Abstract
Peptidoglycan recognition proteins (PGRPs) constitute a family of innate immune recognition molecules. In Drosophila, distinct PGRPs bind to peptidoglycans on gram-positive or gram-negative bacteria and provide essential signals upstream of the Toll and Imd pathways required for immunity against infection. Four PGRPs, PGRP-L, -S, -Iα, and -Iβ, are expressed from three genes in mammals. In this paper, we provide direct evidence that the longest family member, PGRP-L, is a secreted serum protein with the capacity to multimerize. Using gene targeting to create PGRP-L-deficient mice, we demonstrate little contribution by PGRP-L to systemic challenge using gram-negative bacteria (Escherichia coli, slightly less susceptible), Gram-positive bacteria (Staphylococcus aureus), or yeast (Candida albicans). Peritoneal macrophages from PGRP-L-deficient mice produced decreased amounts of the inflammatory cytokines interleukin 6 and tumor necrosis factor alpha when stimulated with E. coli or lipopolysaccharide, but comparable amounts when stimulated with S. aureus, C. albicans, or their cell wall components. Additionally, these cells produced similar amounts of cytokines when challenged with gram-positive or -negative peptidoglycans. In contrast to its critical role in immunity in flies, PGRP-L is largely dispensable for mammalian immunity against bacteria and fungi.
The ability of the host to distinguish between self and nonself remains a central hallmark of innate immunity. Microbial organisms express distinct cell surface molecules, such as peptidoglycan (PGN) and lipoteichoic acid (LTA) on gram-positive bacteria and lipopolysaccharide (LPS) on gram-negative bacteria, that are structurally indispensable components of the cell wall. Recognition of these pathogen-associated molecular patterns, or PAMPs (14), is achieved predominantly by the vertebrate Toll-like receptor (TLR) family proteins, which collectively mediate induction of antimicrobial defensins, cytokines, chemokines, and dendritic cell maturation important to innate and adaptive immune responses.
PGN recognition proteins (PGRPs) were first identified by their ability to bind PGN and complement the prophenoloxidase cascade that induces melanization around pathogens in insects (37). Thirteen PGRP family members are present in Drosophila (36), and each contains a conserved carboxyl-terminal region with homology to bacterial T7 lysozyme (11). Some family members, such as PGRP-SC1b, have enzymatic activity and can digest PGN by hydrolyzing the amide bond between N-acetylmuramic acid and l-alanine (20). The N-acetylmuramyl-l-alanine amidase activity may function to reduce the immunostimulatory effects of PGN (12).
Definitive evidence indicating essential roles for PGRPs in Drosophila innate immunity was revealed in studies of mutants defective in Toll and Imd pathway-mediated antimicrobial responses. Infection of flies by gram-positive bacteria and fungi induces localized production of cleaved Spaetzle, the Toll ligand. Stimulation of Toll by ligand binding activates latent NF-κB-like transcription factors, resulting in expression of the antimicrobial peptide drosomycin. Infection by gram-negative bacteria results in activation of Imd, a Drosophila RIP-like adapter (5), and the Relish pathway, with expression of antibacterial peptides active against these bacteria, including diptericin, attacin, drosocin, and cecropins (11). Drosophila with a mutated PGRP-SA gene failed to activate drosomycin expression and was unable to survive challenge with gram-positive bacteria (21). In contrast, responses to gram-negative bacteria and fungi were not affected. A constitutively active Toll mutant could overcome the PGRP-SA mutation, suggesting that PGRP-SA functions upstream of Toll (21). The capacity of PGRP-SA to bind to PGN, likely in conjunction with gram-negative binding protein (6, 24), must therefore constitute a crucial and nonredundant recognition element upstream of Toll pathway activation in Drosophila.
A similar screen identified PGRP-LC upstream of the Imd pathway (1, 7, 27). Like Imd mutants, PGRP-LC mutants poorly induced antimicrobial peptides, particularly diptericin, and demonstrated increased susceptibility to gram-negative bacteria; responses to gram-positive bacteria and fungi were unaffected. As in the Toll pathway, a constitutively active Imd mutation could rescue antimicrobial peptide induction attenuated by the PGRP-LC mutation (1, 7). Therefore, PGRP-LC plays a crucial and nonredundant role in detecting gram-negative bacteria and leading to activation of the Imd/Relish pathway.
Although LPS is a major component of the gram-negative bacterial cell wall, PGN constitutes a shared cell wall element in both gram-positive and gram-negative bacteria (28). Structurally similar, these cell wall PGNs differ at the third amino acid of the peptide side chain, where lysine (Lys-PGN) and meso-diaminopimelic acid (Dap-PGN) are used in gram-positive and gram-negative organisms, respectively (28). Unlike LPS, Dap-PGN induced gram-negative-like Imd activation in flies, whereas Lys-PGN fully induced the gram-positive-like Toll pathway response (15, 18). Together, these studies suggest that different PGRP proteins in Drosophila mediate direct interactions with distinctive PGN moieties in bacterial cell walls as a proximal mechanism upstream of Toll and Imd activation. Indeed, PGRP-LE binds Dap-PGN but not Lys-PGN in vitro, consistent with this paradigm (31).
PGRP is conserved in mice and humans, where four PGRP proteins are generated from three genes (19). The longest, PGRP-L, is a predicted membrane protein expressed mainly in liver. The shortest, PGRP-S, is a predicted soluble protein expressed in neutrophils. The intermediately sized family members, PGRP-Iα and -Iβ, are splice products from the same gene. PGRP-Iα/Iβ are predicted membrane proteins that are expressed in the esophagus (19). Like the Drosophila PGRPs, mammalian PGRPs have conserved carboxyl-terminal PGRP domains homologous to lysozyme (19). Only PGRP-L, however, has N-acetylmuramyl-l-alanine-amidase activity that digests PGN. The remaining family members have critical amino acid substitutions that abrogate enzymatic activity (4, 35), but all four mammalian proteins retain the ability to bind PGN in vitro (19).
In contrast to the dramatic phenotype of PGRP-SA mutants in Drosophila, the PGRP-S null mouse displayed normal susceptibility to gram-negative and gram-positive challenge with Escherichia coli and Staphylococcus aureus. A nonpathogenic gram-positive Bacillus species was cleared less efficiently, however, and the killing and digestion of gram-positive bacteria by PGRP-SA-deficient neutrophils were compromised in vitro, although phagocytosis remained intact. Further, murine PGRP-S had no role in TLR-2-mediated cytokine induction (3). To define further the function of PGRPs in mammalian innate immunity, we disrupted the mouse PGRP-L gene and examined the role of this protein in host defense against bacterial and fungal challenges.
MATERIALS AND METHODS
Targeting strategy.
The murine PGRP-L genomic sequence was amplified by PCR using 129/SvJ embryonic stem (ES) cell DNA as template. Primers were used as follows: sense (GGGAATTCCGAGTCTGGCTCAGTCTGG) and antisense (GCAAGCTTCATGGTGGGTCTCCAGTTCC) for the 4-kb left arm of the construct; sense (GCGTCGACCACTTGCTTTGTTCAACCCTAATGG) and antisense (GCCTCGAGTCTACCCCTAAGAACCAGTCACATC) for the 4-kb right arm of the construct. The PCR conditions included the first 10 cycles at 94°C for 30 s, 60°C for 30 s, and 68°C for 4 min; the next 20 cycles at 94°C for 30 s, 60°C for 30 s, and 68°C for 4 min plus cycle elongation of 5 s for each cycle. The left arm was subcloned into pgkTK (33) with EcoRI and HindIII, and the right arm was subcloned with SalI and XhoI. An enhanced green fluorescent protein (EGFP) cassette, derived from pIRES-EGFP (Clontech, Palo Alto, Calif.), was inserted in frame with the first PGRP-L ATG codon, using PCR-generated HindIII and ClaI restriction enzyme sites. A loxP-flanked neo cassette from pL2neo2 (8) was ligated into the targeting construct by using ClaI and SalI.
Generation of PGRP-L-deficient mice.
The targeting construct was linearized with NotI and was transfected into PrmCre ES cells, which express the Cre recombinase under control of the protamine promoter (22). After 7 days of selection on 400 μg of G418/ml and 2 μM ganciclovir, resistant ES cell clones were screened for homologous integration by Southern blotting. Three correctly targeted clones were injected into C57BL/6 blastocysts to create chimeric mice. The neomycin resistance cassette was deleted in the male germ line by Cre-mediated recombination after breeding chimeric mice to wild-type mice. Offspring of the heterozygous males and wild-type C57BL/6 females were also screened for the absence of the Cre transgene. Finally, heterozygous animals were interbred to obtain homozygous mice. The absence of PGRP-L expression was determined by reverse transcription (RT)-PCR using the following primers: sense, ACCTGCTAAGAGAGTACTATGG; antisense, TAGTTGCCCACGAAGGCCACAC. The following PCR conditions were used: 94°C for 25 s, 60°C for 25 s, and 72°C for 1 min.
Cell lines and transfections.
HEK 293 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum, l-glutamine, penicillin, and streptomycin. Drosophila Schneider cells were cultured at 29°C with Schneider's Drosophila medium and 10% fetal calf serum. PGRP-L and PGRP-S cDNAs were generated from RNA of mouse hepatocytes and eosinophils, respectively, through RT-PCR. They were subsequently subcloned into the pcDNA 3.1-myc plasmid (Invitrogen, Calif.) by using EcoRI and XhoI to generate PGRP-L-myc and PGRP-S-myc. PGRP-L-V5 was derived from pAC5.1-PGRP-L (described in insect cell protein expression). Two micrograms of each plasmid DNA was transfected into cells plated in six-well plates by using a calcium phosphate protocol. Cells were collected and assayed 48 h after transfection.
Insect cell protein expression and antibody generation.
PGRP-L cDNA and PGRP-S cDNA were subcloned into the pAC 5.1 vector (Invitrogen), using EcoRI and XhoI, to generate pAC5.1-PGRP-L and pAC5.1-PGRP-S. pAC5.1-PGRP-L or -S and pCoBlast (Invitrogen) were cotransfected into Drosophila Schneider cells using the calcium phosphate transfection method to generate blasticidin-resistant cells. The supernatants from the various stably transfected cells were collected, and the insect cell-expressed PGRP-L was partially purified with Ni2+ columns (Invitrogen). The expressed PGRP-L was used to immunize rats for generation of polyclonal rat antiserum according to the Current Protocols in Immunology. Additionally, a peptide (amino acids 75 to 94), KAPSHNTTEPDPHSLSPELQ, was synthesized and used to immunize rabbits for production of rabbit polyclonal antiserum by Research Genetics (Invitrogen).
Coimmunoprecipitation and Western blot analysis.
Transfected 293 cells were lysed with NET-N buffer (150 mM NaCl, 1 mM EDTA, 20 mM Tris-HCl [pH 8.0], 0.5% NP-40, 10% glycerol) and immunoprecipitated with anti-Myc antibodies (Upstate, Lake Placid, N.Y.) at 4°C. After four washes with phosphate-buffered saline (PBS) plus 0.1% NP-40, the immunoprecipitates were subjected to polyacrylamide gel electrophoresis, transferred to polyvinylidene difluoride membranes, and blotted with anti-V5 antibodies (Invitrogen). For serum immunoprecipitation, 50 μl of mouse serum was incubated with rat anti-PGRP-L antibodies at 4°C. PGRP-L was detected by rabbit anti-PGRP-L antibodies. For detection of PGRP-L-myc, anti-Myc antibodies were used.
Challenges of PGRP-L null mice.
Ten sex- and age-matched (6 to 8 weeks old) wild-type and PGRP-L-deficient mice were used in each challenge experiment. LPS of E. coli strain O55:B5 was purchased from Sigma (St. Louis, Mo.). S. aureus (ATCC 33594), E. coli (ATCC 25922), and Candida albicans were grown to log phase and frozen in 17% glycerol. The CFU were counted by plating the frozen stocks overnight, using appropriate plates at 37 or 30°C. Designated amounts of S. aureus and E. coli (shown in the figures) were injected into peritoneal cavities of 10 wild-type or knockout mice. C. albicans was injected into the tail vein. The morbidity of mice after injection was monitored for a 10-day period.
Peritoneal macrophages.
Thioglycolate (1 ml of 3% solution) was injected into the peritoneal cavity of three wild-type or knockout mice. Five days later, the peritoneal cavity was washed with 10 ml of cold PBS. Eluted peritoneal macrophages were counted and plated into 24-well culture plates at a density of 2 × 105 per ml using DMEM plus 1% wild-type or PGRP-L knockout mouse serum for wild-type or knockout macrophages, respectively. After incubation for 2 h at 37°C, nonmacrophage cells were washed away, and different stimulation agents were added with 50 U of gamma interferon (IFN-γ) (R&D Systems, Minneapolis, Minn.)/ml. PGN from S. aureus (Fluka, Milwaukee, Wis.) was used at a concentration of 10 μg/ml, PGN from Bacillus subtilis (Fluka) was used at 10 μg/ml, LPS from E. coli strain 055:B5 (Sigma) was used at 1 μg/ml, LTA (Fluka) was used at 10 μg/ml, zymosan (Fluka) was used at 20 μg/ml, heat-inactivated S. aureus was used at 2 × 107/ml, heat-inactivated E. coli was used at 2 × 107/ml, and heat-inactivated C. albicans was used at 2 × 106/ml. Lysozyme digestion of S. aureus PGN was carried out as described by Mellroth et al. (20). Briefly, 1 mg of PGN/ml was digested with 40 μg of hen egg white lysozyme (Sigma)/ml in PBS for 24 h at room temperature with rocking. Lysozyme was then heat inactivated at 94°C for 10 min. PGN used in all experiments was examined for LPS contamination by using the Limulus amebocyte lysate assay kit (BioWhittaker, Walkersville, Md.). No significant contamination was detected. After challenge, macrophage culture supernatants were collected after 24 h, and interleukin 6 (IL-6) and tumor necrosis factor alpha (TNF-α) levels were determined by enzyme-linked immunosorbent assay (ELISA).
IL-6 and TNF-α ELISA.
IL-6 antibodies (R&D Systems) and TNF-α antibodies (R&D Systems) were coated overnight on 96-well plates at a concentration of 1 μg/ml. After blocking with 5% bovine serum albumin for 1 h, serum samples (1:3 and 1:9 dilutions) or culture supernatants were added. Two hours later, IL-6 antibodies (R&D Systems) or TNF-α antibodies (R&D Systems) were added at 200 ng/ml, respectively. The color reaction was developed using 4-nitrophenyl phosphate as a substrate and measured at an optical density of 495 nm.
RESULTS
PGRP-L is a serum protein.
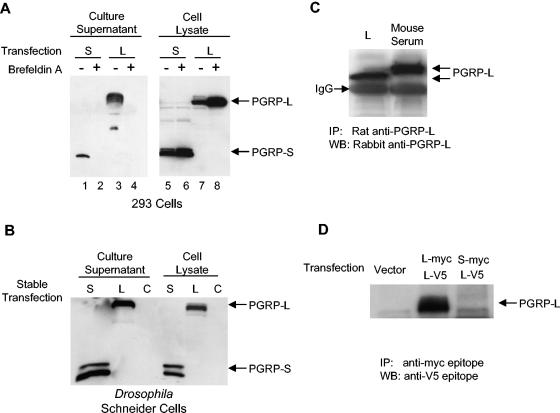
Human PGRP-L is predicted to have two transmembrane domains. In transient-transfection systems, PGRP-L localized intracellularly and on the cell membrane (35). However, human serum amidase, which circulates in blood at 100 μg/ml, has the same 15 N-terminal amino acids as PGRP-L (2) and the same biological activity (35). Similarly, mouse PGRP-L possesses amidase activity, which was first identified in blood (4, 34), raising the possibility that PGRP-L may be a soluble secreted serum protein in vivo. A Myc epitope-tagged murine PGRP-L was created and introduced into HEK 293 cells. PGRP-L was readily detected in the culture supernatants of transfected cells, and its secretion was blocked using brefeldin A, which blocks transport of proteins from the endoplasmic reticulum to the Golgi (Fig. 1A). As a positive control, PGRP-S, a putative secretory protein, was similarly tagged and was readily detected in cell supernatants, as expected. Secretion of PGRP-S was also blocked by brefeldin A, which resulted in the intracellular accumulation of both PGRP-L and PGRP-S (Fig. 1A). Similar results were obtained after expressing Myc-tagged PGRP-L in Drosophila Schneider cells, which express Drosophila PGRP-LC (Fig. 1B). Neither N-terminally tagged nor C-terminally tagged PGRP-L was detected on the cell surface of transfected cells, as assessed using flow cytometry (data not shown). Taken together, these data indicate that mammalian PGRP-L is a secreted protein, consistent with prior studies suggesting that human serum amidase is PGRP-L (2).
FIG. 1.
PGRP-L is a secreted serum protein. (A) HEK 293 cells were transfected with PGRP-L-myc (lanes 3, 4, 7, and 8) or PGRP-S-myc (lanes 1, 2, 5, and 6). Brefeldin A treatment (lanes 2, 4, 6, and 8) was used for 12 h to block protein secretion. Total cell lysates and culture supernatants were blotted with anti-Myc antibodies. (B) Drosophila Schneider cells were stably transfected with pAC5.1PGRP-L (lane L) or pAC5.1PGRP-S (lane S) or were mock transfected (lane C). Total cell lysates and culture supernatants were blotted with anti-V5 antibodies. (C) Mouse serum or PGRP-L-containing Schneider cell culture supernatants (lane L) were subject to immunoprecipitation with rat anti-PGRP-L antibodies. Immunoprecipitates were blotted with rabbit anti-PGRP-L. (D) HEK 293 cells were cotransfected with PGRP-L-myc and PGRP-L-V5 or PGRP-S-myc and PGRP-L-V5 or were mock transfected. Culture supernatants were incubated with anti-Myc antibodies. Immunoprecipitates were blotted with anti-V5 antibodies.
Two polyclonal antibodies recognizing PGRP-L were generated by immunizing rats with PGRP-L expressed from insect cells and by immunizing rabbits with PGRP-L-derived peptides. Both antibodies recognized PGRP-L when tested by ELISA or Western blot analysis and were used in combination to enhance specificity (data not shown). To test whether PGRP-L is a serum protein, coimmunoprecipitation was performed. Indeed, murine PGRP-L was immunoprecipitated from normal mouse serum using rat anti-PGRP-L antibodies (Fig. 1C). The antibody, as expected, immunoprecipitated PGRP-L expressed from insect cells as well, with the different migration properties likely reflecting differences in glycosylation in the different cell types (Fig. 1C). The immunoglobulin G staining was due to the cross-reaction of secondary antibodies used in Western blot detection with the antibodies used in the first step of coimmunoprecipitation. Together, the data presented here and previous findings by others suggest that PGRP-L is a serum amidase.
Human serum amidase is believed to be a homodimeric protein (2). To examine the capacity of PGRP-L to multimerize, we generated Myc and V5 epitopes at the carboxyl terminus of PGRP-L. Both forms were transfected into 293 cells. Myc-tagged PGRP-L was coimmunoprecipitated with the V5-tagged PGRP-L (Fig. 1D). In contrast, Myc-tagged PGRP-S did not coimmunoprecipitate with V5-tagged PGRP-L. Together, these data suggest that PGRP-L is able to form multimers, most likely homodimers, and that it is unlikely that PGRP-L and PGRP-S form heterodimers.
Generation of PGRP-L-deficient mice.
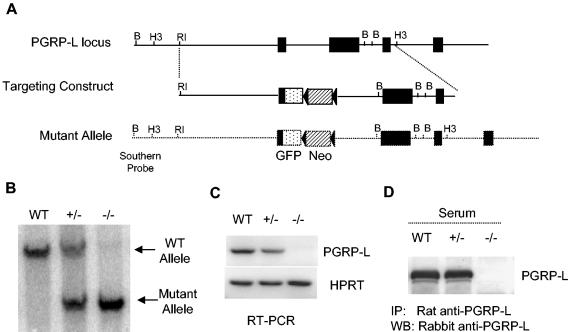
To study the function of PGRP-L in vivo, the PGRP-L gene was disrupted by homologous recombination in embryonic stem cells. A green fluorescent protein (GFP)-neomycin resistance cassette was inserted after the ATG start codon of the gene (Fig. 2A). Although it has been shown that different mRNA splicing variants are present for PGRP-L, all of these variants begin with the first ATG start codon (17). The strategy we used here is, thus, likely to produce a true null mutant. Inclusion of the GFP gene allowed us to identify cell populations that express the PGRP-L gene. Three chimeric mice were obtained, all of which gave germ line transmission of the targeted allele. Interbreeding the heterozygous mutant mice generated homozygous PGRP-L-targeted mice. A probe from the 5′ noncoding region of the gene was used to distinguish the wild-type and targeted alleles. As assessed using Southern blotting, the wild-type alleles generated a single 9.6-kb hybridizing band, whereas the homozygous targeted alleles generated a single 7.5-kb hybridizing band. Heterozygous mice revealed both wild-type and targeted alleles (Fig. 2B).
FIG. 2.
Generation of PGRP-L-deficient mouse. (A) The PGRP-L gene locus is depicted as a single line, with exons represented as filled boxes. Relevant restriction enzyme sites are shown. B, BamHI; H3, HindIII; RI, EcoRI. The organization of the targeting construct and the arrangement of the recombined PGRP-L allele are also shown. The dotted box indicates the GFP cassette that was engineered in-frame with the PGRP-L ATG start codon. The drug selection neomycin cassette (Neo) is depicted as a striped box, with two flanking loxP sites depicted as filled triangles. (B) Genotyping with Southern blot analysis. Tail DNA was subjected to agarose gel electrophoresis. The presence of the mutant allele (7.5 kb) and the wild-type allele (9.6 kb) was determined by Southern blotting, using a probe upstream of the homologous recombined sequence. (C) RT-PCR assay to determine PGRP-L expression. Liver RNA from wild-type, heterozygous, and homozygous gene-targeted mice was extracted and reverse transcribed into cDNA. PGRP-L expression was determined by PCR. The expression of hypoxanthine phosphoribosyltransferase was used as a control. (D) Serum from wild-type, heterozygous, and homozygous gene-targeted mice was immunoprecipitated using rat anti-PGRP-L antibodies. The immunoprecipitates were blotted with rabbit anti-PGRP-L antibodies.
The homozygous PGRP-L-targeted mice were viable without any gross developmental defects. They bred normally and generated normal litter sizes. To verify the absence of PGRP-L expression in the homozygous targeted mice, liver RNA from wild-type, heterozygous, and homozygous mice was reverse transcribed into cDNA and used as a template for PCR assays, using primers covering the 3′ region of the coding sequence. Hypoxanthine phosphoribosyltransferase was used as an amplification control. Using this assay, no PGRP-L expression was observed for the null mice, while expression for heterozygous mice was essentially equivalent to that for wild-type animals (Fig. 2C). There was no PGRP-L expression detected when a separate set of primers that spanned the entire coding region was used (data not shown). Finally, no PGRP-L protein was detected when serum from homozygous targeted mice was immunoprecipitated with anti-PGRP-L antibodies and examined using Western blotting (Fig. 2D). Heterozygous and wild-type animals had readily detected serum PGRP-L of the expected size when assayed similarly. Thus, we have successfully generated PGRP-L-deficient mice.
Pathogen challenges in PGRP-L-deficient mice.
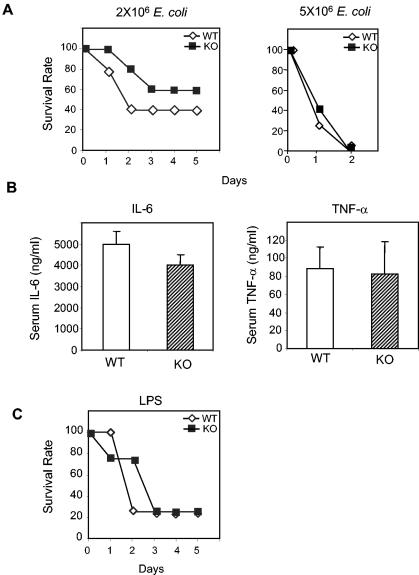
The PGRP family was first identified in innate responses against bacteria in insects (16, 37). In Drosophila, PGRP-LC mutants failed to generate antimicrobial peptides in response to gram-negative bacteria and were rendered susceptible to infection (1, 7, 27). Although lacking amidase activity, Drosophila PGRP-LC is more similar to mouse PGRP-L than to other mammalian PGRPs. Therefore, we challenged the PGRP-L null mice with the gram-negative bacterium, E. coli. Intraperitoneal injection of 2 million viable E. coli bacteria into wild-type mice led to death of 60% of the animals within 2 days, compared to a slightly reduced rate of death (40%) for PGRP-L-deficient mice (Fig. 3A). Animals surviving after 5 days recovered completely in both groups. Using a lethal challenge of E. coli (5 million organisms), both wild-type and PGRP-L-deficient mice died by 2 days (Fig. 3A).
FIG. 3.
E. coli and LPS challenges of PGRP-L-deficient mice. (A) Ten wild-type or PGRP-L-deficient animals were injected intraperitoneally with 2 million (left panel) or 5 million (right panel) live E. coli bacteria and were monitored for 5 days. Survival is plotted against time in days. (B) Three wild-type or PGRP-L-deficient animals were injected intraperitoneally with 2 million E. coli bacteria. Serum IL-6 and TNF-α levels were determined by ELISA 90 min after injection. (C) Ten wild-type or PGRP-L-deficient (KO) mice were injected intraperitoneally with 2 mg of LPS and were monitored for 10 days. Survival is plotted against time in days.
To address further any contributions of PGRP-L to host defense against gram-negative bacteria, we collected sera 90 min after injection of E. coli and analyzed these sera for the inflammatory cytokines TNF-α and IL-6. Although IL-6 levels were consistently diminished in the PGRP-L-deficient mice, this effect was modest. Little difference, if any, was observed on the level of TNF-α (Fig. 3B). As assessed using this challenge, PGRP-L contributes a relatively minor role to gram-negative host defense in mice.
The recognition of gram-negative bacteria in Drosophila is mediated by PGRP-LC-dependent interactions with the Dap-PGN specific to the gram-negative cell wall (18). LPS, the major cell wall component of gram-negative bacteria, plays a minimal role, if any, in activating antimicrobial responses in flies (18). In contrast, LPS directly activates TLR-4-mediated stimulation of the antimicrobial response in mammals (25, 26, 32). To assess whether PGRP-L contributes to LPS recognition in mammals, wild-type and PGRP-L-deficient mice were inoculated intraperitoneally with lethal doses of LPS. Although TLR-4-deficient mice survive the doses used (32), 80% of both wild-type and PGRP-L-null mice died within 2 days (Fig. 3C). Thus, PGRP-L does not seem to affect LPS sensitivity in mammals.
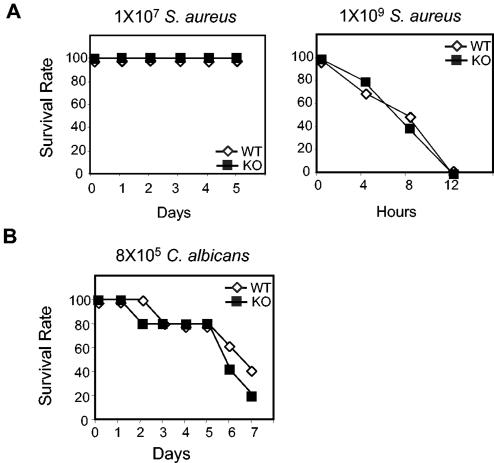
In previous studies, activation of a pathway including TLR-2 and IL-1 receptor-associated kinase 4 (IRAK-4) was necessary to mediate host defense against the gram-positive organism S. aureus, as assessed using a challenge with 10 million bacteria (29). To evaluate the potential role of PGRP-L in this pathway, PGRP-L-deficient mice were challenged with S. aureus and compared with wild-type mice (Fig. 4A). Challenge with 10 million or 100 million S. aureus bacteria caused no lethality in either PGRP-null or wild-type mice (Fig. 4A; also data not shown). Increasing the challenge to 1 billion organisms led to death of all mice in both cohorts within 8 h (Fig. 4A). Thus, in contrast to IRAK-4- and TLR-2-deficient mice, we could discern no differences between wild-type and PGRP-L-deficient mice using challenge with S. aureus in vivo.
FIG. 4.
S. aureus and C. albicans challenges of PGRP-L-deficient mice. (A) Ten wild-type or PGRP-L-deficient mice were injected intraperitoneally with 10 million (left panel) or 1 billion (right panel) live S. aureus bacteria and were monitored for a 10- or 1-day period, respectively. Survival is plotted against time. (B) C. albicans (0.8 million viable organisms) was injected intravenously into 10 wild-type or PGRP-L-deficient mice. Survival was monitored over a 7-day period.
In Drosophila, PGRP has been placed upstream of Toll signaling, which is required to mediate defense against both gram-positive bacterial infections and fungal infections (21). To determine whether mouse PGRP-L is involved in fungal defense, PGRP-L null mice were challenged with C. albicans. Using a tail vein injection of 0.8 million viable yeast cells (13), 60% of both wild-type and PGRP-L-deficient animals succumbed by 7 days (Fig. 4B), suggesting little contribution by PGRP-L to innate fungal immunity.
Macrophage responses to TLR ligands in the absence of PGRP-L.
Macrophages play a crucial role in the innate immune response against infectious organisms. Macrophages express an array of TLRs that are critical in sensing microbial pattern recognition elements and activating the production of inflammatory cytokines, such as IL-6 and TNF-α (14). TLR-2 is involved in responses to PGN and yeast zymosan particles. These distinct responses are mediated by TLR-2 in conjunction with different coreceptors (10, 23), although TLR-2 is required for optimal activation of both pathways and is critical for responses against both gram-positive bacteria and fungi (30). TLR-4 is involved in LPS recognition and is essential for responses against gram-negative bacteria (14). In Drosophila, PGRP-SA functions upstream of Toll (21), raising the possibility that mammalian PGRP-L might be involved upstream of TLR recognition.
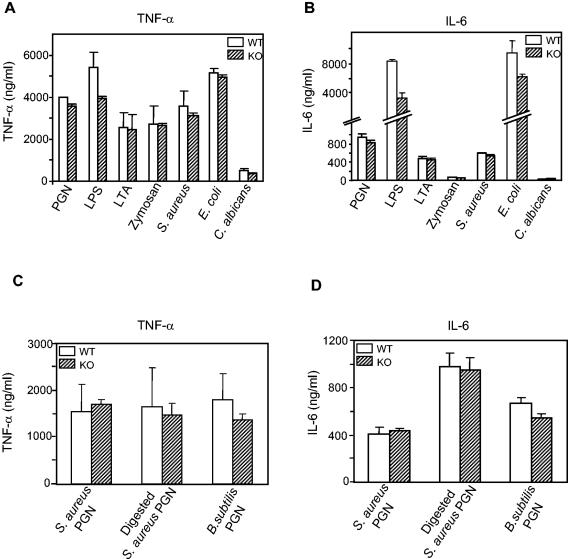
To test this possibility, peritoneal macrophages from wild-type and PGRP-L-deficient littermates were stimulated with various TLR-2 and TLR-4 ligands, and the ability of these macrophages to produce IL-6 and TNF-α was assayed by ELISA. Wild-type or PGRP-L knockout mouse serum was used in the macrophage cultures to avoid any potential interference of PGRP-L present in fetal bovine serum. Over several experiments, PGRP-L-deficient macrophages consistently produced less IL-6 in response to LPS or heat-killed E. coli; TNF-α was also attenuated using LPS, but not with heat-killed E. coli (Fig. 5). Stimulation using PGN, LTA, heat-killed gram-positive bacteria, zymosan, or heat-killed Candida, however, led to comparable generation of inflammatory cytokines by wild-type and PGRP-L-deficient macrophages.
FIG. 5.
Cytokine production by thioglycolate-elicited peritoneal macrophages from PGRP-L-deficient mice. (A) 2 × 105 peritoneal macrophages from wild-type (open bars) or PGRP-L-deficient (striped bars) animals were stimulated in the presence of 50 U of IFN-γ/ml with the various TLR-2 and TLR-4 ligands shown. TNF-α in supernatants was determined by ELISA after 24 h. (B) IL-6 levels in supernatants from peritoneal macrophages prepared as for panel A. (C) Peritoneal macrophages (2 × 105) from wild-type (open bars) or PGRP-L-deficient (striped bars) animals were stimulated in the presence of 50 U of IFN-γ/ml with 10 μg of S. aureus PGN, lysozyme-digested S. aureus PGN, or B. subtilis PGN/ml. TNF-α levels in supernatants were determined by ELISA after 24 h. (D) IL-6 levels in supernatants from peritoneal macrophages prepared as for panel C.
Drosophila distinguishes gram-positive and gram-negative bacterial through recognition of Lys-PGN by PGRP-SA and recognition of Dap-PGN by PGRP-LC, respectively (15, 18). In mammals, PGRP-L optimally recognizes Lys-PGN after it is digested by lysozyme (4). To test whether PGRP-L is involved in recognition of different types of PGN, wild-type and PGRP-L-deficient peritoneal macrophages were stimulated with Lys-PGN from S. aureus, lysozyme-digested PGN from S. aureus, and Dap-PGN from B. subtilis. B. subtilis is a gram-positive bacterium with Dap-PGN on the cell surface (28). PGRP-L-deficient macrophages produced levels of both TNF-α and IL-6 that were similar to those produced by wild-type macrophages (Fig. 5C and D). The increase in IL-6 production by macrophages challenged with lysozyme-digested PGN is most likely due to the easier accessibility of macrophages to smaller PGN particles. Taken together, we cannot exclude a relatively minor role for PGRP-L in Toll-mediated pathogen recognition, at least in response to gram-negative challenge or LPS, but contributions to the TLR-2-mediated signaling pathway appear even less apparent.
DISCUSSION
We present our initial characterization of the role of mammalian PGRP-L, using gene-targeted mice. Although certain Drosophila PGRPs perform nonredundant roles in host defense against both bacteria and fungi, we could define no obvious requirement for PGRP-L after lethal and sublethal challenges with such organisms in vivo. Macrophages isolated from PGRP-L-deficient mice responded to LPS and E. coli with slightly diminished cytokine responses in vitro, but responses to gram-positive organisms, PGN, and yeast were not affected. PGRP-L-deficient macrophages were not attenuated in their capacity to respond to gram positive Lys-PGN or gram-negative Dap-PGN. Additionally, we provide direct evidence that PGRP-L is secreted and multimerizes in vitro and circulates in the serum of mice. Mice deficient in PGRP-L have no obvious phenotypic abnormalities, and further studies will be required to define fully the evolutionary pressures that have maintained this gene in mammals.
Mammalian serum N-acetylmuramyl-l-alanine amidase was purified from human blood almost 10 years ago (2). The enzyme was a 74-kDa protein and could form dimers. The first 15 amino acids were sequenced, and after the cloning of mammalian PGRP-L, it became apparent that the sequences were identical. A discrepancy, however, was the prediction that PGRP-L was a transmembrane rather than a secreted, protein (19). As we demonstrate here in agreement with prior findings (4), mammalian PGRP-L likely represents the previously identified serum amidase. Using tagged PGRP-L, we demonstrate secretion of PGRP-L in two different cell lines. More importantly, PGRP-L antibodies were able to immunoprecipitate PGRP-L in serum from normal mice. Further, as suggested by previous findings (2), PGRP-L could form homodimers. Others, however, using different expression systems, have reported human PGRP-L to be a cell surface protein (35), and it is possible that distinct isoforms may not be secreted. We were unable to detect any surface staining on cells expressing PGRP-L in our studies (data not shown). The expression of PGRP-L in the liver would be consistent with its secretion into serum. Although the liver is the source of acute-phase serum proteins that are increased in response to systemic infections, we were unable to document a substantial increase in hepatic PGRP-L expression in response to systemic challenges that induced typical acute-phase responses in the mouse (data not shown).
Although the range of challenges we used does not rule out some role for PGRP-L in mammalian innate immunity, we think it unlikely that such a role will be major. Mice survived low-dose E. coli challenge slightly better than wild-type littermates and had a slight delay in the time to death following lethal LPS injection. This is in contrast with the substantial impact of TLR-4 deletion on these phenotypes, however (25, 26, 32). Additionally, in contrast to Drosophila, in which Dap-PGN recognition is nonredundantly mediated by PGRP upstream of the Imd/Relish pathway (15, 18), murine cells respond to Dap-PGN in the complete absence of PGRP-L. Thus, detection of LPS instead of Dap-PGN is the major mechanism for mammals to sense gram-negative bacterial infection, and distinguishing Lys-PGN and Dap-PGN may not be necessary with the development of TLR-2-mediated pathways. We did note a diminution of TNF-α and IL-6 production by PGRP-L-deficient macrophages in response to LPS and E. coli in vitro, despite the suggestion that PGRP-L, perhaps via its enzymatic amidase activity, may serve to attenuate proinflammatory responses (12). However, these effects were only partial, and no effects of PGRP-L deficiency could be ascertained in challenges with gram-positive bacteria, gram-positive PGN, yeast, or yeast cell wall constituents in vivo or in vitro in the absence of PGRP-L.
Although expressed as different isoforms, PGRP-L transcripts and proteins were entirely absent from our gene-targeted mice. As we document here and as shown elsewhere (2), PGRP-L can form homodimers, and it is possible that PGRP-L can dimerize with other PGRP proteins. We could find no evidence that PGRP-L and PGRP-S can heterodimerize using in vitro assays. PGRP-Iα/Iβ are also soluble proteins with the capacity to dimerize (9). Thus, heterodimerization may play a critical role in determining the function of PGRPs. Alternatively, other PGRPs might compensate for the lack of PGRP-L. The expression patterns of the three mammalian PGRP genes do not obviously overlap, with PGRP-L predominantly in the liver, spleen and thymus, PGRP-S in neutrophils, and PGRP-Iα/Iβ in the esophagus (19). Considering the fact that they are all soluble, it is not surprising that they can function on the same target tissues. Double- or triple-PGRP knockout mice may be required to rule out definitively a requirement for PGRP family members in host innate immune responses.
Despite the conservation of PGRP and TLR family members from insects to humans, their interactions in innate immune pathways may have diverged. Drosophila contains 13 PGRP genes, 2 of which, PGRP-SA and -LC, are upstream of Toll and Imd, respectively (1, 7, 21, 27). Whereas all of the mammalian TLRs have been implicated in innate immunity, most of the nine Drosophila TLRs have developmental roles. Only Toll has been shown to have a definitive role in innate immune function in the fly (11). The expansion of the role of TLRs in mammalian immunity may have caused a diminution of the role for PGRPs. Another major difference between Drosophila and mammalian TLRs lies in their ligands. In Drosophila, Toll is activated at the end of a serum proteolytic cascade that generates an endogenous ligand, Spaetzle. In mammals, TLRs are believed to act by direct interaction with microbial PAMPs, often in concert with coreceptor(s). The capacity for direct binding to PAMPs by mammalian TLRs may have obviated the need for recognition by soluble binding factors, such as PGRPs. A final consideration that may have lessened the role for PGRPs in innate immunity has been the emergence of adaptive T- and B-cell immune responses. The amplification of immunity acquired through the activities of clonal expansion and antibody production may have contributed to the divergence of the role for PGRPs in vertebrate immunity. Despite the finding that PGRP-L may have a relatively minor role in innate immune functions, the mice we report will be valuable in pursuing possible alternative roles for these proteins in vertebrate biology.
Acknowledgments
We thank N. Flores and D. Winkel for technical support, S. Noble for help with C. albicans, and E. Brown and A. DeFranco for helpful review of the manuscript.
This work was supported by grant AI30663 from the National Institutes of Health and the Howard Hughes Medical Institute. R.M.L. is a Senior Scholar of the Ellison Medical Foundation for Global Infectious Diseases.
REFERENCES
- 1.Choe, K. M., T. Werner, S. Stoven, D. Hultmark, and K. V. Anderson. 2002. Requirement for a peptidoglycan recognition protein (PGRP) in Relish activation and antibacterial immune responses in Drosophila. Science 296:359-362. [DOI] [PubMed] [Google Scholar]
- 2.De Pauw, P., C. Neyt, E. Vanderwinkel, R. Wattiez, and P. Falmagne. 1995. Characterization of human serum N-acetylmuramyl-L-alanine amidase purified by affinity chromatography. Protein Exp. Purif. 6:371-378. [DOI] [PubMed] [Google Scholar]
- 3.Dziarski, R., K. A. Platt, E. Gelius, H. Steiner, and D. Gupta. 2003. Defect in neutrophil killing and increased susceptibility to infection with nonpathogenic gram-positive bacteria in peptidoglycan recognition protein-S (PGRP-S)-deficient mice. Blood 102:689-697. [DOI] [PubMed] [Google Scholar]
- 4.Gelius, E., C. Persson, J. Karlsson, and H. Steiner. 2003. A mammalian peptidoglycan recognition protein with N-acetylmuramoyl-L-alanine amidase activity. Biochem. Biophys. Res. Commun. 306:988-994. [DOI] [PubMed] [Google Scholar]
- 5.Georgel, P., S. Naitza, C. Kappler, D. Ferrandon, D. Zachary, C. Swimmer, C. Kopczynski, G. Duyk, J. M. Reichhart, and J. A. Hoffmann. 2001. Drosophila immune deficiency (IMD) is a death domain protein that activates antibacterial defense and can promote apoptosis. Dev. Cell 1:503-514. [DOI] [PubMed] [Google Scholar]
- 6.Gobert, V., M. Gottar, A. A. Matskevich, S. Rutschmann, J. Royet, M. Belvin, J. A. Hoffmann, and D. Ferrandon. 2003. Dual activation of the Drosophila toll pathway by two pattern recognition receptors. Science 302:2126-2130. [DOI] [PubMed] [Google Scholar]
- 7.Gottar, M., V. Gobert, T. Michel, M. Belvin, G. Duyk, J. A. Hoffmann, D. Ferrandon, and J. Royet. 2002. The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature 416:640-644. [DOI] [PubMed] [Google Scholar]
- 8.Gu, H., Y. R. Zou, and K. Rajewsky. 1993. Independent control of immunoglobulin switch recombination at individual switch regions evidenced through Cre-loxP-mediated gene targeting. Cell 73:1155-1164. [DOI] [PubMed] [Google Scholar]
- 9.Guan, R., E. L. Malchiodi, W. Qian, P. Schuck, and R. A. Mariuzza. 2004. Crystal structure of the C-terminal peptidoglycan-binding domain of human peptidoglycan recognition protein Iα. J. Biol. Chem. 279:31873-31882. [DOI] [PubMed] [Google Scholar]
- 10.Hajjar, A. M., D. S. O'Mahony, A. Ozinsky, D. M. Underhill, A. Aderem, S. J. Klebanoff, and C. B. Wilson. 2001. Cutting edge: functional interactions between toll-like receptor (TLR) 2 and TLR1 or TLR6 in response to phenol-soluble modulin. J. Immunol. 166:15-19. [DOI] [PubMed] [Google Scholar]
- 11.Hoffmann, J. A. 2003. The immune response of Drosophila. Nature 426:33-38. [DOI] [PubMed] [Google Scholar]
- 12.Hoijer, M. A., M. J. Melief, R. Debets, and M. P. Hazenberg. 1997. Inflammatory properties of peptidoglycan are decreased after degradation by human N-acetylmuramyl-L-alanine amidase. Eur. Cytokine Netw. 8:375-381. [PubMed] [Google Scholar]
- 13.Inglis, D. O., and A. D. Johnson. 2002. Ash1 protein, an asymmetrically localized transcriptional regulator, controls filamentous growth and virulence of Candida albicans. Mol. Cell. Biol. 22:8669-8680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janeway, C. A., Jr., and R. Medzhitov. 2002. Innate immune recognition. Annu. Rev. Immunol. 20:197-216. [DOI] [PubMed] [Google Scholar]
- 15.Kaneko, T., W. E. Goldman, P. Mellroth, H. Steiner, K. Fukase, S. Kusumoto, W. Harley, A. Fox, D. Golenbock, and N. Silverman. 2004. Monomeric and polymeric gram-negative peptidoglycan but not purified LPS stimulate the Drosophila IMD pathway. Immunity 20:637-649. [DOI] [PubMed] [Google Scholar]
- 16.Kang, D., G. Liu, A. Lundstrom, E. Gelius, and H. Steiner. 1998. A peptidoglycan recognition protein in innate immunity conserved from insects to humans. Proc. Natl. Acad. Sci. USA 95:10078-10082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kibardin, A. V., I. I. Mirkina, E. V. Baranova, I. R. Zakeyeva, G. P. Georgiev, and S. L. Kiselev. 2003. The differentially spliced mouse tagL gene, homolog of tag7/PGRP gene family in mammals and Drosophila, can recognize Gram-positive and Gram-negative bacterial cell wall independently of T phage lysozyme homology domain. J. Mol. Biol. 326:467-474. [DOI] [PubMed] [Google Scholar]
- 18.Leulier, F., C. Parquet, S. Pili-Floury, J. H. Ryu, M. Caroff, W. J. Lee, D. Mengin-Lecreulx, and B. Lemaitre. 2003. The Drosophila immune system detects bacteria through specific peptidoglycan recognition. Nat. Immunol. 4:478-484. [DOI] [PubMed] [Google Scholar]
- 19.Liu, C., Z. Xu, D. Gupta, and R. Dziarski. 2001. Peptidoglycan recognition proteins: a novel family of four human innate immunity pattern recognition molecules. J. Biol. Chem. 276:34686-34694. [DOI] [PubMed] [Google Scholar]
- 20.Mellroth, P., J. Karlsson, and H. Steiner. 2003. A scavenger function for a Drosophila peptidoglycan recognition protein. J. Biol. Chem. 278:7059-7064. [DOI] [PubMed] [Google Scholar]
- 21.Michel, T., J. M. Reichhart, J. A. Hoffmann, and J. Royet. 2001. Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature 414:756-759. [DOI] [PubMed] [Google Scholar]
- 22.O'Gorman, S., N. A. Dagenais, M. Qian, and Y. Marchuk. 1997. Protamine-Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. Proc. Natl. Acad. Sci. USA 94:14602-14607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ozinsky, A., D. M. Underhill, J. D. Fontenot, A. M. Hajjar, K. D. Smith, C. B. Wilson, L. Schroeder, and A. Aderem. 2000. The repertoire for pattern recognition of pathogens by the innate immune system is defined by cooperation between toll-like receptors. Proc. Natl. Acad. Sci. USA 97:13766-13771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pili-Floury, S., F. Leulier, K. Takahashi, K. Saigo, E. Samain, R. Ueda, and B. Lemaitre. 2004. In vivo RNA interference analysis reveals an unexpected role for GNBP1 in the defense against Gram-positive bacterial infection in Drosophila adults. J. Biol. Chem. 279:12848-12853. [DOI] [PubMed] [Google Scholar]
- 25.Poltorak, A., X. He, I. Smirnova, M. Y. Liu, C. Van Huffel, X. Du, D. Birdwell, E. Alejos, M. Silva, C. Galanos, M. Freudenberg, P. Ricciardi-Castagnoli, B. Layton, and B. Beutler. 1998. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 282:2085-2088. [DOI] [PubMed] [Google Scholar]
- 26.Qureshi, S. T., L. Lariviere, G. Leveque, S. Clermont, K. J. Moore, P. Gros, and D. Malo. 1999. Endotoxin-tolerant mice have mutations in Toll-like receptor 4 (Tlr4). J. Exp. Med. 189:615-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramet, M., P. Manfruelli, A. Pearson, B. Mathey-Prevot, and R. A. Ezekowitz. 2002. Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli. Nature 416:644-648. [DOI] [PubMed] [Google Scholar]
- 28.Schleifer, K. H., and O. Kandler. 1972. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36:407-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzuki, N., S. Suzuki, G. S. Duncan, D. G. Millar, T. Wada, C. Mirtsos, H. Takada, A. Wakeham, A. Itie, S. Li, J. M. Penninger, H. Wesche, P. S. Ohashi, T. W. Mak, and W. C. Yeh. 2002. Severe impairment of interleukin-1 and Toll-like receptor signalling in mice lacking IRAK-4. Nature 416:750-756. [DOI] [PubMed] [Google Scholar]
- 30.Takeda, K., T. Kaisho, and S. Akira. 2003. Toll-like receptors. Annu. Rev. Immunol. 21:335-376. [DOI] [PubMed] [Google Scholar]
- 31.Takehana, A., T. Katsuyama, T. Yano, Y. Oshima, H. Takada, T. Aigaki, and S. Kurata. 2002. Overexpression of a pattern-recognition receptor, peptidoglycan-recognition protein-LE, activates imd/relish-mediated antibacterial defense and the prophenoloxidase cascade in Drosophila larvae. Proc. Natl. Acad. Sci. USA 99:13705-13710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takeuchi, O., K. Hoshino, T. Kawai, H. Sanjo, H. Takada, T. Ogawa, K. Takeda, and S. Akira. 1999. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity 11:443-451. [DOI] [PubMed] [Google Scholar]
- 33.Tybulewicz, V. L., C. E. Crawford, P. K. Jackson, R. T. Bronson, and R. C. Mulligan. 1991. Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl proto-oncogene. Cell 65:1153-1163. [DOI] [PubMed] [Google Scholar]
- 34.Valinger, Z., B. Ladesic, and J. Tomasic. 1982. Partial purification and characterization of N-acetylmuramyl-L-alanine amidase from human and mouse serum. Biochim. Biophys. Acta 701:63-71. [DOI] [PubMed] [Google Scholar]
- 35.Wang, Z. M., X. Li, R. R. Cocklin, M. Wang, K. Fukase, S. Inamura, S. Kusumoto, D. Gupta, and R. Dziarski. 2003. Human peptidoglycan recognition protein-L is an N-acetylmuramoyl-L-alanine amidase. J. Biol. Chem. 278:49044-49052. [DOI] [PubMed] [Google Scholar]
- 36.Werner, T., G. Liu, D. Kang, S. Ekengren, H. Steiner, and D. Hultmark. 2000. A family of peptidoglycan recognition proteins in the fruit fly Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 97:13772-13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshida, H., K. Kinoshita, and M. Ashida. 1996. Purification of a peptidoglycan recognition protein from hemolymph of the silkworm, Bombyx mori. J. Biol. Chem. 271:13854-13860. [DOI] [PubMed] [Google Scholar]