Abstract
To gain a better understanding of the nature of active chromatin in mammals, we have characterized in living cells the various chromatin modification events triggered by the glucocorticoid receptor (GR) at the rat tyrosine aminotransferase gene. GR promotes a local remodeling at a glucocorticoid-responsive unit (GRU) located 2.5 kb upstream of the transcription start site, creating nuclease hypersensitivity that encompasses 450 bp of DNA. Nucleosomes at the GRU occupy multiple frames that are remodeled without nucleosome repositioning, showing that nucleosome positioning is not the key determinant of chromatin accessibility at this locus. Remodeling affects nucleosomes and adjacent linker sequences, enhancing accessibility at both regions. This is associated with decreased interaction of both the linker histone H1 and the core histone H3 with DNA. Thus, our results indicate that nucleosome and linker histone removal rather than nucleosome repositioning is associated with GR-triggered accessibility. Interestingly, GR induces hyperacetylation of histones H3 and H4, but this is not sufficient either for remodeling or for transcriptional activation. Finally, our data favor the coexistence of several chromatin states within the population, which may account for the previously encountered difficulties in characterizing unambiguously the active chromatin structure in living cells.
Modulation of the accessibility of chromatin to transcriptional regulators and to the transcription machinery is key to the regulation of gene expression in eukaryotes. Sequence-specific transcriptional regulators employ a variety of coregulators that act enzymatically on chromatin. Some of the coregulators catalyze covalent modifications of histones, in particular of the histone N-terminal tails. Such modifications may modulate the interaction of the tails with DNA and create binding sites for additional proteins acting on chromatin (20). Other coregulators act on chromatin without catalyzing covalent modifications of the histones. They consist of enzyme complexes that loosen the interaction of DNA with the nucleosome in an ATP-dependent manner, thereby bringing “fluidity” to chromatin (27). They are believed to be responsible for the DNase I-hypersensitive sites that are often found at active regulatory sequences (15). Histone acetyltransferases and ATP-dependent remodeling complexes often cooperate in a context-dependent manner in the regulation of gene expression (27). For some genes, remodeling complexes seem required for subsequent histone acetylation, presumably to unlock a compact chromatin structure (21). For others, histone acetylation appears to precede and to favor the action of chromatin-remodeling complexes by creating a binding site for the bromodomain found in subunits of these complexes (18).
The exact nature of the nucleosomal modifications and the fate of the remodeled nucleosomes are not clear. In vitro, the ATP-dependent remodeling complexes induce a conformational change of the nucleosomes that renders them accessible to nucleases without displacement of histones (references 4 and 27 and references therein). Then, depending on the experimental conditions, these altered nucleosomes can either slide to a nearby position, be transferred to a DNA or protein acceptor, or remain altered at their original position. In vivo analyses of different biological systems using diverse experimental approaches have provided results compatible with either of these possibilities (e.g., see references 4, 10, 22, 31, and 37). It is not clear how the differences in the biological systems and in the experimental approaches contribute to the diversity of the observations. In the case of the PHO5 promoter in yeast, two recent studies combining a multiplicity of experimental approaches have concluded that nucleosomes were completely unfolded upon activation (4, 31). This complete unfolding appears to be responsible for the absence of histone modification detected at the PHO5 promoter using chromatin immunoprecipitation (ChIp) (31). The latter observation could indicate that nucleosome unfolding is not a general outcome of chromatin remodeling, since hyperacetylated histones have been found at the vast majority of active regulatory regions, in particular in higher eukaryotes. However, parallel analyses of a single biological system with a multiplicity of experimental approaches are currently too rare to allow assessment of the generality of the observations.
Our understanding of transcriptional regulation in higher eukaryotes owes a lot to the study of nuclear receptors. These ligand-activated transcriptional regulators have enabled kinetic analyses of the transcriptional regulation process in vitro and in vivo, as well as the identification of transcriptional coregulators in two-hybrid screens (12). They employ both ATP-dependent remodeling complexes and a wide variety of histone acetyltransferases during the transcription activation process, and often trigger the appearance of nuclease-hypersensitive sites at target regulatory sequences. The events occurring at two target genes of one of the nuclear receptors, the glucocorticoid receptor (GR), have been investigated extensively in cultured cells. These targets are the long terminal repeat (LTR) of the mouse mammary tumor virus (MMTV) and the glucocorticoid responsive unit (GRU) of the rat tyrosine aminotransferase gene (Tat) (13, 17). At both of these sites, GR triggers the appearance of a DNase I-hypersensitive site and the parallel recruitment of sequence-specific transcriptional regulators. However, in the MMTV LTR, GR-induced chromatin remodeling takes place at the proximal promoter, whereas in the Tat gene it occurs at a remote enhancer 2.5 kb upstream of the transcription start site (13).
Studies with living cells of the remodeling events taking place at the MMTV LTR and the Tat GRU have led to conflicting results (10, 30, 37). In particular, it is still not clear whether nucleosome positioning contributes to the modulation of transcription factor recruitment and whether nucleosomes are fully displaced upon remodeling. Nucleosome positioning is a popular expectation, since, in principle, it could modulate with precision the access of transcription factors to their sites (22, 29). In contrast, if nucleosomes are not strictly positioned at a regulatory sequence but transcription factor access is still controlled, other types of chromatin structural features must modulate DNA accessibility.
Here, we have investigated the GR-triggered chromatin remodeling events at the Tat GRU, in living cells, to determine nucleosome positioning and displacement and the state and role of histone tail acetylation. We have used a combination of various experimental approaches to circumvent the biases of each individual approach. We have thereby assessed the heterogeneity of the remodeled states and characterized their nature. In particular, we used RNA-fluorescence in situ hybridization (FISH) to quantify the fraction of actively transcribed genes, both low- and high-resolution MNase cleavage analyses, quantitative PCR analyses of MNase digestion, and high-resolution DNase I analyses to monitor nucleosome positioning and remodeling, and finally, chromatin immunoprecipitation to monitor changes in histone H3 and H4 acetylation levels as well as histone H1 and H3 interaction. Our results indicate that nucleosome unfolding rather than repositioning is associated to GR-triggered accessibility.
MATERIALS AND METHODS
Cells, nuclease treatment, and footprinting analyses.
To avoid distortion of the results due to variation in the methylation status of the −2.5 Tat gene GRU, we used H4II rat hepatoma cells where the GRU had been demethylated by a previous 3-day GC treatment, at least a month before the analysis (36). Nuclei were prepared, treated by nucleases, and analyzed as described previously (6, 36). To map MNase cleavages, we used an adaptation of ligation-mediated PCR (LM-PCR) that allows selective detection of the blunt-ended fragments, which presumably allows enrichment for cleavages occurring within the linker region (6, 37). Primers used for amplification and labeling were, respectively the following: set 1, CAGTGTTCTCTATCACAGGGAGAGC and TATCACAGGGAGAGCTGTCAGCCCCTG; set 2, TGGGATAGTTTTCTAGACATAGAACCA and CTAGACATAGAACCACATTCCAGGGGCT; set 3, TAAATAACAGGAAGCCCAAGGTTTAC and GAAGCCCAAGGTTTACCAATCTCTGCTG; set 4, TTCAGATTTTCTATTGCAAATAAAGTAG and TCTATTGCAAATAAAGTAGCTAGAACATCCTG; set 5, TAGCCCTGTAATCCCAGCATTTGG and TGTAATCCCAGCATTTGGGAAGCTGAGGT; and set 6, CACCTCAGCTTCCCAAATGCTGGG and AGCTTCCCAAATGCTGGGATTACAGGGCT.
RNA FISH analysis.
FISH analysis was carried out as described previously, using H4II cells grown on gelatin-coated glass slides (34). The antisense RNA probes used were a 1.5-kb probe spanning introns 6 and 7 and exon 7 (52 bp) of the rat Tat gene and a 2-kb Tat cDNA probe labeled with digoxigenin-11-UTP and biotin-16-UTP, respectively.
ChIp analysis.
Cells were cross-linked and further processed as described previously (7). Sonication conditions were established to shear DNA to an average size of 300 bp. For MNase treatment, nuclei were prepared from scraped cross-linked cells and treated as described previously (6). Nuclei from 1.5 × 107 cells were incubated in 400 μl of 15 mM Tris-HCl (pH 7.5), 60 mM KCl, 15 mM NaCl, 0.5 mM dithiothreitol, 4 mM MgCl2, 1 mM CaCl2, 400 U of MNase, and 40 μg of RNase A for 15 min at 25°C. MNase was stopped by adding four volumes of 20 mM Tris-HCl (pH 8), 165 mM NaCl, 5 mM EDTA, 0.25 mM EGTA, 0.125% sodium dodecyl sulfate, and 1.25% Triton, and the chromatin preparation was further processed as described above. Immunoprecipitation of the lysate from 1.5 × 106 cells was performed in 200 μl of lysis buffer using 5 μl of anti-acetylated H3 or H4 antibodies (Upstate Biotech), anti-H1 (antiserum 355 [35], a kind gift of S. Muller), and anti-green fluorescent protein (GFP) (polyclonal antibody; Clontech). Immunocomplexes were recovered and processed as described previously (7). Real-time quantitative PCR was performed by using a LightCycler and a SYBR-Green-I-containing PCR mix, following the recommendations of the manufacturer (Roche). The immunoprecipitated material was quantified relative to a standard curve of H4II genomic DNA. Primers used were the following: Tat −7, GTCCGAATGGCAGACCTTGTAT and GAGCCAGCCTCTCATCTCTCA; Tat −4.7, ATAAGACCCATAGAGTACCAAGCTGA and CAATTCTTTCTGCCCATGATTAAC; Tat N2, TTTCCCCATGTCCAACAAGACTA and CCCACCTCAGCTTCCCAAAT; Tat N3, AATAACAGGAAGCCCAAGGTTTAC and TGGACATGGGGAAACTTTCAG; Tat N4 (−2.5), TGAAGTCTCTTCTCAGTGTTC and GGCTTCCTGTTATTTATGGATAGTT; Tat N5, ATGGGACAGGGCAGCGAC and GACAGCTCTCCCTGTGATAGAGAA; Tat −1.4, CCACATATCAAGGATCAAAAGTCAA and GCCGGCTGTACATACTTTAGAGTT; Tat promoter, GGGACTTAGTTCTCACTCTCAACCA and TGATGATGAGCTCAGGTGCAGT; globin, TGTAGAGCCACACCCTGGTATTG and GCAAATGTGAGGTGCGACTGA; nucleolin, GTGGAGGTGAGGTCACGAGAA and CCAGCTACAGCTAGCGTCTGAG.
The cell line expressing the GFP-tagged histone H3 was obtained following transfection of H4II cells by the calcium phosphate coprecipitation procedure. We used a vector containing a puromycin selection gene and an H3-GFP fusion (a kind gift of S. Henikoff [2]) controlled by mammalian regulatory sequences (map available upon request). Clones were selected using 80 μg of puromycin/ml and characterized for GFP-tagged histone H3 expression under a microscope and for Tat gene inducibility. Clones that showed homogenous GFP expression within the population and where both basal and induced TAT levels were similar to the parental H4II cells were further analyzed.
RESULTS
Most Tat gene copies are actively engaged in transcription upon a 2-day GC treatment.
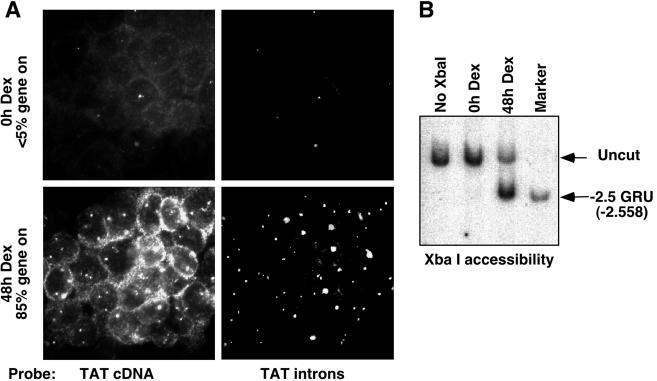
The Tat gene is a single-copy gene located, in the rat genome, on chromosome 19. It is expressed specifically in liver cells, where its transcription is activated by GC through two remote GRUs located 2.5 and 5.5 kb upstream of the transcription start site (14). At the −2.5 GRU, GR induces chromatin remodeling characterized by the appearance of a DNase I-hypersensitive site, leading to the recruitment of several transcription factors (13, 32). Here, we present a detailed analysis with living cells of this chromatin remodeling event occurring at the endogenous gene. We used a well-characterized aneuploid rat hepatoma cell line, H4II, which carries three copies of chromosome 19 and thus of the Tat gene. To ensure that analysis of the GC-induced remodeled state of chromatin at the −2.5 GRU is minimally affected by signals originating from unremodeled copies, we established experimental conditions that allow maximal involvement of Tat gene copies into active transcription complexes. Active foci of Tat gene transcription were visualized using RNA FISH analysis of nascent transcripts (34). Tat mRNA and pre-mRNA were detected in parallel, using an antisense Tat cDNA probe labeled with biotin and revealed with Texas Red and an antisense Tat intron probe labeled with digoxigenin and revealed with fluorescein. The cDNA probe detected both pre-mRNAs and mature mRNAs, whereas the intron probe detected only the pre-mRNAs that accumulated at the site of transcription (Fig. 1A). In H4II cells, cultured in the absence of GCs, most Tat gene copies were not actively transcribed, and RNA-FISH analysis detected only a few foci of transcription, which represented <5% of the Tat gene copies. Kinetic analysis of the GC response revealed that optimal activation was achieved following a 2-day stimulation, with about 80% of the Tat gene copies being actively transcribed (Fig. 1A and data not shown). In some cells, not all Tat gene copies were active, and in others, none were. Still, the accumulation of Tat mRNA in the cytoplasm of cells that had no detectable active foci reveals that the Tat gene had been induced during the 2-day GC treatment.
FIG. 1.
Prolonged GC treatment induces optimal transcriptional induction of the Tat gene and chromatin remodeling at the −2.5 Tat GRU. (A) RNA FISH analysis of nascent and mature Tat transcripts in H4II cells treated with 10−7 M Dex, or left untreated, for 2 days. To quantify the percentage of activated gene copies, the number of transcription foci in about 200 cells was determined. (B) Chromatin accessibility at the −2.5 Tat GRU as assessed by sensitivity to restriction enzyme cleavage of H4II nuclei and indirect end labeling. “Marker” corresponds to genomic DNA cleaved with XbaI in vitro.
The percentage of remodeled GRUs was assessed by using a restriction enzyme accessibility test with a saturating amount of enzyme. Among the various enzymes that cleave within the GRU in nuclei, XbaI appears to be most sensitive to changes in chromatin structure induced by GR (30). Using XbaI as a probe, chromatin accessibility within the GRU paralleled the percentage of active gene copies that were detected by RNA-FISH: <5% of the Tat gene GRUs were cleaved in nuclei of unstimulated H4II cells, whereas about 80% of the GRUs were accessible following a 2-day GC stimulation (Fig. 1B). Thus, upon a 2-day GC treatment, most of the Tat gene copies had remodeled chromatin at the −2.5 GRU and were actively transcribing.
MNase cleavage analyses reveal multiple nucleosomal frames and only partial disruption of nucleosomal organization upon remodeling.
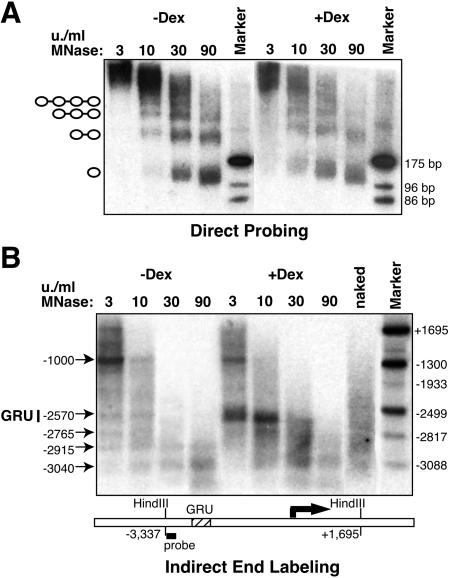
We used MNase to map nucleosomal boundaries and to assess the organization of DNA into chromatin as follows: nuclei of H4II cells treated with GC for 2 days or left untreated were incubated with various amounts of MNase, and the resulting DNA was analyzed at low and high resolutions. First, DNA was separated on an acrylamide gel and probed directly with a ∼200-bp fragment encompassing the −2.5 GRU (Fig. 2A and 3D). In the absence of GCs, the GRU was organized in a nucleosomal array, with both typical and slightly trimmed mononucleosomes being produced at a high concentration of MNase (90 U/ml). Upon GR-induced remodeling, the nucleosomal ladder appeared similar, with no subnucleosomal fragment being produced. However, there was more smear in the nucleosomal ladder, and the overall signal intensity was reduced by 60%, as determined by phosphorimager analysis of several independent experiments. This intensity reduction, as well as smeared DNA, were already apparent using low MNase concentrations (3 U/ml) and were specific for the −2.5 GRU, since they were not observed when either the total genomic DNA population or another Tat 5′-flanking region was analyzed (data not shown). These observations indicate that upon remodeling, the canonical nucleosomal organization has been disrupted at the −2.5 GRU, affecting about 60% of the nucleosomes within the population. This MNase-sensitive fraction is reproducibly smaller than the XbaI-sensitive fraction (80%; Fig. 1B), even though the XbaI site is located within the ∼200-bp fragment used to probe the MNase-treated samples (Fig. 3D). It also differs from a previous analysis, where it was observed that upon a very extensive MNase cleavage, most of the nucleosomal DNA of the GRU could be degraded following remodeling (30). Indeed, a similarly extensive MNase digestion (one order of magnitude harsher) in our system led also to about 80% of the remodeled chromatin being degraded (data not shown).
FIG. 2.
Low-resolution MNase cleavage analysis of the nucleosomal organization at the −2.5 Tat GRU. (A) Direct probing reveals partial GC-induced disruption of the nucleosomal organization. Nuclei were treated with various concentrations of MNase, and the corresponding DNA was separated on a polyacrylamide gel. A −2.5 Tat GRU probe (−2622 to −2417) was used. “Marker” corresponds to genomic DNA cleaved with AluI, giving rise upon probing to 175-, 96-, and 86-bp-long fragments, as indicated. (B) Indirect end labeling does not reveal precise nucleosome positioning at the −2.5 Tat GRU. The MNase-treated genomic DNA was cleaved with HindIII, separated on an agarose gel, and hybridized with a HindIII-EcoRI Tat gene probe (−3337 to −3047), as shown on the diagram below, depicting the end-labeling strategy. “Marker” corresponds to a mixture of genomic DNA doubly digested with HindIII and a second restriction enzyme generating bands at the indicated position. The deduced locations of the preferred MNase cleavage sites are shown on the left.
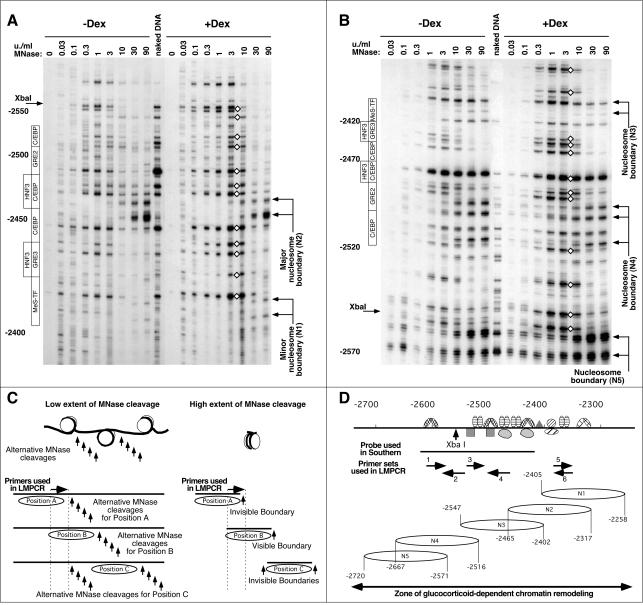
FIG. 3.
High-resolution MNase cleavage analysis reveals multiple nucleosomal frames and partial disruption of nucleosomal organization without nucleosome repositioning upon remodeling. (A) LM-PCR analysis of the MNase cleavage pattern at the −2.5 GRU using primer set no. 6. The locations of the transcription factor binding sites are indicated on the left. The locations of the deduced boundaries of nucleosome N1 and N2 are indicated on the right. Diamonds indicate GC-induced hyperreactivity. (B) LM-PCR analysis of the MNase cleavage pattern at the −2.5 GRU using primer set no. 1. The boundary of nucleosome N3 is only faintly visible here, as primers from set no. 1 hybridize to the corresponding 3′ linker region, but the boundaries of this nucleosome are clearly detected with primer sets no. 3 and no. 4 (see panel D). (C) Schematic interpretation of the cleavage patterns obtained at low and high extents of MNase cleavage for three arbitrary nucleosome positions (A, B, or C). The LM-PCR primers hybridize to a region that is fully protected only by nucleosome B. See the text for a detailed description. (D) Representation of the location of the nucleosome positions detected, as well as depiction of the primer sets used, of the transcription factor binding sites (36), of the probe used in Fig. 2A, and of the area of GC-dependent remodeling as revealed from the high-resolution DNase I analysis shown in Fig. 6C.
Altogether, these results indicate heterogeneity in the chromatin status of the GRU upon remodeling, and it appears possible to group the population in three categories. One fraction behaves as unremodeled chromatin, undistinguishable from that present in the absence of GC treatment, since it is resistant to both MNase and XbaI cleavage. It corresponds to about 20% of the population, a value similar to the fraction of inactive copies detected by RNA-FISH analysis. One fraction appears extensively remodeled, being highly susceptible to XbaI and MNase cleavage. This fraction corresponds to about 60% of the population. A third fraction with more ambiguous behavior can be deduced from the discrepancies in the various quantitative analyses. It would correspond to the fraction of active genes, sensitive to XbaI cleavage but relatively resistant to MNase cleavage. The corresponding DNA could be wrapped around nucleosomes in a particular manner, so that its accessibility depended on the nuclease and enzymatic conditions. Some of the results presented below are also in favor of the existence of this third fraction.
Indirect-end labeling analysis revealed additional features of the chromatin structure at the GRU (Fig. 2B). If nucleosomes were strictly positioned, a regular ladder should be seen. In the absence of GCs, only a faint ladder was detected, indicating some preferred nucleosome positioning, particularly between −3000 and −2570, i.e., upstream the GRU. Bands were weak within a smear (compare with the clear background obtained with the same DNA samples upon direct probing in Fig. 2A), and a clear band was obtained only at −1000, where a DNase I-hypersensitive site is known to be present (14). Upon GC stimulation, chromatin remodeling at the GRU was visible as an MNase-hypersensitive site detectable with low concentrations of MNase, confirming that a large proportion of the −2.5 GRU was highly sensitive to MNase cleavage upon remodeling. No clear nucleosome positioning, nor remodeling-induced repositioning, was detected in this assay.
Next, we sought to establish whether a more subtle nucleosome positioning could be detected using high-resolution LM-PCR analysis. We used the LM-PCR strategy developed by Zaret and collaborators (24), which maps exclusively the MNase cleavage products with blunt ends that are generated preferentially within the linker region (6, 24). To detect any population, irrespective of its sensitivity to MNase cleavage, we used up to eight different MNase concentrations, each concentration differing by a factor of 3 within 4 orders of magnitude (Fig. 3). Figure 3A and B present the data obtained with two representative primer sets hybridizing within the −2.5 GRU. At low concentrations of nuclease, (<3 U/ml), cleavages were evenly distributed throughout the region analyzed, and the pattern is not very different from that of naked DNA. At high concentrations (30 and 90 U/ml, i.e., concentrations that allow production of mononucleosome length fragments within the GRU [Fig. 2 A]), many bands disappeared in favor of a limited number of new bands, which were often absent from the naked DNA control. These bands are thus likely to correspond to trimming of the linker DNA down to the nucleosomal boundary. In support of this interpretation, bands at a distance of 147 ± 2 bp were detected at the same MNase concentrations, using primer sets designed to detect the other end of the nucleosome (see Fig. 3D for the arrangements of the six primer sets used in this study and for the positions of the boundaries detected). In addition, these bands were often seen in doublets, differing by ∼10 bp. Our estimates of the distance that separated the cleavages at the two ends of a nucleosome suggest that the lower bands corresponded to trimming of the first helical turn of the DNA to the entry point of the nucleosome. Importantly, several of the primer sets allowed the detection of fragments generated by low MNase concentrations that were shorter than those visible at high concentration, i.e., corresponding to cleavage within regions that would be identified as nucleosomal DNA based on the extensive MNase digestion pattern (e.g., see Fig. 3A and B). At first glance, this is counterintuitive, but this behavior actually reflects the existence of several nucleosomal positions. The scheme in Fig. 3C presents the interpretation of the cleavage pattern that supports this conclusion. Three arbitrary nucleosomal positions (termed A, B, and C) are represented, as well as the MNase cleavages that could be obtained at either low or high extent of cleavage. The primer set used in this example hybridizes to a region included within nucleosome position B. Cleavage at low MNase concentrations should occur at random positions within the accessible zones, i.e., the linkers, while cleavage at high MNase concentrations should degrade all linker DNA. Thus, at low MNase concentrations, bands corresponding to limited cleavage within either of the linkers corresponding to nucleosome positions A, B, and C should be visible, and bands would be distributed throughout the region downstream of the LM-PCR primers. In contrast, upon extensive MNase cleavage, since the linker would be trimmed on both ends of the nucleosomal DNA, a specific primer set should allow detection solely of the boundaries of the nucleosomes that protect the primer-hybridizing region (nucleosome B in the example represented in Fig. 3C). Consequently, the bands corresponding to cleavage within the linker position A that are shorter than those corresponding to linker position B would disappear. Thus, in the example shown in Fig. 3A, the bands visible at low MNase concentrations that are shorter than those corresponding to nucleosome boundary N2 visible at high concentrations must correspond to the linker of alternatively positioned nucleosome(s) in the population, either the minor position N1 or, possibly, another one that does not protect the primer binding region and thereby is not visible here. We analyzed the extensive MNase digestion experiment with six sets of primers distributed throughout the GRU to validate the various nucleosome positions detected. This allowed us to detect up to five, mutually exclusive nucleosome positions at the −2.5 GRU, as outlined in Fig. 3D. Our analysis provides a minimal estimate of the diversity of positions occupied, since additional primer sets could have allowed detection of additional positions. Note that the analysis of a single MNase concentration with a limited number of primer sets could give the false impression that there was a single position.
Information about the nature of the GC-induced nucleosome remodeling can also be obtained from the high-resolution MNase cleavage pattern (Fig. 3A and B). At high MNase concentrations (90 U/ml), when some nucleosomes were still associated with both the transcriptionally inactive gene copies and a fraction of the active copies (Fig. 2A), the distribution of the nucleosome boundaries was the same as in unremodeled (−Dex) nucleosomes. This finding indicates that there is no modification of the distribution of the nucleosome frames upon remodeling. At low and medium MNase concentrations, the hypersensitivity visible at low resolution (Fig. 2B) was distributed throughout the GRU, as evidenced by multiple bands of increased intensity (Fig. 3 A and B). These bands did not differ from the bands visible at medium MNase concentrations in the absence of hormone (0.3 to 3 U/ml); they were just detected at a lower MNase concentration. The corresponding bands were also obtained upon MNase cleavage of naked DNA in vitro, even though the relative intensity of the bands differed. Thus, the chromatin fraction that was hypersensitive to MNase cleavage resembled naked or linker DNA, indicating that it corresponded rather to DNA loosely bound to chromatin proteins than to a bona fide nucleosome.
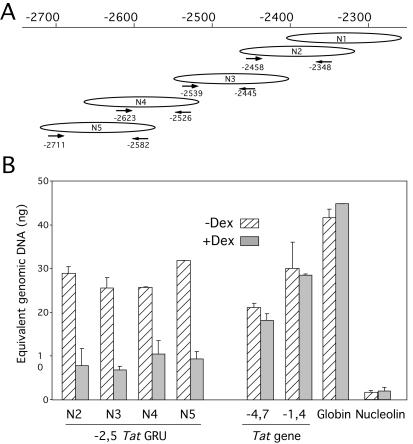
LM-PCR is not quantitative enough to compare the relative proportions of the different nucleosome phases detected. Thus, we established an assay for quantifying the relative proportion of each nucleosome frame, using real-time PCR. Cells were first fixed with formaldehyde to prevent any putative nucleosome redistribution during treatment. Nuclei from these cells were then digested with MNase concentrations that resulted in >95% of the DNA having mononucleosomal length. Using four primer sets that discriminated between nucleosome frames N2 and N4 (nucleosome N1 could not be analyzed using this approach because of repetitive sequences), we quantified each nucleosomal frame relative to genomic DNA (Fig. 4A). External control sequences included two regions flanking the −2.5 GRU within the Tat gene and the promoters of an active housekeeping gene coding for nucleolin and of a silent tissue-specific gene coding for β-globin. This analysis revealed that all four phases were present in comparable proportions, confirming the absence of a preferred nucleosome position within the GRU. Among external control sequences, only the active Nucleolin promoter appeared highly sensitive to MNase digestion. Upon GC-induced remodeling, all nucleosome positions N2 to N5 were lost to a comparable degree (60 to 70% loss), consistent with the direct probing analysis presented in Fig. 2. Thus, nucleosomes occupied multiple positions that were equally susceptible to remodeling.
FIG. 4.
The multiple nucleosomal frames at the −2.5 GRU have similar frequencies and are equally remodeled upon GC stimulation. (A) Location of the primers used to quantify specifically nucleosomal frames N2 to N5. The primers are represented below the specific frame they recognize. (B) Quantification of each nucleosomal frame using real-time PCR. Nuclei from formaldehyde-cross-linked cells were treated with MNase so that >95% of the DNA had mononucleosomal length. Following cross-link reversion and DNA purification, the abundance of each sequence was quantified relative to a standard curve of sonicated genomic DNA.
In conclusion, there was significant heterogeneity in the chromatin organization within the cell population. Nucleosomes occupied multiple positions within the GRU, and there was neither remodeling-induced repositioning nor positions that were preferentially remodeled upon GC treatment. Furthermore, several chromatin states coexisted in the population of GC-treated cells. Within the population, three categories of nucleosomes appear to coexist, i.e., apparently unremodeled, slightly altered, and extensively remodeled nucleosomes, the latter having lost most, if not all, nucleosomal features.
GR induces histone H3 and H4 hyperacetylation at the Tat GRU.
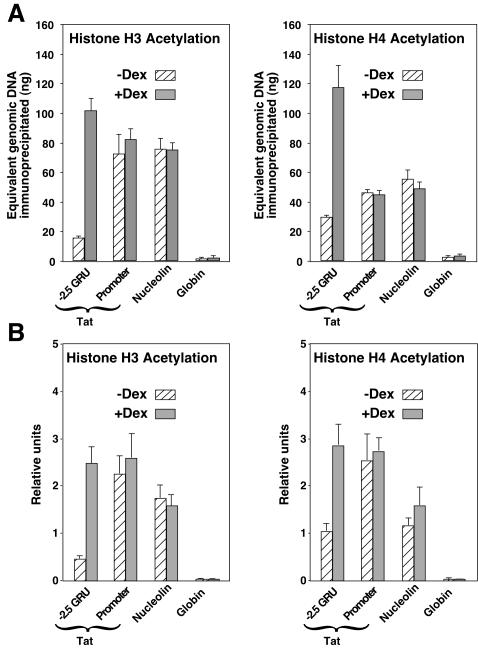
Since nuclear receptors can recruit a variety of histone acetyltransferases, we determined whether GR was promoting any increase in histone acetylation at the Tat GRU. We used ChIp and real-time quantitative PCR to quantify the relative level of acetylated histones H3 and H4 associated with the Tat gene −2.5 GRU and promoter, as well as with the promoters of the active Nucleolin gene and the silent β-globin gene. First, we analyzed cross-linked chromatin that was sonicated to an average size of 300 bp. The analysis revealed that in the absence of GC stimulation, acetylation of both histone H3 and H4 was high at the promoter of both the Tat and Nucleolin genes, low at the silent β-globin promoter, and intermediate at the −2.5 GRU (Fig. 5A). Upon a 2-day GC treatment, a five- to sixfold increase in the acetylation levels of both histone H3 and H4 was observed at the −2.5 GRU. In contrast, no increase in acetylation was visible at the Tat-proximal promoter. The fact that the proximal promoter chromatin was already acetylated before GC induction, even though transcription was inefficient, and that GC induction did not increase its acetylation, reveals that local histone acetylation at the promoter is not sufficient for Tat activation.
FIG. 5.
ChIp analysis reveals GC-induced histone H3 and H4 acetylation of the −2.5 Tat GRU. (A) The amount of immunoprecipitated material from sonicated cross-linked chromatin was quantified using real-time quantitative PCR relatively to a standard curve of genomic DNA. The results shown are means and standard deviations from two immunoprecipitation experiments with duplicate quantification. The −2.5 GRU primers used correspond to nucleosome position N4 in Fig. 4. (B) The amount of immunoprecipitated material from cross-linked chromatin digested with MNase is expressed relative to the input chromatin DNA.
ChIp analysis of MNase-digested chromatin allows assessment of histone modification at the single-nucleosome resolution, specifically of the fraction that is resistant to MNase treatment. Fixed chromatin digested with MNase to mononucleosomal length was analyzed by ChIp and quantified by real-time PCR relative to the input MNase-treated chromatin DNA. The pattern of histone acetylation analyzed in this way was remarkably similar to that obtained with chromatin sheared by sonication (Fig. 5B). At the −2.5 GRU, the MNase-resistant fraction was hyperacetylated upon GC stimulation, showing that at least a subset of the corresponding GR-modified chromatin was hyperacetylated without being fully remodeled.
Histone hyperacetylation induced by inhibition of histone deacetylases neither triggers nor abrogates nucleosome remodeling and HNF-3 recruitment.
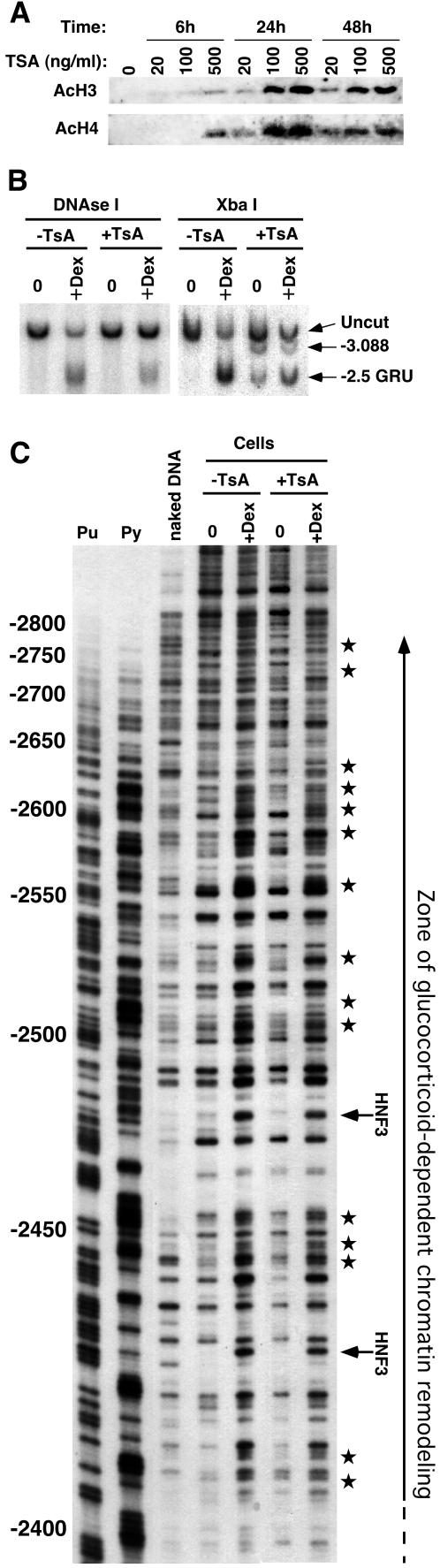
Trichostatin A (TsA) inhibits histone deacetylases, thus inducing histone hyperacetylation. TsA treatment has been shown to provoke nuclease hypersensitivity in several viral promoters (e.g., see references 3 and 38). Since GR induces histone hyperacetylation at the Tat GRU, we tested whether remodeling of the −2.5 GRU could be triggered by TsA-induced histone hyperacetylation. TsA treatment was calibrated for optimal histone H3 and H4 hyperacetylation, as assessed by Western blot analysis with antibodies recognizing the acetylated forms of these histones (Fig. 6A). Neither the optimal TsA treatment nor treatments using different TsA amounts affected the GC-mediated transcriptional induction: Tat gene expression was at most increased twofold, under both the uninduced and induced conditions (data not shown). TsA treatment induced a slight increase in chromatin accessibility, detected as the ability of XbaI to cleave DNA within the −2.5 GRU and at a site further upstream, independent of GCs (Fig. 6B). However, TsA-induced hyperacetylation was not sufficient to promote chromatin remodeling, since it did not increase the reactivity to DNase I at the −2.5 GRU, as seen at both low and high resolutions (Fig. 6B and C). As observed with MNase, the GR-induced DNase I-hypersensitive site visible at low resolution (Fig. 6B) corresponded to enhanced cleavage at multiple positions throughout the GRU when analyzed at high resolution (Fig. 6C). Two of these hyperactive positions are indicative of HNF-3 interaction with its sites within the GRU (32, 33). The other sites of enhanced cleavage were distributed throughout a 450-bp region that included all nucleosome positions mapped herein (Fig. 6C and data not shown; see recapitulation in Fig. 3D). In agreement with the absence of precise nucleosomal phasing observed with MNase, there was also no 10-bp repeat characteristic of rotationally phased nucleosome detected in the absence or presence of hormone. TsA treatment did not substantially modify the unremodeled or remodeled DNase I patterns, indicating that histone hyperacetylation is not the determining event responsible for GC-induced remodeling at the −2.5 GRU.
FIG. 6.
TsA-induced histone hyperacetylation triggers neither chromatin remodeling nor HNF3 recruitment. (A) Western blot analysis of acetylation of histone H3 and H4 induced by different TsA treatments, using antibodies against acetylated histone H3 or H4. (B) Chromatin accessibility at the −2.5 Tat GRU as assessed by sensitivity to XbaI or DNase I cleavage. Cells were treated with 100 ng of TsA/ml for 24 h. (C) High-resolution DNase I cleavage analysis of the GC-induced remodeling, revealing the absence of effect of TsA-induced hyperacetylation. The stars on the right indicate the DNase I hyperreactivities characteristic of the GC-induced remodeling, and the arrows indicate those that are characteristic of HNF3 recruitment. The extent of the zone of GC-induced remodeling is shown.
The interaction of both histone H3 and histone H1 with the −2.5 GRU is reduced upon remodeling.
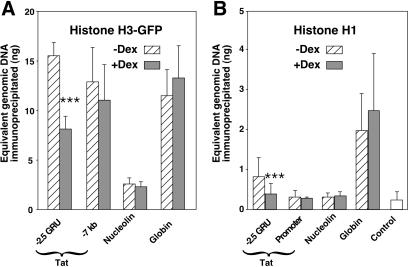
Due to the heterogeneity of the nucleosomal states, it is difficult to deduce unambiguously from the nuclease digestion pattern whether the nuclease-hypersensitive fraction corresponds to structurally altered or fully disrupted nucleosomes. Thus, we sought to quantify directly core histone association irrespective of their modification status by using ChIp. Since efficient antibodies suitable for that purpose are not available, we expressed in H4II cells a GFP-tagged histone H3 to monitor H3 association using antibodies directed against the GFP moiety. In this way, we measured histone H3 association with the −2.5 GRU, with a region of the Tat gene (−7 kb) that is neither remodeled nor in the vicinity of the transcribed region, as well as with the promoters of the active Nucleolin and silent β-globin genes. Similar high levels of tagged histone H3 were found to be associated with the unremodeled −2.5 GRU, the −7 kb Tat region, and the silent β-globin promoter (Fig. 7A). In contrast, the levels of histone H3 found at the active Nucleolin promoter were at least sixfold lower. GC-induced remodeling at the −2.5 GRU led to a twofold decrease of histone H3 association. Thus, histone H3 dissociation appears to parallel MNase sensitivity, as seen in Fig. 4. The amplitude of the differential sensitivity to MNase seems higher than that of histone H3 dissociation. This is clear for the Nucleolin promoter, which was 16-fold more sensitive to MNase than the β-globin promoter, although associated with only sixfold less histone H3. Similarly, even though not as striking, GC-induced remodeling resulted in a threefold increase in MNase sensitivity but in only a twofold decrease in histone H3 association. This suggests that only part of the extensively remodeled fraction was fully depleted of core histones.
FIG. 7.
ChIp analysis reveals local GC-induced dissociation of histones H3 and H1 at the −2.5 GRU. (A) Histone H3 association. An H4II subclone expressing a GFP-tagged histone H3 was analyzed by ChIp using anti-GFP antibodies. Results from three fully independent experiments quantified at least in duplicate were analyzed, and the mean ± standard deviation is represented. For each region, the values obtained from cells treated (or not) with GCs were analyzed with a paired-sample Student t test. Only the −2.5 GRU showed a significant difference (P [two-tail] = 0,0001, indicated by three asterisks), with a twofold GC-induced decrease in the amount of GFP-H3-associated material. (B) Histone H1 association. Results from up to five immunoprecipitation experiments from three independent chromatin preparations were analyzed and are presented as in panel A. Only the −2.5 GRU showed a significant difference with the Student t test (P [two-tail] = 0,001, indicated by three asterisks), with a twofold GC-induced decrease in the amount of H1-associated material.
The inability of the restriction enzyme XbaI to cleave its site (−2558) in the GRU in the absence of hormone reveals that it was inaccessible regardless of whether it is within a nucleosome (e.g., N4; Fig. 3D) or in the linker (e.g., in chromatin with frame N5). Furthermore, the site became accessible upon GC-induced remodeling irrespective of the nucleosomal frame. To gain insight into the possible mechanism that prevents accessibility to the XbaI site, we used ChIp to assess whether the interaction of the linker histone H1 with the −2.5 GRU could be affected by GC treatment. In parallel, we analyzed the interaction of H1 with the Tat gene promoter and with the promoters of the expressed Nucleolin and silent β-globin genes. Immunoprecipitation with the H1 antibody was not very efficient but still allowed us to observe that significantly more H1 was cross-linked to the silent β-globin promoter than to the other regions analyzed (Fig. 7B). Some H1 could be cross-linked at the −2.5 GRU in the absence of GC, whereas the amount of cross-linked H1 was not significantly different from the background level (control in Fig. 7B) at the Tat and Nucleolin promoters. Upon GC induction, H1 interaction with the GRU was decreased back to background levels. These results suggest that the linker histone H1 interacts loosely with particular nucleosomes of poised genes ready to respond to transcription activation, such as Tat, and that this interaction is reduced upon signal-induced local remodeling, leading to transcriptional activation.
Thus, at least part of the GC-induced accessibility within the 450-bp region of the −2.5 GRU results from both core and linker histone depletion occurring irrespective of the nucleosomal frames.
DISCUSSION
The nature of the remodeled chromatin at regulatory sequences in living cells, in particular in higher eukaryotes, remains ambiguous, and it is not yet clear whether nucleosome positioning plays in a general manner a decisive role in the behavior of regulatory sequences. The various studies addressing these questions have led to contradictory conclusions, and the discrepancies could be due as well to differences in the biological systems or in the experimental methods. We have used a combination of varied experimental procedures to characterize the chromatin accessibility induced by the GR at a remote enhancer, the −2.5 GRU of the Tat gene. We analyzed the contribution of nucleosome positioning and histone acetylation in determining chromatin accessibility and studied the nature of nuclease hypersensitivity.
When considering chromatin structure at the mononucleosomal scale, clear differences in DNA accessibility are expected depending on the location of a given sequence with respect to the nucleosome: (i) linker DNA is most accessible; (ii) a sequence at the nucleosomal edge is more accessible than one at the dyad axis; (iii) a sequence facing outward is more accessible than a sequence facing inward. Thus, nucleosome positioning could play a key role in regulating transcription factor access to DNA (29). Indeed, precise nucleosome positioning has been shown to contribute to the tight regulation of the human β-interferon promoter, where it creates a particular requirement for a nucleosome mobilizing activity (22). However, it is not clear whether this level of control is general. Contradictory data about nucleosome positioning have been obtained, to a large extent because assessment of nucleosome positions is dependent on the experimental conditions. MNase is often chosen for positioning studies because it cleaves preferentially within the linker. However, it can also cleave within the nucleosome, thereby necessitating the use of detection methods that enrich for cleavage in the linker DNA (10, 37). Despite these precautions, contradictions remain. This is most striking at the GC-regulated MMTV LTR, where one study has shown precise positioning (37) whereas another one detects multiple frames (10). Chromatin digested to mononucleosomal length is the material of choice for mapping boundaries. However, as shown here, this material can lead to underestimation of the complexity of nucleosomal distribution because only a limited number of nucleosomal frames can be detected with a given primer set. We show that an extended MNase digestion range analyzed with multiple primer sets reveals a much wider complexity of nucleosomal distribution than what would have been detected with the usual analysis of extensively digested chromatin and a limited number of primers. Using quantitative PCR analysis of fixed chromatin, we show that the various nucleosomal frames show up with similar frequencies, thus confirming the heterogeneity of the distribution. Finally, the multiple nucleosome frames detected with MNase are not rotationally phased, in agreement with the absence of a 10-bp repeat in the high-resolution analysis of chromatin digested with DNase I.
Despite this heterogeneity in nucleosome positioning, chromatin accessibility is highly controlled. In the absence of GC-induced remodeling, chromatin is inaccessible to endogenous transcription factors like HNF-3 and COUP-TF (32, 36), as well as to exogenous proteins like the restriction enzyme XbaI used herein, even though their cognate binding sites are located in linker regions in at least parts of the population (see Fig. 3D). Remodeling allows interaction of these factors with DNA, without causing a redistribution of the nucleosomal frames. Thus, nucleosome positioning is not the key factor determining the inaccessibility of these sites in the absence of GR activation. This indicates that chromatin accessibility in vivo cannot be understood readily at the mononucleosomal scale. Most likely, some higher-order folding of the chromatin fiber prevents many DNA-protein interactions, irrespective of the precise nucleosome locations. In favor of this interpretation, using ChIp, we detected linker histone H1 interaction with the −2.5 GRU in the absence of GR-induced remodeling. The corresponding amount of H1 was not as high as that interacting with a silent gene but was significantly above background levels, in contrast to the amount of H1 interacting with the remodeled GRU. Similar observations were made for the MMTV LTR (5, 11), indicating that higher-order folding could contribute relatively frequently to the control of accessibility. It remains to be determined whether histone H1 displacement is a prerequisite for or a consequence of nucleosome remodeling. In vitro, histone H1 inhibits SWI/SNF-catalyzed remodeling (19). However, histone H1 interaction is highly dynamic (26), and it is likely that H1 interaction would be disfavored when the nucleosome is extensively remodeled or unraveled.
Nuclear receptors use histone acetyltransferases as coactivators (12). Accordingly, we observed that GR triggers hyperacetylation of both histone H3 and H4 tails at the −2.5 GRU. This event is presumably not involved per se in the transcriptional activation that takes place at the proximal promoter, since this region is hyperacetylated irrespective of the presence of glucocorticoid and the corresponding activation of transcription. What then could be the role of the GR-triggered hyperacetylation event? Two functions of the acetylation of histone tails have been documented. One function involves neutralization of the positive charges of the tails, thereby loosening their interaction with DNA thus rendering chromatin accessible to transcription factors (16, 28). The other function involves recruitment of bromodomain-containing proteins, in particular ATP-dependent remodeling complexes (1, 20). Both at the MMTV and human immunodeficiency virus LTRs, TsA-induced hyperacetylation can promote local remodeling (3, 38). However, we observed here that TsA treatment does not induce such a remodeling at the −2.5 GRU, even though it facilitates cleavage by the restriction enzyme XbaI. Thus, hyperacetylation is not sufficient to allow the action of the ATP-dependent remodeling complexes at any regulatory sequences. This is not really surprising, since TsA-induced hyperacetylation is widespread and is thus likely to cause a dilution of any targeting effect due to local acetylation. Furthermore, remodeling complex recruitment appears to require both histone acetylation and interaction with factors recruited by DNA-bound proteins (1, 22). Thus, it is more likely that in the situations where TsA treatment elicits chromatin remodeling, its action is indirect. It would first loosen the nucleosomal structure, which would allow access of some transcription factors, promoting the subsequent remodeling events, as observed with the human immunodeficiency virus LTR (28).
The detection of acetylated histones at the remodeled GRU could suggest that histones do not lose contact upon remodeling. However, local chromatin remodeling has long been assumed to correspond to nucleosome displacement, either through sliding along the DNA or through ejection. This view has been based on early electron microscopy and cross-linking analyses (15), but remodeled and still-associated nucleosomes may have escaped these analyses. Indeed, ATP-dependent remodeling complexes can alter the nucleosome architecture in a manner that enhances DNA accessibility without disrupting all histone-DNA contacts (27). In vitro, it has been observed that altered nucleosomes can either remain at the same position, slide to a neighboring position leaving a nucleosome-free region, or be transferred to another molecule, DNA or a chaperone protein (27). The fate of altered nucleosomes in vivo is still unclear. At the human β-interferon gene, a positioned nucleosome appears to slide to a neighboring position upon remodeling (22). At the Pho5 promoter in yeast, the data indicate that the remodeled area is nucleosome free (4, 31). However, it is not clear whether all hypersensitive sites are nucleosome free. It is likely that the nucleosomes remodeled by the SWI/SNF activities are unraveled by other events, in particular transcription factor binding. Thus, it is possible that other remodeled chromatin regions interact with nucleosomes despite being hypersensitive to nucleases due to a low occupancy level by DNA-binding proteins. In contrast to the conclusions of the Pho5 studies, based on the MNase cleavage patterns of the MMTV LTR, it has been proposed that the nucleosome can be altered without being moved sideways or ejected (10, 37). However, heterogeneity within the LTR population was not assessed in these studies, and the analyses were not quantitative.
Here, using a combination of approaches, we have determined the heterogeneity of the −2.5 GRU population when the gene was optimally induced. We observed that depending on the parameter analyzed, the fraction of active or remodeled chromatin differed, suggesting that several conformations coexist. About 80% of the gene copies appeared active at the time of the analyses, as assessed by FISH analyses of transcription, and the chromatin of a similar fraction appeared accessible when analyzed with the restriction enzyme XbaI. A smaller fraction (about 60%) appeared highly accessible to cleavage by MNase. The findings that the MNase-resistant chromatin fraction was hyperacetylated and that TsA-induced hyperacetylation facilitated XbaI cleavage readily account for the discrepancies between the two nuclease sensitivities. An even smaller fraction of the GRU (about 50%) appeared to have lost contact with histone H3. Since we have not observed a corresponding increase in the fraction associated to the replacement histone H3-3 (data not shown) (2), a large part of the MNase-hypersensitive fraction could correspond to unraveled nucleosomes. However, some of our data suggest that this MNase-sensitive fraction could correspond to several populations with both unraveled and remodeled nucleosomes. First, neither the MNase cleavage patterns specifically induced by GC treatment (corresponding to the increased reactivity at intermediate MNase amounts shown in Fig. 3) nor the corresponding DNase I cleavage patterns were exactly identical to those obtained with naked DNA or transcription factor-bound DNA (Fig. 3 and 6) (9; also data not shown). Such a behavior has been observed in vitro with nucleosome remodeled (but not unraveled) by the SWI/SNF complex, where the DNase I cleavage pattern was resembling, but not identical to, that of naked DNA (8). Second, the MNase-sensitive fraction was larger than the fraction that has lost contact with H3. The difference in the sizes of these fractions does not appear very important for the −2.5 GRU, but it is striking when the active Nucleolin and inactive β-globin promoters are compared. Indeed, the Nucleolin promoter was 16-fold more sensitive to MNase than the β-globin promoter, although it was associated with only 6-fold H3 less histone.
Such heterogeneity throughout the population with a coexistence of nucleosomes being unmodified, hyperacetylated, remodeled, or unraveled would account for all our observations. It could also account for the numerous discrepancies found throughout the literature aimed at describing the exact nature of the remodeled chromatin. Indeed, as shown here, different fractions of the chromatin population can be revealed depending on the experimental approaches used. Since the action of nuclear receptors is a dynamic process (23, 25, 32), it is tempting to speculate that the heterogeneity observed herein results from cycling of the chromatin through the various conformations occurring asynchronously in the gene population.
Acknowledgments
This work was supported in part by the CNRS and grants from the Association de Recherche sur le Cancer and the Ligue Nationale contre le Cancer. L.C. was supported by a fellowship from Fondation MEDIC, and C.K. was supported by fellowships from CNRS and the Ligue Nationale contre le Cancer.
We thank S. Muller for the kind gift of anti-H1 antiserum, S. Henikoff for the kind gift of the GFP-histone H3 fusion, and E. M. Geigl, A. Kralli, and A. Prunell for critical reading of the manuscript.
REFERENCES
- 1.Agalioti, T., S. Lomvardas, B. Parekh, J. Yie, T. Maniatis, and D. Thanos. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell 103:667-678. [DOI] [PubMed] [Google Scholar]
- 2.Ahmad, K., and S. Henikoff. 2002. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 9:1191-1200. [DOI] [PubMed] [Google Scholar]
- 3.Bartsch, J., M. Truss, J. Bode, and M. Beato. 1996. Moderate increase in histone acetylation activates the mouse mammary tumor virus promoter and remodels its nucleosome structure. Proc. Natl. Acad. Sci. USA 93:10741-10746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boeger, H., J. Griesenbeck, J. S. Strattan, and R. D. Kornberg. 2003. Nucleosomes unfold completely at a transcriptionally active promoter. Mol. Cell 11:1587-1598. [DOI] [PubMed] [Google Scholar]
- 5.Bresnick, E. H., M. Bustin, V. Marsaud, H. Richard-Foy, and G. L. Hager. 1992. The transcriptionally-active MMTV promoter is depleted of histone H1. Nucleic Acids Res. 20:273-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cappabianca, L., H. Thomassin, R. Pictet, and T. Grange. 1999. Genomic footprinting using nucleases. Methods Mol. Biol. 119:427-442. [DOI] [PubMed] [Google Scholar]
- 7.Chen, H., R. J. Lin, W. Xie, D. Wilpitz, and R. M. Evans. 1999. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell 98:675-686. [DOI] [PubMed] [Google Scholar]
- 8.Cote, J., J. Quinn, J. L. Workman, and C. L. Peterson. 1994. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science 265:53-60. [DOI] [PubMed] [Google Scholar]
- 9.Espinás, M. L., J. Roux, R. Pictet, and T. Grange. 1995. Glucocorticoids and protein kinase A coordinately modulate transcription factor recruitment at a glucocorticoid-responsive unit. Mol. Cell. Biol. 15:5346-5354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fragoso, G., S. John, M. S. Roberts, and G. L. Hager. 1995. Nucleosome positioning on the MMTV LTR results from the frequency-biased occupancy of multiple frames. Genes Dev. 9:1933-1947. [DOI] [PubMed] [Google Scholar]
- 11.Georgel, P. T., T. M. Fletcher, G. L. Hager, and J. C. Hansen. 2003. Formation of higher-order secondary and tertiary chromatin structures by genomic mouse mammary tumor virus promoters. Genes Dev. 17:1617-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glass, C. K., and M. G. Rosenfeld. 2000. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 14:121-141. [PubMed] [Google Scholar]
- 13.Grange, T., L. Cappabianca, M. Flavin, H. Sassi, and H. Thomassin. 2001. “In vivo” analysis of the model tyrosine aminotransferase gene reveals multiple sequential steps in glucocorticoid receptor action. Oncogene 20:3028-3038. [DOI] [PubMed] [Google Scholar]
- 14.Grange, T., J. Roux, G. Rigaud, and R. Pictet. 1989. Two remote glucocorticoid responsive units interact cooperatively to promote glucocorticoid induction of rat tyrosine aminotransferase gene expression. Nucleic Acids Res. 17:8695-8709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gross, D. S., and W. T. Garrard. 1988. Nuclease hypersensitive sites in chromatin. Annu. Rev. Biochem. 57:159-197. [DOI] [PubMed] [Google Scholar]
- 16.Grunstein, M. 1997. Histone acetylation in chromatin structure and transcription. Nature 389:349-352. [DOI] [PubMed] [Google Scholar]
- 17.Hager, G. L. 2001. Understanding nuclear receptor function: from DNA to chromatin to the interphase nucleus. Prog. Nucleic Acid Res. Mol. Biol. 66:279-305. [DOI] [PubMed] [Google Scholar]
- 18.Hassan, A. H., K. E. Neely, and J. L. Workman. 2001. Histone acetyltransferase complexes stabilize SWI/SNF binding to promoter nucleosomes. Cell 104:817-827. [DOI] [PubMed] [Google Scholar]
- 19.Horn, P. J., L. M. Carruthers, C. Logie, D. A. Hill, M. J. Solomon, P. A. Wade, A. N. Imbalzano, J. C. Hansen, and C. L. Peterson. 2002. Phosphorylation of linker histones regulates ATP-dependent chromatin remodeling enzymes. Nat. Struct. Biol. 9:263-267. [DOI] [PubMed] [Google Scholar]
- 20.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 21.Krebs, J. E., C. J. Fry, M. L. Samuels, and C. L. Peterson. 2000. Global role for chromatin remodeling enzymes in mitotic gene expression. Cell 102:587-598. [DOI] [PubMed] [Google Scholar]
- 22.Lomvardas, S., and D. Thanos. 2002. Modifying gene expression programs by altering core promoter chromatin architecture. Cell 110:261-271. [DOI] [PubMed] [Google Scholar]
- 23.McNally, J. G., W. G. Muller, D. Walker, R. Wolford, and G. L. Hager. 2000. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science 287:1262-1265. [DOI] [PubMed] [Google Scholar]
- 24.McPherson, C. E., E.-Y. Shim, D. S. Friedman, and K. S. Zaret. 1993. An active tissue-specific enhancer and bound transcription factors existing in a precisely positioned nucleosomal array. Cell 75:387-398. [DOI] [PubMed] [Google Scholar]
- 25.Metivier, R., G. Penot, M. R. Hubner, G. Reid, H. Brand, M. Kos, and F. Gannon. 2003. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115:751-763. [DOI] [PubMed] [Google Scholar]
- 26.Misteli, T., A. Gunjan, R. Hock, M. Bustin, and D. T. Brown. 2000. Dynamic binding of histone H1 to chromatin in living cells. Nature 408:877-881. [DOI] [PubMed] [Google Scholar]
- 27.Narlikar, G. J., H. Y. Fan, and R. E. Kingston. 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108:475-487. [DOI] [PubMed] [Google Scholar]
- 28.Ng, K. W., P. Ridgway, D. R. Cohen, and D. J. Tremethick. 1997. The binding of a Fos/Jun heterodimer can completely disrupt the structure of a nucleosome. EMBO J. 16:2072-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piña, B., U. Brüggemeier, and M. Beato. 1990. Nucleosome positioning modulates accessibility of regulatory proteins to the mouse mammary tumor virus promoter. Cell 60:719-731. [DOI] [PubMed] [Google Scholar]
- 30.Reik, A., G. Schütz, and A. F. Stewart. 1991. Glucocorticoids are required for establishment and maintenance of an alteration in chromatin structure: induction leads to a reversible disruption of nucleosomes over an enhancer. EMBO J. 10:2569-2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reinke, H., and W. Horz. 2003. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol. Cell 11:1599-1607. [DOI] [PubMed] [Google Scholar]
- 32.Rigaud, G., J. Roux, R. Pictet, and T. Grange. 1991. In vivo footprinting of the rat TAT gene: dynamic interplay between the glucocorticoid receptor and a liver-specific factor. Cell 67:977-986. [DOI] [PubMed] [Google Scholar]
- 33.Roux, J., R. Pictet, and T. Grange. 1995. Hepatocyte nuclear factor 3 determines the amplitude of the glucocorticoid response of rat tyrosine aminotransferase gene. DNA Cell Biol. 14:385-396. [DOI] [PubMed] [Google Scholar]
- 34.Sheardown, S. A., S. M. Duthie, C. M. Johnston, A. E. Newall, E. J. Formstone, R. M. Arkell, T. B. Nesterova, G. C. Alghisi, S. Rastan, and N. Brockdorff. 1997. Stabilization of Xist RNA mediates initiation of X chromosome inactivation. Cell 91:99-107. [DOI] [PubMed] [Google Scholar]
- 35.Stemmer, C., J. P. Briand, and S. Muller. 1994. Mapping of linear epitopes of human histone H1 recognized by rabbit anti-H1/H5 antisera and antibodies from autoimmune patients. Mol. Immunol. 31:1037-1046. [DOI] [PubMed] [Google Scholar]
- 36.Thomassin, H., M. Flavin, M. L. Espinás, and T. Grange. 2001. Glucocorticoid-induced DNA demethylation and gene memory during development. EMBO J. 20:1974-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Truss, M., J. Bartsch, A. Schelbert, R. J. Hache, and M. Beato. 1995. Hormone induces binding of receptors and transcription factors to a rearranged nucleosome on the MMTV promoter in vivo. EMBO J. 14:1737-1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Lint, C., S. Emiliani, M. Ott, and E. Verdin. 1996. Transcriptional activation and chromatin remodeling of the HIV-1 promoter in response to histone acetylation. EMBO J. 15:1112-1120. [PMC free article] [PubMed] [Google Scholar]