Abstract
Nucleolar morphogenesis is a poorly defined process. Here we report that the Saccharomyces cerevisiae nucleolar trimethyl guanosine synthase I (Tgs1p), which specifically selects the m7G cap structure of snRNAs and snoRNAs for m2,2,7G conversion, is required not only for efficient pre-mRNA splicing but also for pre-rRNA processing and small ribosomal subunit synthesis. Mutational analysis indicates that the requirement for Tgs1p in pre-mRNA splicing, but not its involvement in ribosome synthesis, is dependent upon its function in cap trimethylation. In addition, we report that cells lacking Tgs1p showed a striking and unexpected loss of nucleolar structural organization. Tgs1p is not a core component of the snoRNP proteins; however, in vitro, the protein interacts with the KKD/E domain present at the carboxyl-terminal ends of several snoRNP proteins. Strains expressing versions of the snoRNPs lacking the KKD/E domain were also defective for nucleolar morphology and showed a loss of nucleolar compaction. We propose that the transient and functional interactions of Tgs1p with the abundant snoRNPs, through presumed interactions with the KKD/E domain of the snoRNP proteins, contribute substantially to the coalescence of nucleolar components. This conclusion is compatible with a model of self-organization for nucleolar assembly.
The driving forces that govern the dynamic behavior of cellular organelles (i.e., their shape and size and their emergence at specific cellular foci) are unknown. The concept of self-organization that describes the ability of a macromolecular complex, or organelle, to determine its own structure on the basis of the functional and transient interactions of its constituents is particularly attractive (reviewed in reference 17). In a self-organized system, a steady-state structure is generated from highly dynamic components, i.e., “ever-changing partners,” without the need for a rigid architectural framework. Classically, self-organization is opposed to self-assembly, where the physical associations of molecules generate a stable, static structure that reaches a “true equilibrium” (e.g., viruses and phages).
Ribosome synthesis is a dynamic, multistep process that takes place largely in the nucleolus, a specialized region of the nucleus that is highly enriched in RNA-processing factors (RRPs) and further compartmentalized in individual domains, which accounts for the vectorial formation of the ribosomes (reviewed in references 21 and 26). The nucleolus appears as a steady-state compartment rather than a static structure. Indeed, during pre-ribosome assembly, pre-rRNAs, ribosomal proteins (RPs), and several hundred trans-acting factors, including the small nucleolar RNAs (snoRNAs), interact transiently (reviewed in references 2 and 10). Furthermore, major nucleolar antigens, such as fibrillarin and nucleolin are continuously exchanged with the surrounding nucleoplasm (1, 22; reviewed in reference 18).
The spliceosomal RNAs U1, U2, U4, and U5, as well as several snoRNAs, are individually transcribed by RNA polymerase II and synthesized as m7G monomethylated cap precursors. These are selected by trimethyl guanosine synthase 1 (Tgs1p), a recently characterized nucleolar methyl transferase (20), for conversion to m2,2,7G. In metazoans, cap trimethylation is required to promote the nuclear uptake of the snRNPs following their essential cytoplasmic phase of maturation (reviewed in references 7, 15, and 34).
Tgs1p interacts in a two-hybrid assay and in vitro with the C-terminal basic protuberance of SmBp (20). In addition, the protein interacts in vitro with the highly charged carboxyl-terminal KKD/E domain of the snoRNP proteins Nop58p and Cbf5p. However, under physiological conditions, Tgs1p is not found stably associated with its substrates and is neither a core component of the spliceosome nor a core constituent of the snoRNPs. The box C+D snoRNA U3 and the box H+ACA snoRNAs snR10 and snR30 are modified by Tgs1p and are required for early nucleolar pre-rRNA processing at sites A0 to A2, raising the possibility that snoRNA cap trimethylation may be required for pre-rRNA processing.
Here we report that cells with TGS1 deleted are indeed defective for pre-rRNA processing reactions. However, mutational analysis revealed that it is the protein Tgs1p per se, rather than its catalytic activity in cap trimethylation, that is required for efficient small ribosomal subunit synthesis. In addition, we report a novel and unexpected requirement for Tgs1p and the KKD/E extension of the snoRNP proteins in nucleolar morphology that we believe to be fully compatible with a self-organization model for nucleolar assembly.
MATERIALS AND METHODS
Yeast.
Saccharomyces cerevisiae strains were cultivated according to standard procedures.
For the pulse-chase labeling analysis, strains transformed with a URA3 plasmid were grown at 23°C to mid-log phase in synthetic dextrose without uracil, pulse-labeled for 2 min with tritiated uracil, and chased with an excess of cold uracil for the times indicated. For the pre-rRNA processing analysis in tgs1Δ backgrounds, cells were cultivated to mid-log phase in yeast extract-peptone-dextrose at the indicated temperatures (30°C for the XRN1 and xrn1Δ controls). Strains harboring catalytic mutations were grown to mid-log phase at 30°C in SD without uracil and transferred to 16°C for up to 9 h. Conditions of growth for the electron microscopy (EM) analysis were as described for the RNA experiments. The genotypes of the strains used in this work were described previously (5, 19, 20, 27). Strains Nop56p-CFP and Nop56pΔK-CFP were generated as follows. CFP-hph cassettes with appropriate flanking sequences were generated by PCR with oligonucleotides LD382-LD384 and LD383-LD384, respectively, and plasmid pBS4 (a kind gift of Trisha Davis, Yeast Resource Center, University of Washington) and integrated by homologous recombination at the NOP56 locus in YDL401 (8). Transformants were selected on yeast extract-peoptone-dextrose-hygromycin B and screened by PCR on colonies with oligonucleotides LD199-LD200 and fluorescence. The oligonucleotides were as follows (sites of PCR priming are in lowercase): LD199, CTAAAGAAACAAGTTGAGC; LD200, GTGAAAGAGGCATAAGAG; LD382, AAGGAAAAGGAATCCTCAAAGAAGAGAAAATTAGAAGATGATGATGAAggtcgacggatccccgggCCTCAAAGAAGAGAAAATTAGAAGATGATGATGAAGGTCGACGGATCCCCGGG; LD383, AAGAAAGATAAAAAGGAAAAGAAGGATAAAAAGAAGAAAAGTAAGGATggtcgacggatccccgggAAGGAAAAGAAGGATAAAAAGAAGAAAAGTAAGGATGGTCGACGGATCCCCGGG; and LD384, ACAATATACAGCTTCCAAATCCTAAAGAAGAATATTAGAAGGGAATAAatcgatgaattcgagctcgACAATATACAGCTTCCAAATCCTAAAGAAGAATATTAGAAGGGAATAAATCGATGAATTCGAGCTCG.
DNA constructs.
Catalytic mutations in Tgs1p (D103A and W178A) were generated by site-directed mutagenesis (19, 20) and introduced by transformation in a tgs1Δ background. The various Tgs1p alleles were fused to green fluorescent protein (GFP) and expressed from a URA3/CEN-based plasmid. As controls, an isogenic strain transformed with a plasmid-borne wild-type fusion of TGS1 and a empty vector were used. Cyan fluorescent protein (CFP) and yellow fluorescent protein (YFP) fusion versions of Nop1p, Hmo1p, and Nug2p (kind gifts from O. Gadal, Institute Pasteur, Paris, France) were described previously (4).
RNA methods.
Total RNA extraction, pulse-chase labeling, Northern blotting, and primer extension experiments were as described previously (9). For tgs1Δ strains, the pre-rRNA processing defects observed at 16°C were identical to those reported to occur at 23°C (data not shown). Splicing of the box C+D snoRNA U3 was inspected by primer extension as described previously (20). PhosphorImager quantitations were performed with a Typhoon 9400 instrument and the native ImageQuant TL software (Amersham Biosciences). RP-mRNA probes were as follows: anti-RPS9A, GTTGTATACTTTTGTATTTCT; anti-RPS11A, CTTGCTGGTTGCTTAATTT; and anti-RPS11B, TCCCTGGCTTGATACGTT.
EM analysis.
Samples were subjected to an acetylation procedure that specifically enhances the contrast of nuclear components, as described previously (31).
For immunogold labeling, strains were fixed for 16 h at 4°C in 4% formaldehyde in 0.1 M phosphate buffer (0.1 M KH2PO4, 0.1 M K2HPO4 [pH 6.5]) containing 10% sucrose, dehydrated through graded acetone solutions, and embedded in Lowicryl K4 M. Immunogold detection was performed according to classical procedures (28). Nop1p was detected with monoclonal antibody mA66 (a kind gift of J. Aris, University of Florida) diluted 1:20. The secondary antibody was gold-coupled goat anti-mouse immunoglobulin G (10-nm-diameter particles; Amersham Life Sciences) diluted 1:40. No label was seen when the primary antibody was omitted. For an evaluation of the density of labeling, the surface area (Sa) of each compartment studied was first estimated by using a morphometrical approach with the point-counting method (33). After the evaluation of the Sas occupied by the different compartments, the number of gold particles (Ni) present over each of these compartments was counted, and the density of the labeling (Ns) was calculated as Ns = Ni/Sa. In the present study, for the mutant and wild-type strains, 14 and 12 randomly selected micrographs were analyzed and 79 and 52 gold particles were counted, respectively. Values obtained from the resin can be considered background staining. A Student t test (http://www.physics.csbsju.edu/stats/ttest.html; St. John's University, Collegeville, Minn.) was performed to evaluate the statistical relevance of the observations.
Immunofluorescence.
Immunofluorescence analysis was performed according to standard procedures (23). Cells were fixed for 1 h in formaldehyde (3.7%). Nop1p was detected with monoclonal antibody mA66 (a kind gift of J. Aris, University of Florida) at a working dilution of 1:250. The secondary antibody was Alexa Fluor 594 goat anti-mouse immunoglobulin G (heavy plus light chains) (A-11005; Molecular Probes) (working dilution, 1:500). Cells were counterstained with DAPI (4′,6′-diamidino-2-phenylindole). Acquisition was with a Zeiss Axioplan2/KS300 v3.0 microscope equipped with a Plan Neofluar 100×/1.3 objective and a charge-coupled device camera (MicroMax; Princeton Instruments, Inc.). Images were formatted in Adobe Photoshop and Illustrator.
Confocal microscopy.
For live observations, cells were grown to mid-log phase, washed twice in 1× phosphate-buffered saline-5% bovine serum albumin, and embedded in 0.5% agarose. Acquisition was with a Leica DMIRE2/LCS v2.00 microscope equipped with a 100×/Apo/1.4 objective. Quantitations used the native software (LCS v2.00). Files were transferred to and formatted with Adobe PhotoShop and Microsoft Excel.
For stereological quantitations, three-dimensional (3D) reconstructions were performed as follow. Cells were sliced into 20 sections, and a binary image was generated for each section with the segmentation tool (Leica LCII software). Positive voxels in each sections were summed and multiplied by the volume of a single voxel. On average, 20 cells were analyzed for each condition, and a Student t test (http://www.physics.csbsju.edu/stats/t test.html) was performed to evaluate the statistical relevance of the observations.
RESULTS
Tgs1p is required for small ribosomal subunit synthesis.
Besides their well-documented function in pre-RNA 2′-O methylation (box C+D) and pseudouridine formation (box H+ACA), several snoRNA members of both families are required for early nucleolar pre-rRNA processing (reviewed in references 6 and 16) (Fig. 1). Of these, the box C+D snoRNA U3 and the box H+ACA snoRNAs snR10 and snR30 are cap trimethylated by Tgs1p.
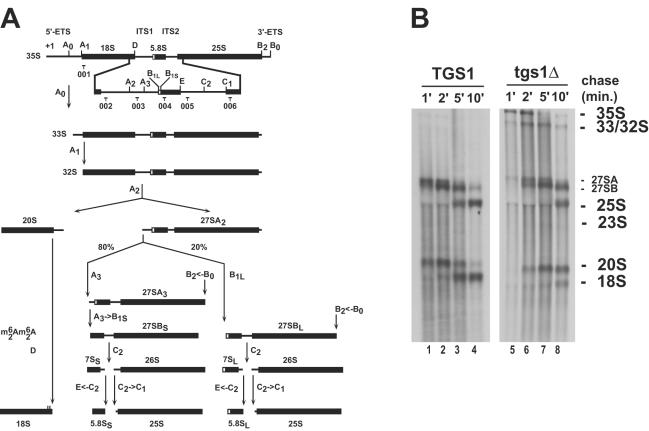
FIG. 1.
Tgs1p is required for small ribosomal subunit synthesis. (A) Yeast pre-rRNA processing pathway. Cleavage sites (A to E) and oligonucleotide probes (001 to 006) used in the Northern blot and primer extension experiments are indicated. See text and reference 10 for a detailed description of the pathway. The box C+D snoRNAs U3 and U14 and the box H+ACA snoRNAs snR10 and snR30 are required for pre-rRNA processing at sites A0 to A2. U3, snR10, and snR30 are cap trimethylated by Tgs1p. ETS, external transcribed spacer. (B) In vivo labeling analysis of pre-rRNA processing in strains with TGS1 deleted (tgs1Δ) and a wild-type isogenic control (TGS1) grown at the semipermissive temperature of 23°C. Samples were collected after 1, 2, 5, and 10 min of chase.
To test whether cap trimethylation is required for ribosome synthesis, rRNA processing was analyzed in a strain with TGS1 deleted (tgs1Δ) and a wild-type isogenic control (TGS1) by pulse-chase labeling, Northern blot hybridization, and primer extension (Fig. 1 and 2). Since the deletion of TGS1 confers a cold-sensitive phenotype for growth (20), these analyses were performed at the physiological temperature of 30°C, as well as at the semipermissive temperature of 23°C (slow growth) and after a prolonged (6- to 9-h) transfer from 30 to 16°C (no residual growth) (Fig. 1 and 2 and data not shown) (see Materials and Methods).
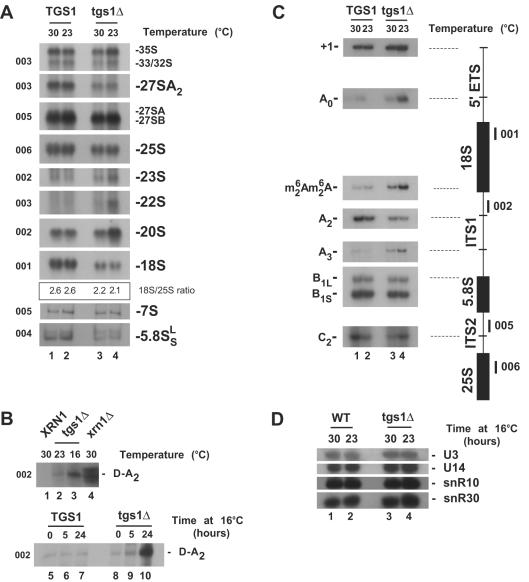
FIG. 2.
Tgs1p is required for pre-rRNA processing. (A to C) Northern blot (A and B) and primer extension (C) analyses of pre-rRNA processing defects in tgs1Δ and isogenic wild-type backgrounds. Temperatures of growth (16, 23, and 30°C) and the oligonucleotides used (001 to 006 [Fig. 1A]) are indicated. The ratio of 18S to 25S was quantitated with a PhosphorImager (Typhoon; Amersham Biosciences) and estimated to be ∼2.6 for the wild type and ∼2.1 to ∼2.2 for the mutant. tgs1Δ cells grown under nonpermissive conditions accumulated the D-A2 spacer fragment that is readily detected in xrn1Δ strains (27) (B). The use of the two alternative pathways of 5.8S-25S synthesis (30) was not affected in tgs1Δ strains, as indicated by the steady-state levels of the two forms of 5.8S rRNA (A) and the conserved ratio of cleavage at site B1S and B1L (C). In wild-type cells, the 18S rRNA is dimethylated at its 3′ end on two adjacent adenosines by Dim1p (9). cDNA strands that extended across this position from oligonucleotide 002 revealed that pre-rRNAs accumulated in tgs1Δ cells are dimethylated (C). (D) Steady-state levels of the box C+D snoRNAs U3 and U14 and the box H+ACA snoRNAs snR10 and snR30 in tgs1Δ cells and a wild-type (WT) isogenic control. Total RNA was extracted at 23 and 30°C. Oligonucleotides specific to the snoRNA species targeted were used to probe a Northern blot membrane (see Materials and Methods).
For the pulse-chase labeling shown in Fig. 1B, cells were grown to mid-log phase at 23°C, pulse-labeled with tritiated uracil for 2 min, and chased with an excess of cold uracil. Total RNA was extracted, separated on agarose-formaldehyde gels, transferred to nylon membranes, and processed for tritium detection.
In wild-type strains, the RNA polymerase I primary transcript (35S) is first cleaved in the 5′ external transcribed spacer at sites A0 and A1 and in the internal transcribed spacer 1 (ITS1) at site A2. This successively generates the 33S, 32S, 27S, and 20S pre-rRNAs (Fig. 1A). The 27S and 20S pre-rRNAs are destined to the small and large ribosomal subunits, respectively (reviewed in references 10 and 30). In tgs1Δ strains, the 35S and 33S-32S pre-rRNAs were readily detected (Fig. 1B). These species are not normally accumulated in wild-type strains due to rapid processing at sites A0 to A2, indicating that cleavage at these sites is delayed in tgs1Δ strains. The 27SA precursor was quantitatively chased into 25S rRNA, albeit with reduced kinetics. However, the overall level of 25S rRNA was only mildly affected. Most striking was the stable accumulation over the time of the chase of the 20S pre-rRNA, which is normally cleaved at site D to provide the 18S rRNA (reviewed in reference 2). As a likely consequence, the 18S rRNA was substantially underaccumulated.
The pre-rRNA processing defect observed in tgs1Δ cells was investigated further by Northern blotting and primer extension (Fig. 2). The delay in cleavage at the early nucleolar processing sites A0 to A2 observed in tgs1Δ cells by pulse-chase labeling was confirmed by Northern blot hybridizations (Fig. 2A), with (i) a significant accumulation of the 35S and 33S-32S pre-rRNAs, (ii) the underaccumulation of the 27SA2 pre-rRNA, and (iii) the accumulation of the aberrant 23S and 22S RNAs. In wild-type strains, a minor fraction of 35S pre-rRNA molecules is directly cleaved in the ITS1 at site A3, generating the 23S RNA; this occurred with a substantially increased incidence in tgs1Δ strains grown at 23°C (Fig. 2A). The 22S RNA that extends from cleavage site A0 to A3 and results from processing of the 33S pre-rRNA at site A3 is not normally detected in wild-type strains (Fig. 2A); a strikingly elevated amount of this RNA was accumulated in the absence of Tgs1p. The 20S pre-rRNA was also substantially accumulated (Fig. 2A); this was unexpected with respect to the inhibition of cleavage at site A2 and suggested a substantial impairment in downstream processing reactions. Cleavage at site D, however, was not fully inhibited, as the D-A2 fragment not only was readily detected but was significantly accumulated in tgs1Δ cells grown under nonpermissive conditions (shown for cells grown at 16°C, cells grown at 23°C, and after a prolonged transfer from 30 to 16°C) (Fig. 2B). The degradation of this spacer fragment, normally carried out by the major cytoplasmic 5′-3′ exoribonuclease Xrn1p (27), was therefore significantly slowed down as well, indicating that this step is closely linked to upstream processing reactions and suggesting a possible “handover” of spacer fragments from the endonucleolytic activity to Xrn1p. These inhibitions led to a severe reduction in the steady-state level of 18S rRNA (Fig. 2A). PhosphorImager quantitation estimated the ratios of 18S to 25S rRNA to be about 2.6 in the wild type and about 2.1 to 2.2 in the mutant (Fig. 2A). After prolonged transfer to 16°C, the 18S/25S ratio remained at 2.1 (data not shown). In contrast, the steady-state levels of the 27SB and 7S pre-rRNAs, as well as the 5.8S and 25S rRNAs, were only marginally affected, indicating that processing reactions downstream from cleavage site B were not greatly affected by the absence of Tgs1p.
Several pre-rRNA species are not easily detected by Northern blotting due to their low abundance, high turnover, or size and require primer extension analysis for their identification. For these species, cDNA strands were extended across the 5′ external transcribed spacer and the ITS1 and ITS2 from oligonucleotides 001, 005, and 006, allowing the detection of the transcription start site (+1) and the cleavage sites A0, A2, A3, B1, and C2, respectively (Fig. 2C). This revealed an increased signal at +1 and A0, a reduction at A2, and, as expected, no inhibition at sites A3, B1, and C2.
The production of mature 18S rRNA involves the modification, by base modification, of two adjacent adenosines at its 3′ end, a modification that is carried out by the dimethyl transferase Dim1p (9). To test for potential functional interactions between Tgs1p and Dim1p, the level of 18S rRNA dimethylation was assessed by primer extension. To this end, primer 002, which is specific to the 5′ end of ITS1, was extended across the 3′ end of 18S rRNA, revealing a stop that corresponds to the modification. Pre-rRNAs that accumulated in the absence of Tgs1p were efficiently methylated (Fig. 2C). The increased signal at the site of modification was well correlated with the accumulation of 20S pre-rRNA (Fig. 2A).
Importantly, the reported pre-rRNA processing inhibitions were not due to decreased amounts of the snoRNAs required for pre-rRNA processing, as Northern blot analysis revealed that the steady-state levels of U3, U14, snR10, and snR30 were not affected (Fig. 2D). This was confirmed for U3 by primer extension (see Fig. 3B).
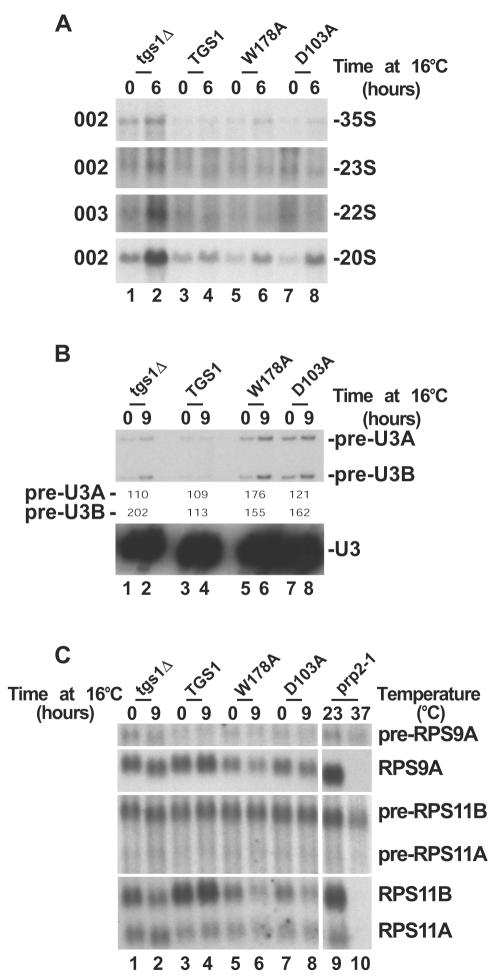
FIG. 3.
Cap trimethylation is not required for pre-rRNA processing. Pre-rRNA processing (A) and splicing analysis (B and C) of Tgs1p catalytic mutants (D103A and W178A), a wild-type isogenic control (TGS1) and a strain with TGS1 deleted (tgs1Δ) grown at 30°C and transferred to 16°C for up to 9 h are shown. (A) Pre-rRNA processing analysis by Northern blotting. The oligonucleotide probes (see Fig. 1A) used are indicated on the left. (B) U3 splicing analysis by primer extension. The splicing defect was estimated by PhosphorImager quantitation (Typhoon; Amersham Biosciences) and expressed as (signal at 16°C/signal at 30°C) × 100. (C) RP pre-mRNA splicing analysis by Northern blotting. A prp2-1 strain grown at 23°C and transferred to 37°C for 90 min was used as a control. See Materials and Methods for the oligonucleotide probes used.
In conclusion, cells deprived of Tgs1p are severely delayed in early nucleolar pre-rRNA processing reactions at sites A0 to A2 and in late cleavage at site D. None of these cleavages is fully inhibited; rather, it is the kinetics of processing that is altered, notably with premature processing in ITS1 at site A3.
Cap trimethylation is not required for pre-rRNA processing.
To test whether the catalytic function of Tgs1p in cap trimethylation is responsible for the pre-rRNA processing defect reported, several conserved residues involved in the methyl transfer reaction were individually substituted for alanines. Comprehensive protein sequence comparisons and phylogenetic analysis allowed the construction by homology modeling of a detailed 3D structural model based on solved structures of well-characterized methyl transferases (19). In this structural model, residues D103 and W178 were singled out as potentially involved in cofactor binding and association with the cap, respectively. D103 is expected to be involved in the water-mediated coordination of the methionine moiety of S-adenosylmethionine; W178 is predicted to be required to stabilize the interaction with the base targeted for modification (i.e., the N7-methylated G of the cap). The predictive value of this model was tested by site-directed mutagenesis. Both residues were individually substituted for alanines and expressed as GFP fusions in tgs1Δ cells (see Materials and Methods). The stability of the catalytic mutant versions of the protein was tested by Western blotting with a GFP-specific antibody; both mutant proteins were expressed at the same level as a wild-type Tgs1p-GFP control construct (data not shown). Furthermore, the nucleolar localization of the catalytic constructs was not affected, as tested by colocalization with an Srp40p-DsRed fusion, a known nucleolar antigen (data not shown). Finally, neither of the two catalytic mutations complemented the slow-growth phenotype associated with tgs1Δ (data not shown). Cap trimethylation was tested on whole-cell extracts by immunoprecipitation with antibodies specific to trimethylated RNAs, and the RNAs recovered in the pellet fractions were tested by Northern blot hybridization. No coprecipitation was observed for the snRNAs and box C+D and H+ACA snoRNAs that are normally modified by Tgs1p, indicating that both mutations led to a complete inhibition of cap trimethylation (data not shown) (19, 20). Importantly, in the D103A and W178A mutants, the methyl transfer activity of Tgs1p is inactivated in a nonconditional fashion; i.e., cap trimethylation is inhibited at all temperatures.
Pre-rRNA processing in the catalytic mutants was analyzed by Northern blot hybridization as described above (Fig. 3A). Total RNAs were extracted from the catalytic mutants, a strain with TGS1 deleted, and a wild-type isogenic control at 30°C and after a transfer from 30 to 16°C that was prolonged for 6 to 9 h (results for 6 h are shown; identical results were observed for 9 h of transfer). Comparison of the steady-state levels of the 35S pre-rRNAs in these strains indicates that the catalytic mutants are not kinetically defective for pre-rRNA processing. Consistently, the 22S and 23S aberrant RNAs were not detected in the catalytic mutants, and the 20S pre-rRNA was accumulated to the same extend as in the wild type. Furthermore, a PhosphorImager quantitation analysis established the 18S/25S ratios to be ∼2.6 and ∼2.2 in the wild type and in tgs1Δ strains, respectively, and ∼2.5 and ∼2.7 in the W178A and D103A mutants, respectively. We concluded that it is not the catalytic activity of Tgs1p in cap trimethylation that is required for efficient pre-rRNA processing.
Many RPs are encoded by intron-containing genes, and strains defective for pre-mRNA splicing are characterized by an imbalanced production of ribosomal components that often results in pre-rRNA processing inhibitions (reviewed in reference 30). Strains with TGS1 deleted are mildly defective for splicing (20) (Fig. 3B and C), and we therefore tested whether the pre-rRNA processing defect reported for tgs1Δ strains is not simply an indirect consequence of the splicing inhibition.
Splicing efficiency was tested for U3 and several pre-mRNAs encoding small ribosomal subunit proteins, by primer extension and Northern blotting, respectively (Fig. 3B and C). Total RNA samples extracted from cells with TGS1 deleted, the catalytic mutants, and a wild-type isogenic control grown at 30°C and transferred to 16°C for 9 h were used for these analyses (Fig. 3B and C).
U3 is encoded by duplicated genes (U3A and U3B) that differ only in the size of their introns. Pre-U3A and pre-U3B precursors could thus easily be detected by primer extension (Fig. 3B). The analysis with U3 confirmed our previous observation of a moderate splicing inhibition (20): (i) the steady-state level of the RNA was unaffected (see also Northern blot hybridization in Fig. 2D), and (ii) low levels of the pre-U3A and pre-U3B precursors were detected in tgs1Δ cells. Significantly, the catalytic strains, which are not kinetically defective for pre-rRNA processing, were at least as defective for splicing as tgs1Δ strains, indicating that the ribosome synthesis defect is not simply a direct consequence of the splicing inhibition.
This conclusion was further substantiated by the analysis of RP pre-mRNA splicing by Northern blot hybridizations. The RP mRNAs selected show weak splicing signals and are expected to be dependent on full splicing efficiency for their maturation. RPS9A, RPS11A, and RSP11B have nonconsensus 5′ splice sites (RPS9A contain GUACGU instead of GUAUGU, while RPS11A and RPS11B contain GUAUGA instead of GUAUGU); RPS9A and RPS11B also lack optimal polypyrimidine tract and branch point region sequences (CACUAAC and GACUAAU, respectively, instead of UACUAAC).
With the exception of the prp2-1 splicing mutant control strain, which did not accumulate any of the mature RNAs at the nonpermissive temperature (37°C), the steady-state levels of the RNAs tested were only marginally affected. The production of RPS11B was more sensitive to tgs1 mutations, but even in this case, catalytic strains were at least as defective as tgs1Δ strains.
Tgs1p is required for intact nucleolar morphology.
Mutational analysis demonstrated that the requirement for Tgs1p in ribosome synthesis is not dependent on cap trimethylation. Additional functions for the protein, which possibly could underlie the ribosome synthesis defect reported for tgs1Δ strains, include a possible involvement in nucleolar structure. Indeed, we reasoned that the transient interactions of the protein with the abundant snoRNPs may contribute to nucleolar morphology.
To test this hypothesis, the nucleolar ultrastructure in tgs1Δ and wild-type control cells was inspected by EM. Wild-type yeast nucleoli typically show two distinct subcompartments, fibrillar strands and granules, that could easily be recognized (Fig. 4A). Cells lacking Tgs1p were quite remarkable in that the nucleus appeared to be nearly entirely homogeneous. A clear nucleolus could not be identified, and the fibrillar and granular subcompartments could not be detected. In stark contrast, both catalytic mutants maintained a conspicuous level of nucleolar organization with distinct fibrils and granules (Fig. 4A).
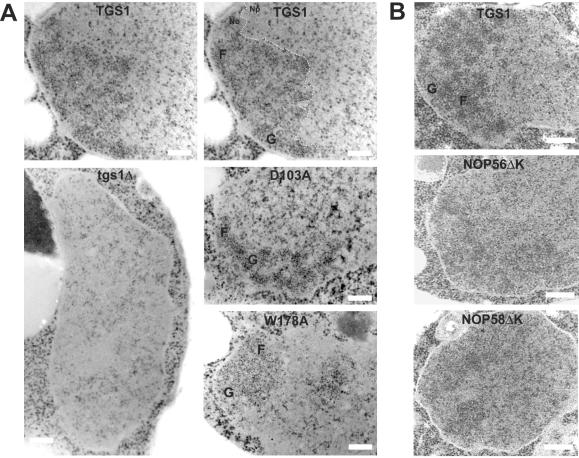
FIG. 4.
Tgs1p and the conserved KKD/E domain of snoRNP proteins are required for intact nucleolar morphology. (A) EM analysis of strains with TGS1 deleted (tgs1Δ), the catalytic mutants (D103A and W178A), and a wild-type isogenic control (TGS1). For convenience, the wild-type panel was duplicated and the nucleolar structure is outlined with a thin dashed line. No, nucleolus; Np, nucleoplasm; F, fibrillar strands; G, granular component. Bars, 0.25 μm. (B) EM analysis of strains lacking the carboxyl-terminal KKD/E domain of Nop56p (NOP56ΔK) or Nop58p (NOP58ΔK) and a wild-type isogenic control. In NOP56ΔK and NOP58ΔK, a residual nucleolar structure can be detected to the left of each cell. Note that the experiments for panel A were performed at 16°C; identical results were obtained with cells grown at 30°C (data not shown).
To ascertain the effects of Tgs1p on nucleolar structure, the distributions of several nucleolar antigens were inspected by epifluorescence, confocal microscopy, and EM, by either indirect immunofluorescence, immunogold labeling, or direct imaging of fluorescent constructs in live cells (Fig. 5 and 6).
FIG. 5.
Nop1p shows a wider distribution in tgs1Δ cells. Indirect immunofluorescence (A) and EM immunogold labeling (B) with a Nop1p-specific antibody are shown. The tgs1Δ strain and a wild-type isogenic control were grown to mid-log phase at 30°C and processed for indirect immunofluorescence or EM immunogold labeling according to standard procedures (see Materials and Methods). (A) The bulk of the DNA was counterstained with DAPI. Cell were observed with a Axioplan2 (Zeiss) microscope and a charge-coupled device-cooled camera. (B) For convenience, the boundary between the nucleolus (No) and the nucleoplasm (Np) in the wild type is outlined with a dotted line and gold particles are circled in red. Bar, 0.25 μm.
FIG. 6.
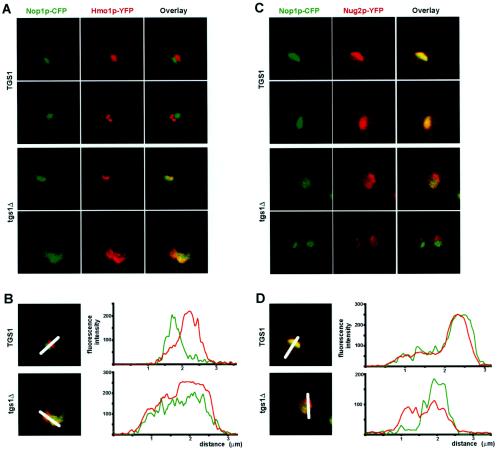
Nucleolar antigens are redistributed in tgs1Δ cells. Nop1p and Hmo1p are colocalized in tgs1Δ cells (A and B); Nop1p and Nug2p are segregated in tgs1Δ cells (C and D). (A) Cells coexpressing a yellow fluorescent version (YFP) of Hmo1p and a cyan fluorescent construct of Nop1p (CFP) were grown to mid-log phase, embedded in agarose, and imaged live with a Leica confocal microscope. The signals detected for the CFP and YFP constructs were artificially colored in green and red, respectively. The overlay (yellow) is also shown. (B) Quantitation was performed with the native LCS v2.00 software (Leica). Color is as in panel A. x axis, distance (micrometers); y axis, fluorescence intensity (arbitrary units). The nucleus always sized on the average of 2.5 μm in diameter. (C) Live cells coexpressing a yellow fluorescent version (YFP) of Nug2p and a cyan fluorescent construct of Nop1p (CFP), artificially colored in red and green, respectively, were observed as described for panel A. (D) The fluorescent signal was quantitated as for panel B.
First, Nop1p, which largely labels the fibrillar component of the nucleolus, was detected by indirect immunofluorescence with a specific monoclonal antibody and imaged by epifluorescence (Fig. 5A). In wild-type cells, Nop1p showed the expected, compact, crescent-like structure facing the bulk of the DNA. By comparison, in tgs1Δ cells the signal was more diffuse and largely overlapped with areas normally occupied by the nucleoplasm (Fig. 5A).
Second, the alteration in the distribution of Nop1p was confirmed at the EM level by immunogold labeling (Fig. 5B). In these experiments, the morphology is not quite as well preserved due to the fixation procedure used (see Materials and Methods). However, the nucleolar territory was easily identified in wild-type cells and quantitatively labeled by Nop1p. Such a distinct territory could not be detected in tgs1Δ cells, where Nop1p lost its confined localization and was scattered throughout the whole nuclear volume (Fig. 5B). This was confirmed by a quantitative analysis that revealed that the density of the gold particles detected in the nucleus increased by a factor of ∼48% (Table 1).
TABLE 1.
Quantitation of gold particles in subcellular compartments by EM-immunogold labeling
| Compartment | Mean no. of particles ± SD (no. of cells)
|
P (Student t test) | |
|---|---|---|---|
| TGS1 (12) | tgs1Δ (14) | ||
| Nucleus (nucleolus + nucleoplasm) | 3.52 ± 1.63 | 5.21 ± 1.91 | 0.024 |
| Nucleolus | 7.04 ± 2.74 | ||
| Nucleoplasm | 0.54 ± 0.98 | ||
| Cytoplasm | 0.04 ± 0.14 | 0.20 ± 0.54 | |
| Resin | 0.03 ± 0.09 | 0 | |
Third, the distribution of nucleolar antigens in live cells was established by confocal microscopy (Fig. 6). For this, Hmo1p and Nug2p, which are specific to distinct nucleolar domains, the ribosomal DNA territory and the granular component of the nucleolus, respectively (3, 4), were fused to a yellow-shifted variant of GFP and coexpressed with a cyan fluorescent version of Nop1p. In wild-type cells, the Hmo1p signal was detected opposite to that of Nop1p, defining the individual ribosomal DNA territory, as previously reported (3). This pattern was lost in tgs1Δ backgrounds, where the two signals largely overlapped (Fig. 6A). The distribution of the intensity of the relative fluorescent signals was quantitated and confirmed our visual inspections (Fig. 6B). The distribution of the Nug2pYFP constructs was analyzed under the same conditions. In wild-type strains, the Nop1p and Nug2p signals overlapped nearly entirely. The overlay was strikingly different in the absence of Tgs1p, with Nug2p distributing more widely, including the Nop1p-positive area (Fig. 6C). This was also confirmed by a quantitative analysis (Fig. 6D).
Finally, in these later experiments (Fig. 6), and in agreement with what we observed by indirect immunofluorescence and immunogold labeling (Fig. 5), the Nop1p-CFP labeling was consistently enlarged and more diffuse in tgs1Δ cells in comparison to wild-type cells. Three-dimensional reconstructions allowed the stereological quantitation of these differences (see Materials and Methods). This analysis revealed that the volume occupied by Nop1p-CFP statistically increased by a factor of ∼122% in the mutant (1.94 μm3 in tgs1Δ cells versus 0.87 μm3 in wild-type cells P ≪ 0.01 by Student's t test) (Table 2).
TABLE 2.
Stereological quantitations of individual nucleolar antigens by confocal analysis
| Antigen (strain) | Mean vol (μm3) ± SD | No. of cells | P (Student t test) |
|---|---|---|---|
| Nop1p-CFP (TGS1) | 0.87 ± 1.01 | 20 | 0.0044 |
| Nop1p-CFP (tgs1Δ) | 1.94 ± 1.21 | 20 | |
| Nop56p-CFP (TGS1) | 0.64 ± 0.45 | 22 | 0.0034 |
| Nop56pΔK-CFP (TGS1) | 1.12 ± 0.59 | 24 |
The conserved KKD/E domain of snoRNP proteins is required for nucleolar compaction.
We previously reported that in vitro Tgs1p interacts specifically with the KKD/E domain (up to 13 repetitions of the corresponding consensus amino acids) found at the carboxyl-terminal ends of the box C+D snoRNP proteins Nop56p and Nop58p and the box H+ACA protein Cbf5p (5, 11, 13, 14, 20, 32, 35). This suggested that in vivo, Tgs1p may also interact with the snoRNPs through these highly charged carboxyl-terminal tails. If so, the KKD/E domains were also expected to contribute to nucleolar morphology.
To test this hypothesis, strains expressing as their unique source of the target protein a carboxyl-truncated version of Nop56p or Nop58p were inspected by EM (Fig. 4B). Importantly, none of these constructs were affected in their interaction with the snoRNAs or in growth (5, 13, 14). In both Nop56pΔK and Nop58pΔK, the nucleus appeared largely homogeneous, with a nucleolus that had lost its compaction and with nucleolar subcompartments that could not be as easily identified as in the wild type (Fig. 4B).
The involvement of the KKD/E domain in nucleolar morphology was quantitated by 3D reconstructions, as described above. For this, a CFP epitope was integrated at the NOP56 locus by PCR, generating either a functional full-length Nop56p-CFP fusion or a version lacking the KKD/E domain (see Materials and Methods). Following 3D reconstructions, the fluorescent signal detected in the Nop56pΔK-CFP strain was compared to that observed in the control full-length Nop56p-CFP strain (see Materials and Methods). In this case, the increase in volume in the mutant was estimated to be about ∼75% (1.12 μm3 for Nop56pΔK-CFP versus 0.64 μm3 for full-length Nop56p-CFP [P ≪ 0.01]) (Table 2).
DISCUSSION
Trimethyl guanosine synthase I (Tgs1p in yeast and PIMT in higher eukaryotes) specifically selects the m7G-monomethylated caps of snRNAs and snoRNAs for m2,2,7G-trimethylation. In metazoans, cap trimethylation is required for the nuclear uptake of snRNPs following their essential cytoplasmic phase of maturation. In yeast, cells with TGS1 deleted are mildly defective for splicing and inhibited for growth at low temperature. We report here that Tgs1p is further required for ribosome synthesis and normal nucleolar morphology.
First, we demonstrate that cells with TGS1 deleted are defective for pre-rRNA processing, primarily at the early nucleolar cleavage sites A0 to A2, and consequently are inhibited for small ribosome subunit synthesis. Surprisingly, while several snoRNA species (U3, snR30, and snR10) are required for pre-rRNA processing at sites A0 to A2 and are cap trimethylated by Tgs1p, we found that alanine substitutions that specifically inhibited the catalytic function of the protein in RNA cap trimethylation did not affect pre-rRNA processing. Importantly, strains harboring catalytically defective alleles of Tgs1p were at least as defective for the splicing of U3 and several pre-mRNAs encoding RPs of the small subunit as were strains with TGS1 deleted, indicating that the ribosome synthesis defect reported for tgs1Δ strains is not simply a direct consequence of the mild splicing defect. We therefore concluded that Tgs1p carries additional functions than in cap trimethylation, functions that are required for efficient ribosome synthesis.
We previously described Dim1p, which selectively selects in the cytoplasm two adjacent adenosines at the 3′ end of the 18S rRNA for m62Am62A conversion; Dim1p is additionally required for nucleolar pre-rRNA processing at cleavage sites A1 and A2 (9). We proposed the existence of a quality control mechanism whereby the association of Dim1p with nucleolar pre-rRNAs is monitored by the processing machinery, resulting in the exclusive maturation at sites A1 and A2 of pre-rRNA molecules that have bound Dim1p and are thus programmed for cytoplasmic methylation (12). Consistently, we know now that Dim1p is indeed detected in the nucleolar 90S preribosomes and remains associated with the pre-40S subunits, within the recently described small ribosomal subunit RRP complex, throughout their final stages of cytoplasmic maturation (25, 29). Tgs1p is not a core component of the snoRNPs; however, its nucleolar localization and its requirement for early pre-rRNA processing suggested that this protein too may be physically associated with known RRPs within nucleolar preribosomes, much in the way that Dim1p is. This was tested extensively by coprecipitation with an epitope-tagged construct (TAP tag [24]) (data not shown). We concluded that Tgs1p is not found in stable association with known RRPs. However, Tgs1p did show significant interactions with uncharacterized proteins, such as Oye2p, Lsp1p, and Asc1p, that in turn are physically associated with and highly connected to several dozen early nucleolar RRPs, components of the U3 small ribosomal subunit processome and 90S preribosomes (G. Colau and D. L. J. Lafontaine, unpublished results).
Several observations indicate that in tgs1Δ strains, the pre-mRNA splicing inhibition contributes more substantially to the reported cold-sensitive phenotype for growth than the pre-rRNA processing defect. First, while the pre-rRNA processing is clearly enhanced at low temperature (there is clear accumulation of the 20S pre-rRNA, the 22S and 23S aberrant RNAs, and the D-A2 fragment [Fig. 2A and B]), the bulk of the processing defect is already detected at 30°C, with, notably, a reduction in 18S rRNA synthesis. This was not increased at the lower temperature of 23 or 16°C (Fig. 2A and data not shown). The accumulation of 35S RNA and the reduction in 27SA2 RNA (best seen in the primer extension and Northern blot hybridization in Fig. 2B and A, respectively) are also nonconditional and already detected at 30°C. Second, the alteration in nucleolar morphology in tgs1Δ cells is not conditional; i.e., it is observed to its full extent at 30°C. Finally, the catalytic mutants that are not defective for pre-rRNA processing are as cold sensitive for growth as tgs1Δ strains.
Additional functions for Tgs1p that could underlie its requirement for ribosome synthesis included a possible involvement in nucleolar morphology, a hypothesis that was tested both at the light microscopy and EM levels. Nucleolar morphology was indeed severely perturbed in tgs1Δ cells, with (i) a loss of structural organization and an apparent loss of compartmentalization (seen in EM analysis on acetylated samples), (ii) a statistically relevant wider distribution of Nop1p (seen in epifluorescence, immunogold labeling, and live observations), and (iii) a drastic alteration in the relative distribution of known antigens (Nop1p, Hmo1p, and Nug2p) (seen in confocal analysis). In contrast, catalytic mutant strains maintained a compact nucleolus with preserved structural organization (seen in EM analysis and acetylation).
In vitro, Tgs1p interacts specifically with the KKD/E domains of the snoRNPs, suggesting that in vivo, the transient interactions of the protein with the snoRNPs may also occur through this domain. Consistently, the nucleolar morphology in strains expressing versions of Nop56p and Nop58p lacking KKD/E was also severely affected, with (i) a loss of compaction and structural organization (seen in EM analysis) and (ii) a statistically relevant wider distribution of a Nop56pΔK fluorescent construct (seen in live observations).
In conclusion, we propose that Tgs1p, through multiple weak and transient interactions with the abundant snoRNPs (presumably through association with the KKD/E domain present in several snoRNPs) contributes significantly to the steady-state dynamics of the nucleolus. This conclusion does not demonstrate but is consistent with the concept that self-organization may be the force at stake in nucleolar morphogenesis.
Archaea strikingly lack KKD/E domains in their snoRNP-like proteins, and it seems quite possible that during eukaryotic evolution, the emergence of a small RNA cap trimethylase was followed by the coevolution of its substrate (i.e., through the acquisition of a carboxyl-terminal charged “landing path”) and that this contributed substantially to the coalescence and local enrichment in RRPs that led to the appearance of an ancestral nucleolus.
Acknowledgments
We acknowledge David Tollervey (Wellcome Trust Centre for Cell Biology, Edinburgh, Scotland) for critical reading of the manuscript, Etienne Pays and Benoit Vanhollebeke (Institut de Biologie et de Médecine Moléculaires, Université Libre de Bruxelles) for helpful discussions, Gabriele Burger (Leica Microsystems Heidelberg GmbH) and Gerard Halloy (Leica Belgium) for assistance with 3D reconstructions and stereological quantitations, and Giovanna Jaramillo-Gutierrez for her initial contribution to this project. We are indebted to Olivier Gadal (Pasteur Institute, Paris, France), Thierry Gautier and Ed Hurt (Biochemie-Zentrum, Heidelberg, Germany), Trisha Davis (Yeast Resource Center, University of Washington), and John Aris (University of California) for kindly providing yeast strains and reagents.
Research in the lab of D.L.J.L. is funded by the Fonds National de la Recherche Scientifique (FNRS), Université Libre de Bruxelles, and the following private charities: Banque Nationale de Belgique, Brachet, Defay, and van Buuren. D.L.J.L. has a permanent position with the FNRS.
REFERENCES
- 1.Chen, D., and S. Huang. 2001. Nucleolar components involved in ribosome biogenesis cycle between the nucleolus and nucleoplasm in interphase cells. J. Cell Biol. 153:169-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fatica, A., and D. Tollervey. 2002. Making ribosomes. Curr. Opin. Cell Biol. 14:313-318. [DOI] [PubMed] [Google Scholar]
- 3.Gadal, O., S. Labarre, C. Boschiero, and P. Thuriaux. 2002. Hmo1, an HMG-box protein, belongs to the yeast ribosomal DNA transcription system. EMBO J. 21:5498-5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gadal, O., D. Strauss, E. Petfalski, P. E. Gleizes, N. Gas, D. Tollervey, and E. Hurt. 2002. Rlp7p is associated with 60S preribosomes, restricted to the granular component of the nucleolus, and required for pre-rRNA processing. J. Cell Biol. 157:941-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gautier, T., T. Berges, D. Tollervey, and E. Hurt. 1997. Nucleolar KKE/D repeat proteins Nop56p and Nop58p interact with Nop1p and are required for ribosome biogenesis. Mol. Cell. Biol. 17:7088-7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kiss, T. 2002. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell 109:145-148. [DOI] [PubMed] [Google Scholar]
- 7.Kuersten, S., M. Ohno, and I. W. Mattaj. 2001. Nucleocytoplasmic transport: Ran, beta and beyond. Trends Cell Biol. 11:497-503. [DOI] [PubMed] [Google Scholar]
- 8.Lafontaine, D., and D. Tollervey. 1996. One-step PCR mediated strategy for the construction of conditionally expressed and epitope tagged yeast proteins. Nucleic Acids Res. 24:3469-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lafontaine, D., J. Vandenhaute, and D. Tollervey. 1995. The 18S rRNA dimethylase Dim1p is required for pre-ribosomal RNA processing in yeast. Genes Dev. 9:2470-2481. [DOI] [PubMed] [Google Scholar]
- 10.Lafontaine, D. L. J. Eukaryotic ribosome synthesis. In K. Nierhaus (ed.), Protein synthesis and ribosome structure, in press. Wiley Interscience, New York, N.Y.
- 11.Lafontaine, D. L. J., C. Bousquet-Antonelli, Y. Henry, M. Caizergues-Ferrer, and D. Tollervey. 1998. The box H + ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev. 12:527-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lafontaine, D. L. J., T. Preiss, and D. Tollervey. 1998. Yeast 18S rRNA dimethylase Dim1p: a quality control mechanism in ribosome synthesis? Mol. Cell. Biol. 18:2360-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lafontaine, D. L. J., and D. Tollervey. 1999. Nop58p is a common component of the box C+D snoRNPs that is required for snoRNA stability. RNA 5:455-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lafontaine, D. L. J., and D. Tollervey. 2000. Synthesis and assembly of the box C+D snoRNAs. Mol. Cell. Biol. 20:2650-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mattaj, I. W., and L. Englmeier. 1998. Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem. 67:265-306. [DOI] [PubMed] [Google Scholar]
- 16.Maxwell, E. S., and M. J. Fournier. 1995. The small nucleolar RNAs. Annu. Rev. Biochem. 64:897-934. [DOI] [PubMed] [Google Scholar]
- 17.Misteli, T. 2001. The concept of self-organization in cellular architecture. J. Cell Biol. 155:181-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Misteli, T. 2001. Protein dynamics: implications for nuclear architecture and gene expression. Science 291:843-847. [DOI] [PubMed] [Google Scholar]
- 19.Mouaikel, J., J. M. Bujnicki, J. Tazi, and R. Bordonné. 2004. Structure-function analysis of Tgs1, the yeast snRNA/snoRNA cap hypermethylase. Nucleic Acids Res. 31:4899-4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mouaikel, J., C. Verheggen, E. Bertrand, J. Tazi, and R. Bordonné. 2002. Hypermethylation of the cap structure of both yeast snRNAs and snoRNAs requires a conserved methyltransferase that is localized to the nucleolus. Mol. Cell 9:891-901. [DOI] [PubMed] [Google Scholar]
- 21.Olson, M. O., M. Dundr, and A. Szebeni. 2000. The nucleolus: an old factory with unexpected capabilities. Trends Cell Biol. 10:189-196. [DOI] [PubMed] [Google Scholar]
- 22.Phair, R. D., and T. Misteli. 2000. High mobility of proteins in the mammalian cell nucleus. Nature 404:604-609. [DOI] [PubMed] [Google Scholar]
- 23.Pringle, J. R., A. E. Adams, D. G. Drubin, and B. K. Haarer. 1991. Immunofluorescence methods for yeast. Methods Enzymol. 194:565-602. [DOI] [PubMed] [Google Scholar]
- 24.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 25.Schafer, T., D. Strauss, E. Petfalski, D. Tollervey, and E. Hurt. 2003. The path from nucleolar 90S to cytoplasmic 40S pre-ribosomes. EMBO J. 22:1370-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scheer, U., and R. Hock. 1999. Structure and function of the nucleolus. Curr. Opin. Cell Biol. 11:385-390. [DOI] [PubMed] [Google Scholar]
- 27.Stevens, A., C. L. Hsu, K. R. Isham, and F. W. Larimer. 1991. Fragments of the internal transcribed spacer 1 of pre-rRNA accumulate in Saccharomyces cerevisiae lacking 5′-3′ exoribonuclease 1. J. Bacteriol. 173:7024-7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thiry, M. 1999. Ultrastructural methods for nucleic acid detection by immunocytology. Prog. Histochem. Cytochem. 34:87-159. [DOI] [PubMed] [Google Scholar]
- 29.Vanrobays, E., J.-P. Gélugne, M. Caizergues-Ferrer, and D. L. J. Lafontaine. 2004. Dim2p, a KH-domain protein required for small ribosomal subunit synthesis. RNA J. 10:645-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Venema, J., and D. Tollervey. 1999. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33:261-311. [DOI] [PubMed] [Google Scholar]
- 31.Verheggen, C., J. Mouaikel, M. Thiry, J. M. Blanchard, D. Tollervey, R. Bordonné, D. L. Lafontaine, and E. Bertrand. 2001. Box C/D small nucleolar RNA trafficking involves small nucleolar RNP proteins, nucleolar factors and a novel nuclear domain. EMBO J. 20:5480-5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watkins, N. J., A. Gottschalk, G. Neubauer, B. Kastner, P. Fabrizio, M. Mann, and R. Luhrmann. 1998. Cbf5p, a potential pseudouridine synthase, and Nhp2p, a putative RNA-binding protein, are present together with Gar1p in all H BOX/ACA-motif snoRNPs and constitute a common bipartite structure. RNA 4:1549-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weibel, E. R. 1969. Stereological principles for morphometry in electron microscopic cytology. Int. Rev. Cytol. 26:235-302. [DOI] [PubMed] [Google Scholar]
- 34.Will, C. L., and R. Luhrmann. 2001. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 13:290-301. [DOI] [PubMed] [Google Scholar]
- 35.Wu, P., J. S. Brockenbrough, A. C. Metcalfe, S. Chen, and J. P. Aris. 1998. Nop5p is a small nucleolar ribonucleoprotein component required for pre-18 S rRNA processing in yeast. J. Biol. Chem. 273:16453-16463. [DOI] [PMC free article] [PubMed] [Google Scholar]