoresis with the spectrally resolvable fluorescent dye Cy3 or Cy5. To normalize for protein abundance differences across multiple 2D gels, a mixed internal standard pool containing equal amounts of the experimental samples was labeled with a Cy2 fluorescent dye. The pooled standard represented the average of all of the samples being compared. Independent Cy2, Cy3, and Cy5 samples were mixed and resolved on the same 2D gels. As a consequence, each gel contained an image with a highly similar spot pattern, simplifying and improving the confidence of inter-gel spot matching and quantification (1). To control for bias in the fluorescent dyes, we reversed the Cy3 and Cy5 dyes used to label the whole-cell extracts in one of the experimental triplicates of the four strains being compared.
In our analysis of in vivo protein levels, we interpreted a 2D gel feature as a protein only if the spot was confirmed with both the Cy3 and Cy5 labels and was present on every gel. The total number of proteins analyzed (approximately 1,500) included multiple isoforms of some individual proteins. Only proteins with ≥1.5-fold differences in abundance were considered significant and included in our analysis. In addition, differences in protein levels between wild-type and asc1Δ null samples were considered statistically significant only if the difference fell within the 95% confidence interval as determined by the Student t test.
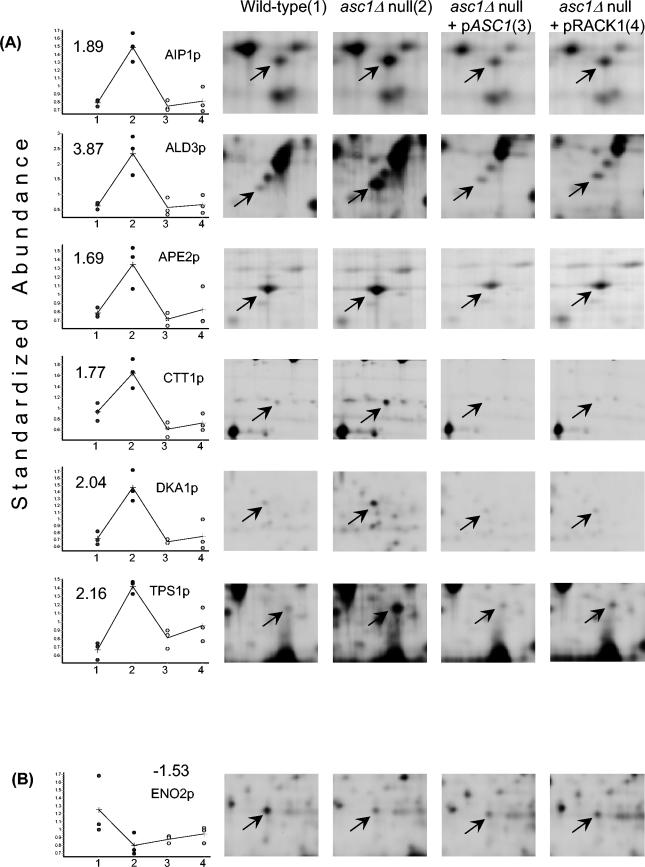
2D DIGE analysis indicated that of the ∼1,500 yeast proteins detected, 27 are ≥1.5-fold more abundant in the asc1Δ null strain than in the wild type (P < 0.05) (Fig. 5A and http://linklab.mc.vanderbilt.edu). Of these 27 elevated proteins, the differences in abundance between the wild-type and asc1Δ null strains ranged from 1.5- to 4.44-fold. Six of the 27 up-regulated had sufficient material for unambiguous identification by MS (Fig. 5A). For AIP1 up-regulated in the asc1Δ null strain, we performed Western analysis with the same extracts used for the 2D DIGE experiments. The relative abundance of Aip1p from the four strains was consistent with results from the 2D DIGE experiments (http://linklab.mc.vanderbilt.edu). Only 3 of the 1,500 proteins demonstrated statistically significant (P < 0.05) down-regulation in the asc1Δ null strain in comparison to the wild-type strain (Fig. 5B and http://linklab.mc.vanderbilt.edu). One of the three proteins had sufficient material for identification by MS (Fig. 5B). Both the asc1Δ null plus pASC1 and asc1Δ null plus pRACK1 strains restored elevated protein levels back to wild-type levels for 24 of 27 of the up-regulated proteins (Fig. 5A and http://linklab.mc.vanderbilt.edu). In contrast, levels of the three down-regulated proteins were not complemented by the asc1Δ null plus pASC1 or the asc1Δ null plus pRACK1 strain (Fig. 5B and http://linklab.mc.vanderbilt.edu). These results are consistent with our in vitro experiments suggesting a repressive role for Asc1p in translation. Additionally, these data provide further evidence that Asc1p and RACK1 are functionally orthologous ribosomal proteins.
FIG.5.
In vivo changes in protein levels in asc1Δ null strains. (A) Up-regulated proteins in the asc1Δ null strain are complemented by either Asc1p or RACK1. Graphs of standardized protein abundance in wild-type (no. 1 on the x axis), asc1Δ null (no. 2 on the x axis), asc1Δ null plus pASC1 (no. 3 on the x axis), and asc1Δ null plus pRACK1 (no. 4 on the x axis) complemented strains are shown. Representative 2D gel images are shown as a reference for protein levels for each strain and show an arrow pointing to the Cy3- or Cy5-labeled protein identified. In the graphs, each dot indicates the standardized protein abundance of a specific protein for a given strain in a single independent experiment. Each dot was calculated by dividing the Cy3 or Cy5 density by the Cy2 density (internal standard) for the respective protein position. Values on the graphs indicate the fold differences in average standardized abundance between the asc1Δ null and wild-type strains. A plus sign marks the average standardized abundance for a given strain and is representative of three independent experiments. The black line connects the average standardized protein abundances with one another. (B) The down-regulated protein in the asc1Δ null strain is not complemented by either Asc1p or RACK1. The graph shows standardized protein abundances in the wild-type (no. 1 on the x axis), asc1Δ null (no. 2 on the x axis), and asc1Δ null plus pASC1 (no. 3 on the x axis) and asc1Δ null plus pRACK1 (no. 4 on the x axis) complemented strains. The graphs and 2D gel images are similar to those in panel A.
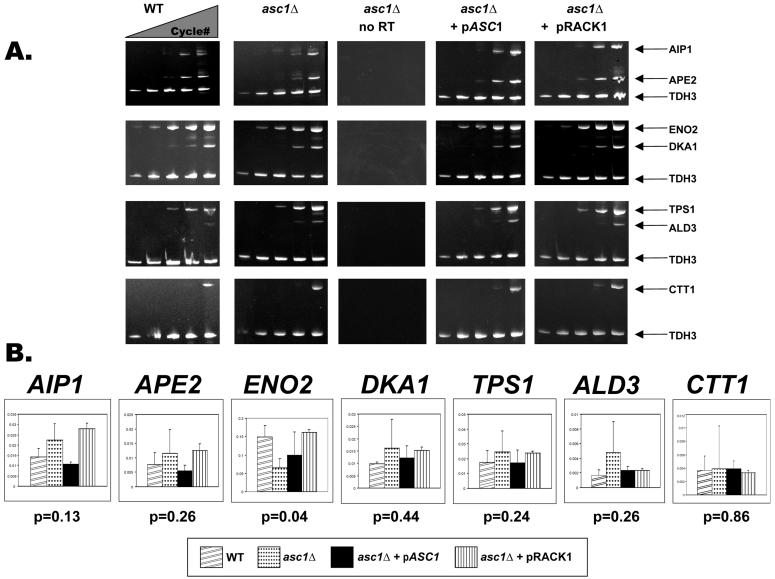
To distinguish between transcriptional and posttranscriptional regulation of gene expression for the seven identified proteins, we compared 2D DIGE protein abundance data with the levels of mRNAs from the four strains. The levels of mRNAs encoding six up-regulated proteins (AIP1p, ALD3p, APE2p, CTT1p, DKA1p, and TPS1p) and one down-regulated protein (ENO2p) were measured in the wild-type, asc1Δ null, and complemented strains by multiplex RT-PCR and real-time RT-PCR. By triplex RT-PCR we observed no apparent difference in transcript levels for any of the seven genes analyzed (Fig. 6A). By real-time quantitative PCR, the average differences in AIP1, ALD3, APE2, CTT1, DKA1, and TPS1 transcript abundance were variable but not statistically significantly different in the asc1Δ null strain relative to those in the wild-type and complemented strains (P < 0.05) (Fig. 6B). However, ENO2 transcript levels were down-regulated in the asc1Δ null strain relative to those in the wild type (P < 0.05) (Fig. 6A). Collectively, these results support a negative role for Asc1p or RACK1 in gene expression.
FIG. 6.
Analysis of mRNA transcript levels in asc1Δ null, asc1Δ null with pASC1, asc1Δ null with pRACK1, and wild-type strains. (A) Triplex RT-PCR of mRNA transcripts in asc1Δ null, asc1Δ null with pASC1, asc1Δ null with pRACK1, and wild-type strains used for 2D DIGE analysis. As a negative control, asc1Δ null RNA was amplified with no reverse transcriptase. AIP1, ALD3, APE2, CTT1, DKA1, ENO2, and TPS1 mRNA transcript abundances were measured by triplex semiquantitative RT-PCR with total RNA prepared from 2D DIGE experiments. cDNA transcripts were coamplified with TDH3 (GAPDH) as the standard. One of three independent samples is shown here. PCR cycles 17, 20, 23, 26, and 30 are shown in order from left to right for each sample. (B) Quantification of mRNA levels in asc1Δ null strains. The graph shows the quantified levels of the mRNAs for the seven genes from the four different strains in panel A. Levels of transcripts for all seven proteins identified by MALDI-TOF were quantified by real-time PCR of cDNA transcripts. As a standard, mRNA transcript levels for each gene were divided by the TDH3 (GAPDH) transcript levels in each sample. Error bars indicate the standard deviation of the mean. P values for each gene were determined through a Student t test comparing the calculated wild-type and asc1Δ null transcript levels.
DISCUSSION
RACK1 or Asc1p is the 33rd ribosomal protein of the small (40S) eukaryotic subunit.
In the classical experiments of Kruiswijk and Planta, proteins were considered core ribosomal components if they remained associated in the presence of 0.5 M KCl (31). Asc1p fulfills this requirement by associating with the ribosome in the presence of 1 M KCl (34). Localization of Asc1p or RACK1 to the 40S, 80S, and polysomal fractions has been observed by several independent studies (2, 4, 6, 26, 34, 50). Here we have demonstrated the localization of Asc1p and RACK1 to polysomes in five different eukaryotic species. Further, we demonstrate that yeast Asc1p and mammalian RACK1 compete for localization to the yeast ribosome. Moreover, we show that RACK1 functionally complements the phenotype of an asc1Δ null mutant. These data, taken together with the results of other studies, strongly suggest that Asc1p and RACK1 are orthologous core ribosomal proteins. Consequently, we propose that a more appropriate name for yeast ASC1 is RPS33, the 33rd ribosomal protein in the S. cerevisiae 40S small subunit. By corollary, RACK1 should be classified as a mammalian 40S core ribosomal protein.
ASC1 has several other features that are common to S. cerevisiae ribosomal genes. Although only a small percentage of S. cerevisiae genes contain an intron, 66% of the yeast ribosomal genes contain an intron. Two previous studies have shown that ASC1 contains an intron (6, 24). The codon adaptation index (CAI) is a measurement of the relative adaptiveness of the codon usage of a gene toward the codon usage of highly expressed genes (28, 48). CAIs range from 0 to 1, with higher values indicating a higher proportion of the most abundant codons. The average CAI for all yeast genes is 0.18, while the average CAI for ribosomal genes is 0.71. ASC1 has a CAI of 0.77, suggesting that it is highly expressed. Several studies have measured the transcriptional expression of ASC1 and found it to be expressed at levels equivalent to those of ribosomal genes (58, 61). A recent study measured the abundances of a large number of yeast proteins (19). The estimated abundances of ribosomal proteins ranged from 4.5 × 103 to 6.02 × 105 molecules per log-phase cell, with an average of 7.0 × 104 molecules per cell. Asc1p was found to be present at approximately 3.33 × 105 molecules per log-phase cell, which is in the same range as other ribosomal proteins (19). These results agreed with an earlier study showing that Asc1p is an abundant yeast protein that is highly enriched in ribosomal fractions (16). Asc1p is present at a concentration equimolar to that of the other ribosomal proteins (34). All of these reports indicate that ASC1 is a highly expressed yeast gene with characteristics similar to those of other ribosomal genes.
ASC1 or RACK1 exerts a repressive effect on protein synthesis.
We found that absence of yeast Asc1p resulted in elevated translational activity. However, this molecular phenotype could not be complemented in vitro by adding recombinant Asc1p. While it is possible the recombinant Asc1p fusion protein is not folded properly or lacks critical modified residues, the fusion protein does complement an asc1Δ null strain. Failure of a core eukaryotic ribosomal protein to complement when added exogenously is not surprising since eukaryotic ribosome assembly is thought to occur as a stepwise process in the nucleolus (14, 57). In contrast, translational initiation factors can be added exogenously to restore activity to wild-type levels in in vitro translation assays (7, 55).
To identify the molecular function of ASC1 and RACK1 in vivo, we used a quantitative global proteomic analysis of the wild-type, asc1Δ null, and yeast pASC1-complemented and mammalian pRACK1-complemented strains. We reasoned that if Asc1p and RACK1 function as repressive ribosomal proteins, we might observe a global up-regulation of protein levels in asc1Δ null strains. Further, this up-regulation should be complemented by expression of yeast pASC1 or mammalian pRACK1 in an asc1Δ null background. Indeed, we found that asc1Δ null strains have elevated levels of some proteins, and this molecular phenotype can be complemented by either yeast pASC1 or mammalian pRACK1. The changes in protein levels appear to be independent of mRNA levels and therefore likely occur through a posttranscriptional mechanism.
The in vitro translation assays with various mRNA templates suggest that translation activity is globally increased for ribosomes lacking Asc1p. However, in vivo assessment of protein levels in asc1Δ null strains shows that only a specific population of proteins (27 of 1,500) is significantly up-regulated. Although in vitro translation assays suggested a 2- to 10-fold increase in translational activity for the asc1Δ null strain ribosomes, in vivo posttranscriptional analysis indicated that specific protein levels were up-regulated between 1.5- and 4.44-fold (compare Fig. 4 and 5). The in vitro translation assays are by nature an artificial environment for translating mRNAs. Therefore, a 1:1 correlation with the in vivo results should not be expected. Despite the observed discrepancy between in vitro and in vivo results, both assays suggest a general trend of increased translational activity for the asc1Δ null strain. Taking the results of both assays into consideration, we propose that Asc1p functions as a translational repressor.
Recent reports suggest that Asc1p and RACK1 function to stimulate eukaryotic translation (4, 50). We speculate that these observations arise from indirect phenomena initiated by the translational up-regulation of regulatory proteins. For example, in this study we observed a reduction of ENO2 protein and mRNA levels in the asc1Δ null strain (Fig. 6A and B). It is conceivable that this reduction in transcript levels arises from the translational up-regulation of a transcriptional repressor.
A previous study observed an increase in specific mRNA levels in asc1Δ null strains relative to those in wild-type strains (24). Interestingly, these up-regulated mRNAs in asc1Δ null strains are regulated by the transcription factor Gcn4p (24). However, in the same study asc1Δ null strains did not show translational up-regulation of GCN4. Here we observed up-regulation of GCN4 translational activity relative to that of the wild type (Fig. 4E). Because a multiplicity of coactivators contributes to GCN4-mediated transcription, it is possible that specific mRNAs are elevated in asc1Δ null strains through the translational up-regulation of specific GCN4 coactivators (53). Our results support a model in which Asc1p and RACK1 are core components of the 40S ribosomal subunit that modulate the translation of mRNAs. Because our results show that average mRNA levels are often higher, but not statistically significantly different, in asc1Δ null strains, it is possible that Asc1p and RACK1 play an indirect role in transcriptional repression.
A number of proteins have been reported to interact with RACK1 in mammalian systems. Although we have not addressed this question, we observe RACK1 in ribosomal fractions when analyzing mammalian species. It is possible that RACK1 dissociates from the ribosome to perform other functions. A precedent for such phenomena has recently been reported for ribosomal protein L13A (35). Several studies have reported an association of RACK1 with the βII isoform of PKC (46, 52). Interactions of RACK1 with signal transduction proteins may direct ribosomes to specific cellular locations where localized translation of proteins is required. Further experiments are required to test these different models.
Acknowledgments
We thank Tracey Fleischer and David Powell for numerous discussions during this project. We thank Tony Weil, Kathy Friedman, and Elizabeth Link for comments during the preparation of the manuscript. We thank Jennifer Jennings for assistance with the MS experiments and Rodney Gabriel for medium preparation. We thank Corbin Williams for assistance with 2D DIGE experiments. We thank Linda Riles and Mark Johnston for yeast lambda clone 5992 (ATCC 70652), John Warner for antibodies to yeast ribosomal protein Rpl3p, David Amberg for antibody to yeast Aip1p, Alan Hinnebusch for GCN4 reporter plasmid p180, Gerhard Braus for plasmid pME1867, Tony Weil for yeast vectors pRS416 and p426GPD, and Alan Sachs for plasmid T3 lucpA and in vitro translation protocols.
We thank the Vanderbilt Academic Venture Capital Fund. This project was supported by NIH grant GM64779 to A.J.L. V.R.G. is supported by NIH training grant T32 CA009385. C.W. is supported by NIH grants GM64779 and HL68744. A.J.L. is supported by NIH grants GM64779, HL68744, NS43952, ES11993, and CA098131.
REFERENCES
- 1.Alban, A., S. O. David, L. Bjorkesten, C. Andersson, E. Sloge, S. Lewis, and I. Currie. 2003. A novel experimental design for comparative two-dimensional gel analysis: two-dimensional difference gel electrophoresis incorporating a pooled internal standard. Proteomics 3:36-44. [DOI] [PubMed]
- 2.Angenstein, F., A. M. Evans, R. E. Settlage, S. T. Moran, S. C. Ling, A. Y. Klintsova, J. Shabanowitz, D. F. Hunt, and W. T. Greenough. 2002. A receptor for activated C kinase is part of messenger ribonucleoprotein complexes associated with polyA-mRNAs in neurons. J. Neurosci. 22:8827-8837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asano, K., L. Phan, T. Krishnamoorthy, G. D. Pavitt, E. Gomez, E. M. Hannig, J. Nika, T. F. Donahue, H. K. Huang, and A. G. Hinnebusch. 2002. Analysis and reconstitution of translation initiation in vitro. Methods Enzymol. 351:221-247. [DOI] [PubMed] [Google Scholar]
- 4.Ceci, M., C. Gaviraghi, C. Gorrini, L. A. Sala, N. Offenhauser, P. Carlo Marchisio, and S. Biffo. 2003. Release of eIF6 [p27(BBP)] from the 60S subunit allows 80S ribosome assembly. Nature 426:579-584. [DOI] [PubMed] [Google Scholar]
- 5.Chang, B. Y., K. B. Conroy, E. M. Machleder, and C. A. Cartwright. 1998. RACK1, a receptor for activated C kinase and a homolog of the β subunit of G proteins, inhibits activity of Src tyrosine kinases and growth of NIH 3T3 cells. Mol. Cell. Biol. 18:3245-3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chantrel, Y., M. Gaisne, C. Lions, and J. Verdiere. 1998. The transcriptional regulator Hap1p (Cyp1p) is essential for anaerobic or heme-deficient growth of Saccharomyces cerevisiae: genetic and molecular characterization of an extragenic suppressor that encodes a WD repeat protein. Genetics 148:559-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi, S. K., J. H. Lee, W. L. Zoll, W. C. Merrick, and T. E. Dever. 1998. Promotion of met-tRNAiMet binding to ribosomes by yIF2, a bacterial IF2 homolog in yeast. Science 280:1757-1760. [DOI] [PubMed] [Google Scholar]
- 8.Christianson, T. W., R. S. Sikorski, M. Dante, J. H. Shero, and P. Hieter. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110:119-122. [DOI] [PubMed] [Google Scholar]
- 9.Cox, E. A., D. Bennin, A. T. Doan, T. O'Toole, and A. Huttenlocher. 2003. RACK1 regulates integrin-mediated adhesion, protrusion, and chemotactic cell migration via its Src-binding site. Mol. Biol. Cell 14:658-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dell, E. J., J. Connor, S. Chen, E. G. Stebbins, N. P. Skiba, D. Mochly-Rosen, and H. E. Hamm. 2002. The βγ subunit of heterotrimeric G proteins interacts with RACK1 and two other WD repeat proteins. J. Biol. Chem. 277:49888-49895. [DOI] [PubMed] [Google Scholar]
- 11.Dever, T. E., L. Feng, R. C. Wek, A. M. Cigan, T. F. Donahue, and A. G. Hinnebusch. 1992. Phosphorylation of initiation factor 2 alpha by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell 68:585-596. [DOI] [PubMed] [Google Scholar]
- 12.Doudna, J. A., and V. L. Rath. 2002. Structure and function of the eukaryotic ribosome: the next frontier. Cell 109:153-156. [DOI] [PubMed] [Google Scholar]
- 13.Friedman, D. B., S. Hill, J. W. Keller, N. B. Merchant, S. E. Levy, R. J. Coffey, and R. M. Caprioli. 2004. Proteome analysis of human colon cancer by two-dimensional difference gel electrophoresis and mass spectrometry. Proteomics 4:793-811. [DOI] [PubMed] [Google Scholar]
- 14.Fromont-Racine, M., B. Senger, C. Saveanu, and F. Fasiolo. 2003. Ribosome assembly in eukaryotes. Gene 313:17-42. [DOI] [PubMed] [Google Scholar]
- 15.Gallina, A., F. Rossi, and G. Milanesi. 2001. Rack1 binds HIV-1 Nef and can act as a Nef-protein kinase C adaptor. Virology 283:7-18. [DOI] [PubMed] [Google Scholar]
- 16.Garrels, J. I., C. S. McLaughlin, J. R. Warner, B. Futcher, G. I. Latter, R. Kobayashi, B. Schwender, T. Volpe, D. S. Anderson, R. Mesquita-Fuentes, and W. E. Payne. 1997. Proteome studies of Saccharomyces cerevisiae: identification and characterization of abundant proteins. Electrophoresis 18:1347-1360. [DOI] [PubMed] [Google Scholar]
- 17.Geijsen, N., M. Spaargaren, J. A. Raaijmakers, J. W. Lammers, L. Koenderman, and P. J. Coffer. 1999. Association of RACK1 and PKCβ with the common beta-chain of the IL-5/IL-3/GM-CSF receptor. Oncogene 18:5126-5130. [DOI] [PubMed] [Google Scholar]
- 18.Gerbasi, V., S. Lutsenko, and E. J. Lewis. 2003. A mutation in the ATP7B copper transporter causes reduced dopamine β-hydroxylase and norepinephrine in mouse adrenal. Neurochem. Res. 28:867-873. [DOI] [PubMed] [Google Scholar]
- 19.Ghaemmaghami, S., W. K. Huh, K. Bower, R. W. Howson, A. Belle, N. Dephoure, E. K. O'Shea, and J. S. Weissman. 2003. Global analysis of protein expression in yeast. Nature 425:737-741. [DOI] [PubMed] [Google Scholar]
- 20.Guillemot, F., A. Billault, and C. Auffray. 1989. Physical linkage of a guanine nucleotide-binding protein-related gene to the chicken major histocompatibility complex. Proc. Natl. Acad. Sci. USA 86:4594-4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hennig, E. E., E. Butruk, and J. Ostrowski. 2001. RACK1 protein interacts with Helicobacter pylori VacA cytotoxin: the yeast two-hybrid approach. Biochem. Biophys. Res. Commun. 289:103-110. [DOI] [PubMed] [Google Scholar]
- 22.Hermanto, U., C. S. Zong, W. Li, and L. H. Wang. 2002. RACK1, an insulin-like growth factor I (IGF-I) receptor-interacting protein, modulates IGF-I-dependent integrin signaling and promotes cell spreading and contact with extracellular matrix. Mol. Cell. Biol. 22:2345-2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hinnebusch, A. G. 1985. A hierarchy of trans-acting factors modulates translation of an activator of amino acid biosynthetic genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 5:2349-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoffmann, B., H. U. Mosch, E. Sattlegger, I. B. Barthelmess, A. Hinnebusch, and G. H. Braus. 1999. The WD protein Cpc2p is required for repression of Gcn4 protein activity in yeast in the absence of amino acid starvation. Mol. Microbiol. 31:807-822. [DOI] [PubMed] [Google Scholar]
- 25.Iizuka, N., L. Najita, A. Franzusoff, and P. Sarnow. 1994. Cap-dependent and Cap-independent translation by internal initiation of mRNAs in cell extracts prepared from Saccharomyces cerevisiae. Mol. Cell. Biol. 14:7322-7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inada, T., E. Winstall, S. Z. Tarun, Jr., J. R. Yates III, D. Schieltz, and A. B. Sachs. 2002. One-step affinity purification of the yeast ribosome and its associated proteins and mRNAs. RNA 8:948-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishikawa, H. 1977. Evolution of ribosomal RNA. Comp. Biochem. Physiol. B. 58:1-7. [DOI] [PubMed] [Google Scholar]
- 28.Jansen, R., H. J. Bussemaker, and M. Gerstein. 2003. Revisiting the codon adaptation index from a whole-genome perspective: analyzing the relationship between gene expression and codon occurrence in yeast using a variety of models. Nucleic Acids Res. 31:2242-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kiely, P. A., A. Sant, and R. O'Connor. 2002. RACK1 is an insulin-like growth factor 1 (IGF-1) receptor-interacting protein that can regulate IGF-1-mediated Akt activation and protection from cell death. J. Biol. Chem. 277:22581-22589. [DOI] [PubMed] [Google Scholar]
- 30.Koehler, J. A., and M. F. Moran. 2001. RACK1, a protein kinase C scaffolding protein, interacts with the PH domain of p120GAP. Biochem. Biophys. Res. Commun. 283:888-895. [DOI] [PubMed] [Google Scholar]
- 31.Kruiswijk, T., and R. J. Planta. 1974. Analysis of the protein composition of yeast ribosomal subunits by two-dimensional polyacrylamide gel electrophoresis. Mol. Biol. Rep. 1:409-415. [DOI] [PubMed] [Google Scholar]
- 32.Kubota, T., N. Yokosawa, S. Yokota, and N. Fujii. 2002. Association of mumps virus V protein with RACK1 results in dissociation of STAT-1 from the alpha interferon receptor complex. J. Virol. 76:12676-12682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liliental, J., and D. D. Chang. 1998. Rack1, a receptor for activated protein kinase C, interacts with integrin beta subunit. J. Biol. Chem. 273:2379-2383. [DOI] [PubMed] [Google Scholar]
- 34.Link, A. J., J. Eng, D. M. Schieltz, E. Carmack, G. J. Mize, D. R. Morris, B. M. Garvik, and J. R. Yates III. 1999. Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol. 17:676-682. [DOI] [PubMed] [Google Scholar]
- 35.Mazumder, B., P. Sampath, V. Seshadri, R. K. Maitra, P. E. DiCorleto, and P. L. Fox. 2003. Regulated release of L13a from the 60S ribosomal subunit as a mechanism of transcript-specific translational control. Cell 115:187-198. [DOI] [PubMed] [Google Scholar]
- 36.McCahill, A., J. Warwicker, G. B. Bolger, M. D. Houslay, and S. J. Yarwood. 2002. The RACK1 scaffold protein: a dynamic cog in cell response mechanisms. Mol. Pharmacol. 62:1261-1273. [DOI] [PubMed] [Google Scholar]
- 37.Mikulits, W., B. Pradet-Balade, B. Habermann, H. Beug, J. A. Garcia-Sanz, and E. W. Mullner. 2000. Isolation of translationally controlled mRNAs by differential screening. FASEB J. 14:1641-1652. [DOI] [PubMed] [Google Scholar]
- 38.Mourton, T., C. B. Hellberg, S. M. Burden-Gulley, J. Hinman, A. Rhee, and S. M. Brady-Kalnay. 2001. The PTPmu protein-tyrosine phosphatase binds and recruits the scaffolding protein RACK1 to cell-cell contacts. J. Biol. Chem. 276:14896-14901. [DOI] [PubMed] [Google Scholar]
- 39.Mumberg, D., R. Muller, and M. Funk. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156:119-122. [DOI] [PubMed] [Google Scholar]
- 40.Olson, M. V., J. E. Dutchik, M. Y. Graham, G. M. Brodeur, C. Helms, M. Frank, M. MacCollin, R. Scheinman, and T. Frank. 1986. Random-clone strategy for genomic restriction mapping in yeast. Proc. Natl. Acad. Sci. USA 83:7826-7830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ozaki, T., K. Watanabe, T. Nakagawa, K. Miyazaki, M. Takahashi, and A. Nakagawara. 2003. Function of p73, not of p53, is inhibited by the physical interaction with RACK1 and its inhibitory effect is counteracted by pRB. Oncogene 22:3231-3242. [DOI] [PubMed] [Google Scholar]
- 42.Reinhardt, J., and T. Wolff. 2000. The influenza A virus M1 protein interacts with the cellular receptor of activated C kinase (RACK) 1 and can be phosphorylated by protein kinase C. Vet. Microbiol. 74:87-100. [DOI] [PubMed] [Google Scholar]
- 43.Rigas, A. C., D. M. Ozanne, D. E. Neal, and C. N. Robson. 2003. The scaffolding protein RACK1 interacts with androgen receptor and promotes cross-talk through a protein kinase C signaling pathway. J. Biol. Chem. 278:46087-46093. [DOI] [PubMed] [Google Scholar]
- 44.Riles, L., J. E. Dutchik, A. Baktha, B. K. McCauley, E. C. Thayer, M. P. Leckie, V. V. Braden, J. E. Depke, and M. V. Olson. 1993. Physical maps of the six smallest chromosomes of Saccharomyces cerevisiae at a resolution of 2.6 kilobase pairs. Genetics 134:81-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodal, A. A., J. W. Tetreault, P. Lappalainen, D. G. Drubin, and D. C. Amberg. 1999. Aip1p interacts with cofilin to disassemble actin filaments. J. Cell Biol. 145:1251-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ron, D., Z. Jiang, L. Yao, A. Vagts, I. Diamond, and A. Gordon. 1999. Coordinated movement of RACK1 with activated βIIPKC. J. Biol. Chem. 274:27039-27046. [DOI] [PubMed] [Google Scholar]
- 47.Sanders, S. L., J. Jennings, A. Canutescu, A. J. Link, and P. A. Weil. 2002. Proteomics of the eukaryotic transcription machinery: identification of proteins associated with components of yeast TFIID by multidimensional mass spectrometry. Mol. Cell. Biol. 22:4723-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sharp, P. M., and W. H. Li. 1987. The codon adaptation index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 15:1281-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sherman, F., G. R. Fink, and J. B. Hicks. 1986. Laboratory course manual for methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 50.Shor, B., J. Calaycay, J. Rushbrook, and M. McLeod. 2003. Cpc2/RACK1 is a ribosome-associated protein that promotes efficient translation in Schizosaccharomyces pombe. J. Biol. Chem. 278:49119-49128. [DOI] [PubMed] [Google Scholar]
- 51.Spahn, C. M., R. Beckmann, N. Eswar, P. A. Penczek, A. Sali, G. Blobel, and J. Frank. 2001. Structure of the 80S ribosome from Saccharomyces cerevisiae—tRNA-ribosome and subunit-subunit interactions. Cell 107:373-386. [DOI] [PubMed] [Google Scholar]
- 52.Stebbins, E. G., and D. Mochly-Rosen. 2001. Binding specificity for RACK1 resides in the V5 region of beta II protein kinase C. J. Biol. Chem. 276:29644-29650. [DOI] [PubMed] [Google Scholar]
- 53.Swanson, M. J., H. Qiu, L. Sumibcay, A. Krueger, S. J. Kim, K. Natarajan, S. Yoon, and A. G. Hinnebusch. 2003. A multiplicity of coactivators is required by Gcn4p at individual promoters in vivo. Mol. Cell. Biol. 23:2800-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tarun, S. Z., Jr., and A. B. Sachs. 1995. A common function for mRNA 5′ and 3′ ends in translation initiation in yeast. Genes Dev. 9:2997-3007. [DOI] [PubMed] [Google Scholar]
- 55.Tarun, S. Z., Jr., S. E. Wells, J. A. Deardorff, and A. B. Sachs. 1997. Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc. Natl. Acad. Sci. USA 94:9046-9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tcherkasowa, A. E., S. Adam-Klages, M. L. Kruse, K. Wiegmann, S. Mathieu, W. Kolanus, M. Kronke, and D. Adam. 2002. Interaction with factor associated with neutral sphingomyelinase activation, a WD motif-containing protein, identifies receptor for activated C-kinase 1 as a novel component of the signaling pathways of the p55 TNF receptor. J. Immunol. 169:5161-5170. [DOI] [PubMed] [Google Scholar]
- 57.Tschochner, H., and E. Hurt. 2003. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 13:255-263. [DOI] [PubMed] [Google Scholar]
- 58.Velculescu, V. E., L. Zhang, W. Zhou, J. Vogelstein, M. A. Basrai, D. E. Bassett, Jr., P. Hieter, B. Vogelstein, and K. W. Kinzler. 1997. Characterization of the yeast transcriptome. Cell 88:243-251. [DOI] [PubMed] [Google Scholar]
- 59.Vilardell, J., and J. R. Warner. 1997. Ribosomal protein L32 of Saccharomyces cerevisiae influences both the splicing of its own transcript and the processing of rRNA. Mol. Cell. Biol. 17:1959-1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, A. M. Chu, C. Connelly, K. Davis, F. Dietrich, S. W. Dow, M. El Bakkoury, F. Foury, S. H. Friend, E. Gentalen, G. Giaever, J. H. Hegemann, T. Jones, M. Laub, H. Liao, R. W. Davis, et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901-906. [DOI] [PubMed] [Google Scholar]
- 61.Wodicka, L., H. Dong, M. Mittmann, M. H. Ho, and D. J. Lockhart. 1997. Genome-wide expression monitoring in Saccharomyces cerevisiae. Nat. Biotechnol. 15:1359-1367. [DOI] [PubMed] [Google Scholar]
- 62.Yarwood, S. J., M. R. Steele, G. Scotland, M. D. Houslay, and G. B. Bolger. 1999. The RACK1 signaling scaffold protein selectively interacts with the cAMP-specific phosphodiesterase PDE4D5 isoform. J. Biol. Chem. 274:14909-14917. [DOI] [PubMed] [Google Scholar]
- 63.Yik, J. H., R. Chen, R. Nishimura, J. L. Jennings, A. J. Link, and Q. Zhou. 2003. Inhibition of P-TEFb (CDK9/cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol. Cell 12:971-982. [DOI] [PubMed] [Google Scholar]