Abstract
Translational initiation of hepatitis C virus (HCV) mRNA occurs by internal entry of ribosomes into an internal ribosomal entry site (IRES) at the 5′ nontranslated region. A region encoding the N-terminal part of the HCV polyprotein has been shown to augment the translation of HCV mRNA. Here we show that a cellular protein, NS1-associated protein 1 (NSAP1), augments HCV mRNA translation through a specific interaction with an adenosine-rich protein-coding region within the HCV mRNA. The overexpression of NSAP1 specifically enhanced HCV IRES-dependent translation, and knockdown of NSAP1 by use of a small interfering RNA specifically inhibited the translation of HCV mRNA. An HCV replicon RNA capable of mimicking the HCV proliferation process in host cells was further used to confirm that NSAP1 enhances the translation of HCV mRNA. These results suggest the existence of a novel mechanism of translational enhancement that acts through the interaction of an RNA-binding protein with a protein coding sequence.
The translation of eukaryotic mRNAs occurs either by cap-dependent scanning or by direct binding of a ribosome to a specialized RNA element called an internal ribosomal entry site (IRES) (12). Although the canonical translation initiation factors function in translation through IRES elements, many RNA-binding proteins have also been shown to play important roles (12, 14). For instance, host cellular proteins such as polypyrimidine tract-binding protein (PTB), the La autoantigen, poly(rC)-binding proteins, and upstream of N-ras (Unr) bind directly to IRESs and enhance the translation of picornaviral mRNAs (12, 14).
Hepatitis C virus (HCV), the major causative agent of virus-related liver cirrhosis and hepatocellular carcinoma in humans, is a positive-sense RNA virus. The HCV RNA contains a 341-nucleotide (nt) 5′ nontranslated region (5′NTR) and a very long open reading frame encoding a polyprotein that is proteolytically processed into 10 or more viral proteins (9, 15). Translation of the HCV polyprotein is directed by an IRES element spanning the 5′NTR (29).
Curiously, while most IRESs require only the 5′NTR for full activity, the HCV IRES depends on the presence of a protein-coding sequence downstream of the initiating AUG (19, 26). An HCV RNA encoding the N-terminal part of the HCV polyprotein was shown to be required for full HCV IRES activity when heterologous reporter genes (influenza NS′ and secreted alkaline phosphatase) were connected to the HCV 5′NTR (26). Moreover, the same region was absolutely required for the generation of a viable chimeric poliovirus that used the HCV IRES element for translation (19). However, several other reports have suggested that the core-coding sequence is not strictly essential for HCV IRES activity (27, 29, 30). Thus, the true molecular basis of translational activation by the HCV core-coding sequence is still a subject of some debate.
Here we investigated the HCV IRES translational initiation mechanism by analyzing proteins that were found to specifically interact with the IRES element. RNA affinity chromatography and peptide mass fingerprinting through matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF) revealed that NS1-associated protein 1 (NSAP1) specifically interacts with a protein-coding region downstream of the HCV polyprotein initiation codon.
NSAP1 was originally identified as a protein capable of interacting with NS1, which is the major nonstructural protein of the mouse minute virus (10). NSAP1 is highly homologous (82.1% identity) to human heterogeneous nuclear ribonucleoprotein R (hnRNP R) (11) and was shown to modulate c-fos mRNA stabilization by forming a protein complex with Unr, PABP, and PAIP1 (6). The protein also interacts with synaptotagmin and with poly(A) RNA, which led to its independent designation as synaptotagmin-binding, cytoplasmic RNA-interacting protein (SYNCRIP) (21).
The role of NSAP1 in HCV IRES-dependent translation was investigated by modifying the cis-acting element and the amounts of NSAP1 in several translation assay systems. Modification of the NSAP1-binding site, which is composed of an adenosine-rich region (hereafter referred to as the ACR), reduced the activity of the HCV IRES in the assay systems and in an HCV replicon system designed to mimic the HCV proliferation process. The expression of NSAP1 enhanced HCV IRES-dependent translation, whereas a knockdown of NSAP1 levels by the use of targeted small interfering RNAs (siRNAs) specifically hampered HCV IRES activity. These results strongly suggest that NSAP1 augments HCV IRES activity through an interaction with the ACR and provide evidence for a novel mode of translational activation occurring through the interaction of an RNA-binding protein with coding sequences downstream of the initiation codon.
MATERIALS AND METHODS
Plasmid construction.
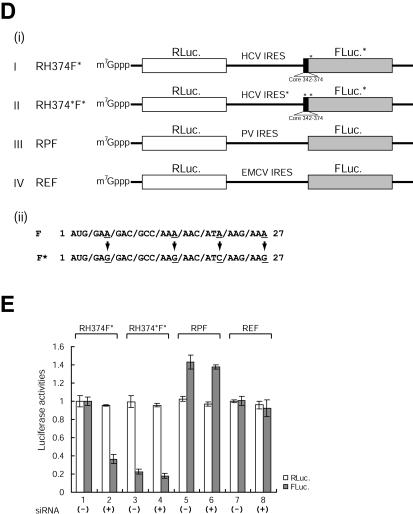
For the generation of pSK(−)/NSAP1, a plasmid containing the full-length coding sequence of murine NSAP1 (pRSETC9-15; kindly provided by C. Astell, University of British Columbia, Vancouver, Canada) was digested with HindIII and XbaI, and the insert was cloned into the pBluescript SK(−) [pSK(−)] vector (Clontech). The mouse and human NSAP1 amino acid sequences differ by only one amino acid (alanine-357 to serine). For the construction of pEGFP-c1/NSAP1, pSK(−)/NSAP1 was treated with EcoRI-Klenow-Asp718, and the DNA fragment was cloned into pEGFP-c1 (Clontech) treated with NheI-Klenow-Asp718. For the generation of pQE31-NSAP1 for recombinant NSAP1 expression, pSK(−)/NSAP1 was treated with BamHI-Klenow-KpnI, and the DNA fragment was cloned into NheI-Klenow-KpnI-treated pQE31 (Qiagen). To obtain the human NSAP1 cDNA clone, we PCR amplified a 1.7-kb fragment spanning the NSAP1 coding region from a Chang's liver cDNA library with the following oligomers: 5′-CCATCGATGCTAGCATGGCTACAGAACATGTTAATGG-3′ and 5′-GCTCTAGACTCGAGTCATTGTAACAGGTCAGGACCG-3′. The amplified DNA was digested with ClaI and XbaI and then inserted into pSK(−). For the generation of pGL3-H(18-402), plasmid pH(18-402)CAT was treated with Asp718-BamHI-Klenow, and the DNA fragment containing the HCV IRES (18-402) was isolated and ligated into an NcoI-Klenow-treated pGL3 control vector (Promega). For the generation of pRH402F, pGL3-H(18-402) was treated with HindIII-XbaI-Klenow, and the DNA fragment containing the HCV IRES and the firefly luciferase reporter was isolated and ligated with XbaI-Klenow-treated pRL-CMV (Promega). For the generation of pRPF, pCATΔ-PCAT (unpublished data), which contains the poliovirus (PV) IRES and pRM531F (17), was treated with SalI-SmaI and then ligated together. For the generation of pREF, the encephalomyocarditis virus (EMCV) IRES fragment was PCR amplified from PMPS1-ECAT (2) by use of the following primers: 5′-CCATCGATACCCCTCTCCCTCCCC-3′ and 5′-TCCCCCCGGGCATGGTTGTGGCCATATTATC-3′. The amplified DNA was treated with AccI-XmaI, and the DNA fragment was cloned into ClaI-XmaI-treated pRM531F. The monocistronic constructs pH(342-402)CAT, pH(18-402)CAT, and pH(18-374)CAT were described previously (8). For the construction of pH(18-374*)CAT, the oligomers 5′-AGACCGTGCATCATGAGCACGAATCCTAAGCCTCAGAGGAAGACCG-3′ and 5′-GATCCGGTCTTCCTCTGAGGCTTAGGATTCGTGCTCATGATGCACGGT-3′ were annealed and inserted into AccI-BamHI-treated pH(18-374)CAT. Construct pRH374F* contains adenine-to-guanosine replacements in the firefly luciferase gene, as shown in Fig. 4D, panel ii, and pRH374*F* contains mutations in both the ACR and the firefly luciferase gene. For the introduction of these mutations, the HCV core-coding region and the N-terminal portion of the firefly luciferase sequence were PCR amplified by use of the following oligomers: 5′-CGGGATCCATGGAGGACGCCAAGAACATCAAGAAGGGCCCGGCG-3′ and 5′-TCCAAAACCGTGATGGAATGG-3′. The DNAs corresponding to the HCV 5′NTRs were isolated from Asp718-BamHI-treated pH(18-374)CAT or pH(18-374*)CAT. The DNA fragments generated by PCR and restriction enzyme digestion were cloned into Asp718-SphI-treated pRH402F by three fragment ligations. To knockdown endogenous NSAP1, we designed a double-stranded RNA corresponding to nt 209 to 231 downstream of the initiation codon of human NSAP1 (GenBank accession no. NM_006372) to generate a siRNA. For the generation of pAVU6+27/NSAP1, oligomers (sense, 5′-TCGACGACGGTGCATTGGCAGTTCTTCAAGAGAGAACTGCCAATGCACCGTCTTTTT-3′; antisense, 5′-CTAGAAAAAGACGGTGCATTGGCAGTTCTCTCTTGAAGAACTGCCAATGCACCGTCG-3′) were annealed and inserted into pAVU6+27 (24) treated with SalI and XbaI. Plasmid pEBV-no promoter was constructed by the self-ligation of pEBV-HisA (Invitrogen) treated with SalI. Plasmids pEBV-U6+27 and pEBV-U6+27/NSAP1 were constructed by ligation of the SalI-Klenow-treated pEBV-no promoter DNA fragment and of pAVU6+27 and pAVU6+27/NSAP1, respectively, treated with BamHI-Klenow-SacII-T4 polymerase. For the construction of plasmids repI374 and repI374*, pRH374F* and pRH374*F* were treated with NcoI-Klenow-NruI, and the resulting DNA fragments were cloned into rep5.1 treated with AscI-Klenow-NruI. All constructs were verified by DNA sequencing.
FIG. 4.
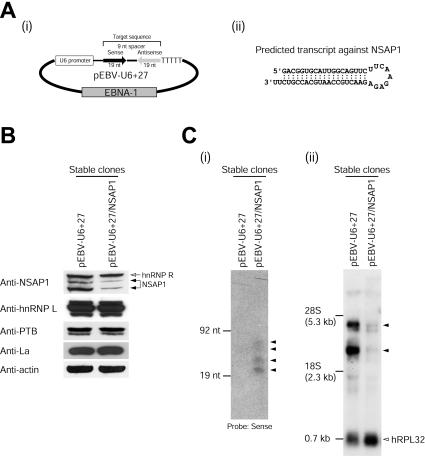
Knockdown of NSAP1 by siRNA reduces HCV IRES activity. (A) (i) Schematic diagram of the pEBV-U6+27 plasmid expressing a tandem-type siRNA against NSAP1. (ii) Predicted siRNA structure generated by pEBV-U6+27/NSAP1. (B) Levels of NSAP1 protein in HeLa cells stably expressing siRNAs against NSAP1 (lane pEBV-U6+27/NSAP1) and in control cells (lane pEBV-U6+27) were monitored by immunoblot analysis. About 1 μg of the pEBV-U6+27 or pEBV-U6+27/NSAP1 plasmid was transfected into HeLa cells by electroporation. After hygromycin (400 μg/ml) selection for 1 month, total cell lysates from pools of hygromycin-resistant colonies were immunoblotted with antibodies against NSAP1, hnRNP L, PTB, La, and actin (ICN Biomedicals). (C) (i) Cellular expression patterns of siRNAs against NSAP1 were monitored by Northern blot analysis. RNA size markers were generated by in vitro transcription. Arrowheads depict siRNAs generated by the transfection of pEBV-U6+27/NSAP1. (ii) Levels of NSAP1 mRNAs in siRNA-expressing HeLa cells were monitored by Northern blot analysis. Arrowheads depict isoforms of NSAP1 transcripts. (D) Schematic diagrams of dicistronic mRNAs used in RNA transfection experiments. (i) The dicistronic plasmids and the β-galactosidase-encoding plasmid pCMV · SPORT-βgal were treated with HpaI and XbaI, respectively. The linearized DNAs were in vitro transcribed with the T7 (dicistronic mRNAs) or SP6 (β-galactosidase mRNA) RNA polymerase in the presence of m7G(5′)pppG(5′). The intercistronic regions of the dicistronic mRNAs RH374F*, RH374*F*, RPF, and REF contained the wild-type HCV IRES (nt 18 to 374), the adenosine-to-guanosine mutant HCV IRES (nt 18 to 374), the PV IRES, and the EMCV IRES, respectively. (ii) Adenosine residues in the adenosine-rich region in the N-terminal coding region of the firefly luciferase gene were mutated to guanosine or cytidine residues to eliminate the adenosine-rich elements in firefly luciferase; the amino acid sequence of the firefly luciferase remained unchanged in both cases. The asterisk denotes the nucleotide changes in the ACR of the firefly luciferase gene. (E) Translation activities of dicistronic mRNAs in siRNA-expressing cells. Dicistronic mRNAs and control β-galactosidase mRNAs were cotransfected into control (pEBV-U6+27) and NSAP1 siRNA-expressing (pEBV-U6+27/NSAP1) cells. At 3 h posttransfection, luciferase activities were measured and normalized against the β-galactosidase activity to control for the transfection efficiency. For IRES-dependent translation, firefly luciferase activity, which was directed by mRNA RH374F*, was arbitrarily set to 1. For cap-dependent translation, Renilla luciferase activity, which was directed by mRNA RH374F*, was arbitrarily set to 1. (−) and (+), control cells and siRNA-expressing cells, respectively. Luciferase activities are shown with bars, and error bars indicate standard deviation values.
RNA affinity chromatography.
Biotinylated RNAs used in affinity chromatography experiments were produced from pH(18-402)CAT linearized with BamHI and T7 polymerase by use of an in vitro transcription kit according to the manufacturer's recommendations (Stratagene). The biotinylated RNAs (15 μg) were conjugated with streptavidin-agarose resin (Pierce) in the presence of incubation buffer (10 mM Tris-Cl [pH 7.5], 150 mM KCl, 1.5 mM MgCl2, 0.5 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, and 0.05% Nonidet P-40). Samples were incubated at 4°C for 2 h, and unbound RNAs were removed by washing two times with incubation buffer. After the washing step, S10 extracts (500 μg) and yeast tRNA (30 μg; Roche) were applied to the resin, which was incubated for 30 min at room temperature and then for 2 h at 4°C. The resin was then washed five times with incubation buffer, and the resin-bound proteins were resolved by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-10% PAGE). The biotinylated RNAs used for pull-down assays were generated as previously described (17). Template DNAs were prepared by AccI digestion of plasmid pH(18-402)CAT and by BamHI digestion of plasmids pH(18-374)CAT, pH(18-374*)CAT, and pH(18-402)CAT.
Purification of recombinant NSAP1.
Escherichia coli (M15) cells were used to produce recombinant NSAP1 from plasmid pQE31-NSAP1. Isopropyl-β-d-thiogalactopyranoside (final concentration, 0.5 mM) was added to induce NSAP1 protein expression at an optical density at 600 nm of 0.5. After incubation at 27°C for 5 h, the cells were harvested, resuspended in lysis buffer (20 mM sodium phosphate [pH 7.6], 300 mM NaCl, 0.5 mM phenylmethylsulfonyl fluoride, 0.5 μg of leupeptin/ml, 1 mM β-mercaptoethanol, and 10% glycerol), and sonicated. The resulting cell extracts were loaded onto a Ni-nitrilotriacetic acid-agarose column (Qiagen) equilibrated with lysis buffer containing 10 mM imidazole. The columns were washed with lysis buffer containing 20 mM imidazole, and bound NSAP1 was eluted with 100 mM imidazole.
Mammalian cell culture and HeLa cell lysate preparation.
293T and HeLa cells were cultivated in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% Fetalclone III (HyClone). S10 cytoplasmic extracts of HeLa S3 cells were prepared as previously described (17).
mRNA purification and Northern blot analysis.
Levels of NSAP1 mRNA and replicon RNAs were analyzed by Northern blotting with 20 μg of total RNA as described by Kim et al. (17). siRNA expression profiles were analyzed as described by Miyagishi and Taira (20), with minor modifications, by the use of a 32P-end-labeled sense-strand probe (5′-GACGGTGCATTGGCAGTTC-3′) and 15 μg of total RNA. For analyses of replicon RNAs, total RNAs were probed with a 32P-labeled negative-sense riboprobe that was complementary to the 3′ end of the HCV RNA. The probe was generated by in vitro transcription with T3 polymerase (Roche) and plasmid rep5.1 treated with BglII.
RESULTS
Identification of a cellular protein that interacts with HCV RNA.
To understand the molecular basis of IRES-dependent translation of HCV mRNA, we used several methods to analyze the cellular factor(s) that interacts with the previously defined HCV IRES element (8, 13, 26). First, a UV cross-linking experiment was performed with micrococcal nuclease-treated S10 cytoplasmic extracts of HeLa S3 cells (S10 extracts) and a 32P-labeled RNA fragment spanning nt 18 to 402 of the genomic HCV RNA. The latter nucleotide (nt 402) corresponds to the 62nd nt of the HCV core-coding sequence (see Fig. 2A, panel i). Four prominent bands, with apparent molecular masses of 45, 65, 100, and 110 kDa, were observed after UV cross-linking (Fig. 1A, lane 1). When homopolymeric ribonucleotides were used as competitors in the UV cross-linking reactions, poly(A) strongly inhibited binding of the 65-kDa protein to the HCV RNA, but poly(C) and poly(U) did not (Fig. 1A, lanes 2 to 4). The specificity of this RNA-binding activity was further investigated by the addition of unlabeled competitor HCV RNAs (nt 18 to 402). Binding of the 45-, 65-, and 110-kDa proteins, but not the 100-kDa protein, was inhibited by the unlabeled competitor RNAs (Fig. 1A, lanes 5 and 6). These results indicate that the 45-, 65-, and 110-kDa proteins specifically interact with the HCV IRES.
FIG. 2.
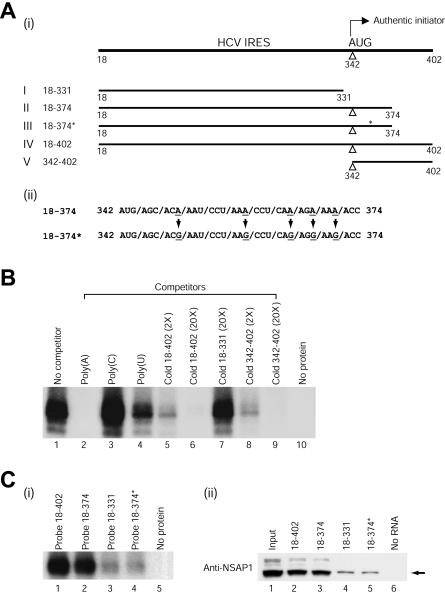
NSAP1 interacts specifically with the ACR in the N-terminal part of the polyprotein. (A) (i) Schematic diagrams of HCV RNA transcripts used for RNA pull-down and UV cross-linking experiments. (ii) Five adenosine residues, at the third nucleotide position of each codon, were replaced with guanosine residues; these changes maintained the same core amino acid sequence. The asterisk in construct 18-374* denotes the nucleotide changes in the ACR. (B) Effects of competitor RNAs on binding of purified NSAP1 to probe IV. The recombinant NSAP1 protein was purified to near homogeneity and then used in UV cross-linking reactions. Recombinant NSAP1 (100 ng) was incubated with RNA probe IV in the UV cross-linking experiments in the presence (lanes 2 to 9) or absence (lane 1) of competitor RNAs. The competitor RNAs consisted of the following: 50 ng of three homopolymeric RNAs (lanes 2 to 4), 2-fold (lane 5) and 20-fold (lane 6) molar excesses of unlabeled transcript IV, a 20-fold molar excess of transcript I (lane 7), and 2-fold (lane 8) and 20-fold (lane 9) molar excesses of transcript V. (C) (i) UV cross-linking experiments were performed with 100 ng of purified NSAP1 protein and with 32P-labeled RNA probes corresponding to HCV wild-type RNAs (18-402, 18-374, 18-331, and 18-402 [lanes 1 to 3 and 5, respectively]) and the mutant RNA 18-374* (lane 4). (ii) In order to investigate whether NSAP1 can form a complex with the HCV IRES, we performed RNA pull-down experiments with S10 extracts and biotinylated RNAs corresponding to various fragments of the wild-type HCV RNAs (18-402, 18-374, and 1-331 [lanes 2 to 4]) and a mutant HCV RNA (18-374* [lane 5]). Immunoblot analysis was performed with an anti-NSAP1 polyclonal antibody.
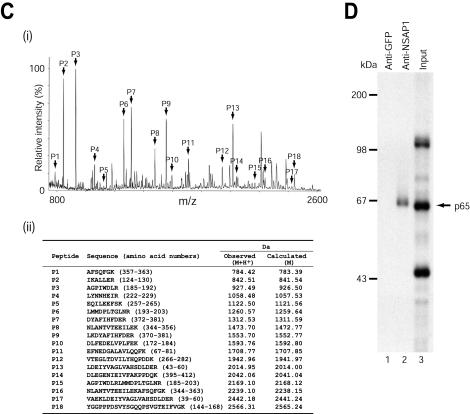
FIG. 1.
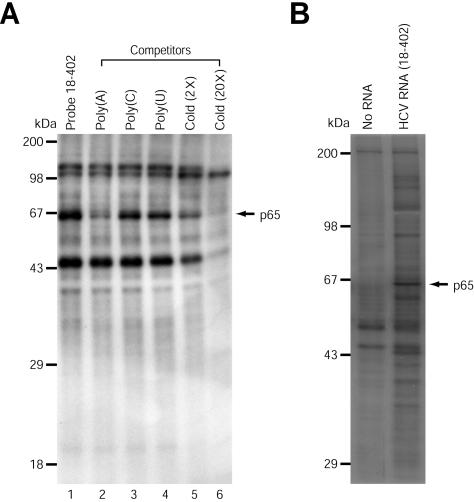
Identification of a cellular protein that interacts with HCV RNA. (A) UV cross-linking experiments using 32P-labeled HCV RNA. S10 extracts of HeLa S3 cells (20 μg) were incubated with 32P-labeled HCV RNA (nt 18 to 402) (3 × 105 cpm) without competitor RNAs (lane 1). Competition experiments were performed with 32P-labeled HCV RNA (nt 18 to 402) in the presence of 50 ng of homopolymeric RNAs [poly(A), poly(C), and poly(U)] (lanes 2 to 4), or 2- and 20-fold molar excesses of unlabeled HCV RNA (nt 18 to 402) (lanes 5 and 6). (B) HCV RNA (nt 18 to 402) affinity chromatography. A biotinylated probe (15 μg) was incubated with S10 extracts (500 μg) and nonspecific tRNAs (30 μg), and then the resin-bound proteins were resolved by SDS-10% PAGE. (C) Identification of the 65-kDa protein as NSAP1. (i) Peptide mass fingerprinting via MALDI-TOF was performed as described previously (2). The peaks labeled with arrows match the estimated tryptic peptide masses of NSAP1 with an error of <50 ppm. (ii) Oligopeptides from mass fingerprinting analysis. (D) Immunoprecipitation of UV cross-linked proteins with a 32P-labeled HCV RNA probe (nt 18 to 402). After RNase treatment, the samples were precleared with protein G-agarose resin and then immunoprecipitated with 1.5 μg of rabbit polyclonal anti-NSAP1 (lane 2) or anti-GFP (lane 1) antibody. Resin-bound proteins were resolved by SDS-10% PAGE. Lane 3, 32P-labeled proteins before immunoprecipitation.
To isolate the cellular proteins interacting with the HCV RNA, we generated a biotinylated HCV RNA (nt 18 to 402) by in vitro transcription and performed RNA affinity chromatography with S10 extracts and the biotinylated RNA. After precipitation and washing of the RNA-protein complex, RNA-bound proteins were analyzed by SDS-PAGE (Fig. 1B), which revealed proteins with apparent molecular masses of 35, 38, 40, 45, 57, 65, and 150 kDa. The 65-kDa band was the most prominent among the purified proteins, and the binding of this protein was specifically inhibited by competitor RNAs, so we focused the following experiments on this protein.
The 65-kDa protein was excised from the gel and analyzed by peptide mass fingerprinting via MALDI-TOF. Figure 1C, panel I, shows the MALDI mass spectrum of the peptides generated by trypsinization of the 65-kDa protein. The obtained masses were compared with those of proteins in the Swiss-Prot database by use of MS-Fit peptide mass search program. As shown in Fig. 1C, panel ii, the peptides exhibited molecular masses that were in good agreement with the theoretically predicted tryptic peptides of NSAP1. The analyzed peptides covered 32% of the human NSAP1 sequence. On the basis of these results, we concluded that the 65-kDa protein is NSAP1. Taken together, these results strongly suggest that NSAP1 interacts with HCV mRNA.
The identity of the 65-kDa protein was further confirmed by immunoprecipitation of proteins that were UV cross-linked with 32P-labeled HCV RNA (nt 18 to 402) by use of a polyclonal antibody against NSAP1 (anti-SYNCRIP-N; kindly provided by A. Mizutani, University of Tokyo, Tokyo, Japan) (Fig. 1D). No bands were detected when a polyclonal antibody against green fluorescent protein (GFP) was used as a negative control (Fig. 1D, lane 1). Immunoprecipitation of the UV cross-linked proteins with the polyclonal anti-NSAP1 antibody successfully detected the 65-kDa protein (Fig. 1D, lane 2). Even though the polyclonal anti-NSAP1 antibody is known to additionally interact with hnRNP R, only the 65-kDa band was detected in the immunoprecipitation results (also see Fig. 3B, panel ii), indicating that NSAP1, but not hnRNP R, interacts with the HCV RNA.
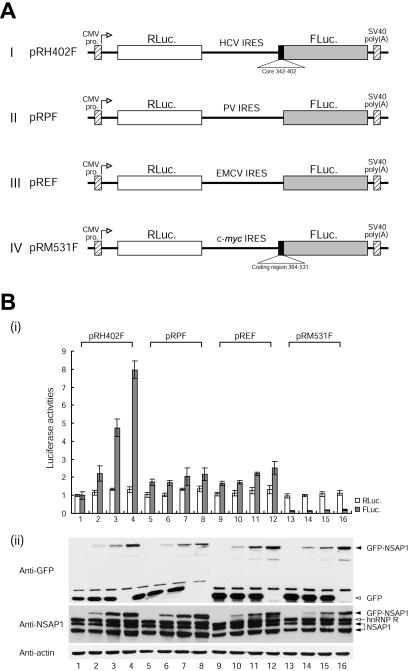
FIG. 3.
NSAP1 augments HCV IRES-dependent translation. (A) Schematic diagrams of dicistronic reporter plasmids used for monitoring efficiency of cap- and IRES-dependent translation in vivo. The vectors contained the cytomegalovirus (CMV) immediate early enhancer-promoter (CMV pro.) to direct transcription in vivo. The transcription start site in the CMV promoter is indicated by open arrows. (B) (i) Translation activities of IRES-containing dicistronic mRNAs in 293T cells. Dicistronic reporter constructs (0.5 μg each of pRH402F [lanes 1 to 4], pRPF [lanes 5 to 8], pREF [lanes 9 to 12], and pRM531F [lanes 13 to 16]) and variable amounts of effecter pEGFP-c1/NSAP1 (2.5 μg [lanes 2, 6, 10, and 14], 5 μg [lanes 3, 7, 11, and 15], and 10 μg [lanes 4, 8, 12, and 16]) and negative control effecter pEGFP-c1 (10 μg [lanes 1, 5, 9, and 13], 7.5 μg [lanes 2, 6, 10, and 14], and 5 μg [lanes 3, 7, 11, and 15]) were cotransfected with the control plasmid pCMV · SPORT-βgal (Invitrogen) to monitor the effect of NSAP1 on IRES activities. Forty-eight hours after transfection by electroporation, the cells were harvested and their luciferase activities were measured. For IRES-dependent translation, firefly luciferase activity, which was directed by pRH402F, was arbitrarily set to 1. For cap-dependent translation, Renilla luciferase activity, which was directed by pRH402F, was arbitrarily set to 1. The transfection efficiency was also determined for each luciferase value relative to that of the β-galactosidase activity of the transfection control. Error bars represent standard deviation values. (ii) Expression profiles of GFP-NSAP1. Immunoblot analysis was performed with antibodies against GFP, NSAP1, and actin.
NSAP1 specifically interacts with an adenosine-rich region encoding the N-terminal part of the HCV polyprotein.
To determine the region of the HCV IRES that interacts with NSAP1, we generated truncated RNAs containing different parts of the HCV IRES (Fig. 2A, panel i) and then performed UV cross-linking experiments. As shown in Fig. 2C, panel i, purified recombinant NSAP1 strongly bound to probes II and IV, which corresponded to nt 18 to 374 and 18 to 402, respectively (Fig. 2C, panel i, lanes 2 and 1, respectively). In contrast, recombinant NSAP1 bound only weakly to probe I (nt 18 to 331) (Fig. 2C, panel i, lane 3). We also found that a biotinylated HCV RNA corresponding to nt 342 to 402 was sufficient for binding to NSAP1 (data not shown). These results were further confirmed by competition assays. The interaction between recombinant NSAP1 and probe IV was strongly inhibited by poly(A) and unlabeled RNA probes IV and V, moderately inhibited by poly(U), and unaffected by poly(C) or unlabeled RNA I (Fig. 2B). The competition patterns for purified recombinant NSAP1 were in good agreement with the binding patterns of the 65-kDa protein to HCV RNA observed by UV cross-linking with S10 extracts (compare Fig. 1A and 2B), suggesting that NSAP1 directly interacts with HCV RNA and that a specific RNA-protein interaction can occur in cells in which numerous proteins exist together with NSAP1.
The RNA-binding data demonstrate that the HCV RNA fragment spanning nt 342 to 374 is necessary and sufficient for the interaction with NSAP1. Moreover, homopolymeric RNA competition data (Fig. 1A and 2B) and a previous report showed that NSAP1 has a strong affinity for adenosine-rich RNAs (21). Interestingly, the 5′-proximal region of the HCV core-coding sequence contains an adenosine-rich region (Fig. 2A, panel ii) that was previously shown to be necessary for the efficient translation of HCV mRNA (26). The ACR is conserved among different HCV isolates and also among HCV-related viruses such as pestiviruses and GB virus B (27). In order to examine whether the ACR participates in the interaction with NSAP1, we generated a mutant construct in which the ACR adenosines were replaced with guanosines (Fig. 2A, panel ii). These site-directed mutagenic changes did not alter the amino acid sequence and were predicted to induce only negligible changes (ΔG = −0.7 kcal/mol) in the secondary RNA structure, since both the wild-type and mutant ACR are predicted to exist in a single-stranded region (according to the mfold RNA folding program). The effect of this ACR mutation on NSAP1 binding was measured by UV cross-linking experiments (Fig. 2C, panel i) and RNA pull-down assays (Fig. 2C, panel ii). Direct binding between NSAP1 and the ACR was assessed by the use of a purified recombinant NSAP1 protein and 32P-labeled HCV RNAs. Purified recombinant NSAP1 interacted strongly with HCV RNAs containing the intact ACR (Fig. 2C, panel i, lanes 1 and 2) but only weakly interacted with mutant HCV RNA (Fig. 2C, panel i, lane 4) and with RNA lacking the core-coding region (Fig. 2C, panel i, lane 3). The weak binding activity of NSAP1 to probes I and III may be attributable to a putative weak binding site in the 5′NTR of HCV mRNA, since the binding activity to probe III is the same as that of probe I and both contain the 5′NTR sequence (nt 18 to 341) of HCV mRNA. The in vitro pull-down experiments were also performed with S10 extracts and biotinylated HCV RNAs. In these experiments, NSAP1 coprecipitated efficiently with wild-type HCV RNAs containing the intact ACR, such as probes II (nt 18 to 374) and IV (nt 18 to 402) (Fig. 2C, panel ii, lanes 3 and 2, respectively), whereas the ACR-mutated HCV RNA showed a dramatically reduced affinity to NSAP1 (Fig. 2C, panel ii, lane 5). The binding affinity of the mutated RNA was the same as that of an HCV RNA entirely lacking the coding region (compare lanes 4 and 5 in Fig. 2C, panel ii). Taken together, these data indicate that some or all of the adenosine residues in the ACR play a key role in the interaction of the HCV RNA with NSAP1.
NSAP1 augments HCV IRES-dependent translation.
To investigate the role of NSAP1 in HCV IRES-dependent translation, we cotransfected 293T cells with a plasmid expressing GFP fused to NSAP1 and plasmid pRH402F, yielding a dicistronic mRNA containing the HCV IRES (nt 18 to 402) in the intercistronic region. Cap- and HCV IRES-dependent translation was monitored by measuring Renilla luciferase and firefly luciferase activities, respectively (Fig. 3A). The cotransfection of full-length NSAP1 augmented HCV IRES-dependent translation in a dose-dependent manner (Fig. 3B, panel i, lanes 1 to 4). In contrast, NSAP1 showed no or moderate effects on translation through the IRES elements of PV, EMCV, and c-myc (Fig. 3B, panel i, lanes 5 to 16). As shown in Fig. 3B, panel ii, the expression profiles of GFP-NSAP1 correlated well with the activity of the HCV IRES element. Immunoblot analysis with a polyclonal anti-NSAP1 antibody (Fig. 3B, panel ii) revealed four bands, with apparent molecular masses of 65, 74, 82, and 90 kDa. As previously reported, the 65- and 74-kDa proteins corresponded to two isoforms of NSAP1 produced from alternatively spliced mRNAs (22). The 82- and 90-kDa proteins corresponded to hnRNP R, homologous to NSAP1, and the GFP-NSAP1 fusion protein expressed from the plasmid pEGFP-c1/NSAP1, respectively.
siRNA-directed knockdown of NSAP1 reduces HCV IRES activity.
To further analyze the role of NSAP1 in HCV IRES-dependent translation, we employed mRNA knockdown by using RNA interference (3). A plasmid generating a siRNA specific for NSAP1, containing the Epstein-Barr virus nuclear antigen 1 coding sequence and the Epstein-Barr virus origin of replication was constructed (Fig. 4A). The siRNA-expressing plasmid was transfected into HeLa cells, and plasmid-containing cells were selected by hygromycin treatment. As shown in Fig. 4B, endogenous NSAP1 levels were dramatically reduced in siRNA-expressing cells (pEBV-U6+27/NSAP1) compared with control cells containing the vector plasmid (pEBV-U6+27) (Fig. 4B). In contrast, the levels of other well-characterized IRES-interacting trans-acting factors (ITAFs; hnRNP L, PTB, and La) and the internal control, actin, were not affected by the siRNAs (Fig. 4B). The expression of the NSAP1 siRNA was monitored on Northern blots probed with a 32P-labeled sense-strand RNA-specific oligomer (Fig. 4C, panel i). Four bands were detected; the smallest RNA (21 nt) was expected to be the completely processed siRNA, whereas the others were likely to be precursors of the siRNA. Similarly, the levels of NSAP1 mRNA were monitored by Northern blot analysis using a 32P-labeled probe corresponding to nt 526 to 1038 of the human NSAP1 mRNA. Multiple bands were detected; these were thought to represent alternatively spliced forms of the NSAP1 mRNA (Fig. 4C, panel ii). The levels of two major NSAP1 mRNA species (∼3.2 and 4.2 kb; indicated by arrowheads) were dramatically reduced in NSAP1 siRNA-expressing cells (Fig. 4C, panel ii), whereas the level of control human ribosomal protein L32 (hRPL32) mRNA was not affected by the presence or absence of the siRNAs (Fig. 4C, panel ii).
The effect of NSAP1 on HCV IRES-dependent translation was further analyzed by the use of dicistronic mRNAs containing wild-type and mutant HCV IRESs (RH374F* and RH374*F*) and control dicistronic mRNAs (RPF and REF). mRNAs were synthesized in vitro with T7 RNA polymerase, and the IRES activities of the four mRNAs in cells were analyzed by RNA transfection experiments.
Three hours after the transfection of these mRNAs into HeLa cells, with or without siRNAs against NSAP1, the IRES activities were monitored by the analysis of luciferase activities. The translation of firefly luciferase by the wild-type HCV IRES was reduced about 2.5-fold after the knockdown of NSAP1 (Fig. 4E, compare lane 2 with lane 1), and the ACR mutation reduced HCV IRES activity 4-fold (Fig. 4E, compare lane 3 with lane 1). However, the activity of the mutant HCV IRES was only marginally reduced by the knockdown of NSAP1 (Fig. 4E, compare lane 4 with lane 3). The activities of the PV and EMCV IRESs were not affected by the reduction in the NSAP1 level (compare lanes 6 and 8 with lanes 5 and 7, respectively). Taken together, these results strongly suggest that NSAP1 augments HCV IRES activity through an interaction with the ACR in the HCV RNA core-coding sequence.
NSAP1 plays a key role in translation of HCV mRNA.
HCV is a positive-sense RNA virus; the genomic RNA in the virus particle serves as an mRNA, and the HCV IRES element directs the translation of a polyprotein that is proteolytically processed into all of the viral proteins. Therefore, IRES activity is critical for efficient HCV proliferation. In order to investigate the effect of NSAP1 on HCV mRNA translation, we constructed HCV replicons that are capable of supporting HCV RNA replication inside cells. The tested replicons contained wild-type HCV IRESs (rep5.1 and repI374) and a mutant IRES (repI374*) in which the ACR adenosines were replaced with guanosines, as shown in Fig. 2A, panel ii (Fig. 5A). In vitro translation of the replicon RNAs in rabbit reticulocyte lysates (RRLs) revealed two major bands. The larger band corresponds to the NS3 protein, which is a proteolytically processed form of the HCV polyprotein. The translation of NS3 is directed by the EMCV IRES. All three mRNAs yielded similar levels of NS3 (Fig. 5B). Curiously, other nonstructural proteins (NS4A to NS5B) in the replicon RNAs were not clearly detected from translation products of replicon RNAs in RRLs. This may be attributed to an imperfect translation of a very long open reading frame of HCV mRNA and to poor proteolytic processing of the partially translated products. The same translation patterns were observed by Friebe et al. (4). In contrast, the translational efficiency of neomycin phosphotransferase II (NPTII) varied among the wild-type and mutant HCV IRESs. Similar levels of NPTII protein were produced from the wild-type mRNAs rep5.1 and repI374 (Fig. 5B), although the rep5.1 mRNA produced a slightly larger NPTII protein, due to rep5.1 containing a coding sequence that is five amino acids longer than that of repI374. In contrast, translation of the NPTII protein by repI374* was about threefold less efficient than that by repI374 (Fig. 5B). The reduction level was similar to that found in our in vivo experiments (Fig. 4E, lanes 1 and 3) and is likely due to an impairment of HCV IRES activity by the ACR mutation, which blocks the binding of NSAP1.
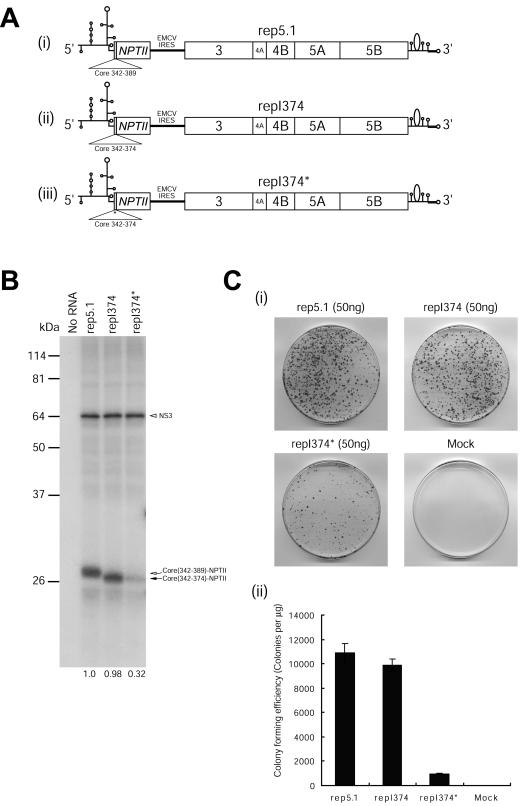
FIG. 5.
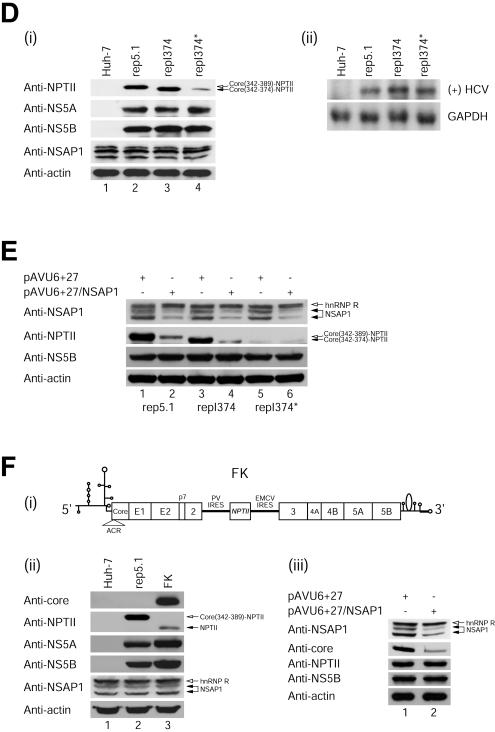
NSAP1 is required for translation of HCV mRNA during HCV proliferation. (A) Schematic diagrams of subgenomic replicons. The NPTII proteins of rep5.1 and repI374 are fused with 16 and 11 amino acids of the N-terminal core-coding sequence, respectively. (B) In vitro translation of replicon RNAs in RRLs. Replicon RNAs (1.0 μg) were in vitro translated for 80 min at 30°C with RRLs (Promega) according to the manufacturer's instructions, and samples were resolved by SDS-12% PAGE. Radioactivities of the core-NPTII fusion and NS3 protein bands were quantified by use of a Fuji phosphorimager. The numbers below the lanes depict the translation levels of the core-NPTII fusion proteins normalized to those of NS3. The translation efficiency obtained with the replicon carrying the HCV IRES (1-389) was set to 1. (C) (i) Colony-forming efficiencies of replicon RNAs. Huh-7 cells containing HCV replicons (rep5.1, repI374, and repI374*) were generated as described by Lohmann et al. (18). After about 2 weeks of G418 (600 μg/ml) selection, the colonies were fixed and stained with crystal violet. (ii) Graphical presentation of the colony-forming efficiency of each replicon. The data shown are means, and error bars show standard deviation values for four independent transfection experiments. (D) (i) Immunoblot analysis using total cell lysates from pooled G418-resistant colonies. Immu- noblot analyses were performed with antibodies against NPTII (anti-NPTII; Upstate), NS5A, NS5B, NSAP1, and actin. The levels of NPTII reflect HCV IRES activities. (ii) Northern blot analysis of HCV RNAs in replicon-containing cells. Total RNAs (20 μg of each) were analyzed with a 32P-labeled riboprobe complementary to the 3′ end of the positive-sense HCV RNA and with a 32P-labeled riboprobe specific for glyceraldehyde-3-phosphate dehydrogenase mRNA. The radioactivities of the HCV and (GAPDH)/GAPDH bands were quantified by use of a phosphorimager. (E) Effect of siRNA against NSAP1 on HCV IRES-dependent translation. The replicon-containing cells were cotransfected with pGL3-control (Promega) and pAVU6+27 or pAVU6+27/NSAP1 by the use of Lipofectamine 2000 (Invitrogen). Forty-eight hours after transfection, an immunoblot analysis was performed with antibodies against NSAP1, NPTII, NS5B, and actin. (F) (i) Schematic diagram of replicon FK containing the full-length HCV polyprotein. The construction procedure for FK will be described elsewhere (S. H. Lee, Y. K. Kim, C. S. Kim, S. K. Seol, J. Kim, S. Cho, Y. L. Song, and S. K. Jang, submitted for publication). Huh-7 cells containing replicon FK was generated as described by Lohmann et al. (18). (ii) Immunoblot analyses were performed with total cell extracts from the pool of G418-resistant colonies and with antibodies against core, NPTII, NS5A, NS5B, NSAP1, and actin. (iii) Effect of siRNA against NSAP1 on HCV IRES-dependent translation. The replicon-containing cells were cotransfected with pGL3-control and pAVU6+27 or pAVU6+27/NSAP1 by the use of Lipofectamine 2000. Forty-eight hours after transfection, an immunoblot analysis was performed with antibodies against NSAP1, core, NPTII, NS5B, and actin.
The importance of the ACR on HCV mRNA translation was also tested by analyzing the colony-forming efficiencies of Huh-7 hepatoma cells containing HCV replicon RNAs programmed to generate NPTII, providing resistance against G418, and viral proteins NS3 to NS5B, which are required for RNA replication. After transfection, RNA replicon-containing cells were selected by the use of G418, which kills cells lacking the replicon-encoded NPTII protein translated in response to the HCV IRES. This allows indirect monitoring of the activities of wild-type and mutant IRESs by measurements of the colony-forming efficiencies of cells containing the replicon RNAs, as shown in Fig. 5C. Cells containing wild-type HCV IRES replicons rep5.1 and repI374 showed similar colony-forming efficiencies, whereas cells containing the mutated ACR replicon showed reduced colony-forming efficiencies (Fig. 5C).
The translational efficiencies of the wild-type and mutant HCV IRESs were then analyzed by immunoblotting cell extracts from the surviving replicon-containing cells with an antibody against NPTII. The translation of NPTII directed by the mutant HCV IRES was significantly reduced compared to that directed by the wild-type HCV IRES (Fig. 5D, panel i, compare lane 4 with lanes 2 and 3 in the anti-NPTII panel). On the other hand, two other viral proteins, NS5A and NS5B, directed by the EMCV IRES were not affected by the mutation (Fig. 5D, panel i, compare lanes 2, 3, and 4 in the anti-NS5A and anti-NS5B panels). The levels of NSAP1 and actin proteins were similar in all tested cells (Fig. 5D, panel i, lanes 1 to 4 in the anti-NSAP1 and anti-actin panels). The levels of replicon RNAs in the cells were also monitored by Northern blotting with a probe complementary to nt 7332 to 8001 of the positive-sense replicon RNA, which revealed the presence of similar levels of positive-sense HCV RNAs in all tested replicon-containing cells (Fig. 5D, panel ii, lanes rep5.1, repI374, and repI374*). This indicates that the observed differences in the anti-NPTII levels may be attributed to differences in IRES activities among the various replicons and also that the ACR plays a key role in the activation of the HCV IRES during HCV proliferation (Fig. 5D, panel i).
The function of NSAP1 in HCV IRES-dependent translation was further analyzed by introducing a siRNA against NSAP1 into cells containing the various HCV replicons. Transfection of the plasmid pAVU6+27/NSAP1 (which produces siRNAs against NSAP1), but not of the control plasmid pAVU6+27, led to reduced NSAP1 protein levels in replicon-containing cells (Fig. 5E, compare lanes 1, 3, and 5 with lanes 2, 4, and 6 in the anti-NSAP1 panel). Note that the levels of hnRNP R, which is also recognized by the anti-NSAP1 antibody, were not changed by the siRNA against NSAP1 (Fig. 5E). The translation levels of NS5B were the same regardless of the existence of the ACR and/or NSAP1 (Fig. 5E). These data suggest that HCV mRNA levels are similar regardless of the existence of the ACR and NSAP1. This in turn indicates that NSAP1 does not affect the stability of HCV mRNA. This was also shown by a Northern blot analysis of replicon RNAs with wild-type and mutant ACRs, with which NSAP1 interacts (Fig. 5D, panel ii).
Wild-type HCV IRES-directed translation of NPTII was greatly reduced by the introduction of the NSAP1-specific siRNA (Fig. 5E, compare lanes 2 and 4 with lanes 1 and 3, respectively, in the anti-NPTII panel). Mutant HCV IRES-directed translation of NPTII was much lower than that directed by the wild-type HCV IRES (Fig. 5E, compare lane 5 with lanes 1 and 3 in the anti-NPTII panel). However, unlike that directed by the wild-type HCV IRES, the translation of NPTII directed from the mutant HCV IRES was not reduced by the siRNA against NSAP1 (Fig. 5E, compare lane 5 with lane 6 in the anti-NPTII panel). EMCV IRES-directed translation of NS5B and cap-dependent translation of actin from a cellular mRNA were not affected by the introduction of the siRNA against NSAP1 (Fig. 5E, anti-NS5B and anti-actin panels).
The effect of NSAP1 on HCV mRNA translation was also tested in the context of the full-length HCV polyprotein. A replicon RNA containing the full-length HCV genome was generated as shown in Fig. 5F, panel i. This replicon is composed of a tricistronic mRNA in which translations of the core to NS2, NPTII, and NS3 to NS5B proteins are directed by the HCV IRES, the PV IRES, and the EMCV IRES, respectively. Viral proteins in the replicon-containing cells were monitored by immunoblotting of cell extracts from the surviving replicon-containing cells. Replicon FK, containing the full-length HCV genome, but not replicon rep5.1, which lacked the structural protein-coding region, produced the core protein (Fig. 5F, panel ii). The NPTII produced from the control rep5.1 replicon was larger than that produced from FK, since NPTII in rep5.1 contains an additional 16 amino acids corresponding to the N-terminal part of the core protein to the authentic NPTII protein. The effect of NSAP1 was monitored by introducing the siRNA against NSAP1 into Huh-7 cells containing the FK replicon (Fig. 5F, panel iii). The siRNA reduced the level of NSAP1 protein in the cells containing replicon FK (Fig. 5F, panel iii, compare lane 1 with lane 2 in the anti-NSAP1 panel). A reduction in core translation, which was directed by the HCV IRES, was observed in the cells containing the siRNA (Fig. 5F, panel iii, compare lane 1 with lane 2 in the anti-core panel). On the other hand, the translation of NPTII, which was directed by the PV IRES, and NS5B, which was directed by the EMCV IRES, was not affected by the siRNA (Fig. 5F, panel iii). This strongly suggests that NSAP1 enhances HCV IRES-dependent translation of the full-length HCV mRNA.
Taken together, these in vitro and in vivo experimental data strongly suggest that the NSAP1 protein augments HCV IRES-dependent translation through an interaction with the ACR at the core-coding region of the HCV mRNA. This also indicates that enhancement of the HCV IRES by NSAP1 plays an important role in the production of viral proteins during HCV infection.
DISCUSSION
To better understand the molecular mechanism of HCV IRES-dependent translation, we analyzed the effects of a cellular protein, NSAP1, on this translation, particularly in terms of its ability to interact with the HCV RNA ACR within the core-coding region. We found that NSAP1 augments HCV IRES-dependent translation. The specificity of the interaction between NSAP1 and the ACR was investigated by UV cross-linking experiments in the presence of competitor RNAs, by RNA affinity chromatography, and by artificial mutagenesis of the ACR. The role of NSAP1 in HCV IRES-dependent translation was investigated by observing the effects of NSAP1 overexpression and knockdown on HCV IRES activity. Moreover, an ACR mutation that inhibits the interaction between NSAP1 and the ACR was examined for its effect on the activity of the HCV IRES in conjunction with the knockdown of NSAP1. The data strongly indicated that NSAP1 augments HCV IRES function through an interaction with the ACR at the core-coding region. This is the first report showing that NSAP1 functions as an ITAF and is the first report of an ITAF augmenting IRES-dependent translation through an interaction with an mRNA coding region.
It may be difficult to envision the NSAP1 ITAF activity as occurring through an interaction with the ACR downstream of the initiation codon, since the ITAF should be removed from the binding site at the elongation stage of translation. Nevertheless, several modes of action of NSAP1 can be speculated. First, NSAP1 may assist the binding of ribosomes to the IRES through a putative interaction. In this respect, it is noteworthy that the 40S ribosomal subunit can bind to the HCV IRES without the assistance of other initiation factors (25). NSAP1 may augment this ribosome-IRES interaction by guiding the 40S ribosomal subunit to the right place through a putative NSAP1-ribosome interaction. It is also possible that NSAP1 shifts the IRES into a structure that makes the IRES more competent to assemble binary IRES-40S complexes. In light of the fact that NSAP1 target sequences need to be unfolded to exert efficient IRES activity (27), NSAP1 might keep the target sequence from exerting a dominant-negative interaction with the IRES. Second, it is possible that NSAP1 recruits a canonical translation factor(s) through a putative protein-protein interaction. eIF-3 is a good candidate for the putative counterpart of NSAP1 because it interacts with stem-loop III of the HCV 5′NTR (28). Third, NSAP1 may activate IRES-dependent translation through interactions with other ITAFs. In this respect, it is noteworthy that many ITAFs are known to interact with each other (7, 16). The RNA-binding protein hnRNP L is known to interact with the HCV RNA N-terminal core-coding sequence, including the ACR (8). Moreover, we observed a protein-protein interaction between NSAP1 and hnRNP L by coimmunoprecipitation (data not shown). However, unlike the NSAP1 situation described here, the enhancing effect of hnRNP L on HCV IRES function was not observed after ectopic expression of hnRNP L by the method described in Fig. 3 (data not shown). Moreover, hnRNP L did not enhance the ITAF activity of NSAP1 on HCV IRES when it was coexpressed with NSAP1 (data not shown). Although these experiments did not reveal an ITAF activity for hnRNP L, the protein might function through protein-protein interactions with both NSAP1, as described here, and PTB, as described by Hahm et al. (8). Further mechanistic studies of the function of NSAP1 within this proposed model are in progress.
Enhancements of biological activities by RNA-binding proteins interacting downstream of active sites have been reported in the cases of RNA transcription and replication. For instance, Tat, a transcriptional activator of human immunodeficiency virus type 1, enhances the transcription of mRNA through an interaction with TAR in an RNA transcript (1). Similarly, the polioviral 3CD protein and poly(rC)-binding proteins activate the replication of positive-sense viral RNAs through an interaction with the clover leaf structure of the replicative intermediate of the positive-strand RNA (5, 23). The difference between translational activation by NSAP1 and transcription by other factors is that NSAP1 should be detached from its binding site after the activation of translation, since the 80S ribosome most likely needs to pass through the binding site. Transcriptional factors, on the other hand, may stay on the binding site after the activation of each round of transcription.
There have been controversial reports on the role of the core-coding region in HCV IRES function. Several reports of experiments with heterologous reporter systems have suggested that the core-coding sequence between nt 354 and 372 plays an important role in HCV IRES function (8, 13, 26). Moreover, the same region was shown to be essential for the proliferation of a chimeric poliovirus by use of a fragment of the HCV IRES spanning the core-coding sequence instead of the polioviral IRES (19). In contrast, several other studies using heterologous reporters such as chloramphenicol acetyltransferase and firefly luciferase have suggested that the core-coding region does not affect HCV IRES activity (27, 30). This discrepancy may be attributed to differences in the employed assay systems and reporters. Considering the NSAP1 function shown in this report, one possible explanation for the successful translation of reporters lacking the core-coding region is that the adenosine-rich regions in the chloramphenicol acetyltransferase and firefly luciferase genes may have functionally replaced the ACR of HCV in these studies. Note that in this work, we intentionally mutated the adenosine-rich region in the luciferase gene to rule out the potential replacement of ACR activity with that by the adenosine-rich region in the luciferase gene. However, further work will be required to fully investigate whether this functional replacement hypothesis accounts for the previous results.
Further investigations into the molecular basis of the role played by NSAP1 in IRES activity will improve our understanding of the mechanisms of IRES-dependent translation. Moreover, this improved understanding may suggest novel therapeutic approaches for the treatment of life-threatening HCV infections based on the inhibition of IRES-dependent translation.
Acknowledgments
We thank A. Mizutani for providing the NSAP1 polyclonal antibody (anti-SYNCRIP-N), C. Astell for providing plasmid pRSETC9-15, D. Engelke for providing plasmid pAVU6+27, and R. Bartenschlager for providing the subgenomic replicon plasmid rep5.1 and antibodies against core, NS5A, and NS5B. We are also grateful to members of our laboratory for helpful discussions and technical assistance.
This work was supported in part by grants M10204000018-03J0000-01410 and M10106000056-03B4500-01010 from MOST, grant 02-PJ2-PG1-CH16-0002 from KHIDI, grant KRF-2003-005-C00011 from KRF, and a grant from POSCO.
REFERENCES
- 1.Berkhout, B., R. H. Silverman, and K. T. Jeang. 1989. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell 59:273-282. [DOI] [PubMed] [Google Scholar]
- 2.Choi, K., J. H. Kim, X. Li, K. Y. Paek, S. H. Ha, S. H. Ryu, E. Wimmer, and S. K. Jang. 2004. Identification of cellular proteins enhancing activities of internal ribosomal entry sites by competition with oligodeoxynucleotides. Nucleic Acids Res. 32:1308-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elbashir, S. M., J. Harborth, K. Weber, and T. Tuschl. 2002. Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods 26:199-213. [DOI] [PubMed] [Google Scholar]
- 4.Friebe, P., V. Lohmann, N. Krieger, and R. Bartenschlager. 2001. Sequences in the 5′ nontranslated region of hepatitis C virus required for RNA replication. J. Virol. 75:12047-12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gamarnik, A. V., and R. Andino. 1998. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 12:2293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grosset, C., C. Y. Chen, N. Xu, N. Sonenberg, H. Jacquemin-Sablon, and A. B. Shyu. 2000. A mechanism for translationally coupled mRNA turnover: interaction between the poly(A) tail and a c-fos RNA coding determinant via a protein complex. Cell 103:29-40. [DOI] [PubMed] [Google Scholar]
- 7.Hahm, B., O. H. Cho, J.-E. Kim, Y. K. Kim, J. H. Kim, Y. L. Oh, and S. K. Jang. 1998. Polypyrimidine tract-binding protein interacts with hnRNP L. FEBS Lett. 425:401-406. [DOI] [PubMed] [Google Scholar]
- 8.Hahm, B., Y. K. Kim, J. H. Kim, T. Y. Kim, and S. K. Jang. 1998. Heterogeneous nuclear ribonucleoprotein L interacts with the 3′ border of the internal ribosomal entry site of hepatitis C virus. J. Virol. 72:8782-8788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han, J. H., V. Shyamala, K. H. Richman, M. J. Brauer, B. Irvine, M. S. Urdea, P. Tekamp-Olson, G. Kuo, Q. L. Choo, and M. Houghton. 1991. Characterization of the terminal regions of hepatitis C viral RNA: identification of conserved sequences in the 5′ untranslated region and poly(A) tails at the 3′ end. Proc. Natl. Acad. Sci. USA 88:1711-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris, C. E., R. A. Boden, and C. R. Astell. 1999. A novel heterogeneous nuclear ribonucleoprotein-like protein interacts with NS1 of the minute virus of mice. J. Virol. 73:72-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hassfeld, W., E. K. Chan, D. A. Mathison, D. Portman, G. Dreyfuss, G. Steiner, and E. M. Tan. 1998. Molecular definition of heterogeneous nuclear ribonucleoprotein R (hnRNP R) using autoimmune antibody: immunological relationship with hnRNP P. Nucleic Acids Res. 26:439-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hellen, C. U., and P. Sarnow. 2001. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 15:1593-1612. [DOI] [PubMed] [Google Scholar]
- 13.Hwang, L. H., C. L. Hsieh, A. Yen, Y. L. Chung, and D. S. Chen. 1998. Involvement of the 5′ proximal coding sequences of hepatitis C virus with internal initiation of viral translation. Biochem. Biophys. Res. Commun. 252:455-460. [DOI] [PubMed] [Google Scholar]
- 14.Jang, S. K., and E. Wimmer. 2001. The role of RNA-binding proteins in IRES-dependent translation, p. 1-33. In K. Sandberg and S. E. Mulroney (ed.), RNA binding proteins: new concepts in gene expression. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 15.Kato, N., M. Hijikata, Y. Ootsuyama, M. Nakagawa, S. Ohkoshi, T. Sugimura, and K. Shimotohno. 1990. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc. Natl. Acad. Sci. USA 87:9524-9528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim, J. H., B. Hahm, Y. K. Kim, M. Choi, and S. K. Jang. 2000. Protein-protein interaction among hnRNPs shuttling between nucleus and cytoplasm. J. Mol. Biol. 298:395-405. [DOI] [PubMed] [Google Scholar]
- 17.Kim, J. H., K. Y. Paek, K. Choi, T. D. Kim, B. Hahm, K. T. Kim, and S. K. Jang. 2003. Heterogeneous nuclear ribonucleoprotein C modulates translation of c-myc mRNA in a cell cycle phase-dependent manner. Mol. Cell. Biol. 23:708-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lohmann, V., F. Korner, A. Dobierzewska, and R. Bartenschlager. 2001. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J. Virol. 75:1437-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu, H. H., and E. Wimmer. 1996. Poliovirus chimeras replicating under the translational control of genetic elements of hepatitis C virus reveal unusual properties of the internal ribosomal entry site of hepatitis C virus. Proc. Natl. Acad. Sci. USA 93:1412-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyagishi, M., and K. Taira. 2002. U6 promoter-driven siRNAs with four uridine 3′ overhangs efficiently suppress targeted gene expression in mammalian cells. Nat. Biotechnol. 20:497-500. [DOI] [PubMed] [Google Scholar]
- 21.Mizutani, A., M. Fukuda, K. Ibata, Y. Shiraishi, and K. Mikoshiba. 2000. SYNCRIP, a cytoplasmic counterpart of heterogeneous nuclear ribonucleoprotein R, interacts with ubiquitous synaptotagmin isoforms. J. Biol. Chem. 275:9823-9831. [DOI] [PubMed] [Google Scholar]
- 22.Mourelatos, Z., L. Abel, J. Yong, N. Kataoka, and G. Dreyfuss. 2001. SMN interacts with a novel family of hnRNP and spliceosomal proteins. EMBO J. 20:5443-5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parsley, T. B., J. S. Towner, L. B. Blyn, E. Ehrenfeld, and B. L. Semler. 1997. Poly(rC) binding protein 2 forms a ternary complex with the 5′-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA 3:1124-1134. [PMC free article] [PubMed] [Google Scholar]
- 24.Paul, C. P., P. D. Good, I. Winer, and D. R. Engelke. 2002. Effective expression of small interfering RNA in human cells. Nat. Biotechnol. 20:505-508. [DOI] [PubMed] [Google Scholar]
- 25.Pestova, T. V., I. N. Shatsky, S. P. Fletcher, R. J. Jackson, and C. U. Hellen. 1998. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 12:67-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reynolds, J. E., A. Kaminski, H. J. Kettinen, K. Grace, B. E. Clarke, A. R. Carroll, D. J. Rowlands, and R. J. Jackson. 1995. Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J. 14:6010-6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rijnbrand, R., P. J. Bredenbeek, P. C. Haasnoot, J. S. Kieft, W. J. Spaan, and S. M. Lemon. 2001. The influence of downstream protein-coding sequence on internal ribosome entry on hepatitis C virus and other flavivirus RNAs. RNA 7:585-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sizova, D. V., V. G. Kolupaeva, T. V. Pestova, I. N. Shatsky, and C. U. Hellen. 1998. Specific interaction of eukaryotic translation initiation factor 3 with the 5′ nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. J. Virol. 72:4775-4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsukiyama-Kohara, K., N. Iizuka, M. Kohara, and A. Nomoto. 1992. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 66:1476-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, C., P. Sarnow, and A. Siddiqui. 1993. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J. Virol. 67:3338-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]