Abstract
Many of the RNAs transcribed from the mitochondrial genome of Physarum polycephalum are edited by the insertion of nonencoded nucleotides, which are added either singly or as dinucleotides. In addition, at least one mRNA is also subject to substitutional editing in which encoded C residues are changed to U residues posttranscriptionally. We have shown previously that the predominant type of editing in these organelles, the insertion of nonencoded single C residues, occurs cotranscriptionally at the growing end of the RNA chain. However, less is known about the timing of dinucleotide addition, and it has been suggested that these insertions occur at a later stage in RNA maturation. Here we examine the addition of both single nucleotides and dinucleotides into nascent RNAs synthesized in vitro and in vivo. The distribution of added nucleotides within individual cloned cDNAs supports the hypothesis that all insertion sites are processed at the same time relative to transcription. In addition, the patterns of partial editing and misediting observed within these nascent RNAs suggest that separate factors may be required at a subset of dinucleotide insertion sites and raise the possibility that in vivo, nucleotides may be added to RNA and then changed posttranscriptionally.
The sequence content of many RNAs in a variety of organisms is completed or diversified by RNA editing, involving the substitution of bases or insertion and deletion of nucleotides during or after transcription. Extensive insertional editing occurs during transcription in Physarum mitochondria, adding hundreds of single C residues, at an average spacing of approximately 25 nucleotides in mRNAs and 40 nucleotides in tRNAs and rRNAs, and a much smaller number of single U residues (6, 11). In addition, there are 19 reported instances of pairs of RNA residues known or likely to be inserted adjacent to each other (6, 12). Physarum mitochondria are distinctive in also performing substitutional editing on a transcript that is insertionally edited: C-to-U conversion occurs at four known sites in the coI mRNA, apparently at a later stage in RNA production (7, 8, 13). Together, these editing events result in the creation of open reading frames encoding proteins involved in oxidative phosphorylation and electron transport, as well as the generation of mature mitochondrial tRNAs and rRNAs.
The factors responsible for single-nucleotide and dinucleotide insertions have yet to be identified. Phylogenetic comparisons indicate that the ability to insert single C nucleotides, single U nucleotides, and dinucleotides arose separately. Horton and Landweber (9) determined the sequence of the mitochondrial coI gene and its transcript among a collection of organisms with various degrees of relatedness to Physarum. On the basis of their analysis, they proposed that U insertion appeared first, followed by C insertion and later dinucleotide additions, perhaps upon the emergence of separate or additional specificity factors (9). It has also been proposed that single and dinucleotide insertions occur via distinct mechanisms in Physarum mitochondria (16). This suggestion was based on the results of experiments in which cytb cDNAs containing unedited and misedited sites were selectively amplified from total mitochondrial RNA pools. Thus, there are a number of possible scenarios regarding the relationship between dinucleotide and mononucleotide insertional editing in Physarum mitochondria. Dinucleotides might be inserted by separate activities which evolved independently or were derived from the mononucleotide insertion apparatus or by the same machinery, potentially with additional or alternative specificity factors.
The insertion of single C nucleotides occurs cotranscriptionally, with the extra nucleotides added to the growing 3′ end of the RNA (5). Three lines of evidence indicate that dinucleotides are also added to nascent transcripts. First, S1 protection studies of labeled run-on transcripts synthesized in isolated mitochondria show that coI mRNAs synthesized in vitro contain added nucleotides at the GU insertion site (13). Second, pulse-chase studies indicate that nascent coI transcripts associated with stalled RNA polymerases contain added nucleotides at the GU site (15). Third, RNA fingerprint analyses indicate that the CU site within the coI mRNA is also edited correctly in transcripts labeled in isolated mitochondria (14). Thus, if a separate activity were responsible for processing of dinucleotide insertion sites, it would have to act shortly after C insertion, unlike the activity which generates the C-to-U substitutions in the same transcript (13).
Our recent analyses of partially edited (2) and misedited (3) transcripts produced during run-on synthesis in mitochondrial transcription elongation complexes (mtTECs) have been informative in regards to the steps required for the insertion of C nucleotides. In these experiments, run-on transcription was performed in partially purified mtTECs, which contain mitochondrial genomes with associated RNA polymerases and nascent transcripts, and individual RNA molecules were characterized by reverse transcription-PCR (RT-PCR), cloning, and sequencing. In this system, the RNA synthesized in vitro is partially edited at C insertion sites, with edited, unedited, and occasional misedited sites interspersed along individual molecules (2-4). This contrasts with RNA synthesized in vivo, which is efficiently and accurately edited at these sites (3); thus, sequences downstream of unedited and misedited sites in run-on transcripts are known to have been extended in vitro. Interestingly, both the addition of only 1 nucleotide at dinucleotide insertion sites (14) and misinsertion of a second residue at C insertion sites (3) have been observed under certain conditions in vitro. These observations, taken together with the biochemical data of Visomirski-Robic and Gott (13-15), led us to investigate whether single nucleotides and dinucleotides may be added at the same time relative to transcription, potentially by the same basic machinery.
Here we use an approach similar to that described above (2, 3) to examine further the relationship between dinucleotide and mononucleotide insertion, providing the first characterization of the sequences of individual nascent RNA molecules containing dinucleotide insertion sites. An interspersion of edited and unedited single and dinucleotide sites is observed, with rather complex patterns of addition at a subset of the dinucleotide sites. In addition, our data raise the intriguing possibility that in vivo, nucleotides may be added to RNA and then changed posttranscriptionally.
MATERIALS AND METHODS
Oligodeoxynucleotides.
The oligodeoxynucleotides used in this study were as follows: 9coI (5′-AACTTCTGGATGGCCAAA-3′), 12coI (5′-GCTGTATTAGTAACTGTG-3′), 18coI (5′-CATAGCATACACCATACC-3′), 44coI (5′-AATCTAGAGTAATTTTAATACATCTTCTCC-3′), 45co1 (5′-ATATCAAGACCAGAATTAGC-3′), 3ssu (5′-ATGGCGTGAGCCTGAGCA-3′), and met2.1 (5′-GAGCCCTGTATGCGAACC-3′).
mtTEC isolation, RNA synthesis, and cDNA cloning.
mtTECs were isolated essentially by the method of Cheng and Gott (4) with minor changes in the dialysis conditions. Run-on transcription reactions (in a volume of 45 to 50 μl) (4) and RNA isolation, RT-PCR, cloning, and sequencing were performed as described previously (2) except where noted. The coI data presented in Fig. 2 were derived from run-on transcription mixtures containing 20 or 500 μM CTP; RT-PCR primers were 12coI and 45coI. The sequences in Fig. 3 were derived from RNA isolated from mtTEC preparations that had not been subjected to run-on transcription. Many of the sequences initially obtained using the 12coI-45coI primer pair for RT-PCR of this mtTEC RNA preparation were found to be edited at the four C-to-U sites, most likely due to copurifying fully processed free RNA. To enrich for cDNAs derived from nascent RNAs, subsequent experiments used oligonucleotide 45coI for priming RT reactions and primers 44coI and 18coI for PCR (see Fig. 3). The upstream primer 44coI preferentially amplifies sequences unedited at the C-to-U sites at editing site 26 (es26), es27, and es28. Recombinant plasmids were then screened for editing of the remaining C-to-U site (es30) by poisoned primer extension of oligonucleotide 9coI. Similar numbers of clones were found to be edited and unedited at this site; those that were unedited (and therefore clearly derived from nascent RNA) were sequenced. The cDNAs depicted in Fig. 4 were obtained using primers 3ssu and met2.1 for RT-PCR of mtTEC RNAs present in the control reactions with no run-on transcription and with no SpeI (run-on transcription with 500 μM concentrations of the four nucleoside triphosphates [NTPs]) in the study of Byrne and Gott (specifically, in the experiment shown in Fig. 6 of reference 2).
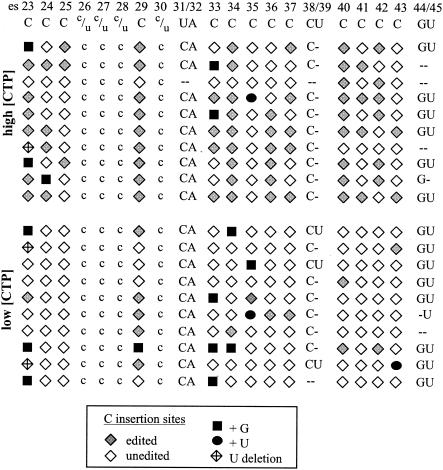
FIG. 2.
Patterns of editing and misediting in run-on transcripts synthesized in vitro from the coI gene. A schematic representation of the editing status of sites within individual cDNA clones is depicted, showing 3 dinucleotide insertion sites, 13 sites of C insertion, and 4 sites of C-to-U substitution, as noted. Insertions at dinucleotide editing sites are indicated by the appropriate uppercase letters, while hyphens indicate no insertion. C insertion sites are indicated by symbols as follows: correctly edited, gray diamonds; G misinsertion, black squares; U misinsertion, black circles. The guanosines at es29 and es34 are misinserted downstream of the encoded C nucleotides adjacent to these sites. Lowercase letters show the status of the four substitutional editing sites (the c-to-u sites at es26 to es28 and es30). cDNA clones were generated from run-on transcripts synthesized under either high nucleotide concentrations (500 μM concentrations of each NTP) or low CTP conditions (20 μM CTP, 500 μM ATP, 500 μM GTP, 500 μM UTP). Note that 2 of the 10 sequences in the high-CTP panel are identical; these were obtained in different transformations using the same ligation reaction. All other clones obtained in these experiments are unique.
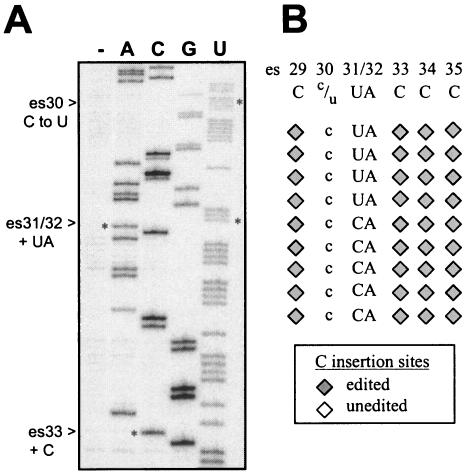
FIG. 3.
Editing patterns among steady-state (A) and nascent (B) coI transcripts synthesized in vivo. (A) Primer extension sequencing of bulk mitochondrial RNA. Lane —, primer extension in the absence of dideoxynucleotides. (B) Schematic representation of the editing status of sites within individual cDNA clones synthesized from nascent RNAs isolated from mtTECs that had not been subjected to run-on transcription. See text for details. Symbols for C insertion sites are explained in the legend to Fig. 2.
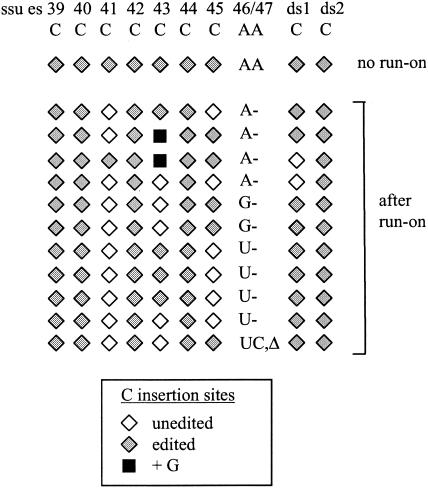
FIG. 4.
Systematic misediting of the AA insertion site (es46/47) in nascent ssu transcripts synthesized in vitro. Patterns of editing at es39 to es47 of the ssu transcript and two downstream C insertion sites (ds1 and ds2) that are present on the same nascent transcript in RNAs isolated from mtTEC before (no run-on; synthesized in vivo) and after run-on transcription in vitro are depicted. Insertions at the dinucleotide site (es46/47) are indicated by the appropriate uppercase letters; -, no insertion; Δ, deletion of the encoded A adjacent to es46/47 (see Fig. 5B for sequence). Symbols for C insertion sites are explained in the legend to Fig. 2. Note that two of the four clones with U inserted at the AA site and the two clones with G inserted at the AA site may not be independent, as they have identical sequences and were generated in the same experiment.
RESULTS
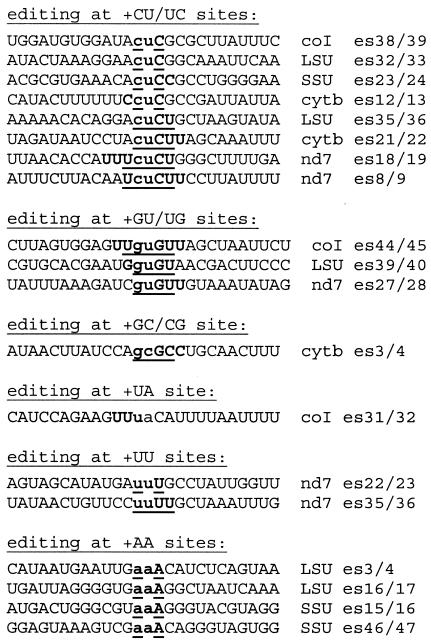
Six different types of dinucleotide insertions have been reported in Physarum mitochondrial RNAs: CU, GU, GC, UA, UU, and AA (7, 10-13, 16). As can be seen in Fig. 1, the exact site of each of these insertions is somewhat ambiguous due to the sequence context of the added nucleotides. The extent of ambiguity at each editing site is shown in boldface type and underlining, with one possible placement of the nontemplated nucleotides indicated in lowercase type for each site. The two nonencoded nucleotides must be inserted adjacent to one another at three of the CU sites listed in Fig. 1 (es32 and es33 [es32/33] and es35/36 in the large rRNA and es38/39 in coI), but in all other cases, the insertions could be either adjacent or separated by one or two encoded residues. For example, the extra cu at es21 and es22 within the cytb RNA could arise in three different ways, assuming that the two residues are added adjacent to each other: a cu could be added prior to the encoded C (AcuCUUA) or between the encoded U's (ACUcuUA), or a uc could be added between the encoded C and U (ACucUUA). A fourth possibility is that the c and u could be added at either side of the downstream encoded U (ACUcUuA). The fact that editing sites have not otherwise been observed closer than 9 nucleotides apart suggests that there are constraints on the proximity of nonencoded residues or the signals that define their location. However, although it seems likely that all of the pairs discussed here involve adjacent addition rather than insertions that are 1 or 2 nucleotides apart, to date there is no direct experimental information on this issue.
FIG. 1.
Dinucleotide insertion site contexts. Potential positions of inserted nucleotides are depicted in lowercase type. Note that the insertion positions are uncertain, since all are flanked by encoded residues of the same type as one or both of the extra nucleotides. The extent of this ambiguity is indicated by showing all residues which could be either regularly encoded or added by editing in bold type. The extent of the uncertainty is reduced to the underlined letters if the added nucleotides are inserted as an adjacent pair. The numbering of the editing sites for the ssu gene differs from that of Mahendran et al. (10), as the individual inserted nucleotides at dinucleotide sites are assigned independent numbers, as for the coI gene. LSU, large-subunit rRNA.
Despite the inherent ambiguities surrounding these “dinucleotide” insertions, sites containing two nonencoded residues (shown in lowercase type) can be loosely categorized into four basic editing patterns as follows: (i) cuC (or Cuc); (ii) xyXY (or XyxY or XYxy), where xy = cu, gu, or gc; (iii) Uua (or uUa); (iv) xxX (or Xxx or xXx), where x = A or U.
Here we examine the extent and pattern of editing and misediting at sites representative of each of these classes in individual cDNA clones derived from nascent and steady-state RNA. As in our previous work examining C insertions within atp mRNAs (2, 3), the newly synthesized transcripts are produced under a variety of different conditions, including both RNAs synthesized in vivo (but still associated with the transcription machinery) and RNAs synthesized in vitro from native templates. We have also examined the effects of altered nucleotide concentrations during RNA synthesis in vitro.
Editing of nascent RNAs at single-nucleotide and dinucleotide sites within the coI mRNA in vitro.
To facilitate comparisons with our previous biochemical data, we examined editing of nascent transcripts within a 534-nucleotide region of the coI mRNA that encompasses 23 editing sites (es23 to es45), including three different types of dinucleotide insertions (CU, GU, and UA sites), 13 single C insertion sites, and 4 sites of C-to-U changes. The latter base alterations serve as independent markers of nascent RNAs: newly synthesized RNAs contain the encoded C residues at C-to-U sites (8, 13), but essentially all steady-state transcripts contain U's at these positions (7). RT-PCR products derived from run-on RNAs synthesized by mtTECs were cloned and sequenced as previously described (2; see Materials and Methods). Because each of these cDNAs is over 500 bp long, editing patterns are represented schematically in Fig. 2, with each editing site indicated by both a number (es23 to es45) and the type of editing event (C, UA, etc.).
The types and patterns of editing in 10 independent clones produced under our standard (or high) (500 μM) nucleotide concentrations are shown at the top of Fig. 2. Each of these clones is clearly derived from nascent RNAs, as they contain a C residue at each of the four C-to-U sites. In addition, unlike the clones isolated from steady-state RNAs (7), none is fully edited, as expected for RNAs synthesized by mtTEC preparations (2, 4). Of the 130 C insertion sites examined in this experiment, 56 (∼43%) were unedited, 67 (∼52%) were edited correctly, and 7 (∼5%) were misedited by the insertion of either a G or U residue or deletion of an adjacent encoded U residue. These results are equivalent to those reported for the C insertion sites within the atp mRNA produced under similar conditions (3).
Interestingly, we observe a number of different patterns at the dinucleotide insertion sites in the same transcripts (Fig. 2, top). At the GU insertion site (es44/45), the extent of editing is roughly similar to that seen at the C insertion sites, with unedited (30%), correctly edited (+GU, 60%), and partially edited (+G, 10%) sites. In contrast, editing at the CU site (es38/39) is much less efficient in vitro. The vast majority of these CU sites are partially edited by the addition of a C residue (+C, 90%), with none of the clones being fully edited or having a misincorporated nucleotide at this site. A third pattern was apparent at the UA site (es31/32), which appears to be systematically misedited, with all nine of the processed sites containing CA rather than UA (+CA, 90%) under these conditions.
The systematic in vitro misinsertion of CA at the UA site was unexpected, given that all instances of misediting observed previously were sporadic (3). mtTECs are clearly capable of inserting U nucleotides, given that U addition is quite efficient at the GU site and that a U is even misincorporated at some C insertion sites (Fig. 2). However, because U insertions were not observed at the CU or UA site in this set of cDNAs, it seemed possible that U addition could be impaired in our mtTEC preparations in certain contexts. If in the mtTEC system, U insertion was somehow compromised at es31 and C was added by default, we would predict that decreasing the concentration of CTP in the reaction mixture might increase the chances of correct editing at this site, given that C addition at single C sites is reduced under these conditions (5). In addition, we previously observed that editing at a given C insertion site increases if the concentration of the encoded nucleotide immediately downstream of that site is reduced (5). Since both the UA and CU sites are followed by an encoded C, editing at these sites might also be enhanced at low CTP concentrations. Thus, by synthesizing RNAs at a reduced CTP concentration, we expected to change the pattern of editing at these two sites.
To examine the effect of lowering the concentration of CTP on editing at both single-nucleotide and dinucleotide insertion sites, we performed run-on transcriptions using mtTECs in the presence of 20 μM CTP, rather than 500 μM CTP, keeping the concentration of the other three nucleotides at 500 μM. Under these conditions (low CTP), the efficiency of single C insertion is reduced about fivefold (Fig. 2), with only 14 of 130 C insertion sites (10.7%) correctly edited. Interestingly, the accuracy of C insertion is also affected by these conditions. Although we see the same types of misediting events as at high CTP concentrations, the frequency of misediting at C insertion sites doubles (14 of 130, or 10.7%), such that half of all added nucleotides are the result of misinsertion.
In contrast, the extents and patterns of editing at the dinucleotide insertion sites varied in their responses to low CTP conditions. All but one of the dinucleotide editing sites in these 20 independent clones are preceded by multiple unedited or misedited C insertion sites, indicating that they were produced in vitro. As expected on the basis of the analysis of labeled run-on transcripts in isolated mitochondria by Visomirski-Robic and Gott (14), at lower CTP concentrations, we observe an increased amount of complete editing at the CU site, which occurs in the context AcuC. Whereas at high CTP concentrations, none of the clones contained CU at the CU site, 3 of the 10 clones contained CU at this site under low CTP conditions (Fig. 2). We also see a slight increase in complete editing at the GU insertion site (8 of 10 correctly edited), which is not adjacent to an encoded C, but this site was completely edited in the majority of clones even under high CTP conditions. Surprisingly, however, lowering the concentration of CTP did not reduce the level of C misinsertion at the UA site, as all 10 independent clones contained CA rather than UA at this position, despite a 25-fold molar excess of UTP over CTP in the reaction mixture.
Editing of nascent coI mRNAs at the UA site in vivo.
Because there are also four instances of C-to-U changes within the coI mRNA (at es26, es27, es28, and es30), it is possible that the UA site is normally edited by the insertion of CA, followed by subsequent deamination of the C to a U. To determine the status of the dinucleotide insertion sites among nascent RNAs produced in vivo, we analyzed both bulk RNA preparations and individual cDNA clones. First, primer extension sequencing was performed on bulk RNA isolated from mitochondria (Fig. 3A). Only U was detected at both es30 (a C-to-U site) and es31 (the first nucleotide of the UA site) in this total mitochondrial RNA preparation. We next sequenced bulk RNA isolated from mtTECs, which should have a higher percentage of nascent RNAs than total mitochondrial RNA pools. Again, the vast majority of RNA contained U at both es30 and es31 (data not shown). Finally, to enrich for nascent coI mRNAs within the in vivo mtTEC RNA pool, we performed RT-PCR using an upstream PCR primer that preferentially anneals to RT products from RNAs having C's at es26, es27, and es28, C-to-U sites that occur in the context UCUUcUccUG (C-to-U sites in lowercase type). The resulting PCR product was then sequenced directly to look at editing within the entire pool (data not shown) and cloned to allow for sequencing of individual cDNAs derived from nascent RNA (Fig. 3B). In both cases, we observed a mixture of C and U at the internal C-to-U site (es30) and the UA site (es31/32).
Sequencing of these individual clones derived from mtTEC RNA in the absence of run-on transcription gave three different patterns. The first group contains a U at both es30 and es31, matching the sequence found in steady-state RNA pools (data not shown). Clones in this group presumably represent fully processed transcripts. The other two sets each have a C at es30 (a C-to-U site) but differ in that the second group contains a C at es31, while the third group has a U at this position. Of the nine clones which had not been substitutionally edited at the upstream C-to-U site (es30) and thus are known to be derived from nascent transcripts, five had a CA and four had a UA (Fig. 3B), while neighboring C insertion sites were fully and correctly edited, as expected on the basis of our previous results for atp mRNA (2). These data argue that at least a portion of the nascent RNA synthesized in vivo has CA rather than UA inserted at es31/32.
Partial editing and misediting at an AA insertion site in vitro.
There are six known cases in which two identical nucleotides are added at adjacent sites: two sites of AA insertion in both the small-subunit (SSU) rRNAs (10) and large-subunit rRNAs (11) and 2 UU nucleotides in the nd7 mRNA (12). Each of these insertions occurs in an ambiguous context (Fig. 1). To determine the in vitro insertion pattern at this type of site, we examined editing at es46/47 (AA inserted) of SSU rRNA, which are the last two editing sites in the ssu sequence. Because our mtTEC preparations contain significant amounts of rRNA synthesized in vivo, a primer complementary to the cotranscribed tRNA2Met gene 3′ of the ssu gene was used for RT and PCR to ensure that we were looking at nascent rRNA rather than the mature rRNA species, which are fully edited. During the course of sequencing these clones, we found two previously unknown sites of single C insertion in the downstream region between the 3′ end of ssu and tRNA2Met; these sites are called ds1 and ds2 and shown in Fig. 4.
The editing status of the last nine insertion sites in the SSU rRNA (es39 to es47) and the two downstream sites in these clones are displayed in Fig. 4. These cDNA clones were derived from nascent RNA associated with mtTECs either before (no run-on) or after run-on transcription in the presence of 500 μM concentrations of the four NTPs. As expected, RNA synthesized in vivo, i.e., present in mtTECs prior to run-on transcription, was completely edited at each of the seven single C insertion sites within this region of the SSU rRNA (es39 to es45) and at the AA site (es46/47) (Fig. 4). Both of the editing sites between the ssu gene and the tRNA2Met gene were also fully edited in these clones. In contrast, although C insertion was relatively efficient in this region (74% correctly edited, 24% unedited, 2% misedited), in the 11 clones derived from RNA synthesized in vitro, none were fully edited at the AA insertion site (Fig. 4). Instead, four independent clones were partially edited, containing a single A at the AA site, and the rest were misedited. Of the clones containing misinsertions, two had a single G, four contained a single U, and the last had a UC added at the AA site as well as a deletion of the adjacent encoded A. We have also sequenced additional clones containing the 3′ portion of the SSU rRNA in experiments examining the effects of sequence context on editing in mtTEC preparations (2). These RT-PCR clones are derived from RNAs synthesized from rearranged templates in which the 5′ portion of the atp gene was joined to the 3′ portion of the ssu gene prior to run-on transcription (2). Interestingly, each of these six independent clones contains a single U at the AA insertion site (data not shown). Thus, during in vitro run-on transcription in mtTEC preparations, it seems that only a single nucleotide is generally added at this AA insertion site, and there appears to be little discrimination in selection of that nucleotide, other than perhaps excluding C.
DISCUSSION
Complex patterns of editing at dinucleotide insertion sites.
We have previously described the editing events that occur in vitro at C insertion sites within individual cDNA clones derived from the atp mRNA (2, 3). Here we extend these observations to include each of the four broad classes of dinucleotide insertion at sites within both the coI mRNA and the small rRNA from Physarum mitochondria. Although these sites are fully edited in cDNA clones derived from steady-state RNAs, editing at these sites is remarkably variable in vitro, ranging from highly efficient editing to systematic misediting.
Our data regarding both the CU and GU insertion sites are similar to what was observed for RNA pools labeled during run-on transcription in isolated mitochondria, confirming at the sequence level what we observed when analyzing bulk RNAs (13-15). Upon in vitro transcription using high nucleotide concentrations, the coI CU site is either not edited or partially edited by C insertion, while at low CTP concentrations, we observe some fully edited molecules. In contrast, editing is fairly efficient at the GU site in vitro under both high and low CTP conditions, although all combinations of editing, partial editing, and no editing are observed. In addition, the fact that either a single G or a single U can be added at the GU site provides further evidence that residues can be added independently at dinucleotide insertion sites.
The systematic misediting observed at the UA site within the coI mRNA and the second AA site in the SSU rRNA was rather surprising, given that previous instances of misediting were relatively rare (∼5% of editing events) and sporadic in nature (3). In the case of the ssu AA (es46/47) site, in vitro we always observe the net addition of a single nucleotide, rather than a dinucleotide (note that in the one case where a dinucleotide is added [UC], an adjacent encoded A residue is deleted [Fig. 4]). In contrast, editing at this site is complete and accurate in both the steady-state RNA pool and in nascent, in vivo transcripts associated with elongating RNA polymerases (Fig. 4 and data not shown). Interestingly, the two A residues that are normally added at this site align with the two A's in the bacterial 16S rRNA that are proposed to be the major determinants of proofreading of the codon-anticodon interaction (17). Thus, correct editing at this site would be predicted to be essential for mitochondrial ribosome function.
Processing of es31/32 of the coI transcript is also more complex than it initially appeared from the sequences of steady-state RNAs. In previous characterizations of the coI mRNA (7, 13), we sequenced 13 different cDNA clones spanning these sites, 8 of which were derived from separate RT-PCR products, using multiple total mitochondrial RNA preparations as the initial template. In each case, a nontemplated UA dinucleotide was present at es31/32. Consistent with these results, primer extension analysis of bulk mitochondrial RNA demonstrates that essentially all steady-state RNAs contain a UA (Fig. 3A), creating a codon for a highly conserved tyrosine residue (UAC). It was unexpected, therefore, to find that the nucleotide at es31 is C, rather than U, in all edited cDNA clones that are derived from RNA synthesized by mtTEC preparations in vitro (Fig. 2) and that a subset of the cDNA clones derived from nascent RNAs synthesized in vivo also contained C at this site (Fig. 3B).
It is possible that either a C or U nucleotide can be inserted at es31 and RNAs containing a C at es31 are preferentially degraded, resulting in a steady-state RNA pool containing only UA. However, given that there are four known sites of C-to-U substitutions within this region of the coI mRNA (7), it seems more likely that C's inserted at es31 are also targeted for deamination in vivo. The fact that we see RNAs synthesized in vivo that contain C at es30 but either U or C at es31 suggests that the C at position 31 is converted to U by substitutional editing before the other C→U sites are processed. From our data, we cannot determine whether a mixture of C and U is inserted at es31 or if C is always added and then changed to U. Thus, the fact that we observe only C's at es31 in cDNA clones derived from RNAs synthesized in vitro could be due to the loss of the ability to insert or extend (discussed below) a U nucleotide at es31 or the lack of specific C-to-U deamination in these experiments. Since the C-to-U changes at coI es26, es27, es28, and es30 do not occur in our in vitro editing systems (13; J. M. Gott, A. Majewski, and Y.-W. Cheng, unpublished data), the latter possibility appears likely.
Although this would be the first known case of C-to-U editing of an added C residue in Physarum, it is likely that at least one other inserted nucleotide in Physarum mitochondria is also modified posttranscriptionally. An additional U is inserted into the T loop of the Physarum mitochondrial tRNAGlu, changing the loop sequence from UCGAUU to UUCGAUU (1). Although the modifications of these tRNAs have not been analyzed directly, presumably this region of the tRNA is modified to TψC. Thus, either the T or the pseudouridine of the final tRNA product is initially derived from a nonencoded U.
Relationship between mononucleotide and dinucleotide insertional editing.
It has been suggested that dinucleotide insertion might proceed via a mechanism separate from the addition of single nucleotides, operating after C insertion sites have been edited (16). However, biochemical data from run-on transcription in isolated mitochondria (13, 14) are consistent with an alternative hypothesis that all insertion sites are processed cotranscriptionally. Here we reexamined this question by looking at individual RNA molecules synthesized in vitro. We find that most dinucleotide insertion sites have been processed by the addition of at least a single nucleotide among sequences produced in vitro in mtTEC preparations, and in fact, it appears that these sites are actually recognized more efficiently than many of the surrounding single C insertion sites in this system. Among individual nascent transcripts synthesized in vivo, full addition was observed at both dinucleotide and C insertion sites (Fig. 3); again, there was no evidence that insertion at the dinucleotide sites was delayed relative to the addition of single C residues.
The responses to lower CTP concentrations at the dinucleotide sites examined are also consistent with the hypothesis that dinucleotides are added at the growing end of the RNA. For example, the stimulation of U insertion at es39 in the coI mRNA (the cuC site) by lower CTP concentrations suggests that addition at this dinucleotide site is in competition with templated C addition. This interpretation is also supported by the observation that unlike what is observed at single C insertion sites, a lower CTP concentration does not decrease C insertion at the CU site. In this scenario, templated incorporation of the encoded C by the transcription apparatus will compete with both editing events, and reducing the rate of transcription elongation at this site by decreasing CTP concentration will increase the probability of editing (5, 14). Similarly, our finding that all cDNA clones are edited at the coI UA-CA site at low CTP concentrations is also consistent with addition of the dinucleotide upstream of the encoded C residue, where, similar to insertion at the CU site, editing would be expected to be enhanced by slowing incorporation of the next templated nucleotide. Thus, our data suggest that C mononucleotide and dinucleotide insertion occur by the same general mechanism but with differences in detail.
However, there may be some systematic differences between the C insertion sites and dinucleotide sites in aspects of the editing or misediting process. First, the dinucleotide sequence contexts are less diverse, as discussed above. Second, we found no evidence of imprecise processing of C insertion sites in nascent RNA synthesized in vivo (3), but here we find that the first position of the coI UA site may actually be naturally processed by nondiscriminating pyrimidine nucleotide insertion in vivo and that there may be subsequent substitutional editing of inserted C's at this site. Third, in the mtTEC run-on system, C insertion sites are sporadically unedited and, at a low frequency, misedited (3). In contrast, we have found no evidence of misediting at the coI CU and GU sites, while, conversely, ssu es46/47 displays a curious mix of partial editing and high-frequency misediting, and if insertion at the coI UA site is split with CA dinucleotides in vivo, it defaults to the CA pattern in vitro.
Effect of CTP concentration on the accuracy of C insertion.
In the course of this work, we examined the effects of lowering the concentration of CTP upon editing at the single C nucleotide and dinucleotide insertion sites in individual molecules of coI mRNA. Editing at C insertion sites was reduced, consistent with what was observed during analysis of pools of labeled run-on transcripts (5). Sequencing individual cDNA clones reveals a low level of misediting at C insertion sites (3), which is not readily detected by direct analysis of labeled transcript pools; here the technique demonstrated that lower CTP levels led to a striking increase in these misediting events. This result is consistent with the idea that decreasing the availability of the correct editing substrate at C insertion sites increases the chance of misbinding and misinserting an alternative nucleotide or enhances the probability of making an error while exiting from the editing mode. Interestingly, use of low concentrations of CTP in the presence of a 25-fold molar excess of UTP did not diminish the frequency of C misinsertion at the UA insertion site within the coI mRNA, making it unlikely that there is a direct competition between insertion of U and C at this site in mtTEC (Fig. 2).
Locations of dinucleotide insertion sites.
The patterns of partial editing and misediting allow us to infer the probable positions at which the dinucleotides are added at sites where assignment is ambiguous due to flanking sequences. At the coI GU insertion site, the fact that we see two consecutive G's in one cDNA clone and three consecutive U's in another implies that if the g and u are added adjacent to each other, they are actually inserted between the G and U of the second encoded GUU as ug (Fig. 5A). Interestingly, the sequence immediately upstream from this insertion is an encoded UG, leaving open the possibility that the nascent RNA might realign with the template prior to extension by the polymerase at this site. A similar scenario could also be envisioned at other dinucleotide sites of the xyxy class as described below. Likewise, the misinserted U and G in the clones in Fig. 4 all appear upstream of the encoded A that flanks the ssu AA (es46/47) site (Fig. 5B), suggesting that at least one of the A's that are normally inserted at this site is likely to be added 5′ of the encoded A. Finally, while the point of insertion of the U at coI es31 is ambiguous due to the presence of adjacent, encoded U's (Fig. 5C), the extra C and A residues present in nascent RNA are clearly added at adjoining sites in one of three possible configurations (UcaCAU, UCacAU, or UCAcaU). Although the data do not unequivocally distinguish between these possibilities, the most likely scenario is that the extra ua or ca is added at the position indicated in Fig. 5C.
FIG. 5.
Likely sites of dinucleotide insertion based on sequences of partially edited and misedited cDNAs. Alignments of unedited, edited, partially edited, and misedited clones encompassing the GU insertion site of the coI mRNA (es44/45) (A), the AA insertion site of the ssu rRNA (es46/47) (B), and the UA insertion site of the coI mRNA (es31/32) (C). See text for details.
Potential editing factors.
Our results suggest that although both single-nucleotide and dinucleotide insertion sites are recognized in our in vitro system, dinucleotide addition might require auxiliary or alternative specificity factors for accurate insertion at a subset of these sites. After nontemplated nucleotide insertion at an editing site, the growing end of the RNA must be juxtaposed correctly with the next incoming substrate nucleotide triphosphate; it is not known how this is achieved. Extension from unpaired dinucleotides might be even more difficult, perhaps requiring the intervention of specific components of the transcription-editing machinery. Intriguingly, most of the dinucleotide insertions occur in contexts that could potentially allow realignment of the nascent RNA with the template, such that upstream templating deoxynucleotides might be used to pair with the extra terminal residue(s) in these contexts. Thus, mixed dinucleotides added at the xyxy class site might be stabilized by 2 bp in this scenario. Even if realignment is used to facilitate extension from added dinucleotides, it seems likely that such a rearrangement would require facilitation by some additional factor.
In vitro, the ssu AA (es46/47) site is recognized efficiently, but insertion is apparently limited to a single extra nucleotide. It is, of course, possible that dinucleotides can be added at the growing end of the RNA here but that they cannot then be extended; observing an incorrect nucleotide pair with omission of the next encoded residue in a clone might be symptomatic of the difficulty extending dinucleotides here. Such a deficit might again be due to loss or inactivation of a factor specifically required for extension of this dinucleotide. It is also possible that incorrect nucleotides are not actually added to the growing end of the transcript at a high frequency but happen to be more easily extended in vitro, leading to the observed bias towards misedited clones at this site. Similar scenarios could be imagined for the systematic insertion of CA at the UA-CA site in vitro. Our mtTEC preparations may, for instance, lack a factor that allows extension of an unpaired dinucleotide at this site, and RNAs with CA at this position might be more efficiently extended than RNAs containing UA. In this case, the biased insertions that we see could be due to an artificial selection process in vitro. The fact that we observed systematic anomalies at two of the dinucleotide sites that we analyzed but relatively normal editing at the other two sites in vitro suggests that different types of insertion sites have distinct factor requirements and that our in vitro system may be depleted for only a subset of these factors.
Taken together, our data indicate that it is likely that dinucleotide insertions, like single-nucleotide insertions, occur cotranscriptionally in Physarum mitochondria. It should be pointed out that the rare, partially edited molecules selected from steady-state RNA pools by Wang et al. (16) are not inconsistent with this conclusion. In addition, our findings are consistent with the hypothesis proposed by Horton and Landweber (9) that all forms of insertional editing in the myxomycetes arose from an ancestral U addition capacity, most likely with the addition of successive specificity factors for C and dinucleotide addition.
Acknowledgments
We thank Angela Stout, Amy Rhee, and Neeta Parimi for technical assistance and Mike Harris, Amy Rhee, Tim Nilsen, Erik Andrulis, and JoAnn Wise for advice and comments on the manuscript.
This work was supported in part by Public Health Service grant GM-54663 to J.M.G. from the National Institutes of Health.
REFERENCES
- 1.Antes, T., H. Costandy, R. Mahendran, M. Spottswood, and D. Miller. 1998. Insertional editing of mitochondrial tRNAs of Physarum polycephalum and Didymium nigripes. Mol. Cell. Biol. 18:7521-7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Byrne, E. M., and J. M. Gott. 2002. Cotranscriptional editing of Physarum mitochondrial RNA requires local features of the native template. RNA 8:1174-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Byrne, E. M., A. Stout, and J. M. Gott. 2002. Editing site recognition and nucleotide insertion are separable processes in Physarum mitochondria. EMBO J. 21:6154-6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng, Y. W., and J. M. Gott. 2000. Transcription and RNA editing in a soluble in vitro system from Physarum mitochondria. Nucleic Acids Res. 28:3695-3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng, Y. W., L. M. Visomirski-Robic, and J. M. Gott. 2001. Non-templated addition of nucleotides to the 3′ end of nascent RNA during RNA editing in Physarum. EMBO J. 20:1405-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gott, J. M. 2001. RNA editing in Physarum polycephalum. Front. Mol. Biol. 34:20-37. [Google Scholar]
- 7.Gott, J. M., L. M. Visomirski, and J. L. Hunter. 1993. Substitutional and insertional RNA editing of the cytochrome c oxidase subunit 1 mRNA of Physarum polycephalum. J. Biol. Chem. 268:25483-25486. [PubMed] [Google Scholar]
- 8.Gott, J. M., and L. M. Visomirski-Robic. 1998. RNA editing in Physarum mitochondria, p. 395-411. In R. Benne (ed.), Modification and editing of RNA. ASM Press, Washington, D.C.
- 9.Horton, T. L., and L. F. Landweber. 2000. Evolution of four types of RNA editing in myxomycetes. RNA 6:1339-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mahendran, R., M. S. Spottswood, A. Ghate, M. L. Ling, K. Jeng, and D. L. Miller. 1994. Editing of the mitochondrial small subunit rRNA in Physarum polycephalum. EMBO J. 13:232-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller, D., R. Mahendran, M. Spottswood, H. Costandy, S. Wang, M. L. Ling, and N. Yang. 1993. Insertional editing in mitochondria of Physarum. Semin. Cell Biol. 4:261-266. [DOI] [PubMed] [Google Scholar]
- 12.Takano, H., T. Abe, R. Sakurai, Y. Moriyama, Y. Miyazawa, H. Nozaki, S. Kawano, N. Sasaki, and T. Kuroiwa. 2001. The complete DNA sequence of the mitochondrial genome of Physarum polycephalum. Mol. Gen. Genet. 264:539-545. [DOI] [PubMed] [Google Scholar]
- 13.Visomirski-Robic, L. M., and J. M. Gott. 1995. Accurate and efficient insertional RNA editing in isolated Physarum mitochondria. RNA 1:681-691. [PMC free article] [PubMed] [Google Scholar]
- 14.Visomirski-Robic, L. M., and J. M. Gott. 1997. Insertional editing in isolated Physarum mitochondria is linked to RNA synthesis. RNA 3:821-837. [PMC free article] [PubMed] [Google Scholar]
- 15.Visomirski-Robic, L. M., and J. M. Gott. 1997. Insertional editing of nascent mitochondrial RNAs in Physarum. Proc. Natl. Acad. Sci. USA 94:4324-4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang, S. S., R. Mahendran, and D. L. Miller. 1999. Editing of cytochrome b mRNA in Physarum mitochondria. J. Biol. Chem. 274:2725-2731. [DOI] [PubMed] [Google Scholar]
- 17.Yoshizawa, S., D. Fourmy, and J. D. Puglisi. 1999. Recognition of the codon-anticodon helix by ribosomal RNA. Science 285:1722-1725. [DOI] [PubMed] [Google Scholar]