Abstract
Mutations in the CSA and CSB genes cause Cockayne syndrome, a rare inherited disorder characterized by UV sensitivity, severe neurological abnormalities, and progeriod symptoms. Both gene products function in the transcription-coupled repair (TCR) subpathway of nucleotide excision repair (NER), providing the cell with a mechanism to remove transcription-blocking lesions from the transcribed strands of actively transcribed genes. Besides a function in TCR of NER lesions, a role of CSB in (transcription-coupled) repair of oxidative DNA damage has been suggested. In this study we used mouse models to compare the effect of a CSA or a CSB defect on oxidative DNA damage sensitivity at the levels of the cell and the intact organism. In contrast to CSB−/− mouse embryonic fibroblasts (MEFs), CSA−/− MEFs are not hypersensitive to gamma-ray or paraquat treatment. Similar results were obtained for keratinocytes. In contrast, both CSB−/− and CSA−/− embryonic stem cells show slight gamma-ray sensitivity. Finally, CSB−/− but not CSA−/− mice fed with food containing di(2-ethylhexyl)phthalate (causing elevated levels of oxidative DNA damage in the liver) show weight reduction. These findings not only uncover a clear difference in oxidative DNA damage sensitivity between CSA- and CSB-deficient cell lines and mice but also show that sensitivity to oxidative DNA damage is not a uniform characteristic of Cockayne syndrome. This difference in the DNA damage response between CSA- and CSB-deficient cells is unexpected, since until now no consistent differences between CSA and CSB patients have been reported. We suggest that the CSA and CSB proteins in part perform separate roles in different DNA damage response pathways.
In order to cope with the continuous attack of endogenous and environmental genotoxic agents on the integrity of their genomes, cells are equipped with a battery of DNA repair systems with partly overlapping substrate specificities. In mammals, chemically and UV-induced helix-distorting lesions are removed through the versatile nucleotide excision repair (NER) pathway. NER functions by excision of the lesion, as an approximately 30-nt oligonucleotide, after which the resulting single-stranded gap is filled in by DNA polymerase and ligase. Recognition of the lesion occurs via two subpathways. In global genome NER (GG-NER), repair of helix-distorting base damage in the entire genome is initiated by recognition of these lesions by the XPC-HR23B-Cen2 complex, facilitated by the XPE dimer (UV-DDB1/2). In transcription-coupled NER (TC-NER), repair of transcriptional blocking lesions is thought to be initiated by an RNA polymerase, which is unable to pass the lesion (for reviews, see references 11, 20, 31, 47, and 48).
Mutations in NER genes can lead to several rare inherited recessive disorders. The prototype NER syndrome is xeroderma pigmentosum (XP), which is characterized by pronounced UV sensitivity, pigmentation abnormalities in sun-exposed areas of the skin, and more than a 1,000-fold-higher risk of developing skin cancer, causing a 30-year life span reduction. In a subpopulation of XP patients, accelerated neurodegeneration occurs, due to early loss of neurons (4, 39). A distinct NER-associated disorder is Cockayne syndrome (CS), which shares with XP the pronounced UV sensitivity but in addition has a wide range of severe physical and mental manifestations. These include postnatal growth failure, chachetic dwarfism, retinal degeneration, deafness, mental retardation associated with neurodemyelination, and skeletal abnormalities such as osteoporosis and a bird-like face (4, 37). Many of these symptoms, together with the average short life span of 12.5 years, point to premature aging.
Complementation analysis by use of cell hybridization studies have shown the involvement of seven genes (XPA through XPG) in the NER-deficient form of XP (4). Mutations in XP genes cause a combined defect in both the TC-NER and GG-NER pathways in five out of seven XP complementation groups. In contrast, mutations in XPC and XPE cause a deficiency in GG-NER only (21, 49, 57, 58). CS is associated with a specific TC-NER defect, caused by mutations in two genes, CSA or CSB (56). Interestingly, mutations in XPB, XPD, or XPG can cause a combination of XP and CS (4).
The notion that mutations in CSA or CSB affect only the TC-NER pathway, while mutations causing XP frequently hamper both TC-NER and GG-NER, is difficult to reconcile with the more severe symptoms observed in CS compared to XP patients. To explain this phenomenon, a role of the CS proteins outside TC-NER has been suggested, such as an auxiliary function in transcription (3, 14, 42) and/or in (transcription-coupled) repair of oxidative DNA damage and other non-NER lesions (12, 13, 27, 30, 38, 46, 52). Similarly, cell lines from XP or CS patients with mutations in XPB, XPD, or XPG show a defect in transcription-coupled repair (TCR) of oxidative DNA damage (9, 30), underscoring the possible involvement of unrepaired oxidative DNA lesions in the CS etiology.
Most studies on the role of CS proteins in processes other than classical TC-NER have been performed with CSB-deficient human cell lines. Since clinical differences between patients belonging to CSA and CSB complementation groups have not been observed, similar responses for CSA- and CSB-deficient cell lines are expected. Indeed, for NER-related assays, there is no evidence for a CSB- or CSA-related difference. Although an early report does not exclude a possible CSA- or CSB-related difference in the cellular response to gamma rays (27), findings obtained with non-NER-related assays in CSB-deficient systems have often been extrapolated to be general CS characteristics. Yet, as clear biochemical differences between CSA and CSB exist (55), a minor variance in the cellular response to genotoxic stress may be present in CSA and CSB cells. In studying such potential subtle differences, isogenic NER-deficient mouse models are highly valuable, as the results obtained are not influenced by differences in genetic background. Previously, using a mouse model for CSB (54), we showed that CSB-deficient cells and animals are sensitive to oxidative DNA damage (12). Recently, we also have generated a mouse model for CSA (53) and have shown that both CS mouse models mimic the human phenotype in terms of the repair defect, retinal degeneration, and manifestation of UV sensitivity of skin and eyes. To determine whether CSA and CSB are truly equivalent in their oxidative DNA damage responses, we systematically compared the sensitivities to oxidative stress in a variety of cell types and in the intact organism in a CSA- and CSB-deficient background.
MATERIALS AND METHODS
Cell lines.
The isolation of primary CSB−/− (FVB/129Ola) and CSA−/− (C57BL6J/129Ola) mouse embryonic fibroblasts (MEFs) and corresponding wild-type cell lines has been described previously (53, 54). Cells were cultured in F10-DMEM (1:1) (Gibco) supplemented with 10% fetal calf serum and 50 μg of penicillin and streptomycin (Gibco) per ml. Spontaneously immortalized cell lines were obtained by continuous subculturing of primary MEFs.
Primary wild-type, CSA−/− and CSB−/− keratinocytes from 2-day-old mice (in a pure C57BL6 genetic background) were isolated as described previously (15, 19). Keratinocytes were cultured on collagen-fibronectin-coated dishes in low-calcium (0.05 mM) Eagle’s minimal essential medium (BioWhittaker) supplemented with 8% fetal calf serum (treated with Chelex 100 [Bio-Rad] to remove Ca2+ ions), 1 ng of keratinocyte growth factor (R&D Systems) per ml, and 50 μg of penicillin and streptomycin (Gibco) per ml. Spontaneously immortalized cell lines were obtained by continuous subculturing of primary keratinocytes.
Isolation of CSB−/− and wild-type embryonic stem (ES) cell lines in a C57BL6 background has been described previously (12). CSA−/− ES cell lines are isolated by the same procedure (12). ES cells were maintained on gelatin-coated dishes in 50% buffalo rat liver cell-conditioned DMEM-50% fresh DMEM supplemented with 15% fetal calf serum, 0.1 mM nonessential amino acids (Gibco), 50 μg of penicillin and streptomycin (Gibco) per ml, 1,000 U of leukemia inhibitory factor (Chemicon) per ml and 0.1 mM 2-mercaptoethanol (Sigma).
Cellular sensitivity studies.
For determination of the gamma-ray sensitivity of immortalized MEFs, keratinocytes, and ES cells, cells were plated in 6-cm-diameter dishes at various dilutions. After 12 to 16 h, cells were irradiated with a single dose in the range of 0 to 8 Gy with a 137Cs source. Cells were grown for 5 to 14 days, fixed, stained, and counted to assess the colony-forming ability. All experiments were performed in triplicate.
This protocol was adapted for determination of UV sensitivity of keratinocytes and ES cells by irradiating the cells with different doses of UV (254 nm; Philips TUV lamp) instead of the gamma ray irradiation.
UV sensitivity of MEFs was determined as described previously (43). Briefly, MEFs were exposed to different doses of UV (254 nm; Philips TUV lamp) and allowed to grow for another 3 to 5 days, before reaching confluency. The number of proliferating cells was estimated by scintillation counting of the radioactivity incorporated during a 3-h pulse with [3H]thymidine (5 μCi/ml; specific activity, 40 to 60 Ci/mmol) (Amersham). Cell survival was expressed as the ratio of 3H incorporations in treated and nontreated cells. This protocol was adapted for paraquat survival by growing MEF cultures for 3 to 5 days in medium containing different concentrations of paraquat, followed by determination of the amount of proliferating cells as described above.
DEHP treatment of mice.
Wild-type, CSB−/−, and CSA−/− female mice in a C57BL6 background were put on a diet containing 6,000 ppm of di(2-ethylhexyl)phthalate (DEHP) (Sigma) or on a regular diet for 4 weeks (untreated, 5 wild-type, 7 CSB−/−, and 10 CSA−/− mice; treated, 8 wild-type, 7 CSB−/−, and 13 CSA−/− mice). Animals were screened daily for discomfort. Animals were weighed at the start of and then weekly during the experiment. The relative weight is calculated as the ratio between the weights of the mouse during the experiment and at the start of the experiment, and the ratio of these relative weights of treated versus untreated animals of the same genotype is plotted. Animal experiments were approved by the local animal ethics committee of the National Institute of Public Health and the Environment, Bilthoven, The Netherlands.
Measurement of 8-oxo-dG in mouse liver.
The 8-oxo-2′-deoxyguanosine (8-oxo-dG) analyses were performed as previously described (41). In short, the DNA from approximately 200 mg of liver or 160 mg of kidney (one kidney) was extracted and precipitated by an NaI-based procedure originally described by Nakae et al. (36) and Asami and Kasai (2). The DNA was resuspended in 10 mM Tris-0.1 mM desferrioxamine prior to enzymatic hydrolysis with nuclease P1 and alkaline phosphatase (Boehringer, Mannheim, Germany). The deoxyribonucleotides were then treated with DOWEX 1 × 8-400 ion-exchange resin (The Dow Chemical Company, Midland, Mich.) to remove I− and finally were filtered through a Micropure-EZ filter (Millipore, Bedford, Mass.). The levels of 8-oxo-dG and α-deoxyguanosine were measured by using a high-pressure liquid chromatography system with electrochemical and UV detection. Peak areas were used for calculations. Calibration curves were run together with each batch of samples.
RESULTS
CSA−/− MEFs lack hypersensitivity to gamma-ray irradiation.
By using [3H]thymidine incorporation assays, primary CSA−/− and CSB −/− MEFs have been shown to be UV sensitive (53, 54). Since gamma-ray sensitivity cannot be determined in this manner but rather requires use of a clonogenic assay, and since primary cells are not suitable for performing clonogenic experiments, we first subcultured MEFs until spontaneous transformation resulted in formation of established cell lines. Similar to the case for primary MEFs, immortalized CSA−/− and CSB−/− MEFs are both UV sensitive (Fig. 1A).
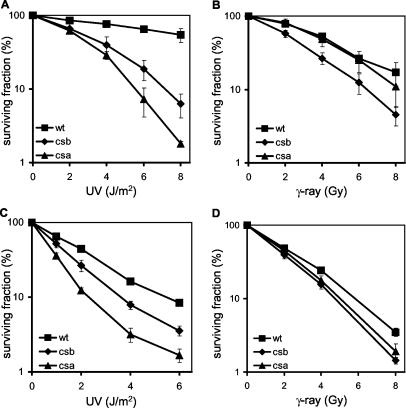
FIG. 1.
Survival of wild-type, CSB−/−, and CSA−/− MEFs after exposure to UV, gamma rays, or paraquat. (A) UV survival of spontaneously immortalized CSB−/−, CSA−/−, and wild-type (wt) MEFs. Experiments were performed at least two times per cell line, with at least two cell lines per genotype as determined by the [3H]thymidine incorporation assay. Shown are representative curves. Error bars indicate the standard errors of the means. (B) Survival of spontaneously immortalized CSB−/−, CSA−/−, and wild-type MEFs after exposure to increasing doses of gamma rays, as determined by the colony assay. Shown are averages for at least two cell lines per genotype, as measured by at least three independent experiments. Error bars indicate the standard errors of the means. (C) Survival of spontaneously immortalized wild-type MEFs in different C57BL6, FVB/129OLA, or C57BL6/129OLA backgrounds after exposure to increasing doses of gamma-rays, as determined by the colony assay. Shown are the averages from at least three independent experiments. Error bars indicate the standard errors of the means. (D) Paraquat survival of spontaneously immortalized CSB−/−, CSA−/−, and wild-type MEFs. Experiments were performed at least two times per cell line, with at least two cell lines per genotype as determined by the [3H]thymidine incorporation assay. Shown are representative curves. Error bars indicate the standard errors of the means.
To critically investigate whether CSA−/− MEFs, like CSB−/− MEFs, display hypersensitivity to ionizing radiation, we performed clonogenic gamma-ray survival experiments with spontaneously transformed wild-type, CSB−/−, and CSA−/− MEFs (at least two independent cell lines per genotype). In accordance with previous experiments (12), we observed that CSB−/− MEFs are approximately twofold more sensitive to gamma-ray irradiation than wild-type MEFs. Surprisingly, however, CSA−/− MEFs show a gamma-ray survival similar to that of wild-type MEFs (Fig. 1B). The observed difference in gamma-ray sensitivity between CSA−/− and CSB−/− MEFs cannot be attributed to differences in genetic backgrounds (FVB/129OLA and C57BL6/129OLA), since wild-type MEFs from these different backgrounds have comparable gamma-ray sensitivities (Fig. 1C).
CSA−/− MEFs are not sensitive to paraquat.
To confirm that CSA−/− MEFs are insensitive to oxidative DNA damage, we next tested the survival of these cell lines following treatment with the herbicide paraquat. Enzymatic reduction converts paraquat into radicals that react with molecular oxygen and thereby produce superoxide anions, giving rise to hydrogen peroxide (1). As shown previously, CSB−/− MEFs are sensitive to paraquat exposure (12). In marked contrast, CSA−/− MEFs possess paraquat sensitivity in the wild-type range (Fig. 1D). On the basis of the observed insensitivity of CSA−/− MEFs to both gamma rays and paraquat exposure, we conclude that CSA−/− MEFs are not sensitive to oxidative DNA damage.
Cell type-specific differences in gamma-ray sensitivity in CSA−/− and CSB−/− cells.
To determine whether the insensitivity to oxidative damage of the CSA−/− MEFs is a general feature, we extended our study to other cell types. To this end, we isolated keratinocytes from wild-type, CSA−/−, and CSB−/− newborn mice, all in a genetically identical C57BL6 background to avoid any influence of genetic background. Similar to the case for MEFs, spontaneously transformed CSA−/− and CSB−/− keratinocytes are UV sensitive, as determined by clonogenic assays (Fig. 2A). Next, we performed clonogenic gamma-ray survival experiments with wild-type, CSA−/−, and CSB−/− keratinocytes. In line with our observations of MEFs, we showed that a CSB deficiency causes hypersensitivity of keratinocytes to gamma-ray irradiation, whereas a deficiency of CSA does not make these cells more sensitive to gamma rays (Fig. 2B).
FIG. 2.
UV and gamma-ray sensitivities of CSB−/−, CSA−/−, and wild-type (wt) keratinocytes and ES cells. (A and B) UV (A) and gamma-ray (B) survival of spontaneously immortalized CSB−/−, CSA−/−, and wild-type keratinocytes, as determined by the colony assay. The wild-type curve represents the average from wild-type, CSA+/−, and CSB+/− cell lines, as determined in at least three experiments. The CSB−/− curve is the average from two cell lines, as measured by three independent experiments. The CSA−/− curve is the average from at least two independent experiments. Error bars indicate the standard errors of the means. (C and D) UV (C) and gamma-ray (D) survival of CSB−/−, CSA−/−, and wild-type ES cells, as determined by the colony assay. Shown are the averages for at least two cell lines per genotype, as measured by at least three independent experiments. Error bars indicate the standard errors of the means.
Previously, we have demonstrated that CSB−/− ES cells have a slight gamma-ray sensitivity (12). To extend this study to CSA−/− ES cells, we isolated pluripotent ES cells from blastocysts derived from intercrosses between CSA−/− animals in a C57BL6 background. We first analyzed the UV sensitivity of these ES lines and observed that CSA−/−, and to a somewhat lesser extent CSB−/−, ES lines are UV sensitive (Fig. 2C). Subsequently, we performed a clonogenic gamma-ray assay on wild-type, CSA−/−, and CSB−/− ES cells. Unexpectedly, since CSA−/− MEFs and keratinocytes display a wild-type gamma-ray sensitivity, we found a slight gamma-ray sensitivity in three independent CSA−/− ES cell lines, comparable to the mild gamma-ray sensitivity in CSB−/− ES cells (Fig. 2D). A fourth CSA−/− ES cell line exhibited, for unknown reasons, wild-type sensitivity. A possible explanation might be a loss of pluripotency in this ES cell line.
We conclude that a CSB deficiency causes cellular sensitivity to gamma-ray irradiation in MEFs, keratinocytes, and ES cells but with a significant difference in magnitude. A deficiency of CSA has no effect on cellular sensitivity to gamma rays in MEFs and keratinocytes, whereas in ES cells a CSA deficiency causes a slight hypersensitive phenotype.
To investigate whether a combined CSA-CSB deficiency would act either synergistically or epistatically, we generated double mutant CSA−/− CSB−/− mice. Double mutant animals appear normal and do not display any overt phenotype up age 18 months. (A detailed comparative study of the phenotypes of CSA−/−, CSB−/−, and CSA−/− CSB−/− mice is under way.) Next, we generated established CSA−/− CSB−/− MEFs and showed that they do not display an increased sensitivity to UV or gamma-ray irradiation, compared to the most sensitive single mutant. These findings indicate that both proteins function in the same pathway and are epistatic (data not shown).
Different responses of CSB−/− and CSA−/− mice to a DEHP-containing diet.
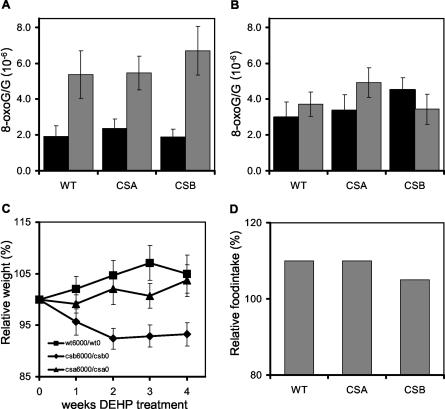
It is not known how the observed cell type- and genotype-specific gamma-ray response in cultured wild-type, CSB−/−, and CSA−/− cells can be extrapolated to cells in the context of the whole animal. Therefore, we aimed at investigating the sensitivity of the intact animal to oxidative DNA damage. As a potential oxidative damage inducing agent, we used the plasticizer DEHP, which causes proliferation of peroxisomes in the liver by activation of peroxisome proliferator-activated receptor alpha (60). This is believed to induce higher oxidative stress in the liver, which in turn leads to the induction of a wide range of DNA lesions. To test whether DEHP indeed could be used as a suitable method to induce oxidative stress in mice, we administrated wild-type, CSA−/−, and CSB−/− female mice (n ≥ 5) food containing 6,000 ppm of DEHP for 4 weeks. The control group received unmodified food. Induction of oxidative DNA damage was assessed by double-blind measurement of the 8-oxo-dG damage level in the DNA of treated versus untreated animals. We observed that DEHP-treated mice showed a 2.5-times-higher 8-oxo-dG content in the liver than untreated animals (Fig. 3A), whereas no significant induction of 8-oxo-dG in kidneys was observed in any genotype tested (Fig. 3B). This result confirms that DEHP is a liver-specific toxic compound, inducing oxidative DNA damage. Interestingly, there seems to be no difference in accumulation of 8-oxo-dG between wild-type, CSA−/−, and CSB−/− mice, suggesting that, in line with the findings of Osterod and coworkers (38), bulk repair of this type of lesion in the mouse does not require CSA or CSB.
FIG. 3.
DEHP sensitivities of CSB−/−, CSA−/−, and wild-type (WT) mice. (A and B) Average 8-oxo-dG/G ratios in livers (A) and kidneys (B) of CSB−/−, CSA−/−, and wild-type mice after 4 weeks of a DEHP-containing diet (grey bars) or a control diet (black bars), as measured by using a high-pressure liquid chromatography system with electrochemical and UV detection. Shown are the average 8-oxo-dG/G ratios from at least five animals per group. Error bars indicate the standard errors of the means. (C) Relative weights of CSB−/−, CSA−/−, and wild-type mice fed with food containing 6,000 ppm of DEHP versus animals on a regular diet. Shown are the average weight ratios for at least five animals per group. (D) Relative food intakes of CSB−/−, CSA−/−, and wild-type mice. Shown are the ratios of average food intake per week over the whole treatment for mice fed with food containing 6,000 ppm of DEHP versus regular a diet.
As a read-out of the DEHP sensitivity of the mouse, we used the overall condition, as determined by body weight. The weight of every mouse was compared to the original weight of the mouse before treatment. Plotted in Fig. 3C is the relative weight of treated animals divided by that of untreated animals (times 100), showing a clear weight loss in CSB−/− mice compared to wild-type and CSA−/− mice (n ≥ 5). Surprisingly, wild-type and CSA−/− (although to a somewhat lesser extent) animals fed with a DEHP-containing diet gained more weight than untreated animals of the same genotype. This phenomenon is probably due to an increased food intake in treated animals (Fig. 3D). We suggest that CSB−/− mice at the organismal level are sensitive to oxidative DNA damage, reflecting the observed hypersensitivity to oxidative DNA damage in various cultured cell types. CSA−/− mice are far less sensitive for oxidative damage caused by DEHP treatment, which is in agreement with the observed insensitivity of CSA−/− keratinocytes and MEFs to oxidative DNA damage.
DISCUSSION
Sensitivity of CSB−/− cells and mice to oxidative stress.
Using genetically homogeneous mouse models, we investigated the effect of a CSB deficiency on oxidative DNA damage sensitivity in various cell types and at the level of the intact organism. As shown previously (12), cellular gamma-ray sensitivity in general markedly depends on cell type. For instance, wild-type ES cells are significantly more sensitive to gamma-ray irradiation than either MEFs or keratinocytes. In line with previous findings (12, 27), we showed oxidative DNA damage sensitivity in CSB-deficient fibroblasts, keratinocytes, and ES cells, although to different relative extents. Previously we showed that the sensitivity to oxidative DNA damage in CSB−/− MEFs can be corrected by introducing hCSB cDNA into these cells (12), arguing that indeed the CSB deficiency accounts for the observed sensitivity. Moreover, at the level of the intact animal, we also could demonstrate a significant effect of CSB deficiency on sensitivity to oxidative DNA damage, as illustrated by the DEHP experiment. This finding is in line with the previously observed tendency of CSB−/− mice to be more sensitive to the toxic or killing effects of gamma rays than wild-type mice (12). In light of the recent discussion about the validity of TCR of oxidative DNA lesions (10, 16, 26, 26a), these data clearly show that CSB deficiency causes sensitivity to oxidative DNA damage in various cell types and on the organismal level.
A cell type-specific CSA effect on gamma-ray response.
Previously, one primary human fibroblast cell line derived from a CSA patient was investigated and was reported to display a slight gamma-ray sensitivity (27). However, because of the large genetic variation in the human population on the one hand and the only minor difference in sensitivity on the other hand, it is difficult to draw firm conclusions about the potential link between a CSA deficiency and gamma-ray sensitivity on the basis of the human fibroblast studies. This study even suggests that the CSA and CSB proteins might have different functions in TCR. Surprisingly, we failed to show an increased sensitivity to oxidative DNA damage in CSA−/− MEFs and keratinocytes. DEHP-treated CSA−/− mice, in contrast to CSB−/− mice, failed to show a pronounced reduction in body weight compared to untreated mice, indicating that CSA−/− animals are barely sensitive to the two- to threefold-higher levels of 8-oxo-dG lesions. Despite the absence of oxidative damage sensitivity in CSA−/− MEFs, keratinocytes, and animals, we could still demonstrate a slight gamma-ray sensitivity in CSA−/− ES cells, which compares well to that observed in CSB−/− ES cells.
These findings suggest that in a wide range of cell types, CSA is dispensable for the response to oxidative DNA damage. Yet, given the slight gamma-ray sensitivity of CSA−/− ES cells and the tendency for somewhat less growth in DEHP-treated CSA−/− mice than in wild-type mice, its function might be needed in specific types of cells. To our knowledge, these data provide the first critical cell biological evidence that the CSA and CSB proteins have separable functions.
Different functions of CSA and CSB?
These observed biological differences between CSA- and CSB-deficient cells and mice, as uncovered by the divergence in oxidative damage sensitivity, seem to be in contrast to the widely accepted notion that both CSA and CSB function in the same subpathway of TCR (27, 30, 56). However, biochemical analysis of the CSA and CSB proteins has revealed marked differences that are suggestive of a potential difference in function. The CSA protein resides in a 420-kDa complex, whereas CSB is part of a >700-kDa complex (55). While CSA was found to be a constituent of a complex containing DDB1, cullin 4A, Roc1, and the COP9 signalosome (17), CSB was found to interact with RNA polymerase II (50, 55), XPA, XPG, TFIIE, and TFIIH (23, 42) and with several splicing factors. Moreover, a role of CSB, but most likely not of CSA, in RNA polymerase I transcription has been suggested (5). Also, a CSB deficiency rather than a CSA deficiency might cause metaphase fragility for genes encoding specific highly structured transcripts (61).
In contrast, arguments that CSA and CSB in some way might function together are provided by the reported in vitro interaction between CSA and CSB (18). Moreover, a recent study shows that genotoxic stress-mediated translocation of CSA to the nuclear matrix is hampered in CSB-deficient cell lines (24). Most importantly, there is no evidence for a difference in the clinical appearances of CSA and CSB patients (37, 44). Similarly, the phenotypes of CSA −/− and CSB−/− mice fail to reveal obvious differences, since both mouse models are UV sensitive, lack TCR (53, 54), show photoreceptor loss upon aging (53; T. G. M. F. Gorgels, personal communication) and die before weaning when combined with an XPA or XPC deficiency (35, 53; I. van der Pluijm, personal communication). Finally, our CSA CSB double mutant cells and mice provide genetic evidence for an epistatic relationship and involvement in the same pathway.
In conclusion, arguments both in favor of and against differences between CSA and CSB functions exist.
Possible functions of CSA and CSB in different biological processes.
To find an explanation for the observed differences in gamma-ray sensitivity between CSA- and CSB-deficient cells, we list the possible functions of CSA and CSB in response to oxidative DNA damage.
(i) Transcriptional bypass.
It has been shown that oxidative DNA lesions (such as 8-oxo-dG) can block RNA polymerase II, although far less efficiently than UV-induced lesions do (25). Escherichia coli RNA polymerase can bypass 8-oxo-dG by putting either adenine or cytosine opposite the 8-oxo-dG (6, 59). For the yeast counterpart of CSB, Rad26, strong indications for a role in transcriptional bypass of methyl methanesulfonate-induced DNA damage have been found (29). Recent reports show that human RNA polymerase II also is able to bypass oxidative DNA lesions in vitro (25, 51). Since CSB has already been associated with transcription elongation (especially of damaged templates, pause sites, and highly structured RNAs), these data suggest a possible role of CSB in transcriptional bypass of some oxidative DNA lesions. This function may require the reported chromatin-remodeling activity of the CSB protein (8). Transcriptional bypass might be CSA independent, since, for example, CSA appears not to be involved in stimulation of transcription of genes for highly structured RNAs (61). This function of CSB may not be relevant to UV-induced damage, as photolesions form strong RNA polymerase blocks and are therefore not subject to transcriptional bypass.
(ii) TCR.
TCR of UV-induced lesions, as well as recovery of RNA synthesis after UV treatment, has been demonstrated to depend on both CSA and CSB (33, 56). Although bypass of 8-oxo-dG is possible, the absence of the mfd protein in E. coli (which is required for TCR) causes higher bypass rates, suggesting that TCR acts on 8-oxo-dG (6). Also, several reports point to a function of mammalian CSB in TCR of oxidative DNA lesions (reference 12 and references therein). The influence of CSA on TCR of oxidative DNA lesions is unknown.
(iii) Ubiquitination of RNA polymerase II.
Upon UV treatment, stalled RNA polymerase can be ubiquitinated in a CSA- and CSB-dependent manner (7, 28, 40). This ubiquitination might be needed for TCR and/or the degradation of the stalled polymerase. The latter event would allow access of the repair machinery to the lesion and subsequent recovery of RNA synthesis (34). The potential link between CSA and ubiquitination of RNA polymerase II may lie in the facts that CSA is known to regulate the ubiquitin ligase activity of the complex containing DDB1, cullin 4A, Roc1, and the COP9 signalosome (17) and that this complex could, in some way, be involved in the ubiquitination of the polymerase. However, CSA and CSB proteins are not prerequisites for the breakdown of RNA polymerase II, as CSA- and CSB-deficient cells can still degrade the polymerase (32). Ubiquitination of RNA polymerase II has been also been shown to occur after exposure of cells to oxidative DNA damage (22). However, the mechanism of ubiquitination differs from that provoked by UV light and is not CSA or CSB dependent (22).
(iv) Other repair pathways.
Evidence for an indirect role of CSB in BER-mediated global genome repair and mitochondrial DNA repair of oxidative DNA lesions has been reported (13, 38, 45, 46, 52).
Explanation of CSA- and CSB-related differences in oxidative damage response.
Taking into account the possible functions of CSA and CSB, we discuss two scenarios that might explain our findings.
(i) Oxidative DNA damage does not have a major impact on the onset of CS features. Although the effect of CSA on the response to oxidative DNA damage is poorly investigated, we do not favor this explanation, since this would argue against a large body of evidence supporting a function of CSB in repair of oxidative DNA damage and the general importance of oxygen radicals (12, 13, 27, 30, 38, 45, 46, 52).
(ii) The differences in oxidative stress sensitivity between CSB−/− and CSA−/− cells and animals (as observed during cellular survival experiments and in vivo exposure studies) are the consequence of the acute response of a heavily challenged system. Evidently, the conditions used in such studies are not representative of the processes in a patient that cause the CS manifestations. Although in acute (high-dose) experiments CSA- and CSB-deficient cells differ in sensitivity, under physiological conditions a deficiency of CSA or CSB may cause a similar effect upon exposure to low but constitutive levels of oxidative DNA damage. The hypersensitivity of CSB−/− cells and mice could be due to other functions of CSB. For example, CSB−/− cells could be unable to perform adequate transcription bypass, which might be CSA independent. In the presence of large amounts of oxidative DNA damage, inefficient bypass in CSB−/− cells might lead to cell death. The absence of a significant RNA synthesis block after high gamma-ray doses suggests that the majority of oxidative DNA lesions are bypassed. In contrast, since the major UV lesions are probably not bypassed during transcription, UV sensitivity of CSA- and CSB-deficient cell lines mainly reflects the inability of these cells to perform TCR and RNA synthesis recovery. These processes depend on both CSA and CSB for removal of the stalled RNA polymerase.
The second explanation suggests that the gamma-ray sensitivity in CSB−/− cells could be partly attributable to processes other than a TCR defect. Consequently, the lack of gamma-ray sensitivity in CSA−/− cells does not per se exclude a role of CSA in TCR of oxidative DNA damages. Moreover, the observed gamma-ray sensitivity in CSA−/− ES cells suggests some function of CSA in response to oxidative DNA damage. Therefore, a deficiency in TCR in specific cell types could still underlie CS symptoms.
Concluding remarks.
Comparison of the responses of isogenic cells of different tissues to ionizing radiation has demonstrated significant variation in sensitivity and in dependence on CS proteins. This finding suggests the use of different genome-caretaking strategies by different cell types and tissues. In addition, our study reveals that the absence of CSA or CSB has different impacts on the response to oxidative DNA damage and consequently that the two proteins are not functionally equivalent. However, this appears not to be reflected in the CS phenotypes of patients and mice. The increased sensitivity to ionizing radiation due to CSB inactivation is consistent with a role of CSB protein in cellular resistance to oxidative damage. The notion that the TCR defect for UV lesions in both CSA and CSB mutants is the same suggests that the sensitivity of CSB-deficient cells to ionizing radiation is due to some extra function of the CSB protein. However, this extra function does not significantly influence the clinical outcome. The findings described above also highlight a hitherto-unanticipated functional dissimilarity of the main TCR factors CSA and CSB.
Acknowledgments
This research was supported by The Netherlands Organization for Scientific Research (NWO) through the foundation of the Research Institute on Diseases of the Elderly, as well as by grants from the NIH (AG17242-02 and RFA-ES-00-005), the EC (QRTL-1999-02002 and QRLT-CT-1999-00181), the Dutch Cancer Society (EUR-1774 and EUR-2004), and the Danish Medical Research Council.
REFERENCES
- 1.Ali, S., S. K. Jain, M. Abdulla, and M. Athar. 1996. Paraquat induced DNA damage by reactive oxygen species. Biochem. Mol. Biol. Int. 39:63-67. [DOI] [PubMed] [Google Scholar]
- 2.Asami, S., and H. Kasai. 2000. 8-OH-dG: extraction/enzyme treatment/ measurement of 8-OH-dG, p. 224-228. In N. Taniguchi and J. M. Gutteridge (ed.), Experimental protocols for reactive oxygen and nitrogen species. Oxford University Press, Oxford, United Kingdom.
- 3.Balajee, A. S., A. May, G. L. Dianov, E. C. Friedberg, and V. A. Bohr. 1997. Reduced RNA polymerase II transcription in intact and permeabilized Cockayne syndrome group B cells. Proc. Natl. Acad. Sci. USA 94:4306-4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bootsma, D., K. H. Kraemer, J. E. Cleaver, and J. H. J. Hoeijmakers. 2001. Nucleotide excision repair syndromes: xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy, 8th ed., p. 677-703. McGraw-Hill, New York, N.Y.
- 5.Bradsher, J., J. Auriol, L. Proietti de Santis, S. Iben, J. L. Vonesch, I. Grummt, and J. M. Egly. 2002. CSB is a component of RNA pol I transcription. Mol. Cell 10:819-829. [DOI] [PubMed] [Google Scholar]
- 6.Bregeon, D., Z. A. Doddridge, H. J. You, B. Weiss, and P. W. Doetsch. 2003. Transcriptional mutagenesis induced by uracil and 8-oxoguanine in Escherichia coli. Mol. Cell 12:959-970. [DOI] [PubMed] [Google Scholar]
- 7.Bregman, D. B., R. Halaban, A. J. van Gool, K. A. Henning, E. C. Friedberg, and S. L. Warren. 1996. UV-induced ubiquitination of RNA polymerase II: a novel modification deficient in Cockayne syndrome cells. Proc. Natl. Acad. Sci. USA 93:11586-11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Citterio, E., V. Van Den Boom, G. Schnitzler, R. Kanaar, E. Bonte, R. E. Kingston, J. H. Hoeijmakers, and W. Vermeulen. 2000. ATP-dependent chromatin remodeling by the Cockayne syndrome B DNA repair-transcription-coupling factor. Mol. Cell. Biol. 20:7643-7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooper, P. K., T. Nouspikel, S. G. Clarkson, and S. A. Leadon. 1997. Defective transcription-coupled repair of oxidative base damage in Cockayne syndrome patients from XP group G. Science 275:990-993. [DOI] [PubMed] [Google Scholar]
- 10.Cozzarelli, N. R. 2003. Editorial expression of concern. Proc. Natl. Acad. Sci. USA 100:11816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Laat, W. L., N. G. Jaspers, and J. H. Hoeijmakers. 1999. Molecular mechanism of nucleotide excision repair. Genes Dev. 13:768-785. [DOI] [PubMed] [Google Scholar]
- 12.de Waard, H., J. de Wit, T. G. Gorgels, G. van den Aardweg, J. O. Andressoo, M. Vermeij, H. van Steeg, J. H. Hoeijmakers, and G. T. van der Horst. 2003. Cell type-specific hypersensitivity to oxidative damage in CSB and XPA mice. DNA Repair 2:13-25. [DOI] [PubMed] [Google Scholar]
- 13.Dianov, G., C. Bischoff, M. Sunesen, and V. A. Bohr. 1999. Repair of 8-oxoguanine in DNA is deficient in Cockayne syndrome group B cells. Nucleic Acids Res. 27:1365-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dianov, G. L., J. F. Houle, N. Iyer, V. A. Bohr, and E. C. Friedberg. 1997. Reduced RNA polymerase II transcription in extracts of Cockayne syndrome and xeroderma pigmentosum/Cockayne syndrome cells. Nucleic Acids Res. 25:3636-3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dlugosz, A. A., A. B. Glick, T. Tennenbaum, W. C. Weinberg, and S. H. Yuspa. 1995. Isolation and utilization of epidermal keratinocytes for oncogene research. Methods Enzymol. 254:3-20. [DOI] [PubMed] [Google Scholar]
- 16.Gowen, L. C., A. V. Avrutskaya, A. M. Latour, B. H. Koller, and S. A. Leadon. 1998. BRCA1 required for transcription-coupled repair of oxidative DNA damage. Science 281:1009-1112. (Retraction, 300:1657, 2003.) [DOI] [PubMed] [Google Scholar]
- 17.Groisman, R., J. Polanowska, I. Kuraoka, J. Sawada, M. Saijo, R. Drapkin, A. F. Kisselev, K. Tanaka, and Y. Nakatani. 2003. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell 113:357-367. [DOI] [PubMed] [Google Scholar]
- 18.Henning, K. A., L. Li, N. Iyer, L. D. McDaniel, M. S. Reagan, R. Legerski, R. A. Schultz, M. Stefanini, A. R. Lehmann, L. V. Mayne, et al. 1995. The Cockayne syndrome group A gene encodes a WD repeat protein that interacts with CSB protein and a subunit of RNA polymerase II TFIIH. Cell 82:555-564. [DOI] [PubMed] [Google Scholar]
- 19.Hennings, H., K. Holbrook, P. Steinert, and S. Yuspa. 1980. Growth and differentiation of mouse epidermal cells in culture: effects of extracellular calcium. Curr. Probl. Dermatol. 10:3-25. [DOI] [PubMed] [Google Scholar]
- 20.Hoeijmakers, J. H. 2001. Genome maintenance mechanisms for preventing cancer. Nature 411:366-374. [DOI] [PubMed] [Google Scholar]
- 21.Hwang, B. J., J. M. Ford, P. C. Hanawalt, and G. Chu. 1999. Expression of the p48 xeroderma pigmentosum gene is p53-dependent and is involved in global genomic repair. Proc. Natl. Acad. Sci. USA 96:424-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inukai, N., Y. Yamaguchi, I. Kuraoka, T. Yamada, S. Kamijo, J. Kato, K. Tanaka, and H. Handa. 2004. A novel hydrogen peroxide-induced phosphorylation and ubiquitination pathway leading to RNA polymerase II proteolysis. J. Biol. Chem. 279:8190-8195. [DOI] [PubMed] [Google Scholar]
- 23.Iyer, N., M. S. Reagan, K. J. Wu, B. Canagarajah, and E. C. Friedberg. 1996. Interactions involving the human RNA polymerase II transcription/nucleotide excision repair complex TFIIH, the nucleotide excision repair protein XPG, and Cockayne syndrome group B (CSB) protein. Biochemistry 35:2157-2167. [DOI] [PubMed] [Google Scholar]
- 24.Kamiuchi, S., M. Saijo, E. Citterio, M. de Jager, J. H. Hoeijmakers, and K. Tanaka. 2002. Translocation of Cockayne syndrome group A protein to the nuclear matrix: possible relevance to transcription-coupled DNA repair. Proc. Natl. Acad. Sci. USA 99:201-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuraoka, I., M. Endou, Y. Yamaguchi, T. Wada, H. Handa, and K. Tanaka. 2003. Effects of endogenous DNA base lesions on transcription elongation by mammalian RNA polymerase II. Implications for transcription-coupled DNA repair and transcriptional mutagenesis. J. Biol. Chem. 278:7294-7299. [DOI] [PubMed] [Google Scholar]
- 26.Leadon, S. A. 2003. Retraction. DNA Repair 2:361. [DOI] [PubMed] [Google Scholar]
- 26a.Leadon, S. A., and A. V. Avrutskaya. 1998. Requirement for DNA mismatch repair proteins in the transcription-coupled repair of thymine glycols in Saccharomyces cerevisiae. Mutat. Res. 407:177-187. (See also reference 26.) [DOI] [PubMed] [Google Scholar]
- 27.Leadon, S. A., and P. K. Cooper. 1993. Preferential repair of ionizing radiation-induced damage in the transcribed strand of an active human gene is defective in Cockayne syndrome. Proc. Natl. Acad. Sci. USA 90:10499-10503. (See also reference 10.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, K. B., D. Wang, S. J. Lippard, and P. A. Sharp. 2002. Transcription-coupled and DNA damage-dependent ubiquitination of RNA polymerase II in vitro. Proc. Natl. Acad. Sci. USA 99:4239-4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, S. K., S. L. Yu, L. Prakash, and S. Prakash. 2002. Yeast RAD26, a homolog of the human CSB gene, functions independently of nucleotide excision repair and base excision repair in promoting transcription through damaged bases. Mol. Cell. Biol. 22:4383-4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Le Page, F., E. E. Kwoh, A. Avrutskaya, A. Gentil, S. A. Leadon, A. Sarasin, and P. K. Cooper. 2000. Transcription-coupled repair of 8-oxoguanine: requirement for XPG, TFIIH, and CSB and implications for Cockayne syndrome. Cell 101:159-171. [DOI] [PubMed] [Google Scholar]
- 31.Lindahl, T., and R. D. Wood. 1999. Quality control by DNA repair. Science 286:1897-1905. [DOI] [PubMed] [Google Scholar]
- 32.Luo, Z., J. Zheng, Y. Lu, and D. B. Bregman. 2001. Ultraviolet radiation alters the phosphorylation of RNA polymerase II large subunit and accelerates its proteasome-dependent degradation. Mutat. Res. 486:259-274. [DOI] [PubMed] [Google Scholar]
- 33.Mayne, L. V., and A. R. Lehmann. 1982. Failure of RNA synthesis to recover after UV irradiation: an early defect in cells from individuals with Cockayne's syndrome and xeroderma pigmentosum. Cancer Res. 42:1473-1478. [PubMed] [Google Scholar]
- 34.McKay, B. C., F. Chen, S. T. Clarke, H. E. Wiggin, L. M. Harley, and M. Ljungman. 2001. UV light-induced degradation of RNA polymerase II is dependent on the Cockayne's syndrome A and B proteins but not p53 or MLH1. Mutat. Res. 485:93-105. [DOI] [PubMed] [Google Scholar]
- 35.Murai, M., Y. Enokido, N. Inamura, M. Yoshino, Y. Nakatsu, G. T. van der Horst, J. H. Hoeijmakers, K. Tanaka, and H. Hatanaka. 2001. Early postnatal ataxia and abnormal cerebellar development in mice lacking Xeroderma pigmentosum group A and Cockayne syndrome Group B DNA repair genes. Proc. Natl. Acad. Sci. USA 98:13379-13384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakae, D., Y. Mizumoto, E. Kobayashi, O. Noguchi, and Y. Konishi. 1995. Improved genomic/nuclear DNA extraction for 8-hydroxydeoxyguanosine analysis of small amounts of rat liver tissue. Cancer Lett. 97:233-239. [DOI] [PubMed] [Google Scholar]
- 37.Nance, M. A., and S. A. Berry. 1992. Cockayne syndrome: review of 140 cases. Am. J. Med. Genet. 42:68-84. [DOI] [PubMed] [Google Scholar]
- 38.Osterod, M., E. Larsen, F. Le Page, J. G. Hengstler, G. T. Van Der Horst, S. Boiteux, A. Klungland, and B. Epe. 2002. A global DNA repair mechanism involving the Cockayne syndrome B (CSB) gene product can prevent the in vivo accumulation of endogenous oxidative DNA base damage. Oncogene 21:8232-8239. [DOI] [PubMed] [Google Scholar]
- 39.Rapin, I., Y. Lindenbaum, D. W. Dickson, K. H. Kraemer, and J. H. Robbins. 2000. Cockayne syndrome and xeroderma pigmentosum. Neurology 55:1442-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ratner, J. N., B. Balasubramanian, J. Corden, S. L. Warren, and D. B. Bregman. 1998. Ultraviolet radiation-induced ubiquitination and proteasomal degradation of the large subunit of RNA polymerase II. Implications for transcription-coupled DNA repair. J. Biol. Chem. 273:5184-5189. [DOI] [PubMed] [Google Scholar]
- 41.Riis, B., L. Risom, S. Loft, and H. E. Poulsen. 2002. Increased rOGG1 expression in regenerating rat liver tissue without a corresponding increase in incision activity. DNA Repair 1:419-424. [DOI] [PubMed] [Google Scholar]
- 42.Selby, C. P., and A. Sancar. 1997. Cockayne syndrome group B protein enhances elongation by RNA polymerase II. Proc. Natl. Acad. Sci. USA 94:11205-11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sijbers, A. M., W. L. de Laat, R. R. Ariza, M. Biggerstaff, Y. F. Wei, J. G. Moggs, K. C. Carter, B. K. Shell, E. Evans, M. C. de Jong, S. Rademakers, J. de Rooij, N. G. Jaspers, J. H. Hoeijmakers, and R. D. Wood. 1996. Xeroderma pigmentosum group F caused by a defect in a structure-specific DNA repair endonuclease. Cell 86:811-822. [DOI] [PubMed] [Google Scholar]
- 44.Stefanini, M., H. Fawcett, E. Botta, T. Nardo, and A. R. Lehmann. 1996. Genetic analysis of twenty-two patients with Cockayne syndrome. Hum. Genet. 97:418-423. [DOI] [PubMed] [Google Scholar]
- 45.Stevnsner, T., S. Nyaga, N. C. de Souza-Pinto, G. T. van der Horst, T. G. Gorgels, B. A. Hogue, T. Thorslund, and V. A. Bohr. 2002. Mitochondrial repair of 8-oxoguanine is deficient in Cockayne syndrome group B. Oncogene 21:8675-8682. [DOI] [PubMed] [Google Scholar]
- 46.Sunesen, M., T. Stevnsner, R. M. Brosh, Jr., G. L. Dianov, and V. A. Bohr. 2002. Global genome repair of 8-oxoG in hamster cells requires a functional CSB gene product. Oncogene 21:3571-3578. [DOI] [PubMed] [Google Scholar]
- 47.Svejstrup, J. Q. 2002. Mechanisms of transcription-coupled DNA repair. Nat. Rev. Mol. Cell Biol. 3:21-29. [DOI] [PubMed] [Google Scholar]
- 48.Svejstrup, J. Q. 2003. Rescue of arrested RNA polymerase II complexes. J. Cell Sci. 116:447-451. [DOI] [PubMed] [Google Scholar]
- 49.Tang, J. Y., B. J. Hwang, J. M. Ford, P. C. Hanawalt, and G. Chu. 2000. Xeroderma pigmentosum p48 gene enhances global genomic repair and suppresses UV-induced mutagenesis. Mol. Cell 5:737-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tantin, D., A. Kansal, and M. Carey. 1997. Recruitment of the putative transcription-repair coupling factor CSB/ERCC6 to RNA polymerase II elongation complexes. Mol. Cell. Biol. 17:6803-6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tornaletti, S., L. S. Maeda, D. R. Lloyd, D. Reines, and P. C. Hanawalt. 2001. Effect of thymine glycol on transcription elongation by T7 RNA polymerase and mammalian RNA polymerase II. J. Biol. Chem. 276:45367-45371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tuo, J., M. Muftuoglu, C. Chen, P. Jaruga, R. R. Selzer, R. M. Brosh, Jr., H. Rodriguez, M. Dizdaroglu, and V. A. Bohr. 2001. The Cockayne syndrome group B gene product is involved in general genome base excision repair of 8-hydroxyguanine in DNA. J. Biol. Chem. 276:45772-45779. [DOI] [PubMed] [Google Scholar]
- 53.van der Horst, G. T., L. Meira, T. G. Gorgels, J. de Wit, S. Velasco-Miguel, J. A. Richardson, Y. Kamp, M. P. Vreeswijk, B. Smit, D. Bootsma, J. H. Hoeijmakers, and E. C. Friedberg. 2002. UVB radiation-induced cancer predisposition in Cockayne syndrome group A (Csa) mutant mice. DNA Repair 1:143-157. [DOI] [PubMed] [Google Scholar]
- 54.van der Horst, G. T., H. van Steeg, R. J. Berg, A. J. van Gool, J. de Wit, G. Weeda, H. Morreau, R. B. Beems, C. F. van Kreijl, F. R. de Gruijl, D. Bootsma, and J. H. Hoeijmakers. 1997. Defective transcription-coupled repair in Cockayne syndrome B mice is associated with skin cancer predisposition. Cell 89:425-435. [DOI] [PubMed] [Google Scholar]
- 55.van Gool, A. J., E. Citterio, S. Rademakers, R. van Os, W. Vermeulen, A. Constantinou, J. M. Egly, D. Bootsma, and J. H. Hoeijmakers. 1997. The Cockayne syndrome B protein, involved in transcription-coupled DNA repair, resides in an RNA polymerase II-containing complex. EMBO J. 16:5955-5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Hoffen, A., A. T. Natarajan, L. V. Mayne, A. A. van Zeeland, L. H. Mullenders, and J. Venema. 1993. Deficient repair of the transcribed strand of active genes in Cockayne's syndrome cells. Nucleic Acids Res. 21:5890-5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Venema, J., A. van Hoffen, V. Karcagi, A. T. Natarajan, A. A. van Zeeland, and L. H. Mullenders. 1991. Xeroderma pigmentosum complementation group C cells remove pyrimidine dimers selectively from the transcribed strand of active genes. Mol. Cell. Biol. 11:4128-4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Venema, J., A. van Hoffen, A. T. Natarajan, A. A. van Zeeland, and L. H. Mullenders. 1990. The residual repair capacity of xeroderma pigmentosum complementation group C fibroblasts is highly specific for transcriptionally active DNA. Nucleic Acids Res. 18:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Viswanathan, A., and P. W. Doetsch. 1998. Effects of nonbulky DNA base damages on Escherichia coli RNA polymerase-mediated elongation and promoter clearance. J. Biol. Chem. 273:21276-21281. [DOI] [PubMed] [Google Scholar]
- 60.Ward, J. M., J. M. Peters, C. M. Perella, and F. J. Gonzalez. 1998. Receptor and nonreceptor-mediated organ-specific toxicity of di(2-ethylhexyl)phthalate (DEHP) in peroxisome proliferator-activated receptor alpha-null mice. Toxicol. Pathol. 26:240-246. [DOI] [PubMed] [Google Scholar]
- 61.Yu, A., H. Y. Fan, D. Liao, A. D. Bailey, and A. M. Weiner. 2000. Activation of p53 or loss of the Cockayne syndrome group B repair protein causes metaphase fragility of human U1, U2, and 5S genes. Mol. Cell 5:801-810. [DOI] [PubMed] [Google Scholar]