Abstract
The heterochromatic domains of Drosophila melanogaster (pericentric heterochromatin, telomeres, and the fourth chromosome) are characterized by histone hypoacetylation, high levels of histone H3 methylated on lysine 9 (H3-mK9), and association with heterochromatin protein 1 (HP1). While the specific interaction of HP1 with both H3-mK9 and histone methyltransferases suggests a mechanism for the maintenance of heterochromatin, it leaves open the question of how heterochromatin formation is targeted to specific domains. Expression characteristics of reporter transgenes inserted at different sites in the fourth chromosome define a minimum of three euchromatic and three heterochromatic domains, interspersed. Here we searched for cis-acting DNA sequence determinants that specify heterochromatic domains. Genetic screens for a switch in phenotype demonstrate that local deletions or duplications of 5 to 80 kb of DNA flanking a transposon reporter can lead to the loss or acquisition of variegation, pointing to short-range cis-acting determinants for silencing. This silencing is dependent on HP1. A switch in transgene expression correlates with a switch in chromatin structure, judged by nuclease accessibility. Mapping data implicate the 1360 transposon as a target for heterochromatin formation. We propose that heterochromatin formation is initiated at dispersed repetitive elements along the fourth chromosome and spreads for ∼10 kb or until encountering competition from a euchromatic determinant.
The chromosomes of higher eukaryotes are organized into multiple domains with distinct properties. The partitioning of chromosomes into zones of condensed heterochromatin and dispersed euchromatin is readily seen in interphase nuclei. Heterochromatin has the property of silencing most genes that are normally packaged in euchromatin; this silencing is strikingly evident where euchromatic genes are abnormally juxtaposed to heterochromatic domains by chromosome rearrangement or transposition, resulting in position-effect variegation (PEV) (16). Work with multiple systems has led to a model of heterochromatin formation based on a specific combination of biochemical marks. Heterochromatic regions are characterized by histone hypoacetylation and methylation of histone H3 at lysine 9 (producing H3-mK9), in most cases accompanied by binding of heterochromatin protein 1 (HP1) and in some cases associated with methylation of the DNA (31). In Drosophila melanogaster, mutations in HP1 [Su(var)2-5] and in an H3-K9 methyltransferase [Su(var)3-9] cause a loss of heterochromatin-induced silencing. A remarkable property of heterochromatin is the ability to spread in cis in response to the loss of boundary constraints or to changes in the dosage or activity of chromatin components (24, 38). The interaction of HP1 with both the modified histone H3-mK9 and the modifying enzyme SU(VAR)3-9 suggests a plausible model for the maintenance and spreading of heterochromatin (reviewed in reference 16).
In most higher eukaryotes, domains of constitutive heterochromatin are normally limited to pericentric and telomeric DNA. An apparent exception to this is the small fourth chromosome of Drosophila melanogaster (22), which exhibits characteristics of both heterochromatin and euchromatin. Chromosome 4 does not undergo detectable meiotic recombination (6, 8) and the chromosome as a whole is late replicating (5), which are well-established characteristics of heterochromatic regions. However, the distal 20 to 25% of chromosome 4 is amplified in polytene nuclei to an extent similar to that of other euchromatic regions, and the gene density there (82 genes in 1.2 Mb) is similar to that found in the other euchromatic regions (15). Immunofluorescent staining of the polytene chromosomes shows that HP1, primarily localized in the pericentric heterochromatin and known to play a role in heterochromatin-induced silencing, is distributed in a banded pattern across this region (14, 19), suggesting that there are interspersed heterochromatic and euchromatic domains.
The transposable P element P[hsp26-pt, hsp70-w], containing an hsp70-driven white (w) gene and hsp26 fused with a fragment of plant (pt) DNA (Fig. 1), is a useful reporter of chromatin packaging. Transposition into pericentric or telomeric heterochromatin results in variegated expression of white, reduced nuclease accessibility in the hsp26 regulatory region, and a shift to a more regularly spaced nucleosome array across the transgene (11, 37, 40). Insertion of this element at various sites in the fourth chromosome has identified a minimum of three euchromatic domains (resulting in a red eye) interspersed with heterochromatic domains (inducing a variegating phenotype) (36). Mapping of the site of insertion of each P element, reported here, generated a surprising result: in many of the lines showing a variegating phenotype, the P element is inserted close to or within a gene. Thus, heterochromatic domains are not limited to tandem clusters of repetitious DNA or satellite sequences.
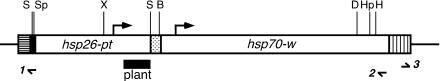
FIG. 1.
A map of the P[hsp26-pt, hsp70-w] construct drawn to scale (40). The hsp26 sequences from −1917 to +490 were fused to a 740-bp fragment of barley cDNA (plant probe), followed by hsp70 transcription termination sequences (dotted box). This fusion gene was cloned into the P element vector A412 possessing an hsp70-white gene. Transcription start sites for hsp26-pt and hsp70-w are marked by bent arrows. The 3′P end is on the left side (horizontally striped box), and the 5′P end is on the right side (vertically striped box). Restriction sites used in the analysis are the following: S, SalI; Sp, SpeI; X, XbaI; B, BglII; D, DpnII; Hp, HpaII; H, HhaI. Primers used for inverse or direct PCR to identify flanking DNA (small half-arrows below the diagram) are the following: 1, 5′-AACTCGAGGCCTCGAGGT-3′; 2, 5′-GACGAAATGACCCACTCGG-3′; 3, 5′-GCTTCGGCTATCGACGGGACCACC-3′.
To identify the DNA sequence elements that might determine heterochromatic or euchromatic status, we mobilized the reporter transgene and screened for a switch from one phenotype to the other. We show here that the deletion or duplication of 5 to 80 kb of DNA flanking specific P element reporters can lead not only to the conversion of a red-eye phenotype to a variegating phenotype (as in PEV) but also to the conversion of a variegating phenotype to a red-eye phenotype. The variegating phenotype observed as a result of a deletion or duplication is dependent on HP1, as a loss of silencing results from crossing lines with these traits with a line carrying Su(var)2-502, a hypomorphic allele of the gene for HP1. A change in the nuclease accessibility of the reporter gene in tested lines indicates that the underlying mechanism is a change in the chromatin structure. Analysis of the end points of the deletions and duplications recovered implicates the 1360 repetitive element (a.k.a. hoppel) as an initiation site for heterochromatin formation in the region of the fourth chromosome we have examined in detail. Heterochromatin packaging appears to extend for ca. 10 kb from the 1360 target, with no evidence of a boundary. Recent studies of fungi and plants provide evidence that heterochromatin formation is targeted to repetitious elements through an RNA interference (RNAi) mechanism (17, 39), and we have observed that mutations in components of the RNAi system lead to suppression of PEV (loss of silencing) in Drosophila (29). The results reported here suggest that RNAi uses the 1360 element to target heterochromatin formation in discrete regions of the fourth chromosome.
MATERIALS AND METHODS
Screens for Drosophila lines with the P element on the fourth chromosome.
All Drosophila stocks were raised on cornmeal sucrose-based medium (35). All crosses were performed at 25°C. Flies similar in age were used for all comparisons of eye phenotype. In the original screen (screen 0) to recover lines with a variegating phenotype carrying the P element P[hsp26-pt, hsp70-w], females from two lines homozygous for the P element insertion on the X chromosome (lines 39C-X and 118E-X) were crossed to w/Y, Sb Δ2-3/TM6 males, with Δ2-3 serving as a genomic source of transposase (32). The male progeny carrying the P element and the Sb Δ2-3 chromosome were crossed to females of the y w67c23 mutant stock. Male progeny showing PEV of hsp70-white expression and lacking the Sb Δ2-3 chromosome were made homozygous, and the site of insertion was determined by in situ hybridization (40). These homozygous lines are designated by 39C- or 118E- followed by a numeral. One line (1-M707) showing a red-eye phenotype was recovered from a similar screen (screen 1) starting with females from line 39C-X, using the y w67c23; net; sbd; spapol line to ascertain the segregation of the P element with the fourth chromosome.
To improve the rate of recovery of lines with a red-eye phenotype, a screen (screen 2) was executed, using females carrying the P element on the fourth chromosome and searching for a switch in phenotype from variegating eyes to red eyes; subsequently, similar screens were executed to look for a switch from red eyes to variegating eyes or to look for recombination by using a fourth chromosome marked with spapol. Characterization of the new lines revealed not only local transposition events but also local deletions and duplications, reflecting mobilization of only one end of the P element. New lines are designated by a numeral indicating the screen number (1 through 6) followed by M and a numeral. New lines were characterized to determine whether or not there had been a change in the P element itself and to monitor changes in flanking genomic DNA. The starting female line, selection phenotype, and types of lines recovered in screens 0 through 6 are shown in Table 1. In screen 2, females from line 39C-12 (with a transgene at position 102B on the fourth chromosome and a variegating phenotype) were crossed to w/Y, Sb Δ2-3/TM6, spapol/spapol males; male progeny that were Sb Δ2-3; P[hsp26-pt, hsp70-w]/spapol were independently crossed to females of the y w67c23; net; sbd; spapol line, and the resulting male progeny with red eyes were screened (using the same female stock) for segregation of the P element with the fourth chromosome. Initial results from experiments using lines recovered in screen 2 have been previously reported (36), assuming that these lines represented transposition events; new tests used here (see below) have shown that while red-eyed lines 2-M390R, 2-M371R, and 2-M1021R are the results of transposition events, lines 2-M59A.R and 2-M010R have switched phenotype as a consequence of a flanking deletion (see Fig. 3). To obtain a more detailed map, we carried out several additional screens using this same protocol, starting with w/Y, Sb Δ2-3/TM6 males; screen 4 used females from line 39C-12 as the source of the P element, searching for conversion from a variegating to a red-eye phenotype, and screen 5 used females from line 2-M59A.R as the source of the P element, searching for conversion from a red-eye to a variegating phenotype. Screens 3 and 6 were carried out in the same manner but started from lines carrying the P element on a fourth chromosome marked with spapol and selected for recombination (loss of the spapol marker).
TABLE 1.
Screens leading to the recovery of lines characterized here
| Screen no. | Starting line(s); eye phenotypea | Selected eye phenotypea | No. of chromosomes screened | Total no. of lines recovered | Classb
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | U | |||||
| 0 | 39C-X; R | V | 2,971 | 24 | 24 | |||||||
| 118E-X; R | V | |||||||||||
| 1 | 39C-X; R | R | 828 | 1 | 1 | |||||||
| 2 | 39C-12; V | R | 1,143 | 8 | 3 | 2 | 2 | 1 | ||||
| 3 | 2-M529; R | R or V, not spapol | 836 | 2 | 1 | 1 | ||||||
| 4 | 39C-12; V | R | 1,396 | 56 | 4 | 12 | 5 | 23 | 7 | 1 | 4 | |
| 5 | 2-M59A.R; R | V | 3,000 | 64 | 5 | 33 | 5 | 2 | 13 | 3 | 1 | 2 |
| 6 | 2-M530, 2-M802, 2-M823; R | R or V, not spapol | 542 | 17 | 2 | 2 | 9 | 4 | ||||
R, red-eye phenotype; V, variegating phenotype.
The recovered lines are grouped into phenotypic classes based on molecular characterization. Columns 1 to 7 give the number of lines recovered of each type. Class 1, transposition (single P element at a new locus); class 2, nonspecific duplication (more than one partial or full copies of the P element); class 3, duplication ([3′P to 3′P] P element duplication); class 4, modified P element (single damaged P element); class 5, rightward change (single P element in its original position adjacent to a change in genomic DNA at the 5′P [right] end), class 6, leftward change (single P element in its original position adjacent to a change in genomic DNA at its 3′P [left] end), class 7, rearrangements (single P element with one end at its original position while the other end maps to a position not on the fourth chromosome or to a position on the fourth chromosome that suggests a local inversion); class U, unclassified or other.
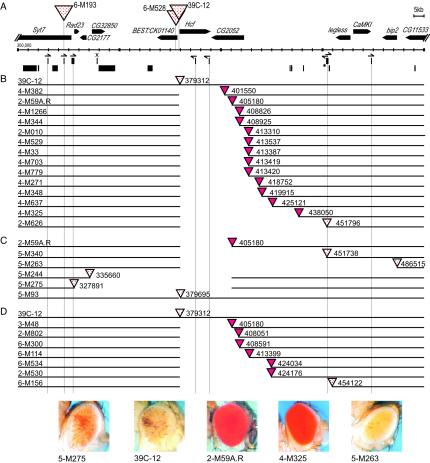
FIG. 3.
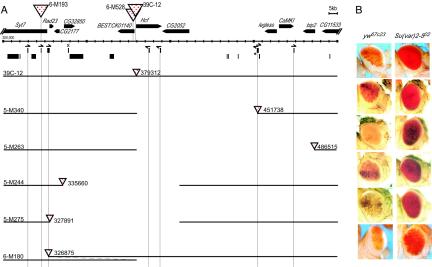
Local deletions can induce a change in eye phenotype. (A) A high-resolution map of a ∼200-kb region from the fourth chromosome. Scale divisions are 5 kb. Genes are shown as thick horizontal arrows, with the start of transcription at the blunt end. The solid blocks below the scale bar indicate fragments of known repetitive elements, with 1360 elements in the upper row and others in the lower row. Half-arrows over the 1360 elements indicate the orientation suggested by homology (>90% over at least 50 bp) to the beginning of transcripts from the 1360 element within the Su(Ste) repeat locus (3). A PCR was used to verify the presence of the 1360 fragments shown in all lines used in this study. The copy marked with an asterisk is present in the y w67c23 and 39C-12 lines but not in the Sb Δ2-3/TM6 or y w67c23; net; sbd; spapol lines. The presence of this element was verified for individual derived lines as needed; it is present in lines 2-M626, 4-M325, and 5-M340, those lines where the P element lies close to this position. The smallest fragment of 1360 is marked with an X; see the text for discussion. In parts B to D, genomic DNA is shown as a solid black line. The number after the triangle gives the insertion site or deletion end point on the 1.2-Mb map of chromosome 4 (15). (B) Rightward deletions generated from P element mobilization in line 39C-12. (C) Rightward and leftward deletions generated from P element mobilization in line 2-M59A.R. (D) Rightward deletions generated from P element mobilization in lines derived from 39C-12, screened for a recombination event rather than a change in phenotype. Shown below are photographs of representative eyes from the starting Drosophila line 39C-12 (variegating phenotype); lines 2-M59A.R and 4-M325, derived from 39C-12 by local deletion (full red expression); and lines 5-M275 and 5-M263, derived from 2-M59A.R by local deletion (variegating phenotype). Together, the results indicate the presence of a 45- to 50-kb euchromatic domain flanked by heterochromatic domains.
Molecular characterization of P element inserts.
Genomic DNA prepared from 100 flies was digested with either SalI, XbaI, or BglII and used for Southern blot analysis, hybridizing with the radiolabeled plant (pt) probe to detect the P element DNA. DNA from each derivative line was compared with that from its progenitor line. A modified, multiple, or missing SalI fragment indicates a modified (damaged or duplicated) P element. A change in size of the XbaI and/or BglII band indicates a change in the flanking DNA, while a multiplied or missing band indicates a modified P element (Fig. 1). A SpeI-SphII double digestion (where the SphII site is in the flanking genomic DNA) was used to resolve head-to-head duplications. Only lines shown by these assays to have a single undamaged P element were analyzed further. Primer 3 (from the 5′P end of the P element) and a primer from genomic DNA near the parental line P element insert site (5′-CCTCACACACGCAATTACT-3′ for screen 4 and 5′-GCTCGGATGAAATAAGTGT-3′ for screen 5) were used to PCR amplify DNA from single adult flies. For lines derived from 39C-12 (screens 2 and 4) or its derivatives (screens 3, 5, and 6), PCR at the 3′P end was carried out using primer 1 from the P element sequence and L-850 (5′-CTCAGCGCCACAGTTATTTA-3′) from the genomic sequence. The absence of products at both the 5′P and 3′P ends indicates a transposition, while the absence of product in one case or the other indicates a change in the flanking DNA. Where a rightward change occurred, genomic DNA was digested with DpnII, HpaII, or HhaI, circularized, and amplified by inverse PCR (with primers 2 and 3; Fig. 1) (10). PCR products were purified with either ExoSAP or gel isolation and directly sequenced. The genomic DNA sequence recovered was mapped by a BLAST search. The end points of selected leftward 3′P-adjacent deletions were mapped by a BLAST of a sequence from the BglII fragment cloned in a pLITMUS28 vector. (The P element lacks sufficient novel DNA sequence to allow inverse PCR from primers close to the 3′P end.) Map positions were confirmed by generating PCR products by using one primer from the P element and one primer from the predicted flanking genomic DNA.
Mapping of 1360 elements in different Drosophila stocks.
The maps shown for chromosome 4 in Fig. 2, 3, 4, and 7 are based on the Gbrowser map at FlyBase release 3 (15); the positions of repetitious elements are based on a previous work (20). Since repetitious elements can be unstable, we used PCRs (with oligos based on the genomic DNA) to confirm the presence of the 1360 copies in the stocks used here. One copy reported at FlyBase coordinates 476495.477610 is not present in any of our stocks and so is not shown on the figures. A second copy, at FlyBase coordinates 451822.452927 (see Fig. 3, 4, and 7), is present in the y w67c23 and 39C-12 lines but not in the Sb Δ2-3/TM6 or y w67c23; net; sbd; spapol lines. The presence of this element was verified for individual derived lines as needed; it is present in lines 2-M626, 4-M325, and 5-M340, those lines where the P element lies close to this position. All other copies of 1360 shown on the map in Fig. 3, 4, and 7 were verified in all of the stocks used here.
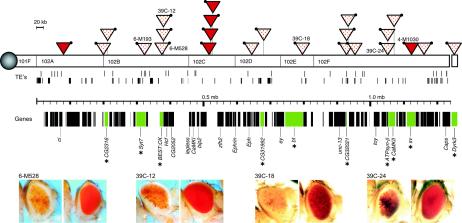
FIG. 2.
Heterochromatin is not limited to tandem repetitious sequences. The phenotype of P[hsp26-pt, hsp70-w] transgenes indicates that while domains of euchromatin are present, the fourth chromosome is largely heterochromatic. The map of the fourth chromosome (top) (regions 101F to 102F) shows the positions of known and predicted genes and of transposable elements (upper row, 1360 elements; lower row, others) (15). Solid triangles and dotted triangles mark P element insertion sites that result in a solid red eye or induce a variegating phenotype, respectively. The black dot indicates the 5′ end of the P element. See Materials and Methods for detailed descriptions of the screens used to recover these lines and of the techniques used to confirm the presence of a single intact P element in each line and to map its position. In cases where a line has been recovered with a P element inserted within a gene, the gene (in green) is indicated by an asterisk. In all cases tested, the variegating phenotype is suppressed by a mutation in HP1; representative examples are shown in the pictures at the bottom, with the line crossed to the y w67c23 line (the left-hand picture of each pair) compared to the line crossed to the y w67c23, Su(var)2-502 line (the right-hand picture of each pair).
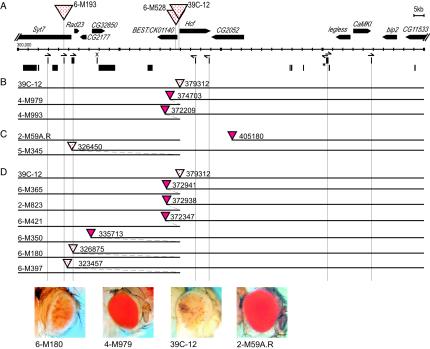
FIG. 4.
Local duplications can induce a change in eye phenotype. (A) A high-resolution map of a ∼200-kb region from the fourth chromosome; symbols are the same as those described in the legend to Fig. 3. (B) Duplications generated from P element mobilization in line 39C-12. (C) A duplication generated from P element mobilization in line 2-M59A.R. (D) Duplications generated from P element mobilization in spapol-marked lines derived from 39C-12, screened for a recombination event rather than a change in phenotype. In each case, the extent of the sequence that is duplicated (from the proximal side of 39C-12) is shown by the overlap in the horizontal lines indicating genomic DNA. Shown below are photographs of representative eyes from the starting Drosophila lines 39C-12 (variegating phenotype) and 2-M59A.R (red-eye phenotype) and lines carrying a duplication, 6-M180 (variegating phenotype) and 4-M979 (red-eye phenotype).
FIG. 7.
Silencing as a consequence of local deletions or duplications is dependent on HP1. (A) A high-resolution map of a ∼200-kb region from the fourth chromosome; symbols are those described in the legend to Fig. 3. (B) The variegating phenotype displayed by flies carrying the P element within 10 kb of a 1360 element is suppressed by mutation in HP1; representative examples are shown, with the line crossed to the y w67c23 line (left) compared to the line crossed to the y w67c23, Su(var)2-502 line (right). Shown are the following lines: 39C-12, a transposition; 5-M340, 5-M263, 5-M244, and 5-M275, deletions; and 6-M180, a duplication. The P element in line 5-M263 does not lie close to a 1360 element but may be affected by a nearby F element (see the text).
Assessing change in chromatin structure.
Previous mapping of hsp26 has identified two DNase I hypersensitivity sites in the 5′ regulatory region; fortuitously, each has an XbaI site (25). Quantitative assessment of changes in chromatin structure can be obtained by digestion of nuclei isolated from third-instar larvae using that restriction enzyme (9). Following XbaI digestion of the chromatin, the DNA was purified, digested with SalI, and size separated by gel electrophoresis, and a Southern blot was probed with the pt DNA fragment isolated from plasmid pGH19 and labeled with [32P]dCTP. The accessibility of the proximal XbaI site was calculated as the ratio of the signal intensity from the resulting gel band relative to that of the sum of all three bands obtained. The values were then normalized to that from the control line 39C-X (P element inserted into euchromatin), which is set at 100%.
Assessing eye phenotype.
Males were collected from cleared vials 2 days after eclosion and photographed 3 days after collection (3 to 5 days posteclosion). Lines such as 39C-12, showing strong PEV (little expression), show a very consistent phenotype. Lines such as 39C-34, which shows weak PEV, exhibit more variation in phenotype on visual inspection. To measure eye pigmentation, four to five samples from five males each (3 to 5 days posteclosion) were homogenized in 0.5 ml of 0.01 M HCl in ethanol. The homogenate was kept at 4°C overnight, warmed at 50°C for 5 min, and clarified by centrifugation. The supernatant was recovered, and the optical density at 480 nm was determined (21). Mean values and standard errors compared with the value for the 39C-X control stock (P element inserted in euchromatin), set at 100%, are reported.
Assessing suppression of PEV by mutations in HP1.
Lines to be tested were crossed with the y w67c23, Su(var)2-502/CyO line; progeny with straight wings were compared to progeny of a control cross with the y w67c23 line. (Note that the CyO chromosome carries a suppressor of PEV, making a comparison of sibs inappropriate.) Males were collected from cleared vials 2 days after eclosion and photographed 3 days after collection (3 to 5 days posteclosion). Similar experiments to examine the impact of a duplication of the HP1 gene were carried out using the y w67c23, Dp(2;2)P90/CyO line (41).
RESULTS
Heterochromatic domains on the fourth chromosome encompass genes.
To test the domain organization of the fourth chromosome, the transposable P element P[hsp26-pt, hsp70-w] was previously used as a phenotypic probe, identifying lines with the P element on the fourth chromosome showing either a variegating or red-eye phenotype (36). We have now approximately doubled the number of such characterized lines with a single transposon inserted at different sites within the fourth chromosome region 101F-102F, increasing the resolution of the map. (See Fig. 1 for a map of the P element used and Materials and Methods for a description of the screens.) Insertion sites have now been precisely mapped onto the published D. melanogaster genome sequence (15) by using genomic DNA sequences flanking the 5′ P element end, recovered by inverse PCR. Figure 2 (top) shows the distribution of insertion sites resulting in a variegating or red-eye phenotype relative to the positions of genes and transposable elements. The pattern of reporter gene expression reveals three euchromatic domains in a largely heterochromatic chromosome. However, additional small euchromatic domains may well be present (see Fig. 3). Note that in practice, it is much easier to recover fourth-chromosome P element insertion events from among lines with a variegating phenotype than from lines with a red-eye phenotype, as the bulk of the red-eye lines represents insertion into the major euchromatic domains on the other chromosomes; thus, the present map is most likely not saturated for euchromatic domains. While one might anticipate that variegating inserts would be associated with gene-poor regions of the chromosome, potentially containing tandem arrays of repetitious elements, this proved not to be the case. In fact, most (17 of 18) of the variegating P elements lie within ca. 2 kb of a gene, with 11 lying within the transcribed portions of 10 different genes (exact positions are given in Table 2). In all cases tested, the variegating phenotype is suppressed (loss of silencing) by a mutation in HP1 (Fig. 2, bottom) (36).
TABLE 2.
Precise insertion sites of the reporter transgenesa
| Line | Phenotype | GenBank accession no.: coordinate | Assembled chromosome coordinate | P position relative to nearest gene | Distance to gene (kb) |
|---|---|---|---|---|---|
| 2-M1021 | R; viable | AE003845: 79754 | 79754 (+) | Right of ci (−) | 2.087 |
| 118E-3 | V; viable | AE003844: 26903 | 200718 (−) | Left of CG2219 (+) | 1.114 |
| 39C-34 | V; viable | AE003844: 39223 | 213037 (+) | Inside CG2316 (−) | 0 |
| 6-M193 | V; viable | AE003844: 149625 | 323400 (−) | Inside 1360 within Syt7 (−) | 0 |
| 6-M528 | V; lethal | AE003844: 204553 | 378368 (+) | Inside BEST:CK01140 (−) | 0 |
| 39C-12 | V; viable | AE003844: 205498 | 379312 (+) | Left of Hcf (+) | 0.397 |
| 5-M29 | V; viable | AE003844: 205837 | 379651 (+) | Left of Hcf (+) | 0.040 |
| 4-M1285 | R; lethal | AE003843: 31713 | 521313 (−) | Right of CG11533 (+) | 12.403 |
| 2-M371 | R; lethal | AE003843: 32114 | 521714 (−) | Right of CG11533 (+) | 12.803 |
| 1-M707 | R; lethal | AE003843: 32792 | 522392 (+) | Right of CG11533 (+) | 13.481 |
| 2-M390 | R; viable | AE003843: 38248 | 527848 (−) | Left of zfh2 (+) | 13.364 |
| 39C-33 | V; viable | AE003843: 190621 | 680221 (−) | Inside CG31992 (−) | 0 |
| 39C-52 | V; viable | AE003843: 190746 | 680346 (−) | Right of CG31992 (−) | 0.011 |
| 39C-18 | V; lethal | AE003843: 305620 | 795220 (−) | Inside bt (+) | 0 |
| 118E-19 | V; viable | AE003846: 2362 | 927509 (−) | Inside CG32021 (−) | 0 |
| 118E-25-5 | V; viable | AE003846: 3435 | 928582 (+) | Inside CG32021 (−) | 0 |
| 5-M359 | V; lethal | AE003846: 7800 | 932947 (+) | Left of elF-4G (−) | 1.926 |
| 39C-42 | V; viable | AE003846: 29117 | 954264 (−) | Left of Glu-RA (+) | 2.166 |
| 39C-28 | V; lethal | AE003846: 126063 | 1051210 (−) | Right of CG11076 (−) | 0.074 |
| 39C-24 | V; viable | AE003846: 126166 | 1051313 (−) | Inside ATPsyn-beta (+) | 0 |
| 118E-9 | V; viable | AE003846: 145671 | 1070818 (+) | Inside CaMKII (−) | 0 |
| 4-M1030 | R; lethal | AE003846: 193104 | 1118250 (−) | Inside sv (+) | 0 |
| 5-M72 | V; viable | AE003846: 235373 | 1160520 (+) | Inside CG32017 (−) | 0 |
| 118E-15 | V; viable | AE003163: 42799 | Unassembled sequence | Inside Dyrk3 | 0 |
Gene positions are derived from the release 3.1 annotation by using the FlyBase Gbrowser online (http://flybase.org); mapping in Dyrk3 is based on a personal communication from L. L. Wallrath and D. Cryderman. The coordinate of each insertion site on the assembled chromosome is given, with + and − indicating the orientation of the P element; + and − are also used to indicate the orientation of the closest gene. R, red-eye phenotype; V, variegating phenotype.
Taken together, the banded pattern of HP1 distribution and the position-dependent expression of the P element reporter inserted at different sites across the fourth chromosome strongly support a model of interspersed euchromatic and heterochromatic domains. However, rather than finding the fourth chromosome genes restricted to euchromatic domains, the experimental results described above point to the conclusion that many fourth-chromosome genes lie in heterochromatic domains, defined as regions inducing a variegating white phenotype, dependent on HP1. Given that euchromatic and heterochromatic domains are interspersed on the fourth chromosome, heterochromatin formation clearly is not dictated simply by proximity to the chromocenter, nor is it limited to regions of tandem repeats. A more subtle signaling mechanism must be at work.
Small deletions and duplications reveal cis-acting determinants of heterochromatin formation.
The interspersion of heterochromatic and euchromatic domains on the fourth chromosome presents a unique opportunity to identify and dissect determinants of heterochromatin and euchromatin structure. If specific sequence elements create initiation sites for the assembly of heterochromatin and/or euchromatin, deletions of DNA flanking a transposon reporter that remove such a determinant would switch the phenotype of the reporter. We have used the mobilization of P elements in region 102B to generate nested deletions and duplications of DNA flanking the transposon insertion sites (1, 30).
A large set of nested deletions was generated in the 102B-C interval using each of three distinct genetic screens: (i) mobilization of the variegating insert in line 39C-12 and selection for red-eyed flies (Fig. 3B), (ii) mobilization of the transposon in red-eyed line 2-M59A.R and selection for flies with the variegating phenotype (Fig. 3C), and (iii) P element-catalyzed recombination between the 39C-12 insert and spapol (Fig. 3D). Rightward deletions that switch the variegated phenotype to a red-eye one range in size from 20 to 60 kb. The switch in phenotype indicates the presence of a euchromatic domain in the region beyond the deletion. A deletion (line 2-M626) that extends ca. 73 kb (selected because it showed a more extreme phenotype than 39C-12) retains a variegating phenotype, indicating that another heterochromatic domain lies beyond the more proximal euchromatic domain. Additional deletions confirm the presence of heterochromatic domains to the right and left of the euchromatic domain in 102B.
In addition to local deletions, nine lines carrying local duplications were also recovered in these screens (Fig. 4). These nine duplications consist of variable amounts of DNA sequence found proximal to the initial P element insertion site that are now duplicated in tandem with the second copy on the distal side of the element. For six of these duplications, the variegated phenotype has been converted to a red-eye phenotype. The results suggest that the duplicated material either (i) increases the distance between the P element and a heterochromatin-initiating element, switching a variegated phenotype to a red-eye phenotype (e.g., line 4-M979) or (ii) brings a heterochromatin-initiating element into close apposition to the P element, switching a red-eye phenotype to a variegating-eye phenotype (e.g., line 5-M345).
Sequence analysis indicates that 1360 serves as an initiator of heterochromatin formation.
The deletion data shown in Fig. 3 indicate that there are two transition regions between heterochromatic and euchromatic domains, the ∼20-kb interval between the 39C-12 insertion point and the 4-M382 end point and the ∼25 kb to the right of the 4-M325 end point. A pairwise comparison identified copies of terminal inverted repeat (TIR) element 1360 (a.k.a. hoppel) within each of these intervals as the only significant shared sequence. Two copies of this element are lost in the smallest deletion leading to a switch from a variegating to a red-eye phenotype; the variegating phenotype is reestablished in deletions that place the P element close to copies of 1360 on the proximal or distal side of the 39C-12 insertion site (Fig. 3). Additional DNA inserted on the distal side of the 39C-12 transposon (by local duplication) that increases the distance between the P element and the two naturally occurring copies of the 1360 transposable element distal to 39C-12 results in a shift from a variegating to a red-eye phenotype. However, the three largest duplications we identified result in a copy of 1360 that lies between Syt7 and Rad23, being placed even closer to the initial P element; lines with these duplications either switch to (5-M345) or maintain (6-M180 and 6-M397) a variegating phenotype.
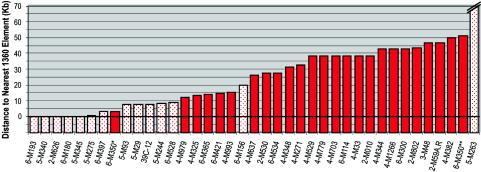
Figure 5 presents a bar diagram indicating the distance between the P element and the nearest 1360 element for all lines examined in the interval shown in Fig. 3 and 4. The results suggest that 1360 serves as an initiator of heterochromatin formation and that heterochromatin assembly spreads for ca. 10 kb. Line 6-M350 is presented twice, first in relation to a 1360 fragment lacking the terminal repeats and known transcription initiation sites (marked with an X in Fig. 3A and 4A) and second in relation to a 1360 fragment with such sites (described in reference 3). The results are consistent with other observations if the transcription initiation sites and/or terminal repeats are required for 1360 to serve as an initiator of heterochromatin formation. All of the 1360 elements shown in Fig. 3A and 4A satisfy this criterion except the X fragment. The X fragment, at 205 bp, is also the smallest in this interval identified as homologous to 1360, with the others ranging from 453 to 1,113 bp. Lines 6-M156 and 5-M263, with variegating phenotypes, also appear anomalous. The variegating P element in 6-M156 lies ∼20 kb from the nearest 1360; this result might be explained if the associated deletion had removed a cis-acting signal promoting euchromatin formation (Fig. 3D). There are no copies of 1360 close to the P element in 5-M263, but the single F element in this region is 8.8 kb away, suggesting that F elements might also play a role (see Discussion).
FIG. 5.
Silencing is correlated with proximity to transposon 1360 fragments. The distance between the P element insertion site and the nearest 1360 element is shown for the different Drosophila lines described in the legends to Fig. 3 and 4, including those generated by independent insertion events, local deletions, and local duplications. The bar for each line starts below 0 to allow visualization of the cases where the P element is within or very close to a 1360 element. A switch from a variegating phenotype (dotted bar) to a red-eye phenotype (solid bar) is observed when the distance is greater than ca. 10 kb. See the text for a discussion of the apparent exceptions to this observation, particularly line 6-M350, marked with asterisks.
Conversion of phenotype reflects an alteration in local chromatin structure.
To test whether changes in the local chromatin structure have occurred, we assayed the accessibility of the hsp26-pt transgene at the proximal XbaI site (within the 5′ regulatory DNase I hypersensitive site) in deletion lines 4-M382, 4-M33, 4-M348, and 4-M325, all having a red-eye phenotype. Nuclei were isolated from third-instar larvae of each heterozygous line (all of the deletions are homozygous lethal) and treated with an excess of XbaI enzyme (9). Accessibility was determined as the percentage of the chromatin cut at the proximal XbaI site normalized to results obtained with line 39C-X (set as 100%), where the P element is in a euchromatic site on the X chromosome. All four lines showed an obvious increase in XbaI accessibility (63 to 81%) compared to line 39C-12 (42%), the starting variegating line (Fig. 6A). The shift in eye phenotype was confirmed by quantitative assessment of pigment extracted from adult males (Fig. 6B). Pigment levels were found to be over 80% of that of line 39C-X, while the amount of pigment from the variegating eyes of line 39C-12 is about 10% of that of line 39C-X. The results demonstrate that the deletion-induced shift in eye phenotype reflects a shift in the underlying chromatin structure.
FIG. 6.
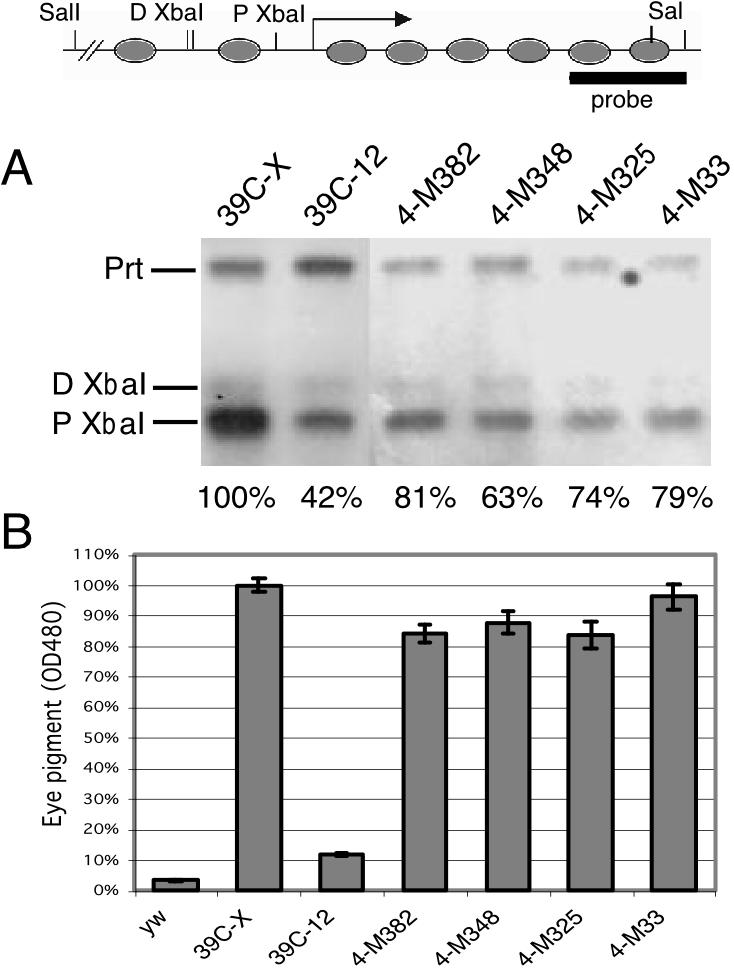
A switch in eye phenotype reflects a switch in chromatin structure. (A) Changes in chromatin structure were assessed by digestion of nuclei isolated from third-instar larvae using the restriction enzyme XbaI, as previously described (9). The DNA was then purified, digested with SalI, and size separated by gel electrophoresis, and a Southern blot was probed with the pt DNA fragment isolated from plasmid pGH19 and labeled with [32P]dCTP. Prt, parental SalI fragment (copies not cleaved by XbaI); D XbaI and P Xba, products of cleavage at the distal and proximal sites, respectively (see map above panel A). Fly lines used as the source of nuclei are indicated above each lane. The accessibility of the proximal XbaI site was calculated as the ratio of the signal intensity from the resulting gel band relative to the sum of all three bands obtained. The percent cleavage of the proximal XbaI site, normalized to that of line 39C-X (where the P element is in euchromatin) is shown below each lane. (B) Quantitative assessment of the eye phenotype was obtained by measuring eye pigmentation. Eye pigments were extracted from adult male fly heads using acidified ethanol, and the optical density was measured at 480 nm. The mean (bar) and standard error of four or more samples (five males each) are indicated. The switch in chromatin structure is associated with restoration to 80% or more of pigment levels (normalized to 39C-X).
Variegation of fourth-chromosome inserts is suppressed by mutation in HP1.
Heterochromatin formation is generally associated with the presence of HP1. To test whether HP1 mediates silencing of these fourth-chromosome P element reporters, we crossed representative stocks to flies carrying the Su(var)2-502 allele of the gene encoding HP1. HP1 activity at the sites in question can be inferred by the observation that the variegating phenotype is suppressed (loss of silencing) in the presence of a mutation in HP1. This is observed whether the variegating line was generated as the consequence of transposition, local deletion, or local duplication (Fig. 7). Conversely, one might ask whether or not duplication of the HP1 gene might lead to a switch from a red-eye phenotype to variegation, as the Dp(2;2)P90 duplication is known to enhance the PEV induced by pericentric heterochromatin (41). However, tests using several different red-eyed lines found no effect (data not shown).
DISCUSSION
Reporter phenotypes on the fourth chromosome indicate a mosaic of heterochromatic and euchromatic domains and show that many genes lie within HP1-associated, heterochromatic domains.
The published DNA sequence of the fourth chromosome (15) shows a fairly uniform distribution of genes across region 101F-102F at a normal gene density (Fig. 2). However, this region is enriched in repetitious sequences compared to similar intervals on the other euchromatic chromosome arms (20, 23, 26). The average transposable element density is 10 to 15 per Mb in the major chromosome arms but over 82 per Mb for chromosome 4 due to an order-of-magnitude increase in remnants of long interspersed element (LINE)-like and TIR elements (elements that transpose via a DNA intermediate, flanked by short inverted repeats). The most abundant TIR in chromosome 4 is 1360, a repetitive sequence that is abundant in the chromocenter, pericentric heterochromatin, and the telomeres of the major chromosome arms (20). 1360 is the only transposable element found across the whole of the fourth chromosome, including the centromere and telomere (15).
While there appear to be some “hot spots” for P element insertion, heterochromatic domains detected by our assay are also distributed across the chromosome. Variegating inserts are not restricted to juxtaposition with repetitious DNA or even to gene-free regions. In fact, most (17 of 18) of the variegating P elements lie within 2 kb of a gene, and 10 variegating P elements lie within the transcribed portion of nine different genes (Table 2). Thus, the heterochromatic domains are not restricted to tandem repeat arrays; rather, the local pattern of dispersed repetitious elements, particularly 1360 in the region examined, appears to be critical for heterochromatin formation.
HP1, a consistent marker of heterochromatic domains, is prominently and extensively associated with the fourth chromosome, as shown by immunofluorescent staining of the polytene chromosomes (19). All of the insertion lines from this study showing a variegating phenotype that have been examined directly (Fig. 2) show a loss of silencing as a consequence of the introduction of a mutation (a hypomorph) in the gene for HP1. This group includes lines with the P element inserted into the genes BEST:CK01140, bt, and ATPsyn-beta. One can infer that a significant number of fourth-chromosome genes are packaged with HP1. While much of the Drosophila heterochromatin at centromeres and telomeres is made up of tandem repeats, classical genetics have identified several genes within the pericentric heterochromatin, and data from genome sequencing suggest that several hundred genes may reside in these regions in D. melanogaster (18). Many of the genes on the fourth chromosome have specific developmental functions and must have some developmental regulation superimposed on the effects of domain packaging. It will be of interest to determine how these genes function within a heterochromatic environment.
cis-Acting determinants of heterochromatin formation.
The switch in phenotype of the 2-M59A.R reporter transgene from a red-eye to a variegating phenotype (Fig. 3C) resembles the classical phenomenon of PEV, in which a euchromatic white gene is brought into juxtaposition with heterochromatin by a chromosomal rearrangement. In a current model, PEV reflects the spread of heterochromatin across the rearrangement breakpoint, with the euchromatic reporter gene being packaged into heterochromatin in a stochastic process. The model assumes the presence of initiators of heterochromatin formation within each heterochromatic domain and the presence of a barrier to the spread of heterochromatin, removed by rearrangement, normally separating the domains. HP1 appears designed to play a central role in such spreading, being able to recognize both a key histone modification, methylation of lysine 9 in histone H3, and histone H3-K9 methyltransferases, including SU(VAR)3-9 (33; reviewed in reference 16). This model is supported by recent findings for Schizosaccharomyces pombe showing that the spreading of heterochromatin upon removal of a putative boundary to the silent mating type region requires the yeast homologue of HP1 and the H3-K9 methyltransferase (17). In the analysis presented here, in contrast to classical PEV, the switch can be related to the loss of a relatively small fragment of DNA, pointing to local cis-acting determinants controlling heterochromatin spreading. As observed in other cases, the silencing is dependent on HP1, as shown by the loss of silencing upon the loss of HP1 in lines with small deletions (Fig. 7).
The results shown here also reveal the reciprocal effect, the conversion of a variegating to a red-eye phenotype upon the deletion of DNA flanking the reporter transgene (Fig. 3B). The underlying mechanism for the switch in phenotype involves a shift in the chromatin structure of the transgenes, shown by changes in XbaI sensitivity of the hsp26-pt gene (Fig. 6). This reciprocal local position effect suggests a model of competitive equilibrium between the two types of chromatin, rather than supporting the common perception that heterochromatin is a dominant form. In this model, the balance between heterochromatin and euchromatin may be determined by the presence and/or strength of nearby initiator elements for each form of chromatin, presumably acting to determine the modification state of the histone cores (Fig. 8). The effect of euchromatin initiator elements might explain some of the discrepancies in Fig. 5 and will need to be taken into account in developing a detailed model of chromatin packaging. The idea of a competitive equilibrium is supported by recent experiments demonstrating that an increase in transcription factor for a variegating reporter gene can antagonize heterochromatin silencing (2).
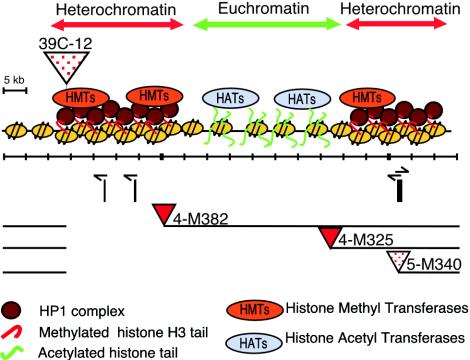
FIG. 8.
A model for the determination of chromatin structure. A map of the 102B region is shown, with the smallest and largest rightward deletions that result in a switch from the variegating 39C-12 transgene to a red-eye phenotype indicated as demarcating a euchromatic domain. Heterochromatic domains appear to be initiated from 1360 elements (shown as bars below the map), presumably targeted by the RNAi mechanism, localizing HP1 association with concomitant H3 modification for K9 methylation. Euchromatic domains may be initiated from regions having a high concentration of histone acetyltransferase activity. Both configurations can spread until stopped by competition.
The model cited above suggests that the switch in phenotype occurs upon the removal of a barrier normally constraining the heterochromatin from spreading. However, we have observed that a switch in phenotype can also occur as the consequence of a local duplication on the fourth chromosome. This seems unlikely to be due to the loss of a heterochromatin and euchromatin barrier, as no sequences have been lost (Fig. 4). In particular, we observe that the duplication of DNA fragments that appear to lie in heterochromatic domains of the genome can cause the switch from a variegating to a red-eye phenotype, indicating a local change in chromatin packaging as a consequence of the rearrangement (see lines 4-M979 and others in Fig. 4). The results are most readily explained by a model in which heterochromatin packaging is the result of proximity to an initiator of heterochromatin formation, which in this region of the genome is the 1360 element, and where packaging in that form spreads for about 10 kb (Fig. 5). This distance is similar to that reported for spreading of heterochromatin in S. pombe (17, 34). Deletions that lead to a loss of 1360 elements can result in a switch from a variegating to a red-eye phenotype (Fig. 3B and D), while those that position the P element close to a copy of 1360 result in a switch from a red eye to a variegating eye (Fig. 3C) or result in the maintenance of a variegating eye (Fig. 3D). Similarly, duplications that increase the distance between the P element and a copy of 1360 can switch a variegating eye to a red eye (Fig. 4B and D), while duplications that move a P element close to a copy of 1360 can switch a red eye to a variegating eye (Fig. 4C) or maintain a variegating eye (Fig. 4D). Note that while 1360 appears to play a dominant role in this region (ca. 200 kb), both the results obtained here and an examination of the fourth chromosome map we have generated (Fig. 2) suggest that other repetitious elements are also involved in targeting heterochromatin formation. There are relatively few 1360 elements in the distal portion of 102F; several lines carrying a variegating P element inserted in this region have been recovered. An investigation of this region, using the strategy reported above, is being undertaken. At the same time, the results presented here argue that not all fragments of repetitious elements can serve as initiators of heterochromatin formation.
Recent reports have indicated that numerous proteins, particularly those involved in chromatin modification to achieve the active form, such as subunits of Swi/Snf, Mediator, TFIID, and other complexes, have efficient barrier function. These activities, which result in histone acetylation and chromatin remodeling, have been shown in yeast to block the spread of silencing (28). The switch to a red-eye phenotype observed here as a consequence of a local deletion (the removal of two 1360 elements) is associated with chromatin remodeling (Fig. 6), presumably reflecting a change in the histone modification state (7). In a competitive system, transcription regulatory elements associated with high levels of histone acetylation might serve as initiators of euchromatin formation, creating barriers to the spread of heterochromatin (13, 27, 42), while sites that target HP1 association and methylation of H3-K9 would serve as initiators of heterochromatin formation. In this region of the genome, regulatory elements associated with gene CG2052 might be initiators of euchromatin formation.
How might the 1360 element target heterochromatin formation? Repetitious elements have been implicated as nucleation sites for heterochromatin formation in fungi and plants via an RNAi mechanism (12, 17, 39). Recent studies have demonstrated that heterochromatic silencing in Drosophila, including that found in line 39C-12, is dependent on the RNAi machinery. The silencing is lost as a result of mutations in piwi, abergine, or spindle-E (homeless), which encode RNAi components. The loss of silencing is concomitant with a loss in histone H3-mK9 and delocalization of HP1, suggesting a need for RNAi to target heterochromatin assembly (29). Interestingly, a 1360 element has recently been implicated in double-stranded RNA-mediated posttranscriptional silencing of Stellate via the Su(Ste) locus (3). Most of the 1360 elements present on the fourth chromosome (7 of 8 in Fig. 3, not including that marked with an X) are fragments that include the same 1360 transcription start site and thus are presumably competent to produce transcripts targeting double-stranded RNA-mediated silencing to the region of chromatin in which they are embedded. 1360 sequences have been identified among the small RNAs identified in Drosophila embryos, where heterochromatin structure is established (4). (Most other known repetitious elements are similarly identified in this pool [4] and may participate as well.) As an initial hypothesis, we propose that the 1360 elements can serve as drivers of heterochromatin formation along the fourth chromosome, most likely through an RNAi-based mechanism. Several tests of this hypothesis are under way.
Acknowledgments
We thank Lori L. Wallrath (University of Iowa) for important contributions to the early stages of this work and for communication (with Diane Cryderman) of unpublished results; Jeremy Buhler (Department of Computer Science and Engineering, Washington University) for assistance with the analysis of repeats; and Elizabeth Slawson, Anne Beckert, and Peter Weitzel for assistance with the genetic screens.
This work was supported by National Institutes of Health grants HD23844 and GM068388 (to S.C.R.E.) and GM57005 (to J.C.E.) and by institutional support from the Friedrich Meischer Institute (to F.-L.S.). K.H. is supported in part by a Chancellor's Fellowship from Washington University, and C.L.S. was supported in part by a Summer Undergraduate Research Fellowship under a grant from the Howard Hughes Medical Institute to Washington University.
REFERENCES
- 1.Adams, M. D., and J. J. Sekelsky. 2002. From sequence to phenotype: reverse genetics in Drosophila melanogaster. Nat. Rev. Genet. 3:189-198. [DOI] [PubMed] [Google Scholar]
- 2.Ahmad, K., and S. Henikoff. 2001. Modulation of a transcription factor counteracts heterochromatic gene silencing in Drosophila. Cell 104:839-847. [DOI] [PubMed] [Google Scholar]
- 3.Aravin, A. A., N. A. Naumova, A. V. Tulin, V. V. Vagin, Y. M. Rozosky, and V. A. Gvozdev. 2001. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol. 11:1017-1027. [DOI] [PubMed] [Google Scholar]
- 4.Aravin, A. A., M. Lagos-Quintana, A. Yalcin, M. Zavolan, D. Marks, B. Snyder, T. Gaasterland, J. Meyer, and T. Tuschl. 2003. The small RNA profile during Drosophila melanogaster development. Dev. Cell 5:337-350. [DOI] [PubMed] [Google Scholar]
- 5.Barigozzi, C., S. Dolfini, M. Fracacaro, G. Rezzonico-Raimondi, and L. Tiepolo. 1966. In vitro study of the DNA replication patterns of somatic chromosomes of Drosophila melanogaster. Exp. Cell Res. 43:231-234. [DOI] [PubMed] [Google Scholar]
- 6.Bartolome, C., X. Maside, and B. Charlesworth. 2002. On the abundance and distribution of transposable elements in the genome of Drosophila melanogaster. Mol. Biol. Evol. 19:926-937. [DOI] [PubMed] [Google Scholar]
- 7.Berger, S. 2002. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12:142-148. [DOI] [PubMed] [Google Scholar]
- 8.Bridges, C. B. 1935. The mutants and linkage data of chromosome four of Drosophila melanogaster. Biol. Zh. 4:401-420. [Google Scholar]
- 9.Cartwright, I. L., D. E. Cryderman, D. S. Gilmour, L. A. Pile, L. L. Wallrath, J. A. Weber, and S. C. R. Elgin. 1999. Analysis of Drosophila chromatin structure in vivo. Methods Enzymol. 304:462-496. [DOI] [PubMed] [Google Scholar]
- 10.Cryderman, D. E., M. H. Cuaycong, S. C. R. Elgin, and L. L. Wallrath. 1998. Characterization of sequences associated with position-effect variegation at pericentric sites in Drosophila heterochromatin. Chromosoma 107:277-285. [DOI] [PubMed] [Google Scholar]
- 11.Cryderman, D. E., E. J. Morris, H. Biessman, S. C. R. Elgin, and L. L. Wallrath. 1999. Silencing at Drosophila telomeres: nuclear organization and chromatin structure play critical roles. EMBO J. 18:3724-3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dernburg, A. F., and G. H. Karpen. 2002. A chromosome RNAissance. Cell 111:159-162. [DOI] [PubMed] [Google Scholar]
- 13.Donze, D., C. R. Adams, J. Rine, and R. T. Kamakaka. 1999. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev. 13:698-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eissenberg, J. C., and S. C. R. Elgin. 2000. The HP1 protein family: getting a grip on chromatin. Curr. Opin. Genet. Dev. 10:204-210. [DOI] [PubMed] [Google Scholar]
- 15.FlyBase Consortium. 2003. The FlyBase database of the Drosophila genome projects and community literature. Nucleic Acids Res. 31:172-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grewal, S., and S. C. R. Elgin. 2002. Heterochromatin: new possibilities for the inheritance of structure. Curr. Opin. Genet. Dev. 12:178-187. [DOI] [PubMed] [Google Scholar]
- 17.Hall, I. H., G. D. Shankaranarayana, K. Noma, N. Ayoub, A. Cohen, and S. I. Grewal. 2002. Establishment and maintenance of a heterochromatin domain. Science 297:2232-2237. [DOI] [PubMed] [Google Scholar]
- 18.Hoskins, R. A., C. D. Smith, J. W. Carlson, A. B. Carvalho, A. Halpern, J. S. Kaminker, C. Kennedy, C. J. Mungall, B. A. Sullivan, G. G. Sutton, J. C. Yasuhara, B. T. Wakimoto, E. W. Myers, S. E. Celniker, G. M. Rubin, and G. H. Karpen. 31 December 2002, posting date. Heterochromatic sequences in a Drosophila whole-genome shotgun assembly. Genome Biol. 3:research0085.1-0085.16. [Online.] http://genomebiology.com/2002/3/12/RESEARCH/0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.James, T. C., J. C. Eissenberg, C. Craig, V. Dietrich, A. Hobson, and S. C. R. Elgin. 1989. Distribution patterns of HP1, a heterochromatin-associated nonhistone chromosomal protein of Drosophila. Eur. J. Cell Biol. 50:170-180. [PubMed] [Google Scholar]
- 20.Kaminker, J. S., C. M. Bergman, B. Kronmileer, J. Carlson, R. Svirskas, S. Patel, E. Frise, D. A. Wheeler, S. Lewis, G. M. Rubin, M. Ashburner, and S. E. Celniker. 23 December 2002, posting date. The transposable elements of the Drosophila melanogaster euchromatin: a genomics perspective. Genome Biol. 3:research0084.1-0084.20. [Online.] http://genomebiology.com/2002/3/12/research/0084. [DOI] [PMC free article] [PubMed]
- 21.Khesin, R. B., and B. A. Leibovitch. 1978. Influence of deficiency of the histone gene-containing 38B-40 region on X-chromosome template activity and the white gene position effect variegation in Drosophila melanogaster. Mol. Gen. Genet. 162:323-328. [DOI] [PubMed] [Google Scholar]
- 22.Locke, J., and H. McDermid. 1993. Analysis of Drosophila chromosome 4 using pulsed field gel electrophoresis. Chromosoma 102:718-723. [DOI] [PubMed] [Google Scholar]
- 23.Locke, J., L. Podemski, K. Roy, D. Pilgrim, and R. Hodgetts. 1999. Analysis of two cosmid clones from chromosome 4 of Drosophila melanogaster reveals two new genes amid an unusual arrangement of repeated sequences. Genome Res. 9:137-149. [PMC free article] [PubMed] [Google Scholar]
- 24.Lu, B. Y., J. Ma, and J. C. Eissenberg. 1998. Developmental regulation of heterochromatin-mediated gene silencing in Drosophila. Development 125:2223-2234. [DOI] [PubMed] [Google Scholar]
- 25.Lu, Q., L. L. Wallrath, B. D. Allan, R. L. Glaser, J. T. Lis, and S. C. R. Elgin. 1992. Promoter sequence containing (CT)n.(GA)n repeats is critical for the formation of the DNase I hypersensitive sites in the Drosophila hsp26 gene. J. Mol. Biol. 225:985-998. [DOI] [PubMed] [Google Scholar]
- 26.Miklos, G. L. G., M.-T. Yamamoto, J. Davies, and V. Pirrotta. 1988. Microcloning reveals a high frequency of repetitive sequences characteristic of chromosome 4 and the β-heterochromatin of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 85:2051-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mutskov, V. J., C. M. Farrell, P. A. Wade, A. P. Wolffe, and G. Felsenfeld. 2002. The barrier function of an insulator couples high histone acetylation levels with specific protection of promoter DNA from methylation. Genes Dev. 16:1540-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oki, M., L. Valenzuela, T. Chiba, T. Ito, and R. T. Kamakaka. 2004. Barrier proteins remodel and modify chromatin to restrict silenced domains. Mol. Cell. Biol. 24:1956-1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pal-Bhadra, M., B. S. Leibovitch, S. G. Gandhi, M. Rao, U. Bhadra, J. A. Birchler, and S. C. R. Elgin. 2004. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science 303:669-672. [DOI] [PubMed] [Google Scholar]
- 30.Preston, C. R., J. A. Sved, and W. R. Engels. 1996. Flanking duplications and deletions associated with P-induced male recombination in Drosophila. Genetics 144:1623-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richards, E. J., and S. C. R. Elgin. 2002. Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell 108:489-500. [DOI] [PubMed] [Google Scholar]
- 32.Robertson, H. M., C. R. Preston, R. W. Phillis, D. M. Johnson-Schlitz, W. K. Benz, and W. R. Engels. 1988. A stable genomic source of P element transposase in Drosophila melanogaster. Genetics 118:461-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schotta, G., A. Ebert, V. Krauss, A. Fischer, J. Hoffmann, S. Rea, T. Jenuwein, R. Dorn, and G. Reuter. 2002. Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J. 21:1121-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schramke, V., and R. Allshire. 2003. Hairpin RNAs and retrotransposon LTRs effect RNAi and chromatin-based gene silencing. Science 301:1069-1074. [DOI] [PubMed] [Google Scholar]
- 35.Shaffer, C. D., J. M. Wuller, and S. C. R. Elgin. 1994. Raising large quantities of Drosophila for biochemical experiments. Methods Cell Biol. 44:99-108. [DOI] [PubMed] [Google Scholar]
- 36.Sun, F.-L., M. H. Cuaycong, C. A. Criag, L. L. Wallrath, J. Locke, and S. C. R. Elgin. 2000. The fourth chromosome of Drosophila melanogaster: interspersed euchromatic and heterochromatic domains. Proc. Natl. Acad. Sci. USA 97:5340-5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun, F.-L., M. H. Cuaycong, and S. C. R. Elgin. 2001. Long-range nucleosome ordering is associated with gene silencing in Drosophila melanogaster pericentric heterochromatin. Mol. Cell. Biol. 21:2867-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Leeuwen, F., and D. E. Gottschling. 2003. The histone minority report: the variant shall not be silenced. Cell 112:591-593. [DOI] [PubMed] [Google Scholar]
- 39.Volpe, T. A., C. Kidner, I. M. Hall, G. Teng, S. I. Grewal, and R. A. Martienssen. 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297:1833-1837. [DOI] [PubMed] [Google Scholar]
- 40.Wallrath, L. L., and S. C. R. Elgin. 1995. Position effect variegation in Drosophila is associated with an altered chromatin structure. Genes Dev. 9:1263-1277. [DOI] [PubMed] [Google Scholar]
- 41.Wustmann, G., J. Szidonya, H. Taubert, and G. Reuter. 1989. The genetics of position-effect variegation modifying loci in Drosophila melanogaster. Mol. Gen. Genet. 217:520-527. [DOI] [PubMed] [Google Scholar]
- 42.Yu, Q., R. Qui, T. B. Foland, D. Griesen, C. S. Galloway, Y.-H. Chiu, J. Sandmeier, J. R. Broach, and X. Bi. 2003. Rap1p and other transcriptional regulators can function in defining distinct domains of gene expression. Nucleic Acids Res. 31:1224-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]