Abstract
Recent data in invertebrates demonstrated that huntingtin (htt) is essential for fast axonal trafficking. Here, we provide direct and functional evidence that htt is involved in fast axonal trafficking in mammals. Moreover, expression of full-length mutant htt (mhtt) impairs vesicular and mitochondrial trafficking in mammalian neurons in vitro and in whole animals in vivo. Particularly, mitochondria become progressively immobilized and stop more frequently in neurons from transgenic animals. These defects occurred early in development prior to the onset of measurable neurological or mitochondrial abnormalities. Consistent with a progressive loss of function, wild-type htt, trafficking motors, and mitochondrial components were selectively sequestered by mhtt in human Huntington's disease-affected brain. Data provide a model for how loss of htt function causes toxicity; mhtt-mediated aggregation sequesters htt and components of trafficking machinery leading to loss of mitochondrial motility and eventual mitochondrial dysfunction.
Huntington's disease (HD) is a progressive neurodegenerative disorder caused by a CAG repeat expansion mutation in the coding region of a novel gene. The mechanism of HD is unknown. However, most data suggest that polyglutamine-mediated aggregation contributes to the pathology (32). Studies of human brain (14), mouse models (48), and cells (8, 28) demonstrate that mutant huntingtin (mhtt) binds and sequesters its normal counterpart as well as many cellular proteins (41). But whether pathophysiology results from a loss of normal function or a gain of a new function in adult neurons is not well understood.
A major gap in our understanding of the disease mechanism is the absence of a known function for normal huntingtin (htt). Emerging evidence suggests that htt is likely to be a multifunctional protein that can mediate transactions in both the nucleus and the cytoplasm. Transcriptional dysfunction caused by mhtt has been proposed to lead to toxicity. The mutation in full-length htt prevents its normal ability to bind and sequester a repressor of brain-derived neurotrophic factor expression, reducing the availability of brain-derived neurotrophic factor to striatal neurons (54). The N-terminal, truncated form of mhtt can bind to and interfere with nuclear factors such as CREB (51), CREB binding protein (30, 39), corepressor (22), and transcriptional activator Sp1 (12, 23).
Cytoplasmic dysfunction has also been implicated as a toxic mechanism. Recently, novel data obtained with Drosophila (17) and isolated squid axoplasm (42) have provided direct evidence that htt is an essential protein involved in fast axonal trafficking. Additionally, these data demonstrate that the mutation in htt causes trafficking abnormalities. Reduction of htt expression in Drosophila caused axonal transport defects in larval nerves and the same neurodegenerative phenotype in adult eyes as expression of mutant dynein or p150Glued (17). In invertebrate models for HD, expression of truncated proteins with an expanded glutamine tract impairs vesicle movement and promotes vesicle accumulation in axons, though the detailed molecular mechanism remains illusive (17, 42). These data are consistent with many reports linking htt to trafficking proteins and motors (9, 15, 19, 21, 24, 25, 44). Therefore, a possible scenario of trafficking dysfunction could include mhtt-dependent titration of soluble motor proteins into aggregates (17). Intriguingly, the presence of truncated htt or androgen receptor with expanded polyglutamine regions caused similar defects in organelle trafficking in the isolated squid axoplasm in the absence of the nucleus, protein synthesis, and detectable aggregation (42).
It is now critical to establish whether mhtt-related trafficking defects observed in invertebrates or in vitro systems also occur in mammals. Moreover, trafficking defects were measured in invertebrate models after expression of artificially truncated proteins. To exclude any assumptions about cleavage and N-terminal fragment generation, it is also important to measure trafficking in models expressing full-length mhtt in vivo. This is a crucial point since pathological features of the protein can change with truncation (52). Short N-terminal fragments are known to be toxic. However, recent data from mice, cells, and humans suggest that the full-length protein initiates cell death before the generation of N-terminal fragments in the cytoplasm (14, 20, 43).
We have previously been demonstrated that expression of full-length mhtt initiates toxicity in the cytoplasm of primary striatal neurons (43). To test whether htt is an essential protein for fast axonal trafficking in mammals, we investigated the effect of reduced htt expression or expression of mhtt on vesicular dynamics in whole animals and primary striatal neurons from mice. Mouse models were chosen to test both effects of a pathogenic length of glutamine tract and the dosage of expressed htt. Our data from both mouse neurons and human HD-affected brain suggest that trafficking defects are caused by abnormal protein interactions of mhtt that lead to titration of wild-type htt and motor proteins from soluble pools. These effects not only inhibited motility of vesicles but also of organelles, notably mitochondria. Taken together, the data provide a testable model for toxicity; mhtt-mediated aggregation causes loss of vesicular trafficking and progressive immobilization of mitochondria.
MATERIALS AND METHODS
Human brain tissue extraction.
Brain tissue was obtained from the Mayo Clinic (Rochester, Minn.) and the Harvard Brain Tissue Resource Center (McLean Hospital, Belmont, Mass.). Tissue was extracted based on solubility by utilization of stronger denaturing conditions (2% sodium dodecyl sulfate [SDS], 3 M urea, and 1 mM dithiothreitol) as previously described (14). The resulting supernatants, S1 and S3, represented soluble and poorly soluble fractions, respectively.
Size exclusion chromatography.
Protein from brain extractions was quantified by the Bradford assay using bovine serum albumin as a standard. An equal amount of total protein (16 mg) was resolved on a 2.5-cm by 100-cm column of Sepharose CL-2B (linear fractionation range, 70,000 to 40,000,000 Da) at a flow rate of 0.5 ml/min in 50 mM Tris (pH 8.0), 150 mM NaCl, and 1 mM EDTA at 4°C. The column was extensively washed (3 column volumes) in buffer after each run. The column was precalibrated with high-range molecular weight (MW) markers (Pharmacia, Piscataway, N.J.). Calibration standards included dextran blue (∼2,000 kDa), thyroglobulin (698 kDa), ferritin (418 kDa), catalase (206 kDa), and aldolase (167 kDa), as previously described (14). Due to the large fractionation range of the column, dextran blue (∼2,000 kDa) was resolved. Therefore, the void volume of 100 ml was determined by the elution profile that contained aggregates of an estimated size of 3 × 108 Da. To determine the fractionation profiles of select proteins, an equal volume (80 μl) of each fraction was analyzed by blotting with the following antibodies: monoclonal anti-htt 2166 (1:1,200; Chemicon, Temecula, Calif.), kinesin MC44 (1:5,000; M. McNiven, Mayo Foundation, Rochester, Minn.), dynactin p150Glued (1:250; BD Transduction Labs, San Diego, Calif.), α-tubulin Tu-01 (1:1,000; Zymed, San Francisco, Calif.), βIII-tubulin 5G8 (1:20,000; Promega, Madison, Wis.), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (1:6,000; Chemicon), actin (1:200; Santa Cruz, Santa Cruz, Calif.), and Ran C-20 (1:200; Santa Cruz). Monomeric htt refers to the elution volume at which a monomeric form of htt is expected based on the calibration standards and htt's predicted MW of 350,000, based on its primary amino acid sequence (3,144 amino acids).
For some experiments, equal total protein amounts (20 to 30 μg) of the S1 and S3 extracts were analyzed by direct immunoblotting without prior size exclusion chromatography. This analysis reveals the enrichment of a specific protein in the soluble (i.e., S1) or poorly soluble (i.e., S3) pool.
Immunoprecipitation.
htt antibody 2166 (Chemicon) was conjugated to Dynabeads protein G (Dynal Inc., Lake Success, N.Y.). Ascites fluid (100 μl at 2 to 3 mg/ml of immunoglobulin G) was mixed with 450 μl of protein G beads that were previously washed three times with 1 ml of 0.1 M sodium phosphate buffer, pH 7.0, and incubated at room temperature for 20 min. Following capture, the beads were washed three times with 1 ml of 0.1 M sodium phosphate buffer, pH 7.0. Beads were prepared for cross-linking by washing three times in 1 ml of 0.2 M triethanolamine, pH 8.2, and resuspended in 1 ml of 20 mM dimethyl pimelimidate and dihydrochloride in 0.2 M triethanolamine, pH 8.2. The suspension was incubated at room temperature for 30 min. The beads were washed for 15 min with rocking in 1 ml of 50 mM Tris, pH 7.5, and then they were washed three times with 1 ml of 0.1% Tween 20 in phosphate-buffered saline (PBS) and resuspended in PBS containing 0.1% Tween 20 and 0.02% sodium azide. For immunoprecipitation, 10 μl of beads (either unconjugated or conjugated to 2166) were combined with 450 μg of S1 extract in 1 ml of radioimmunoprecipitation assay buffer or with 1 ml of gel filtration fractions (reconstituted to radioimmunoprecipitation assay buffer conditions) and incubated at 4°C overnight with rocking. Beads were washed three times with 1 ml of ice-cold TNT buffer (10 mM Tris, pH 8.0, 140 mM NaCl, 0.1% Triton X-100). Beads were washed once with PBS, and proteins were eluted in 50 μl of SDS-polyacrylamide gel electrophoresis sample buffer (58.3 mM Tris, pH 6.8, 1.67% SDS, 5% glycerol, 2.5% 2-mercaptoethanol, 0.002% bromophenol blue) by heating 1 h at 56°C.
Neuronal cell cultures and mouse models.
All procedures involving animals were approved by the Institutional Animal Care and Use Committee. The following mouse models were used: control FVB/N model (3) with seven glutamines in the mouse endogenous htt homologue, and the homozygous transgenic model with full-length human HD cDNA containing 16 (HD16) (33) or 72 (HD72) (20) CAG repeats (both models were constructed using the FVB/N mouse strain). We also used a mouse model with conditional knockout (KO) of the murine htt gene, R1ag5 (10). We have established two colonies of transgenic mice, Hdhflox/flox and R1ag5-cre/+; Hdh+/−, that allow the conditional elimination of htt. The Hdhflox/flox mice contain an engineered Hdh gene flanked by two 34-bp loxP sites that permit cleavage by a recombinase from bacteriophage P1 (Cre). The R1ag5-cre/+; Hdh+/− mice contain a single allele of the mouse endogenous htt gene that allows mice to develop normally. However, these mice also contain an engineered Cre recombinase that catalyzes the deletion of an intervening DNA sequence located between the two loxP sites in the htt gene. Cre expression is under the control of the calmodulin kinase 2a promoter. Loss of htt is limited to sites of the Camk2a expression that occurs throughout the neurons in the forebrain and in the cerebellum. Crossing of the Hdhflox/flox and R1ag5-cre/+; Hdh+/− mice will produce progeny in which the single endogenous mouse gene containing the lox sites is cleaved and deleted, and this occurs when the Cre recombinase becomes active near birth. Consequently, the htt protein expression is normal in the brains of progeny from zygote formation until embryonic day 15 (E15) but is increasingly diminished as the mouse Hdh homologue is inactivated due to Cre expression. Mutant mice (Hdhflox/−; cre/+, hereafter referred to as KO mice) were screened for the presence of Hdhflox allele, Hdh null allele, and cre transgene according to previously published methods (10). All experiments were performed in neurons from mice that were originally heterozygous for the htt allele (Hdhflox/−; cre/+). Thus, at E17, when the experiments with KO neurons were performed, the levels of htt expression must be reduced by more than 50% due to initiation of cre/lox recombination. Littermates Hdhflox/−; Hdh+/− (hereafter referred to as Flox/−) were used as a control. Estimation of htt loss in striatum of KO mice at E17 was done by Western blotting of striatal extracts and showed the reduction in protein levels below 50%.
Striatal neurons were plated according to the protocol published previously (43). Briefly, mice were anesthetized with ether on gestational day 17, and fetuses were rapidly removed. Fetal brains were extracted and placed in sterile HEPES- buffered saline (HBS) (pH 7.3). The ventral part of the medial ganglionic eminence (the developmental precursor to the striatum) was dissected under a microscope in sterile conditions. Tissue was minced and placed in 10 μl of 0.3-mg/ml trypsin (type XII-S from bovine pancreas) in HBS for 30 min at room temperature. After two washes in HBS, the dissociated tissue was triturated in Dulbecco's modified Eagle's medium containing 10% Ham's F-12 with glutamine (Gibco/BRL, Grand Island, N.Y.), 10% heat-inactivated fetal calf serum (HyClone Laboratories, Logan, Utah), and 1× pen/strep antibiotic mixture. Cells were counted and diluted to 3 × 105 cells/ml, and 2 ml of this stock was placed in each well of a six-well dish containing glass coverslips coated with poly-l-ornithine (1 mg in 2 ml of sterile borate buffer, pH 8.4). Plated cells were maintained in an incubator with 5% CO2 at 37°C. After 72 h in culture medium, containing serum was replaced with a serum-free Neurobasal-based medium (without glutamine; Gibco/BRL) containing 1× pen/strep antibiotic mixture and 1× B27 supplement (Gibco/BRL) to reduce the proliferation of astrocytes. All experiments were performed 6 to 7 days after plating. For experiments with KO mice, neurons were plated from individual pups, and tissue was used for DNA extraction and genotyping as described above.
Differential interference contrast (DIC) microscopy.
Coverslips with plated neurons were mounted in a chamber containing serum-free Ham's F-12K medium (Sigma, St. Louis, Mo.) and placed on a heated microscope stage (37°C). Cells were imaged using a Zeiss Axiophot microscope (Carl Zeiss Inc., Thornwood, N.Y.) with a 63× Plan-APO DIC lens (1.4 numeric aperture [n.a.]) and a 4× extender. Vesicular movement within the neurites was recorded for 30 min using a Hamamatsu Newvicon camera (Hamamatsu Phototonics K.K., Hamamatsu, Japan) at 24-h mode and played at 2-h mode. The frame capture rate was five per 2 s. Images were collected on an Apple Desktop G3/300 Minitower with a Targa 2000 PCI video card. Video images were analyzed using Adobe Premiere Raid array software. Particles were visualized by adjusting the prism to achieve the desired shadowing. Analysis was performed by a blind investigator; the movies were encoded by numbers not by phenotype. For every neuron, the amount of visible particles was calculated and sorted depending on shape (round or elongated) and pattern of motion. For every particle observed, we recorded the time it spent in motion or stationary period along with direction of motion (anterograde [A] or retrograde [R]). Depending on the pattern of motion, the particle was considered to move smoothly if it did not stop over the distance of 50 μm (the average length of the neurite observed in the movie was about 60 μm). The particle was considered stationary if it did not move over the time of the whole movie. A particle that moved back and forth without net displacement was counted as saltatory. Many particles underwent stop-and-go motion where they moved, stopped, and then resumed movement often changing the direction. For both round and elongated particles, velocities were estimated by measuring distance and the time the particle spent in motion. If the same particle performed multiple short-term movements in both directions (stop-and-go), velocities in A and R directions were calculated independently and averaged.
Identification of axons and dendrites in neuronal preparations.
Typically axons were chosen for motility measurements. However, real-time imaging required presence of living cells that could not be fixed and probed until after the experiment. To distinguish between axons and dendrites in living neurons, we first identified axons based upon known morphological characteristics such as uniform thickness and lack of branching. After the morphological selection, neurons were postfixed and immunostained with anti-MAP2 antibody (Upstate Biotechnology, Lake Placid, N.Y.) or Tau (C-17) antibody (Santa Cruz) as described earlier (43). We calculated the percentage of correct identifications in three separate experiments. Morphological characteristics of neurites yielded a correct assignment of an axon in roughly 85% of cases.
Mitochondria staining in neurons.
For experiments with mitochondria trafficking in living neurons, cells were treated for 5 min with tetramethylrhodamine methyl ester (TMRM) (Molecular Probes, Eugene, Oreg.) (10 nM in medium). The experiments were performed using a LSM 510 confocal microscope (Carl Zeiss Inc., Thornwood, N.Y.) with a Plan-Apochromat 100× (1.4 n.a.) oil objective. However, the confocal aperture was opened to allow the fluorescence from the entire thickness of the cell to be collected. Utilization of a laser scanning microscope allowed photo bleaching of the sample to be avoided. During imaging, cells were kept at low excitation intensity (laser power was 25% and transmission was 0.1%). Cells were incubated at 37°C during time of recording. All recordings were started 5 min after the coverslip was placed on the microscopic stage to allow equilibration of the sample. The Ar/Kr laser was set up to 568 nm for excitation; emission was set up to 585 nm and greater. TMRM was washed away with fresh F-12K medium prior to imaging, and images of mitochondria were taken every 1 s at the highest scan speed (0.9 sec) for 10 min. A total of 600 images was recorded per cell with frame capturing being one per second.
Movies were analyzed using LSM 510 software that allowed animation of 600 images into a movie. Analysis of mitochondria movement was performed by four blind investigators. Each mitochondrion was traced from the first frame of the movie to the last. The time and the distance that mitochondrion traveled in axon or spent in stationary state was recorded. For every organelle, the average speed that it moved in A or R direction was estimated. Data were averaged for each phenotype. We also created the pattern of motion for randomly selected mitochondria moving in smooth or stop-and-go mode where time that mitochondrion spent in motion or stationary state was plotted versus distance.
Statistical analysis.
Vesicles observed by DIC optics were evaluated for net transport in one direction using a chi-square test. Data were compared to an expected distribution of 50:50 for A to R movement and round to elongated particles, and statistically significant differences reflected net transport in one direction. Comparison of mean velocities was done using a multicomparison Bonferroni t test; comparison of fluoro-gold (FG) uptake was done using an unpaired two-tailed t test. In all cases, differences were considered statistically significant where P was <0.05.
Immunostaining.
Immunostaining was performed as previously described (43). Primary antibodies were polyclonal goat anti-htt antibodies N18 and C20 (1:50; Santa Cruz), kinesin heavy chain MC44 polyclonal rabbit antibody (1:1,000; M. McNiven, Mayo Foundation), monoclonal mouse p150Glued (1:750; Transduction Laboratories, San Diego, Calif.), mouse monoclonal anti-GAPDH (1:6,000; Chemicon), and polyclonal rabbit α-synapsin (1:500; kind gift of P. McPherson). Secondary antibodies were rabbit anti-goat Cy5 (1:120; Zymed); goat anti-mouse Cy5 (1:1,500; Amersham), goat anti-mouse tetramethylmodamine (TMR) (1:400), and goat anti-rabbit TMR (1:50; Molecular Probes).
Cell death measurement.
Before and after the recording of the vesicular trafficking, neurons were cytochemically labeled with the DNA dye Hoechst 33258 (2.5 μg/ml, 5 min), and neurons were scored as healthy or apoptotic by morphological criteria. Neurons were scored as apoptotic only when they had a pyknotic and/or fragmented nuclei and degenerated or absent neuritis.
Confocal imaging.
For colocalization studies, cells were imaged using a LSM 510 confocal laser scanning microscope (Carl Zeiss Inc.) with a 100× oil objective (1.4 n.a.) with optical section set to ∼0.5 μm.
Electron microscopy.
For conventional electron microscopy, cells were fixed using cacodylate, sucrose, and glutaraldehyde mixture. Cells were incubated in 1% osmium tetroxide, dehydrated in a graded series of ethanol, and embedded in Quetol 651 (Ted Pella, Inc). Thin sections (0.09 to 0.1 μm) were cut parallel to the ventral surface of the cells using a diamond knife (Diatome US) and an Ultracut E microtome (Reichert-Jung, Vienna, Austria). Sections were collected on copper grids, poststained with lead citrate, and viewed at ∼60 kV with a JEOL 1200 transmission electron microscope (JEOL USA).
Estimation of mitochondrial and nuclear DNA damage.
Quantitative PCR was utilized to determine the DNA damage in neurons (2). This technique enables the quantitative measurement of both nuclear and mitochondrial DNA in the extracts from the brains of control and transgenic mice. The technique is based on the premise that DNA lesions, including oxidative damage such as strand breaks, base modifications, and AP sites, will block the progression of the polymerase resulting in the decreased amplification of the target sequence. DNA from striatal tissue of control (FVB/N), HD16, and HD72 mice of 8 to 9 weeks of age was extracted with a genomic DNA extraction kit (QIAGEN, Chatsworth, Va.) using the protocol supplied with the kit. DNA estimation was done using a PicoGreen double-stranded DNA (dsDNA) quantitation kit (Molecular Probes). Free PicoGreen dye is essentially nonfluorescent and exhibits >1,000-fold fluorescence enhancement upon binding to dsDNA at excitation and emission wavelengths of 480 and 530 nm, respectively. The assay displays a linear correlation between dsDNA quantity and fluorescence, being extremely sensitive (detection range extending from 25 pg/ml to 1 μg/ml). Quantitative PCR was done using a protocol described previously (2) except that the quantitation of PCR products was done using PicoGreen dye. The primer sequences used were as follows: for the 6.5-kb nuclear gene, β-polymerase, 5′-TAT CTC TCT TCC TCT TCA CTT CTC CCC TGG-3′ and 5′-CGT GAT GCC GCC GTT GAG GGT CTC CTG-3′; for the 10-kb mitochondrial genome, 5′-GAG AGA TTT TAT GGG TGT AAT GCG G-3′ and 5′-GCC AGC CTG ACC CAT AGC CAT AAT AT-3′; and for the 117-bp small mitochondrial fragment, 5′-CCC AGC TAC TAC CAT CAT TCA AGT-3′ and 5′-GAT GGT TTG GGA GAT TGG TTG ATG-3′.
DNA lesion frequencies were calculated as described previously (2). Briefly, the amplification of damaged samples (AD) was normalized to the amplification of a nondamaged control (AO) resulting in a relative amplification ratio. Assuming a random distribution of lesions and using the Poisson equation [f(x) = e−λλx/x!, where λ is the average lesion frequency for the nondamaged template (i.e., the zero class; x = 0)], the average lesion per DNA strand was determined by the following equation: λ = −ln AD/A0.
ATP measurements.
Plated neurons were lysed with ice-cold perchloric acid (5%). The clarified supernatant was neutralized, and ATP was quantified using Sigma's lucigenin/luciferase ATP detection kit. The total amount of ATP extracted was correlated with the protein amount.
FG neurotracer experiments.
Control and HD72 5-month-old mice were injected with 0.2 μl of 4% fluorogold (FG) in water over a 4-min time period. Mice were deeply anesthetized with injection of 10 μl of 8-mg/ml ketamine-1-mg/ml xylazine per g of the body weight. FG was injected into both hemispheres in the substantia nigra using a stereotaxic device. Injections were performed with a Hamilton gastight 10-μl syringe. The anterior-posterior, medial lateral, and dorsoventral coordinates relative to the bregma were as follows: anterior-posterior, −3.2 mm; medial lateral, ±1.5 mm; dorsoventral, −3.8 mm. Animals were cardioperfused with 4% paraformaldehyde solution after 24, 48, or 72 h postinjection. Brains were removed and placed overnight in freshly prepared 4% paraformaldehyde. Sections were cut through the caudate-putamen region and through the sight of injection in the frontal plane at 30 μm thick with an Oxford Vibratome. The sections were mounted on glass slides and air dried over night. FG was detected using UV illumination (360- to 370-nm excitation, 420- to 460-nm emission) with an Olympus AX70 microscope under 40× or 10× magnification. We first identified the accuracy of injection. If injection was placed correctly, analysis of FG staining in 10 consecutive striatal slices was performed. As a reference point we took the slice where lateral ventricles joined the dorsal third ventricle that appears as a “flap” on the slice. Five consecutive slices before and after the reference slice were compared to one another regarding intensity of FG staining in the striatum. Thus, intensity of staining was compared in control and HD72 mice in brain slices that precisely represented the same brain regions, were of the same thickness, and allowed observation of most of the caudate putamen (striatum) region. For each slice imaged under 40× magnification, the total number of neurons accumulating FG in their cell bodies was counted in the same area in the striatum. Numbers were collected from 10 slices and averaged per mouse. The mean number of the cell bodies accumulating FG was calculated for five control and five HD72 mice for 1- and 3-day time points.
Mass spectrometry.
S3 extracts were prepared from the caudate-putamen region of human HD-affected brain 3882 by a method previously described (14). Gel filtration fractions from S3 extracts were pooled, concentrated and desalted by phenol-ether (36), separated by SDS-polyacrylamide gel electrophoresis, and stained with Coomassie blue. The gel lane was then cut horizontally into 37 even-sized gel slices that were processed for trypsin digestion as described previously (46). Extracted, trypsin-digested peptides were separated on a BioBasic C18 reversed-phase column and analyzed on a CapLC Q-TOF mass spectrometer (MicroMass) as described previously (46). The MS/MS data was peaklisted (MassLynx; MicroMass) and submitted to Mascot (MatrixScience) software for database search analysis against the National Center for Biotechnology Information nonredundant database. Only peptides with a confidence level of 95% or greater (as defined by Mascot software) were accepted. Peptides at the low end of the confidence interval were subjected to a second round of analysis by manual inspection.
RESULTS
Vesicles are progressively immobilized in neurons from transgenic mice expressing mhtt with increasing polyglutamine length.
To evaluate whether expression of mhtt alters fast axonal trafficking, we performed real-time imaging in primary embryonic (E17) striatal neurons isolated from control and transgenic mice expressing either a high level of htt with 16 glutamines (HD16) (33) or a low level of mhtt with 72 glutamines (HD72) (20) (Fig. 1). The use of mouse models offered several advantages. First, trafficking could be monitored in primary neurons from brain regions that are most affected in HD. Further, these mice expressed the full-length mhtt as is found in vivo. Therefore, we made no assumptions as to what cleavage products, if any, were present in the cells at the time of measurements. In these experiments, all mice were generated in identical genetic backgrounds allowing accurate comparison.
FIG. 1.
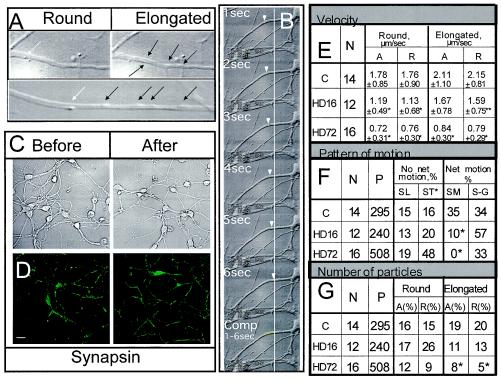
mhtt expression causes inhibition of trafficking in neurons of transgenic mice as visualized by DIC optics. (A) Examples of round and elongated particles as they appear using DIC optics. Images are shown at different magnification (top, 63× with 4× extender; bottom, 100× with 4× extender and 2× zoom). White arrows indicate round particles, black arrows indicate elongated particles. (B) Time-lapse DIC images of particle motility taken 1 s apart. A round vesicle moving towards the cell body is indicated by an arrow. The line is an arbitrary point of reference. Technical details are described in Materials and Methods. (C) DIC imaging does not affect neuronal health. Phase images (63× oil DIC, 1.4 n.a.) of primary striatal neuronal cultures before and after 30 min of DIC imaging. Scale bar, 20 μm. (D) DIC imaging does not affect synaptic activity. Primary striatal neurons immunostained with synapsin antibody before and after 30 min of DIC imaging (63× oil DIC, 1.4 n.a.). Scale bar, 20 μm. Analysis of speed (panel E), pattern of motion (panel F), and number of moving round and elongated particles (panel G) in striatal neurons from control (C) and transgenic mice expressing htt with 16 (HD16) or 72 (HD72) glutamines obtained using DIC optics. (E) Velocity of particles in micrometers per second. N, number of neurites observed; *, P < 0.0001; **, P < 0.01. Thirty to 50 particles were randomly selected for evaluation in every group. (F) Analysis of pattern of motion includes all particles observed (round and elongated). SL, saltatory (back and forth without net direction); ST, stationary (no motion); SM, smooth (without stops over the range of 50 μm); S-G, stop-and-go (the particle stops for a period of time before resuming motion); P, number of particles evaluated; *, P < 0.0001. (G) The percentage of particles moving in neurites in the A or R direction is significantly lower in neurons from HD72 mice. *, P < 0.0001. Statistical analysis in panels F and G was done by chi-square test.
Vesicle motility was measured using DIC optics allowing real-time imaging of individual cells. Particles were detected only by optical shadowing in the absence of any mechanical or chemical treatment (Fig. 1A and B; also unpublished data). In all experiments, neurons were maintained under physiological conditions during the time of recording (30 min). Imaging was primarily performed in neurites that, by known morphological criteria (e.g., lack of branching and uniform thickness), were defined as axons. Neurite origin after DIC recording was, in random cases, confirmed by immunostaining with tau antibody.
We observed two types of moving particles that were defined by size and morphology (Fig. 1A). The dimensions of small, round particles were consistent with endocytic vesicles (0.6 to 1.8 μm) while the larger elongated particles were most likely mitochondria (1.9 to 4.5 μm). Optical shadowing alone could not unambiguously identify them. Therefore, we analyzed the motility of both.
In neurons from HD72 mice, the speed of both round and elongated particles dropped relative to control neurons by nearly 60% in the A as well as R directions (Fig. 1E). The drop in speed was accompanied by a distinctly different pattern of motion. We found that particles could be divided into two broad classes: those that moved with net direction and those that moved without net direction (Fig. 1F). Particles that moved with net direction did so either by smooth, rapid progress along the neurite or by jerky stop-and-go motion (Fig. 1F). Particles without net direction were either stationary or displayed saltatory (back and forth) movement (Fig. 1F) without net displacement. By observing 300 to 500 individual vesicles in 12 to 16 different neurons, we found that the number of particles with net motion significantly diminished from roughly 69 to 33% as the glutamine tract in htt increased from 7 to 72 (Fig. 1F). Concomitantly, the percentage of particles without net displacement increased from 30 to 67%.
In neurons from HD16 mice, an increasing number of particles displayed stop-and-go rather than smooth motility (Fig. 1F). No particles moved smoothly in neurons from HD72 mice. Rather, particles in HD72 neurons displayed exclusively stop-and-go motion, and the number of stationary vesicles increased threefold compared to control neurons (Fig. 1F). Together, these data suggested particles in neurons were increasingly immobilized in the presence of mhtt or had a more difficult time moving along the neurites. For round particles, the number of moving ones dropped modestly in both the A and R directions as the glutamine length increased from 7 (control neurons) to 72 (HD72) (Fig. 1G). However, the number of moving elongated particles was substantially affected in both directions in transgenic animals relative to control (Fig. 1G). Thus, there was a direct correlation between the reduction in number of moving particles and the length of the polyglutamine tract primarily for elongated particles.
Alterations in vesicular dynamics were not due to experimental conditions. After imaging, we observed no changes in neuronal appearance (Fig. 1C), nuclei staining (data not shown), or synaptic activity, as measured by staining with synapsin antibody (Fig. 1D). Thus, trafficking defects were not an artifact of cell health status.
Trafficking is impaired in vivo in living animals expressing mhtt.
To test whether expression of mhtt also causes trafficking dysfunction in vivo, we measured the uptake of the neurotracer, FG, in whole animals (Fig. 2). FG freely crosses the axolemma and becomes trapped in lysosomes, which are subsequently transported to the cell bodies in neurons (47). FG labeling is a sensitive measure of trafficking efficiency and neuronal integrity since accumulation of the tracer in cell bodies depends on R transport and intact axons (38).
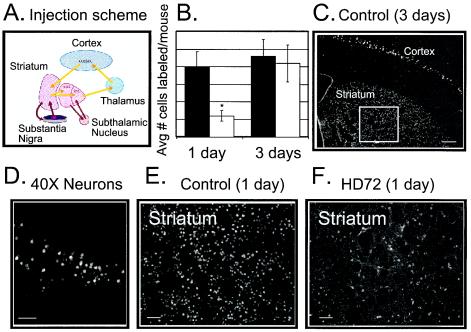
FIG. 2.
Retrograde transport of FG into striatum is slow in mice expressing mhtt. (A) Injection scheme for FG uptake. FG was injected into substantia nigra, and uptake in striatum was monitored with time. (B) Quantification of the number of neurons that accumulate FG in the cell bodies in control and HD72 mice 1 and 3 days after the injection. Horizontal lines represent 0, 50, 100, etc. cells labeled/mouse. Black bars, control FVB/N mice; white bars, HD72 mice; *, P < 0.01 (t test). (C) Retrograde uptake of FG in striatum of control mice 3 days after the injection. The box indicates the area in the striatum that was used for quantification in panel B. Scale bar, 200 μm. (D) View of cortical neurons with FG accumulation in the cell bodies. Scale bar, 50 μm. (E, F) FG uptake in the striatum of control (E) and HD72 (F) mice 1 day after the injection. Neuronal cell bodies labeled with FG are seen as bright circular structures in tissue. Scale bar, 50 μm.
In presymptomatic HD patients, striatal neurons projecting to substantia nigra are preferentially affected (1, 34). Therefore, we injected FG into the substantia nigra of adult control and HD72 mice and measured uptake into the striatum of 5-month-old animals (Fig. 2). At this age, HD72 mice do not display any movement disorder and are indistinguishable from control animals (20). After the injection, mice were allowed to recover and were sacrificed at 1 to 3 days. Trafficking was measured by the intensity of staining in the neuronal cell bodies in the striatum with time (Fig. 2B to F).
We found striking differences in striatal staining between control and HD72 mice at 1 day after injection in all animals tested (n = 10) (Fig. 2B, E, and F). At this time, the number of neurons that accumulated FG in the cell body in the striatum of HD72 mice was less than 25% of that for control animals. By 3 days, differences in staining intensity between control and HD72 mice were not significant. These data provided direct evidence that expression of mhtt slows vesicular trafficking not only in embryonic neurons in vitro but also in adult animals in vivo. Moreover, impairment of trafficking occurred before measurable onset of symptoms associated with neuronal loss in HD72 mice (20).
Mitochondrial movement is impaired in neurons from transgenic mice expressing mhtt.
Analysis of data obtained in DIC experiments indicated that the motility of elongated particles was most affected in neurons expressing mhtt. To test whether the motility of organelles was also affected by the expression of mhtt, we identified mitochondria by specific staining with the cationic dye TMRM and examined their dynamics using fluorescence imaging (6) (Fig. 3A and B). The dye itself does not alter mitochondrial function or motility (6), and the rates obtained for elongated particles in DIC experiments were similar to ones obtained using TMRM.
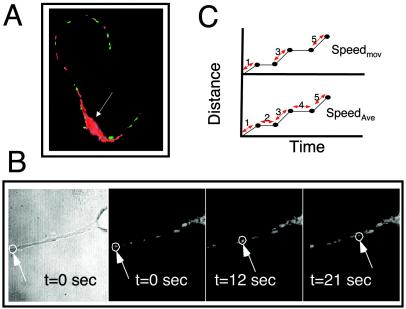
FIG. 3.
Analysis of mitochondrial motion in neurons from control, transgenic, and htt KO mice visualized by TMRM staining. (A) Image of striatal neuron stained with TMRM; red are stationary mitochondria and green are moving mitochondria identified by computer and appropriately colored. Most moving mitochondria are in the neurites. The arrow indicates the cell body. (B) An example of time-lapse recording of a TMRM-stained mitochondrion moving in the R direction along the axon in an E17 striatal neuron. Phase and fluorescence images are shown. The arrow and circle indicate the mitochondrion being traced. (C) Schematic diagram of speed calculations used in the analysis of mitochondrial movement. Speedmov (speed when moving) was calculated by dividing the distance over time for every step in which mitochondria moved (double arrows), and data were averaged. In this example, time equals the sum of steps 1, 3, and 5. No time for stationary phases was included. SpeedAve (average speed) was calculated by dividing distance by total time that includes both moving and stationary phases (sum of steps 1, 2, 3, 4, and 5).
We found that the majority of mitochondria (roughly 70%) in neurons were stationary and tended to be located near the cell body (Fig. 3A). Therefore, we restricted measurements and analyses to mitochondria moving in neurites (Fig. 3A and B; also unpublished data). Neurons were incubated with TMRM and imaged every second over 10 to 15 min on the heated microscopic stage. Indeed, we found that trafficking of mitochondria was impaired in primary neurons from HD72 mice compared to control.
The speed of mitochondria was calculated using two methods. In method 1, the average speed was measured by dividing the total distance by the total time of mitochondrial movement during the observation period (Fig. 3C, bottom). In method 2, we calculated the speed only when the mitochondrion moved (Fig. 3C, top).
Quantification of vesicular dynamics of 300 to 530 mitochondria by method 1 confirmed that they moved with diminished average speed in HD72 mice relative to control (Table 1). The average speed of mitochondria progressively decreased in both directions (up to 70%) as the glutamine tract increased from 7 to 72 (Table 1). Similarly, with method 2, we found that when mitochondria moved, their actual speed was 50% slower in HD72 mice compared to control (Table 1). Thus, mitochondria had an increasingly difficult time moving along neurites in the presence of mhtt, and the degree of diminished motility directly correlated with the glutamine length.
TABLE 1.
Dynamics of mitochondrial motility in neurons of control, transgenic, and KO micea
| Lineb | Direction of motion | Moving speed (μm/s)c | Average speed (μm/s)d | Fme | No. of stopsf |
|---|---|---|---|---|---|
| Cnt | A | 1.91 ± 1.11 | 1.19 ± 0.50 | 0.72 ± 0.14 | 0.9 |
| Cnt | R | 1.74 ± 0.91 | 1.27 ± 0.29 | 0.65 ± 0.16 | 0.5 |
| HD16 | A | 1.33 ± 0.61 | 1.06 ± 0.60 | 0.66 ± 0.28 | 1.1 |
| HD16 | R | 1.28 ± 0.59 | 0.75 ± 0.46* | 0.66 ± 0.19 | 1.8 |
| HD72 | A | 0.92 ± 0.36* | 0.45 ± 0.22* | 0.56 ± 0.22* | 5.7* |
| HD72 | R | 0.82 ± 0.34* | 0.40 ± 0.30* | 0.46 ± 0.19* | 5.1* |
| Flox/− | A | 0.63 ± 0.27 | 0.30 ± 0.16 | 0.71 ± 0.18 | 2.8 |
| Flox/− | R | 0.80 ± 0.35 | 0.34 ± 0.11 | 0.45 ± 0.17 | 5.0 |
| KO | A | 0.53 ± 0.22* | 0.21 ± 0.14* | 0.51 ± 0.22* | 7.4* |
| KO | R | 0.53 ± 0.20* | 0.21 ± 0.12* | 0.42 ± 0.19 | 7.5* |
Mitochondrial speed progressively declines as the glutamine tract in htt increases or htt is lost.
Data were generated in primary striatal embryonic neurons (E17) from control FVB/N (Cnt), transgenic mice with full-length htt with 16 (HD16) and 72 (HD72) glutamines, and mice with 50% loss of wild-type htt (Flox/−) that are littermates of htt conditional KO mice expressing less than 50% endogenous htt.
Speed when moving represents the total distance divided by the time in motion. Here times does not include stationary phases. Data were calculated using 92 to 355 mitochondria from 10 to 16 neurons. *, P < 0.0001.
Average speed represents the total distance traveled by the mitochondria divided by the total time. This involves time both in movement and in stationary phases. *, P < 0.0001.
The Fm is the fraction of time that the mitochondrion is moving. *, P < 0.05.
Number of stops represents the average number of pauses by a mitochondrion. *, P < 0.0001. Data presented in the last three columns were generated from 10 randomly selected organelles observed in five random neurons.
Mitochondria not only moved slowly in HD72 neurons, they also stopped more frequently. We found that the number of stops increased with the glutamine length (Table 1). This is also reflected in the fraction of time mitochondrial spent in motion, which was significantly smaller in neurons from HD72 mice (Table 1). Thus, the inhibition of mitochondria speed in HD16 and HD72 mice was accompanied by a distinctly different pattern of motion (Fig. 4). Mitochondria in control mice moved smoothly in both directions with individual organelles covering a relatively long distance between stops (Fig. 4). In neurons from either HD16 or HD72 mice, mitochondrial motility was more stop-and-go (Fig. 4). The most striking result was a reduction in distance covered by mitochondria between stops in HD72 mice (Fig. 4). Mitochondria in these animals traveled roughly 50% of the distance in the A direction and less than 30% in the R direction relative to control mice (Fig. 4). Trafficking defects were not related to the neuronal health since DNA staining with Hoechst did not reveal any signs of apoptosis before and after the experiment. Thus, motility of mitochondria in HD72 mice was significantly impaired by expression of mhtt.
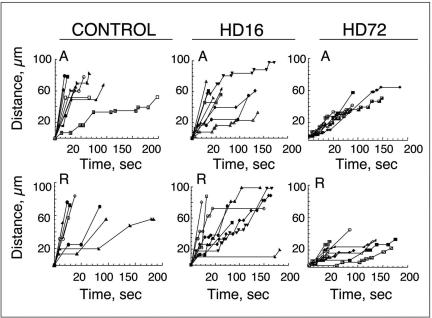
FIG. 4.
Pattern of mitochondria motion in neurites of control and HD-affected mice. The majority of mitochondria in control neurons cover a longer distance in a shorter time and stop less frequently than mitochondria in HD-affected mice. The pattern of motion was recorded for randomly selected individual mitochondria from 10 to 15 neurons that could be traced for over 150 s or moved over the distance of 50 μm. Stationary and mobile phases were plotted over time.
The defect imparted by mhtt expression occurred at the level of trafficking since neither the number, integrity, or the morphology of mitochondria in embryonic striatal neurons was significantly different from controls (Fig. 5A to C). Mitochondria moved slower in embryonic neurons from HD72 mice but were able to synthesize ATP as well as in control neurons, and lactate, a signature of mitochondrial dysfunction, was not elevated (Fig. 5D).
FIG. 5.
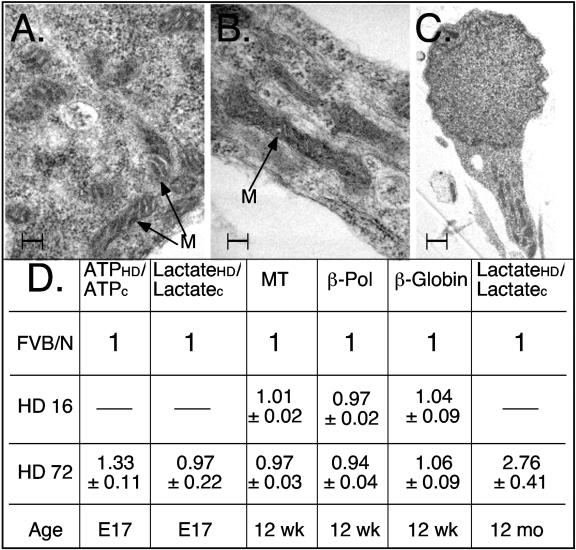
Ultrastructure and function of mitochondria from control and HD-affected mice. Mitochondria morphology was not altered in embryonic striatal neurons (E17) in control (A) or HD72 (B) mice. Scale bar, 10 nm. There was also no detection of abnormal mitochondria accumulation in neuritis in HD72 neurons (C) compared to control (not shown). Scale bar, 1 μm. M, mitochondrion. (D) Trafficking defects precede mitochondrial dysfunction in HD72 mice. Mitochondria function (ATP synthesis and lactate accumulation) was not altered in embryonic (E17) striatal neurons in HD72 mice compared to control despite detected abnormalities in mitochondrial trafficking. At the same time, increased lactate accumulation was measured in adult HD72 mice (12 months). There was also no difference in mitochondrial DNA damage in striatal tissue from control, HD16, and HD72 mice at 12 weeks. Estimated mitochondrial (MT) and nuclear (β-polymerase and β-globin) DNA damage was measured by quantitative PCR in striatal tissues (see Materials and Methods). ATPHD/ATPC is the ratio of ATP generated within E17 cultures of HD16 or HD72 animals (ATPHD) to that of control neurons (ATPC). LactateHD/LactateC is the ratio of lactate measured in E17 striatal cultures or striatal tissue from HD72 mice (lactateHD) to that of control mice (lactateC). Age is indicated.
Mitochondria become increasingly dysfunctional with age.
Loss of mitochondrial motility might diminish ATP production and increase oxygen radical formation with age, both of which can be toxic to neurons. To measure mitochondrial function with age, flash frozen brain tissue from HD72 mice at different ages were tested for ATP content, lactate formation, and oxidative damage in mitochondrial DNA. We detected no mitochondrial dysfunction in HD16 or HD72 at 12 weeks as measured by oxidative lesions of either mitochondrial or nuclear (β-globin, β-polymerase) DNA (Fig. 5D). Oxidative damage is a sensitive marker of mitochondrial dysfunction since oxygen radicals can arise from damaged mitochondria and even a small number of DNA lesions can be precisely measured (2). Thus, impairment of mitochondrial motility observed in embryonic HD72 neurons preceded signs of mitochondrial dysfunction. In contrast to embryonic neurons, lactate accumulation in the striatum from adult HD72 mice (12 months) was significantly increased (Fig. 5D). Mitochondrial dysfunction occurred between 12 weeks and 12 months, much later than the initial movement defects observed in embryonic neurons. Thus, mitochondria became dysfunctional as the animals aged.
Expression of mhtt and loss of htt cause similar trafficking defects in neurons.
We next addressed whether the trafficking defect imparted by mhtt was due to a loss of normal htt function or a gain of a new function. In Drosophila htt was shown to be essential for fast axonal trafficking (17). If htt has the same function in mammalian neurons and mutation in htt inhibits that function, then trafficking defects observed in HD72 mice should also be observed when htt is reduced or lost. Trafficking of mitochondria was visualized in embryonic (E17) striatal neurons from mice genetically engineered to express less than 50% of htt normal levels upon cre-mediated recombination (KO) (10). The results were compared to data obtained in mice heterozygous for expression of htt (Flox/−) that do not have the HD phenotype. The analysis of mitochondrial rates, patterns of motion, and vesicular dynamics was performed using TMRM staining and the protocol identical to the one used for HD72 neurons. Indeed, we found that reduction of wild-type htt below 50% of normal levels decreased speed of mitochondria in neurons from KO mice (see Flox/− and KO speed data in Table 1), increased number of stops (see data for stops in Table 1), decreased time that mitochondria spent in motion (see Fm data in Table 1), and decreased distances mitochondria covered between stops (data not shown). Thus, reduction of htt in the neurons altered mitochondrial dynamics in both the A and R directions. Trafficking defects were significant in KO neurons compared to Flox/−, suggesting a dose-dependent effect. These data demonstrate that normal htt plays a direct role in mitochondrial trafficking. In embryonic neurons, loss of wild-type htt caused the same trafficking defects as expression of mhtt in cells that were otherwise normal.
mhtt titrates motor proteins from soluble pool to poorly soluble protofibrillar complexes in human HD-affected brain.
mhtt is prone to aggregation and forms inclusions in human, mouse, and cellular systems (27). Thus, mhtt may impair vesicular trafficking by sequestering normal htt and/or trafficking machinery. To test whether such sequestration occurs in humans, we isolated extracts from human control and HD-infected brain and tested whether trafficking-related proteins were associated with mhtt in disease tissue. The mutation of htt is known to alter interaction with several partners (18). One of them is htt-associated protein 1 (HAP1) that interacts with the key trafficking proteins dynactin p150Glued, a kinesin heavy chain homologue, and tubulin (15, 24). By using a differential extraction procedure (14) in combination with size exclusion chromatography and mass spectrometry, we addressed whether soluble aggregates from human HD-affected brain contained components of vesicular trafficking.
We first characterized the composition of aggregated complexes using mass spectrometry. To examine htt-containing complexes from brain tissue, we developed and applied an extraction method that allows the isolation of htt-containing complexes from control and HD-affected brain tissue based on their solubility (Fig. 6A) (14). By using this method, we have previously shown that wild-type htt is found in soluble (S1) extracts while mhtt is primarily found in poorly soluble (S3) protofibrillar complexes. Therefore, mass spectrometry was performed on the S3 fraction from HD-affected brain. The S3 extract from control brain contained virtually no htt. Therefore, htt-containing complexes were not present in the S3 of control brain and we could not perform a comparative analysis.
FIG. 6.
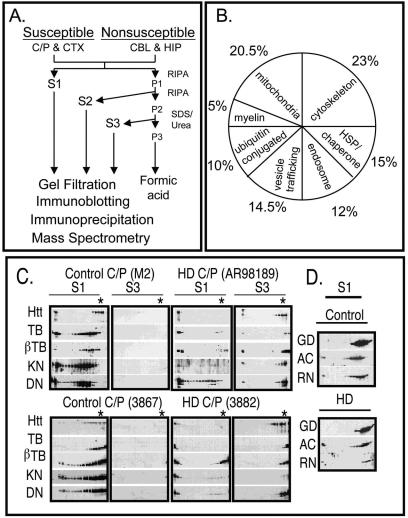
Components of the trafficking machinery are sequestered in human HD-affected brain. (A) Extraction scheme used to resolve proteins based on their solubility. Age- and gender-matched human brain tissues in HD-affected (C/P, CTX) and spared (CBL, HIP) regions were prepared as described in Materials and Methods. The S1 fraction contains proteins that are the most soluble, S2 contains proteins of intermediate solubility, and S3 represents the least soluble proteins that are specific to the HD-affected brain. CTX, cerebral cortex; CBL, cerebellar cortex; C/P, caudate and putamen; HIP, hippocampus; RIPA, radioimmunoprecipitation assay buffer. (B) Distribution of cytoplasmic, nonmetabolic proteins identified by mass spectrometry in gel filtration fractions of S3 extracts of HD-affected caudate and putamen. Note that mitochondrial and cytoskeletal proteins constitute the largest portion of this pool. (C) Redistribution of cytoskeleton and trafficking motors from S1 to S3 in HD-affected brain. Gel filtration profiles of htt, α-tubulin (TB), neuronal specific βIII-tubulin (βTB), kinesin (KN), and dynactin p150Glued (DN) in the S1 and S3 extracts of HD-affected caudate and putamen. Equal total protein amounts were resolved by size exclusion chromatography, and then an equal volume of each fraction was analyzed by immunoblotting. Data from two human control brains (M2 and 3867), age- and gender-matched with two human HD-affected brains (AR98189 and 3882), are shown. The asterisk indicates the predicted migration of a htt monomer. (D) Migration of nontrafficking proteins, actin (AC), GAPDH (GD), and Ran (RN) was unaffected in HD-affected compared to control brain.
Mass spectrometry sequencing revealed that the S3 aggregate fraction from HD-affected brain contained both nuclear (∼8%) and cytoplasmic (∼92%) proteins (unpublished data). Nuclear proteins included histones, leucine acidic nuclear protein, transportins, elongation factors, and Sir12 histone deacetylase. Sixty percent of the cytoplasmic proteins were metabolic enzymes or proteins of unknown function (Fig. 6B). However, the remaining 40% primarily comprised trafficking motors, mitochondrial proteins, endosomal proteins, and cytoskeletal components (Fig. 6B) Overall, these data suggest that components of the vesicular trafficking machinery and cargo were well represented in aggregated fractions from human HD-affected brain.
We next tested whether key components of the trafficking machinery codistributed into S3 extracts together with mhtt in human HD-affected brain. To test this hypothesis, the total protein amount from the soluble (S1) and poorly soluble (S3) extracts from control and HD-affected brain were resolved by size exclusion chromatography. An equal volume of the eluted fractions was analyzed by Western analysis to determine the presence and size distribution of the proteins of interest.
As previously shown, htt was extracted as a soluble protein (S1) in control brain, while in the HD-affected brain, it was consistently depleted in the S1 fraction and redistributed into the poorly soluble S3 fraction (Fig. 6C) (13). Most of the htt reactivity appeared to migrate near its monomer MW range (Fig. 6C). htt was present in control S1 but consistently absent in comparable extracts for HD S1. Instead, normal htt was redistributed into the poorly soluble S3 fraction of HD-affected brain (Fig. 6C, compare the migration from S1 and S3 for htt).
We next tested whether proteins involved in vesicular trafficking were also redistributed into the S3 fraction together with mhtt. Similar to htt, we found that α-tubulin, neuron-specific βIII-tubulin, kinesin, and dynactin p150Glued were extracted in the soluble S1 fraction from control caudate putamen region but were redistributed to the S3 extract from the HD-affected brain (Fig. 6C). Gel filtration analysis confirmed that the loss of tubulin, kinesin, and dynactin p150Glued in the S1 extract of HD-affected caudate-putamen region was due to redistribution into S3 rather than simply a shift in the association state (Fig. 6C). The shift in solubility of the trafficking proteins occurred in both the caudate and the cerebral cortex, which are the two major sites of HD pathology (Fig. 7A and C). The abundance of tubulin, kinesin, and dynactin p150Glued increased in fractions with decreased solubility (S2 and S3) in either affected brain region (Fig. 7C). In contrast, GAPDH was unaffected by the presence of mhtt and primarily extracted in the S1 fraction (Fig. 7A and C).
FIG. 7.
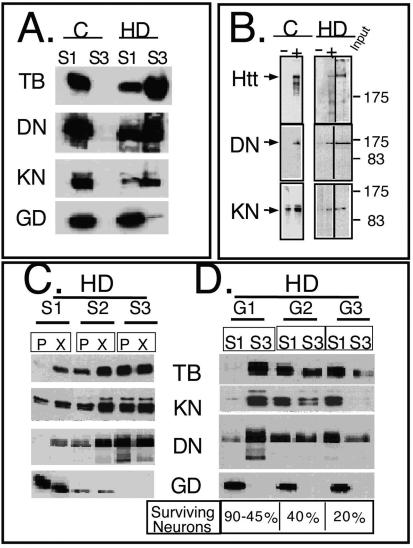
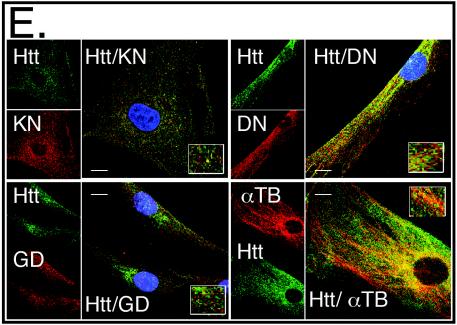
mhtt causes redistribution of soluble htt, cytoskeletal proteins, and trafficking motors to poorly soluble S3 extracts of human HD-affected brain. (A) An equal amount of total protein was analyzed for solubility by comparing S1 and S3 extracts by immunoblotting. Trafficking proteins α-tubulin (TB), dynactin (DN), and kinesin (KN) are present only in the S1 extract of control brain but are redistributed to the S3 extract in the HD-affected human brain. Control (C) and HD-affected brain extracts are from the caudate and putamen regions. GAPDH (GD) remains in the S1 of both control and HD-affected brain. (B) Trafficking proteins interact with both normal and mhtt in human brain. Immunoprecipitation of htt, dynactin, and kinesin from an S1 extract of control brain or from S3 gel filtration fractions of HD-affected brain. Since no htt was present in the S3 extract of control brain, it was not analyzed. Immunoprecipitation utilized the 2166 htt antibody; −, protein G beads alone; +, protein G beads cross-linked with antibody. Input is 8% of the protein sample in the immunoprecipitation reactions. (C) Reduced solubility of α-tubulin, kinesin, and dynactin in the brain regions vulnerable to HD. An equal amount of total protein from the S1, S2, and S3 extracts of grade 1 caudate-putamen (P) or cerebral cortex (X) was analyzed by immunoblotting. The motor-associated proteins are enriched in the poorly soluble S3 extract. The control protein GAPDH is found predominantly in the soluble S1 extract. (D) Solubility of trafficking proteins as a function of disease severity. An equal amount of total protein from S1 and S3 extracts from the caudate and putamen of three HD-affected brains of grade 1 (G1), 2 (G2), and 3 (G3) were probed for α-tubulin, kinesin, dynactin, and GAPDH. The level of α-tubulin, kinesin, and dynactin in S1 increases with grade and decreases with the average number of surviving neurons characteristic of each grade as previously described (44). The level of proteins in S3 shows the inverse pattern. The control protein GAPDH remains totally soluble in both control and HD-affected brains. (E) Colocalization of htt and trafficking proteins in fibroblasts from control individuals and HD patients. Note colocalization of htt with kinesin, α-tubulin, and dynactin but not with GAPDH. In all panels, htt is green; kinesin, dynactin, GAPDH, and α-tubulin are red; blue indicates the nucleus. Overlays are indicated. Confocal images were taken with 100× oil DIC (1.4 n.a.), thickness of the slices was ∼0.5 μm. Scale bar, 10 μm.
We found that the distribution and solubility of Ran GTPase, actin, or GAPDH, all of which are unrelated to trafficking, were equally present in the S1 extracts of either control or HD-affected brain (Fig. 6D) and were not found in the S3 extracts of either tissue (see GAPDH data in Fig. 7A). Column elution profiles for the latter group were nearly identical in normal and HD-affected brain demonstrating that their solubility and association state were not influenced by the presence of mhtt (Fig. 6D). Thus, htt-associated trafficking proteins and motors specifically codistributed with mhtt in human diseased brain and displayed properties expected of soluble aggregates.
If trafficking proteins are selectively sequestered in HD-affected brain, they should physically associate with htt. Therefore, we used immunoprecipitation to determine whether redistribution of trafficking proteins into S3 depended on interaction with mhtt. Since the S3 fraction did not contain htt, the S1 of normal tissue was evaluated as a control. S3 fractions from the exclusion chromatography were used as a source of the relevant proteins. We found that in fractions from both S1 control and S3 HD-affected brain, dynactin p150Glued and kinesin could be pulled down by immunoprecipitation using the 2166 htt antibody (Fig. 7B). Thus, it appeared that trafficking proteins found in aggregates in HD-affected brain normally interact with htt. However, these proteins are sequestered into poorly soluble aggregates due to interaction with mhtt.
Trafficking proteins not only coprecipitated with htt, but they also colocalized in both human control and HD-affected patient fibroblasts (Fig. 7E). Colocalization of htt with kinesin or dynactin was specific, since an equally abundant protein GAPDH showed only minimal colocalization. At the same time, colocalization with α-tubulin, a known partner of htt (21, 44), was pronounced (Fig. 7E). Thus, components of the vesicular trafficking machinery were redistributed into poorly soluble complexes by specific association with htt.
If selective sequestration of trafficking proteins contributes to HD, then the degree of sequestration should correlate with the progression of the disease. The severity of HD is determined by the extent of neuronal degeneration in the caudate and putamen. Five grades (grade 0 to 4) are recognized, with grade 4 being the most severely affected (45). Grade 3 and 4 tissues typically have fewer than 20% of the medium spiny neurons normally present in the caudate (45). Thus, with disease progression, the number of neurons decline and were substituted with glial cells. Indeed, we found that sequestration of trafficking proteins was inversely associated with disease severity and corresponded to the amount of surviving neurons (45) (Fig. 7D).
In grade 1 tissue, tubulin, kinesin, and dynactin p150Glued were most abundant in the S3 fraction (Fig. 7D). In grade 2 tissue, about equal amounts of protein were present in S1 and S3, while in grade 3 tissue most of the material was extracted in the S1 fraction. Under the same conditions, GAPDH (which is not sequestered in disease) was always present in the S1 extract (Fig. 7D). The data indicated that as fewer neurons survived and the tissue mass was increasingly composed of glial cells, the extraction profiles became more reflective of normal tissue. Thus, mhtt-mediated sequestration of trafficking motors appeared to be associated with neurons.
DISCUSSION
Here, we provide direct and functional evidence that expression of full-length mhtt impairs vesicular and mitochondrial trafficking in mammalian neurons in vitro and in vivo. Impairment of vesicular and mitochondrial motility increased with the length of the polyglutamine tract and occurred in the absence of cellular toxicity. Embryonic neurons expressing mhtt showed no morphological changes, no alterations in synapse formation, and no signs of nuclear fragmentation under any of the experimental conditions. In isolated neurons from mice expressing mhtt, trafficking defects in the brain were observed early in development prior to the onset of neurological symptoms. Trafficking defects were also observed in mice prior to onset of disease. Mice develop disease when their expressed level of htt falls below 50%, but haploinsufficient animals are healthy (10, 13, 53). We find that trafficking defects in KO animals that have the HD phenotype were significantly worse compared to animals with 50% loss of htt (Flox/−), suggesting a dose-dependent effect. Thus, trafficking defects occurred in animals and in embryonic neurons prior to the onset of disease. Inhibition of trafficking by mhtt is likely to be a direct effect since impairment of trafficking by mutant polyglutamine protein can occur in a cell-free system (i.e., squid axoplasm), which lacks a nucleus and in which protein synthesis does not occur (42).
Data presented here also suggest a normal role for htt in trafficking. We find that mice expressing less than 50% of htt display a severe trafficking defect, suggesting that normal trafficking requires the presence of htt. In fact, the trafficking defects in these animals are more extensive than those expressing mhtt. These results are consistent with the predictions of earlier genetics studies. Both White et al. (49) and Nasir et al. (29) demonstrated that mice lacking htt die early in embryogenesis but that behavioral and developmental defects can be partially rescued by one allele of mhtt (49). Consistent with these data, we find that expression of mhtt impairs trafficking relative to control mice but partially restores the defect in the conditional KO animals. Thus, the impairment of trafficking that we observe cannot be a simple loss of htt function. Progressive alteration in vesicular dynamics is likely to arise from two sources. mhtt may functionally inactivate trafficking motors as well as inhibit a normal function of htt in trafficking. Such a model is supported by data from human brain. We find that mhtt redistributes htt and soluble components of vesicular trafficking machinery, such as kinesin and the dynein-associated protein dynactin p150Glued, into poorly soluble aggregates.
These data raise the issue of how toxicity might arise from a trafficking defect. Our data as well as others suggest that neurite outgrowth and maintenance may be affected by impaired trafficking. Szebenyi and colleagues (42) have shown that truncated, mutant androgen receptor inhibits full neurite outgrowth in SH-SY5Y cells induced to differentiate to a neuronal phenotype with retinoic acid and brain-derived neurotrophic factor. These events preceded signs of cellular toxicity, indicating that trafficking impairment and dendritic effects are primary events in pathogenesis. Indeed, we have previously shown that full-length mhtt resides in the cytoplasm and initiates toxicity there (43). Expression of full-length mhtt causes neurite retraction, collapse of cytoskeleton, and commitment to cell death before nuclear entry.
We demonstrate that trafficking of mitochondria is particularly affected by expression of mhtt. Therefore, the morphological defects in dendrites observed in HD could be the result of inefficient delivery of mitochondria to the growth cone during neurite formation or to the periphery of mature neurites. Although the previous studies in the invertebrate systems did not analyze mitochondria per se, we demonstrate that mitochondrial trafficking is slower and less fluid in either embryonic neurons from htt KO mice or those from transgenic animals expressing full-length mhtt. Additionally, recent data in the Drosophila model have identified a protein required for mitochondrial transport to the synapse (40). This protein, Milton, has a high homology to HAP1, a protein known to interact more strongly with mhtt than with wild-type htt (25). We also find that mitochondrial proteins succinate dehydrogenase, glutaminase, ATP synthase β- and γ-chain, and outer mitochondrial membrane translocase were among those identified by mass spectrometry sequencing of mhtt-containing complexes from human HD-affected brains.
ATP production is an essential component of cell survival. Thus, impairment of mitochondrial function could deplete energy stores in neurons. Further, mitochondria are required to support synaptic connections and formation of growth cones. The loss of motility might prevent mitochondria from being in the right place at the right time. While trafficking defects occurred in the embryo prior to any sign of neuropathology or mitochondrial dysfunction, mitochondrial dysfunction was detected between 12 weeks and 12 months of age. These data provide a link between a defined function of htt and a long-standing model for mitochondrial involvement in HD (26). In human HD patients, magnetic resonance imaging confirms that creatine (a free radical scavenger, a substrate for the enzyme creatine kinase, and a precursor for ATP) was depleted and lactate was increased (35). In rats, systemic administration of the mitochondrial complex II inhibitor, 3-nitropropionic acid, causes neurobehavioral and pathological abnormalities consistent with HD (4). In HD patients, the caudate has severe deficiencies in mitochondrial complexes II and III (5, 16). Mitochondria from human HD lymphoblasts are more sensitive to stress (31, 37). Thus, disruption of mitochondrial function is consistent with HD pathophysiology in both animals and humans. Vesicular trafficking defects are likely to be a general hallmark of aggregation disorders. Mouse and fly models of Alzheimer's disease (11) and amyotrophic lateral sclerosis have declines in intracellular transport and abnormal accumulation of neurofilaments, actin, and tubulin (7, 50).
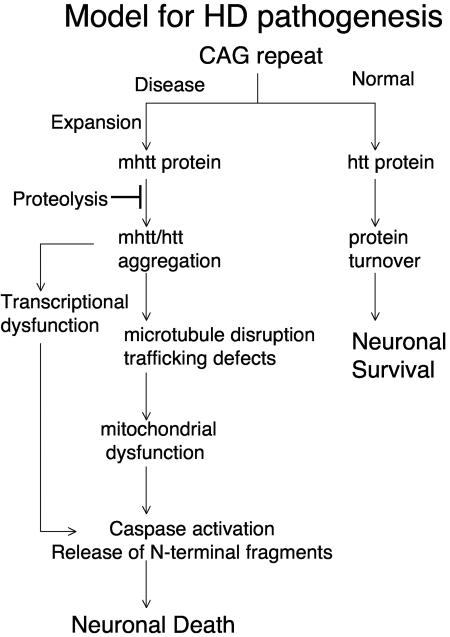
Taken together, these data provide a model as to how cytoplasmic dysfunction and loss of a normal htt trafficking function could lead to toxicity. mhtt redistributes htt and soluble trafficking components into protofibrillar complexes that impair motility of vesicles and organelles. With time, sequestration of mitochondrial proteins along with defective trafficking might lead to failure of ATP synthesis, energy depletion, and ultimately cell death (Fig. 8).
FIG. 8.
Proposed mechanism of neuronal dysfunction in HD. Model for HD pathogenesis that includes inhibition of mhtt proteolysis and initiation of aggregation that involves specific cellular targets along with wild-type htt. Sequestration leads to disruption of cytoskeleton, vesicular and organelle trafficking, transcriptional dysfunction, and eventually causes mitochondrial dysfunction and neuronal death.
Acknowledgments
This work was supported by the Mayo Foundation, National Institutes of Health, grants NS40738 and DK 43694-01; National Science Foundation grant IBN 9728120 (to C.T.M.); Canadian Institutes of Health Research grant mitochondria-15396 (to P.S.M.; the Valorisation Recherche Quebec, Genome Quebec and Genome Canada); and The Norwegian Research Council (FUGE). Human brain tissue was generously provided by the Harvard Brain Tissue Resource Center, which is supported in part by PHS grant MH/NS 31862.
We thank Line Roy, Nathalie Hamel, Daniel Boismenu, Alex Bell, and John Bergeron of the Montreal Proteomics Centre for help with sequencing, Eugene Krueger for technical assistance in DIC imaging, Alexey A. Leontovich for help with statistical analysis, and Timothy Farnham for help with manuscript preparation.
REFERENCES
- 1.Albin, R. L., A. Reiner, K. D. Anderson, L. S. Dure IV, B. Handelin, R. Balfour, W. O. Whetsell, Jr., J. B. Penney, and A. B. Young. 1992. Preferential loss of striato-external pallidal projection neurons in presymptomatic Huntington's disease. Ann. Neurol. 31:425-430. [DOI] [PubMed] [Google Scholar]
- 2.Ayala-Torres, S., Y. Chen, T. Svoboda, J. Rosenblatt, and B. Van Houten. 2000. Analysis of gene-specific DNA damage and repair using quantitative PCR. Methods Companion Methods Enzymol. 22:135-147. [DOI] [PubMed] [Google Scholar]
- 3.Barnes, G. T., M. P. Duyao, C. M. Ambrose, S. McNeil, F. Persichetti, J. Srinidhi, J. F. Gusella, and M. E. MacDonald. 1994. Mouse Huntington's disease gene homolog (Hdh). Somat. Cell Mol. Genet. 20:87-97. [DOI] [PubMed] [Google Scholar]
- 4.Beal, M. F., R. J. Ferrante, K. J. Swartz, and N. W. Kowall. 1991. Chronic quinolinic acid lesions in rats closely resemble Huntington's disease. J. Neurosci. 11:1649-1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brennan, W. A., Jr., E. D. Bird, and J. R. Aprille. 1985. Regional mitochondrial respiratory activity in Huntington's disease brain. J. Neurochem. 44:1948-1950. [DOI] [PubMed] [Google Scholar]
- 6.Chen, L. B. 1988. Mitochondrial membrane potential in living cells. Annu. Rev. Cell Biol. 4:155-181. [DOI] [PubMed] [Google Scholar]
- 7.Collard, J. F., F. Cote, and J. P. Julien. 1995. Defective axonal transport in a transgenic mouse model of amyotrophic lateral sclerosis. Nature 375:61-64. [DOI] [PubMed] [Google Scholar]
- 8.De Cristofaro, T., A. Affaitati, E. V. Feliciello, A. Avvedimento, and S. Varrone. 2000. Polyglutamine-mediated aggregation and cell death. Biochem. Biophys. Res. Commun. 272:816-821. [DOI] [PubMed] [Google Scholar]
- 9.DiFiglia, M., E. Sapp, K. Chase, C. Schwarz, A. Meloni, C. Young, E. Martin, J. P. Vonsattel, R. Carraway, S. A. Reeves, F. M. Boyce, and N. Aronin. 1995. Huntingtin is a cytoplasmic protein associated with vesicles in human and rat brain neurons. Neuron 14:1075-1081. [DOI] [PubMed] [Google Scholar]
- 10.Dragatsis, I., M. S. Levine, and S. Zeitlin. 2000. Inactivation of Hdh in the brain and testis results in progressive neurodegeneration and sterility in mice. Nat. Genet. 26:300-306. [DOI] [PubMed] [Google Scholar]
- 11.Drouet, B., M. Pincon-Raymond, J. Chambaz, and T. Pillot. 2000. Molecular basis of Alzheimer's disease. Cell. Mol. Life Sci. 57:705-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunah, A. W., H. Jeong, A. Griffin, Y. M. Kim, D. G. Standaert, S. M. Hersch, M. M. Mouradian, A. B. Young, N. Tanese, and D. Krainc. 2002. Sp1 and TAFII130 transcriptional activity disrupted in early Huntington's disease. Science 296:2238-2243. [DOI] [PubMed] [Google Scholar]
- 13.Duyao, M. P., A. B. Auerbach, A. Ryan, F. Persichetti, G. T. Barnes, S. M. McNeil, P. Ge, J.-P. Vonsattel, J. F. Gusella, A. L. Joyner, and M. E. MacDonald. 1995. Homozygous inactivation of the mouse Hdh gene does not produce a Huntington's disease-like phenotype. Science 269:407-410. [DOI] [PubMed] [Google Scholar]
- 14.Dyer, R. B., and C. T. McMurray. 2001. Mutant protein in Huntington's disease is resistant to proteolysis in affected brain. Nat. Genet. 29:270-278. [DOI] [PubMed] [Google Scholar]
- 15.Engelender, S., A. H. Sharp, V. Colomer, M. K. Tokito, A. Lanahan, P. Worley, E. L. Holzbaur, and C. A. Ross. 1997. Huntingtin-associated protein 1 (HAP1) interacts with the p150Glued subunit of dynactin. Hum. Mol. Genet. 13:2205-2212. [DOI] [PubMed] [Google Scholar]
- 16.Gu, M., M. T. Gash, V. M. Mann, F. Javoy-Agid, J. M. Cooper, and A. H. Schapira. 1996. Mitochondrial defect in Huntington's disease caudate nucleus. Ann. Neurol. 39:385-389. [DOI] [PubMed] [Google Scholar]
- 17.Gunawardena, S., L. S. Her, R. G. Brusch, R. A., Laymon, I. R. Niesman, B. Gordesky-Gold, L. Sintasath, N. M. Bonini, and L. S. B. Goldstein. 2003. Disruption of axonal transport by loss of huntingtin or expression of pathogenic polyQ proteins in Drosophila. Neuron 40:25-40. [DOI] [PubMed] [Google Scholar]
- 18.Harjes, P., and E. E. Wanker. 2003. The hunt for huntingtin function: interaction partners tell many different stories. Trends Biochem. Sci. 28:425-433. [DOI] [PubMed] [Google Scholar]
- 19.Hilditch-Maguire, P., F. Trettel, L. A. Passani, A. Auerbach, F. Persichetti, and M. E. MacDonald. 2000. Huntingtin: an iron-regulated protein essential for normal nuclear and perinuclear organelles. Hum. Mol. Genet. 9:2789-2797. [DOI] [PubMed] [Google Scholar]
- 20.Hodgson, J. G., N. Agopyan, C.-A. Gutekunst, B. R. Leavitt, F. LePiane, R. Singararja, D. J. Smith, N. Bissada, K. McCutcheon, J. Nasir, L. Jamot, X.-J. Li, M. E. Stevens, E. Rosemond, J. C. Roder, A. G. Phillips, E. M. Rubin, S. M. Hersch, and M. R. Hayden. 1999. A YAC mouse model for Huntington's disease with full-length mutant huntingtin, cytoplasmic toxicity, and selective striatal neurodegeneration. Neuron 23:181-192. [DOI] [PubMed] [Google Scholar]
- 21.Hoffner, G., P. Kahlem, and P. Djian. 2002. Perinuclear localization of huntingtin as a consequence of its binding to microtubules through an interaction with β-tubulin: relevance to Huntington's disease. J. Cell Sci. 115:941-948. [DOI] [PubMed] [Google Scholar]
- 22.Kegel, K. B., A. R. Meloni, Y. Yi, Y. J. Kim, E. Doyle, B. G. Cuiffo, E. Sapp, Y. Wang, Z. H. Qin, J. D. Chen, J. R. Nevins, N. Aronin, and M. DiFiglia. 2002. Huntingtin is present in the nucleus, interacts with the transcriptional corepressor C-terminal binding protein, and represses transcription. J. Biol. Chem. 277:7466-7476. [DOI] [PubMed] [Google Scholar]
- 23.Li, S.-H., A. L. Cheng, H. Zhou, S. Lam, M. Rao, H. Li, and X.-J. Li. 2002. Interaction of Huntington disease protein with transcriptional activator Sp1. Mol. Cell. Biol. 22:1277-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, S. H., C. A. Gutekunst, S. M. Hersch, and X. J. Li. 1998. Interaction of huntingtin-associated protein with dynactin p150Glued. J. Neurosci. 18:1261-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, X. J., S. H. Li, A. H. Sharp, F. C. Nucifora, Jr., G. Schilling, A. Lanahan, P. Worley, S. H. Snyder, and C. A. Ross. 1995. A huntingtin-associated protein enriched in brain with implications for pathology. Nature 378:398-402. [DOI] [PubMed] [Google Scholar]
- 26.Manfredi, G., and M. F. Beal. 2000. The role of mitochondria in the pathogenesis of neurodegenerative diseases. Brain Pathol. 10:462-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Michalik, A., and C. Van Broeckhoven. 2003. Pathogenesis of polyglutamine disorders: aggregation revisited. Hum. Mol. Genet. 12:R173-R186. [DOI] [PubMed] [Google Scholar]
- 28.Narain, Y., A. Wyttenbach, J. Rankin, R. A. Furlong, and D. C. Rubinsztein. 1999. A molecular investigation of true dominance in Huntington's disease. J. Med. Genet. 36:739-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nasir, J., S. B. Floresco, J. R. O'Kusky, V. M. Diewert, J. M. Richman, J. Zeisler, A. Borowski, J. D. Marth, A. G. Phillips, and M. R. Hayden. 1995. Targeted disruption of the Huntington's disease gene results in embryonic lethality and behavioral and morphological changes in heterozygotes. Cell 81:811-823. [DOI] [PubMed] [Google Scholar]
- 30.Nucifora, F. C., Jr., M. Sasaki, M. F. Peters, H. Huang, J. K. Cooper, M. Yamada, H. Takahashi, S. Tsuji, J. Troncoso, V. L. Dawson, T. M. Dawson, and C. A. Ross. 2001. Interference by huntingtin and atrophin-1 with cbp-mediated transcription leading to cellular toxicity. Science 291:2423-2428. [DOI] [PubMed] [Google Scholar]
- 31.Panov, A. V., C. A. Gutekunst, B. R. Leavitt, M. R. Hayden, J. R. Burke, W. J. Strittmatter, and J. T. Greenamyre. 2002. Early mitochondrial calcium defects in Huntington's disease are a direct effect of polyglutamines. Nat. Neurosci. 5:731-736. [DOI] [PubMed] [Google Scholar]
- 32.Perutz, M. F., and A. H. Windle. 2001. Cause of neural death in neurodegenerative diseases attributable to expansion of glutamine repeats. Nature 412:143-144. [DOI] [PubMed] [Google Scholar]
- 33.Reddy, P. H., M. Williams, V. Charles, L. Garrett, L. Pike-Buchanan, W. O. Whetsell, Jr., G. Miller, and D. A. Tagle. 1998. Behavioral abnormalities and selective neuronal loss in HD transgenic mice expressing mutated full-length HD cDNA. Nat. Genet. 20:198-202. [DOI] [PubMed] [Google Scholar]
- 34.Reiner, A., R. L. Albin, K. D. Anderson, C. J. D'Amato, J. B. Penney, and A. B. Young. 1988. Differential loss of striatal projection neurons in Huntington disease. Proc. Natl. Acad. Sci. USA 85:5733-5737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanchez-Pernaute, R., J. M. Garcia-Segura, A. del Barrio Alba, J. Viano, and J. G. de Yebenes. 1999. Clinical correlates of striatal 1H MRS in Huntington's disease. Neurology 53:806-812. [DOI] [PubMed] [Google Scholar]
- 36.Sauvé, D. M., D. T. Ho, and M. Roberge. 1995. Concentration of dilute protein for gel electrophoresis. Anal. Biochem. 10:382-383. [DOI] [PubMed] [Google Scholar]
- 37.Sawa, A., G. W. Wiegand, J. Cooper, R. L. Margolis, A. H. Sharp, J. F. Lawler, Jr., J. T. Greenamyre, S. H. Snyder, and C. A. Ross. 1999. Increased apoptosis of Huntington disease lymphoblasts associated with repeat length-dependent mitochondrial depolarization. Nat. Med. 5:1194-1198. [DOI] [PubMed] [Google Scholar]
- 38.Schmued, L. 1990. Fluoro-gold and 4-acetamido-4′-isothiocyanostilbene-2,2′-disulfonic acid: use of substituted stilbenes in neuroanatomical studies, p. 317-330. In Methods in neuroscience, vol. 3. Academic Press, Inc., New York, N.Y.
- 39.Steffan, J. S., A. Kazantsev, O. Spasic-Boskovic, M. Greenwald, Y. Z. Zhu, H. Gohler, E. E. Wanker, G. P. Bates, D. E. Housman, and L. M. Thompson. 2000. The Huntington's disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc. Natl. Acad. Sci. USA 97:6763-6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stowers, R. S., L. J. Megeath, J. Gorska-Andrzejak, I. A. Meinertzhagen, and T. L. Schwarz. 2002. Axonal transport of mitochondria to synapses depends on milton, a novel Drosophila protein. Neuron 36:1063-1077. [DOI] [PubMed] [Google Scholar]
- 41.Suhr, S. T., M. C. Senut, J. P. Whitelegge, K. F. Faull, D. B. Cuizon, and F. H. Gage. 2001. Identities of sequestered proteins in aggregates from cells with induced polyglutamine expression. J. Cell Biol. 153:283-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Szebenyi, G., G. A. Morfini, A. Babcock, M. Gould, K. Selkoe, D. L. Stenoien, M. Young, P. W. Faber, M. E. MacDonald, M. J. McPhaul, and S. T. Brady. 2003. Neuropathogenic forms of huntingtin and androgen receptor inhibit fast axonal transport. Neuron 40:41-52. [DOI] [PubMed] [Google Scholar]
- 43.Trushina, E., M. P. Heldebrant, C. M. Perez-Terzic, R. Bortolon, I. V. Kovtun, J. D. Badger II, A. Terzic, A. Estévez, A. J. Windebank, R. B. Dyer, J. Yao, and C. T. McMurray. 2003. Microtubule destabilization and nuclear entry are sequential steps leading to toxicity in Huntington's disease. Proc. Natl. Acad. Sci. USA 100:12171-12176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tukamoto, T., N. Nukina, K. Ide, and I. Kanazawa. 1997. Huntington's disease gene product, huntingtin, associates with microtubules in vitro. Mol. Brain Res. 51:8-14. [DOI] [PubMed] [Google Scholar]
- 45.Vonsattel, J. P., R. H. Myers, T. J. Stevens, R. J. Ferrante, E. D. Bird, and E. P. Richardson, Jr. 1985. Neuropathological classification of Huntington's disease. J. Neuropathol. Exp. Neurol. 44:559-577. [DOI] [PubMed] [Google Scholar]
- 46.Wasiak, S., V. Legendre-Guillemin, R. Puertollano, F. Blondeau, M. Girard, E. de Heuvel, D. Boismenu, A. W. Bell, J. S. Bonifacino, and P. S. McPherson. 2002. Enthoprotin: a novel clathrin-associated protein identified through subcellular proteomics. J. Cell Biol. 158:855-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wessendorf, M. W. 1991. Fluoro-gold: composition and mechanism of uptake. Brain Res. 553:35-148. [DOI] [PubMed] [Google Scholar]
- 48.Wheeler, V. C., J. K. White, C. A. Gutekunst, V. Vrbanac, M. Weaver, X. J. Li, S. H. Li, H. Yi, J. P. Vonsattel, J. F. Gusella, S. Hersch, W. Auerbach, A. L. Joyner, and M. E. MacDonald. 2000. Long glutamine tracts cause nuclear localization of a novel form of huntingtin in medium spiny striatal neurons in HdhQ92 and HdhQ111 knock-in mice. Hum. Mol. Genet. 9:503-513. [DOI] [PubMed] [Google Scholar]
- 49.White, J. K., W. Auerbach, M. P. Duyao, J. P. Vonsattel, J. F. Gusella, A. L. Joyner, and M. E. MacDonald. 1997. Huntingtin is required for neurogenesis and is not impaired by the Huntington's disease CAG expansion. Nat. Genet. 17:404-410. [DOI] [PubMed] [Google Scholar]
- 50.Williamson, T. L., and D. W. Cleveland. 1999. Slowing of axonal transport is a very early event in the toxicity of ALS-linked SOD1 mutants to motor neurons. Nat. Neurosci. 2:50-56. [DOI] [PubMed] [Google Scholar]
- 51.Wyttenbach, A., J. Swartz, H. Kita, T. Thykjaer, J. Carmichael, J. Bradley, R. Brown, M. Maxwell, A. Schapira, T. F. Orntoft, K. Kato, and D. C. Rubinsztein. 2001. Polyglutamine expansions cause decreased CRE-mediated transcription and early gene expression changes prior to cell death in an inducible cell model of Huntington's disease. Hum. Mol. Genet. 10:1829-1845. [DOI] [PubMed] [Google Scholar]
- 52.Yu, Z. X., S. H. Li, J. Evans, A. Pillarisetti, H. Li, and X. J. Li. 2003. Mutant huntingtin causes context-dependent neurodegeneration in mice with Huntington's disease. J. Neurosci. 23:2193-2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zeitlin, S., J. P. Liu, D. L. Chapman, V. E. Papaioannou, and A. Efstratiadis. 1995. Increased apoptosis and early embryonic lethality in mice nullizygous for the Huntington's disease gene homologue. Nat. Genet. 11:155-163. [DOI] [PubMed] [Google Scholar]
- 54.Zuccato, C., M. Tartari, A. Crotti, D. Goffredo, M. Valenza, L. Conti, T. Cataudella, B. R. Leavitt, M. R. Hayden, T. Timmusk, D. Rigamonti, and E. Cattaneo. 2003. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat. Genet. 35:13-24. [DOI] [PubMed] [Google Scholar]