Abstract
The Swi/Snf chromatin remodeling complex has been previously demonstrated to be required for transcriptional activation and repression of a subset of genes in Saccharomyces cerevisiae. In this work we demonstrate that Swi/Snf is also required for repression of RNA polymerase II-dependent transcription in the ribosomal DNA (rDNA) locus (rDNA silencing). This repression appears to be independent of both Sir2 and Set1, two factors known to be required for rDNA silencing. In contrast to many other rDNA silencing mutants that have elevated levels of rDNA recombination, snf2Δ mutants have a significantly decreased level of rDNA recombination. Additional studies have demonstrated that Swi/Snf is also required for silencing of genes near telomeres while having no detectable effect on silencing of HML or HMR.
The Saccharomyces cerevisiae Swi/Snf complex is an ATP-dependent chromatin remodeling complex that can activate or repress transcription (see references 3, 34, and 44 for recent reviews). Swi/Snf contains 11 different subunits, including Snf2, a highly conserved ATPase. The Snf2 subunit is the catalytic core of Swi/Snf; single amino acid changes in the DNA-dependent ATPase domain eliminate both the ATPase activity and the chromatin remodeling activity of Swi/Snf (13, 53). While Swi/Snf binds to both DNA and nucleosomes, it does not do so in a site-specific manner (12, 52). Rather, sequence-specific transcriptional activators and repressors have been shown to target Swi/Snf to specific promoters (for examples, see references 16, 46-48, 51, and 72; see reference 23 for a review).
Gene expression microarray analysis has shown that the mRNA levels of a small subset of S. cerevisiae genes are significantly affected by the loss of Swi/Snf activity (25, 65). This apparent specificity of Swi/Snf control is likely caused by several factors, including its recruitment by particular transcriptional regulators. In addition, there is strong evidence that Swi/Snf is redundant with other transcription complexes in vivo and may therefore play a wider role than is indicated by microarray analysis (4, 50, 54, 64). A third factor is that Swi/Snf may be required only at promoters with a particular chromatin structure. Indeed, one study has suggested that different chromatin structures can determine the dependency upon Swi/Snf (9).
Since chromosomal context can influence chromatin structure, we wanted to test whether genomic position might affect the Swi/Snf dependence of a gene. To address this issue, we randomly integrated the SUC2 gene, which is strongly Swi/Snf dependent (69), into the yeast genome to identify locations where SUC2 expression becomes independent of Swi/Snf. Surprisingly, we discovered that when SUC2 is integrated into the ribosomal DNA (rDNA) locus (RDN1, hereafter referred to as rDNA), its dependence on Swi/Snf is reversed. That is, when SUC2 is located in the rDNA, SUC2 transcription is repressed rather than activated by Swi/Snf.
The S. cerevisiae rDNA consists of a tandem array of 9.1-kb units repeated 100 to 200 times on chromosome XII (49) (Fig. 1). The rDNA is located in the nucleolus in an arrangement reminiscent of the heterochromatin of higher eukaryotes (reviewed in references 45 and 57). Each rDNA repeat unit includes the 5S rRNA gene, transcribed by RNA polymerase III, and a 35S precursor rRNA gene, transcribed by RNA polymerase I. About half of the tandemly repeated rRNA genes are transcriptionally active; the active rRNA gene copies are randomly distributed along the ribosomal rRNA gene locus (14). Unlike the results seen with rRNA genes, the expression of several different Pol II-transcribed genes, when integrated into various regions of the rDNA, is repressed (7, 18, 60). rDNA silencing also represses recombination, believed to play an important role in preventing rDNA loss (21).
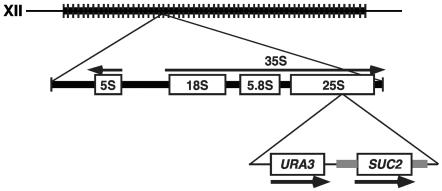
FIG. 1.
Diagram of the SUC2 insertion in the rDNA. The top line represents the rDNA array on the right arm of chromosome XII. A representative rDNA repeat, indicating the position of the integration of the SUC2 URA3 cassette (see text), is shown below. The thick gray line flanking SUC2 represents DNA that normally flanks the SUC2 gene.
Many trans-acting proteins required for rDNA silencing of Pol II transcription have been identified (6-8, 60, 62, 63, 66). These include Sir2, a member of a highly conserved family of NAD-dependent protein and histone deacetylases (30, 40, 61), and Set1, a histone methyltransferase (6, 8). The experiments presented in this paper identify another factor required for rDNA silencing, the Swi/Snf complex. Our results strongly suggest that Swi/Snf-mediated silencing occurs by a mechanism independent of Sir2 and Set1. Additional experiments show that Swi/Snf is also required for silencing at telomeres but not at silent-mating-type cassettes.
MATERIALS AND METHODS
Yeast strains, genetic methods, and plasmids.
All S. cerevisiae strains used in this study (Table 1) are derivatives of a GAL2+ S288C strain (70). Standard strain construction methods and medium recipes were as described previously (55). Deletion of SUC2 was achieved by replacing the open reading frame with the PCR-amplified KanMX4 gene from plasmid pRS400 (5). The snf2Δ::LEU2 (10), snf2-798 (K-to-A change of amino acid 798) (33), and snf5Δ2 (64) alleles have been described previously. Strains used for assaying telomeric silencing were previously described (1). Strains with mURA3-LEU2 integrated in the rDNA and at the leu2Δ1 locus were generated by a cross to strains previously described, JS215-10 and JS210-1 (60). Plasmids were constructed and isolated from Escherichia coli by standard methods (2). Plasmid pVD1 is a derivative of plasmid pRS406 (59) that contains a BglII fragment with SUC2 sequences from −1187 to +2076 with respect to the SUC2 ATG.
TABLE 1.
S. cerevisiae strains
| Strain | Genotype |
|---|---|
| FY49 | MATaura3-52 lys2-128Δ snf1Δ |
| FY78 | MATahis3Δ200 |
| FY328 | MATα his3Δ200 his4-917δ lys2-173R2 snf2Δ1::HIS3 |
| FY1658 | MATahis3Δ200 ura3-52 lys2-128δ snf5Δ2 |
| FY1856 | MATα his3Δ200 leu2Δ0 lys2-128Δ ura3Δ0 |
| FY2084 | MATaura3Δ0 snf2-798 |
| FY2310 | MATahis3Δ200 ura3Δ0 lys2-128δ suc2Δ::KanMX4 snf5-51 |
| FY2311 | MATα lys2-128δ leu2Δ0 his3Δ200 ura3Δ0 suc2Δ::KanMX4 |
| FY2312 | MATα lys2-128δ leu2Δ0 his3Δ200 ura3Δ0 suc2Δ::KanMX4 rDNA::URA3-SUC2 |
| FY2313 | MATα lys2-128δ leu2Δ0 his3Δ200 ura3Δ0 suc2Δ::KanMX4 rDNA::URA3-SUC2 |
| FY2314 | MATα ura3Δ0 lys2-128δ leu2Δ0 snf1Δ suc2Δ::KanMX4 rDNA::URA3-SUC2 |
| FY2315 | MATaura3-52 arg4-12 leu2Δ0 snf2Δ::LEU2 suc2Δ::KanMX4 rDNA::URA3-SUC2 |
| FY2316 | MATaura3Δ0 lys2-128δ leu2Δ1 snf2Δ::LEU2 suc2Δ::KanMX4 rDNA::URA3-SUC2 |
| FY2317 | MATα ura3Δ0 lys2-128δ leu2Δ0 trp1Δ63 snf2Δ::LEU2 suc2Δ::KanMX4 rDNA::URA3-SUC2 |
| FY2318 | MATaleu2Δ0 his3Δ0 ura3-52 his4-912δ lys2-128δ suc2Δ::KanMX4 set1Δ::KanMX4 rDNA::URA3-SUC2 |
| FY2319 | MATaleu2Δ0 arg4-12 ura3-52 lys2-128δ snf2Δ::LEU2 suc2Δ::KanMX4 set1Δ::KanMX4 rDNA::URA3-SUC2 |
| FY2320 | MATα leu2Δ0 ura3Δ0 his3Δ200 lys2-128δ suc2Δ::KanMX4 met15::URA3-SUC2 |
| L1075 | MATaleu2Δ0 his3Δ0 orΔ200 ura3Δ0 lys2-128δ suc2Δ::KanMX4 sir2Δ::KanMX4 rDNA::URA3-SUC2 |
| FY2321 | MATaleu2Δ0 ura3Δ0 his3Δ200 snf2-798 suc2Δ::KanMX4 rDNA::URA3-SUC2 |
| L1076 | MAT? his3Δ0 leu2Δ0 ura3-52 suc2Δ::KanMX4 sir2Δ::KanMX4 snf2Δ::LEU2 rDNA::URA3-SUC2 (mating type not known due to sir2 mutation) |
| L1077 | MATα arg4-12 his3Δ0 leu2Δ0 ura3-52 suc2Δ::KanMX4 sir2Δ::KanMX4 snf2Δ::LEU2 rDNA::URA3-SUC2 |
| L1078 | MATα ura3-52 leu2Δ0 snf2Δ::LEU2 suc2Δ::KanMX4 met15::URA3-SUC2 |
| L1079 | MATα leu2Δ1 ura3-52 his3Δ200 met15Δ0 trp1Δ63 snf2Δ::LEU2 rDNA::mURA3-LEU2 |
| L1080 | MATα leu2Δ1Δ0 ura3Δ0 his3Δ200 lys2Δ0 snf2Δ::LEU2 rDNA::mURA3-LEU2 |
| L1081 | MATα leu2Δ0 ura3-52 his3Δ200 lys2Δ0 met15Δ0 trp1Δ63 rDNA::mURA3-LEU2 |
| L1082 | MATaleu2Δ1 ura3Δ0 his3Δ200 lys2Δ0 met15Δ0 trp1Δ63 leu2Δ1::mURA3-LEU2 |
| L1083 | MATα leu2Δ0 ura3Δ0 his3Δ200 met15Δ0 trp1Δ63 leu2Δ1::mURA3-LEU2 |
| L1084 | MATaleu2Δ0 ura3Δ0 his3Δ200 rDNA::mURA3-LEU2 |
| L1085 | MATaleu2Δ1 ura3Δ0 his3Δ200 met15Δ0 lys2Δ0 snf2Δ::LEU2 leu2Δ1::mURA3-LEU2 |
| L1086 | MATα leu2Δ1 ura3Δ0 his3Δ200 snf2Δ::LEU2 leu2Δ1::mURA3-LEU2 |
| L1087 | MATα his3 ura3 leu2Δ1 TEL-VR::URA3 |
| L1088 | MATaura3Δ0 leu2Δ0 TEL-VR::URA3 |
| L1089 | MATaura3-52 leu2Δ0 snf2Δ::LEU2 TEL-VR::URA3 |
| L1090 | MATα his3 ura3Δ0 leu2Δ1 snf2Δ::LEU2 TEL-VR::URA3 |
| L1091 | MATahis3 ura3Δ0 leu2Δ1 met15Δ0 sir2Δ::KanMX4 TEL-VR::URA3 |
| L1092 | MATahis3 ura3-52 leu2Δ0 met15Δ0 sir2Δ::KanMX4 TEL-VR::URA3 |
Isolation of transformants with SUC2 integrated at random locations and in the rDNA.
To randomly integrate the plasmid pVD1 into the yeast genome, the restriction enzyme-mediated transformation method (43, 58) was used. The plasmid pVD1 was linearized with the restriction enzyme SacI, and 10 μg of the plasmid was used to transform the strain FY2310 in the presence of 100 units of the restriction enzyme BglII. Strain FY2310 contains no homology to pVD1 and also contains the snf5-51 mutation, a temperature-sensitive mutation in SNF5, which encodes a component of Swi/Snf. Transformants were selected on synthetic complete (SC) plates lacking uracil. To determine the site of integration of plasmid pVD1, genomic DNA was extracted and digested with BamHI, which digests only once in pVD1, and the DNA was self-ligated under dilute conditions. This DNA was then used to transform E. coli strain DH5α. The resulting plasmids, which contained genomic DNA flanking the site of plasmid integration, were then isolated from the bacteria and sequenced. To directly integrate a URA3-SUC2 cassette into the same position within the rDNA, URA3 and SUC2 from plasmid pVD1 were amplified by PCR, using the primers F (5′ GGC TTG GCA GAA TCA GCG GGG AAA GAA GAC CCT GTT GAG GAT GCC GGG AGC AGA CAA GC 3′) and R (5′ ACA CCC TCT ATG TCT CTT CAC AAT GTC AAA CTA GAG TCA CAA AAG CTG GAG CTC CAC CG 3′). This 5.2-kb URA3-SUC2 PCR fragment was then used to transform strain FY2310 to Ura+.
Northern hybridization analysis.
For measurement of SUC2 mRNA levels, strains were grown in yeast extract-peptone-dextrose (YPD) to approximately 107 cells/ml, washed in water, resuspended in either YPD (repressed sample) or yeast extract-peptone (YEP) plus 0.05% glucose (derepressed sample), and grown for 2 h 45 min. For measurement of URA3 mRNA levels to assay telomeric silencing, strains were grown in SC medium containing 100 mg of uracil/liter (1). RNA was prepared and analyzed as described previously (67). The SUC2 probe was synthesized by PCR amplification of 603 bp of plasmid pRB58 (11) corresponding to positions +949 to +1552 of the SUC2 open reading frame. The ACT1 probe was synthesized by PCR amplification of 190 bases from +532 to +722 of the ACT1 open reading frame. The URA3 probe was synthesized by PCR amplification of 474 bases extended from +206 to +680. The α1 probe was synthesized by PCR amplification of 328 bases extended from +40 to +369. All probes were radiolabeled with [α-32P]dATP by random priming (2). Quantitation of relative levels of mRNA was performed by using a PhosphorImager (Molecular Dynamics).
Analysis of chromatin structure by MNase.
S. cerevisiae strains were grown in YPD medium to 107 cells/ml and then shifted to derepressing conditions as described for Northern blot analyses. Spheroplasts were isolated and subjected to micrococcal nuclease (MNase) digestion as adapted from previously described methods (31, 32). Approximately 1.2 × 109 cells were incubated with 2 mg of Zymolyase (ICN)/ml (100,000 units/g) for 2 min. Spheroplasts from 2 × 108 cells were aliquoted and digested with 0, 0.625, 1.25, 2.5, or 5 units of MNase at 37°C for 4 min. Purified genomic DNA from an equivalent amount of cells was digested using either 0.5 or 0.75 units of MNase at 37°C for 1 min to serve as naked DNA controls. The DNA from the MNase-treated chromatin samples was purified and then digested completely with HinfI, separated on a 1% agarose gel, and analyzed by indirect end labeling (24). A 156-bp PCR product corresponding to base pairs +140 to +296 (+1 = ATG) of the SUC2 open reading frame was synthesized by PCR, radiolabeled by random priming (2), and used as the probe to detect SUC2 DNA. A 1-kb DNA ladder was used as a size standard to calculate positions of MNase cleavage.
ChIP.
The procedure for chromatin immunoprecipitation (ChIP) was adapted from previously described methods (17, 39). Briefly, cells from 200-ml YPD cultures were cross-linked by adding formaldehyde to achieve a final concentration of 1%. Chromatin was prepared in fluorescent-antibody lysis buffer containing 140 mM NaCl and no sodium dodecyl sulfate. Cross-linked chromatin was sonicated to an average length of 500 bp, with a size range from 200 to 1,200 bp. Sir2 was immunoprecipitated from 1/10 of the cross-linked chromatin by a two-step method (22) using rabbit polyclonal anti-Sir2 antibody (26) followed by immunoglobulin G-Sepharose beads (Pharmacia). Dilutions of input DNA (1/200 and 1/400) and immunoprecipitated DNA (1/10 and 1/20) were subjected to quantitative radioactive PCR as described previously (41), and the products were separated on a 7.5% nondenaturing polyacrylamide gel. Primers of 20-nucleotide oligonucleotides for amplification of products of 250 bp for the rDNA were as previously described (28). Specific binding of Sir2 to DNA amplified by each primer set was evaluated by calculating the ratio of the percentage of immunoprecipitation (IP) of the primer set to the percentage of IP of a control region of the genome (36). The control region used amplifies bp 9716 to 9863 of chromosome V, a region devoid of transcription by RNA polymerase II (36). Levels of H3 K4 methylation were measured as previously described (8) by the use of the same control region used for the Sir2 ChIP experiments.
Spot tests to assay expression of mURA3-LEU2.
SNF2 and snf2Δ strains, containing the mURA3-LEU2 marker in the rDNA or at the leu2Δ1 locus, were grown in 10 ml of YPD cultures to saturation at 30°C. Tenfold serial dilutions of each culture were made in sterile water, and 5 μl of each dilution was spotted onto YPD and 5-FOA solid medium. Plates were photographed after 2 days of incubation at 30°C.
Mitotic stability of the URA3 gene in the rDNA.
The mitotic stability of the URA3 gene was assayed as described previously (7). Briefly, single Ura+ colonies were inoculated into 10 ml of YPD medium and grown overnight at 30°C. Cultures were diluted 1:10,000 in fresh YPD and grown to saturation. Appropriate dilutions of the ninth serial culture were spread on YPD solid medium to obtain 50 to 200 cells per plate. After growth, colonies were counted and the plates were replica plated to SC medium lacking uracil. Recombination frequencies were calculated by counting the number of colonies that failed to grow on medium lacking uracil and dividing that number by the total number of colonies that grew on YPD. This procedure was performed three times for each strain.
RESULTS
Integration of SUC2 at random locations in the S. cerevisiae genome.
Our studies began with the goal of testing whether the genomic position of a gene might affect its control by the Swi/Snf chromatin remodeling complex. To do this, we integrated the Swi/Snf-dependent gene SUC2 at several random locations in the S. cerevisiae genome and then tested whether its expression was still dependent upon Swi/Snf. Random integrants were obtained under conditions in which there is no homology between the transforming DNA and the genome (58) (see Materials and Methods). Briefly, a linearized plasmid (pVD1) containing SUC2 and URA3 was used to transform S. cerevisiae strain FY2310 (Table 1), which contains a temperature-sensitive allele of SNF5 (snf5-51) (20) and lacks the complete SUC2 and URA3 genes. We selected for Ura+ transformants and then screened them for the dependence of SUC2 expression on Swi/Snf at both permissive (30°C) and nonpermissive (37°C) temperatures for snf5-51. This was done by screening the transformants for growth on YEP raffinose medium, which is dependent upon SUC2 expression.
Of 18 Ura+ transformants, we identified 3 candidates that were Raf+ at 37°C. For each of the three candidates, the site of integration was determined as described in Materials and Methods. For the first two cases, the apparent Swi/Snf independence was likely due to multiple copies of SUC2 DNA. In one, the plasmid had integrated into the 2μm circle plasmid; in the other, several tandem copies had integrated into the NUM1 gene, which contains tandem repeats (37). In the third transformant, however, a single copy of SUC2 DNA had integrated into the 35S region of rDNA. As this was our only Raf+ isolate in which SUC2 was present in single copy, we focused our studies on expression of SUC2 in this genomic position. To retest the phenotype and to simplify our subsequent analyses, we first constructed a new URA3-SUC2 cassette with fewer plasmid sequences and integrated it into the same position within the 35S of the rDNA repeat as was identified for the original isolate (Fig. 1) (see Materials and Methods). By pulsed-field gel electrophoresis and Southern blot analyses, we verified that seven of eight URA3-SUC2 cassette transformants were present in single copy in the rDNA. Three of these were mapped to different repeats within the array; however, all behaved similarly with respect to SUC2 transcription in wild-type and snf2Δ mutants (described below and data not shown). Two of these transformants were used for the remainder of the experiments described in later sections.
SUC2 transcription is repressed by the Swi/Snf complex when SUC2 is in the rDNA.
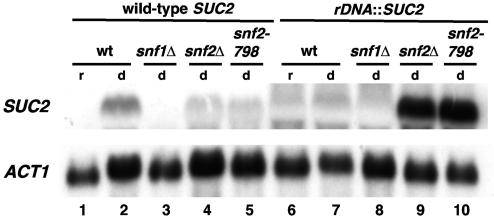
Our initial characterization had suggested that when SUC2 is located in the rDNA its expression is Swi/Snf independent. To characterize the effect of Swi/Snf on expression of SUC2 in the rDNA in greater detail, we measured SUC2 mRNA levels in a wild-type (SNF2) strain and in two different snf2 mutants, snf2Δ and snf2-798. The snf2-798 mutation encodes a K798A amino acid change, which impairs the Snf2 ATPase activity (33), which is critical for Swi/Snf chromatin remodeling activity. This analysis revealed two aspects of SUC2 regulation in the rDNA. First, in a wild-type genetic background, when cells are grown under conditions derepressing for SUC2 transcription, SUC2 mRNA levels are significantly reduced when SUC2 is in the rDNA compared to when SUC2 is at its natural location (Fig. 2; compare lanes 2 and 7). This result is consistent with previous studies demonstrating silencing of RNA polymerase II-dependent transcription in the rDNA (45, 57). Second, in both snf2 mutants tested, the rDNA silencing of SUC2 is abolished, as SUC2 mRNA is present at a high level (Fig. 2; compare lane 7 to lanes 9 and 10). The finding that the snf2-798 mutation causes a silencing defect strongly suggests that the Swi/Snf remodeling activity is required for rDNA silencing of SUC2. A similar derepression was observed in a snf5Δ mutant (data not shown). In contrast, Snf1, a protein kinase that activates SUC2 transcription independently from Swi/Snf (68), does not play any role in rDNA silencing of SUC2 (Fig. 2, lane 8). Taken together, these data suggest that Swi/Snf represses SUC2 transcription in the rDNA, the opposite of its role in activation of SUC2 at its natural location.
FIG. 2.
Swi/Snf represses transcription of SUC2 in the rDNA. Northern analysis of SUC2 mRNA levels was performed on strains that contain SUC2 in its normal genomic location (lanes 1 to 5) or in the rDNA (lanes 6 to 10). Strains were grown in YPD (repressing conditions; lanes r) and then shifted to YEP plus 0.05% glucose (derepressing conditions; lanes d) for 2 h 45 min. ACT1 served as a loading control. The strains analyzed are as follows: lanes 1 and 2, FY78; lane 3, FY49; lane 4, FY328; lane 5, FY2084; lanes 6 and 7, FY2313; lane 8, FY2314; lane 9, FY2316; and lane 10, FY2321. wt, wild type.
Analysis of the chromatin structure of the SUC2 promoter in rDNA compared to its normal position.
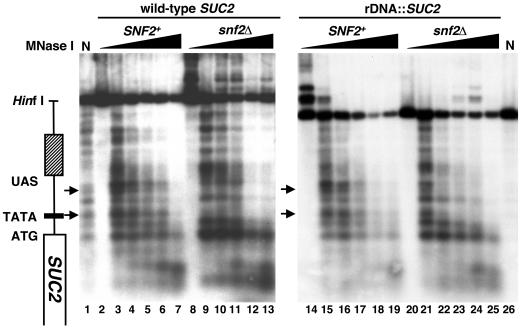
Previous studies showed that under derepressing conditions, a snf2Δ mutation causes changes in chromatin structure over the SUC2 promoter. These studies demonstrated that in wild-type strains, the SUC2 promoter region is generally sensitive to digestion by MNase. However, in snf2Δ mutants, MNase digestion is more inhibited in particular regions of the promoter, strongly suggesting the presence of nucleosomes over the TATA and the region between the TATA and upstream activation sequence (UAS) (19, 24, 71) (Fig. 3). To determine whether snf2Δ also causes changes in SUC2 chromatin structure when SUC2 is in the rDNA, we performed indirect end-labeling analysis of MNase-digested chromatin for cells grown under conditions derepressing for SUC2 transcription (Materials and Methods). In contrast to what was found for SUC2 at its natural location, our results revealed that a snf2Δ mutation causes little if any detectable effect on SUC2 chromatin structure in the rDNA. In this location, the SUC2 MNase cleavage pattern is the same in both wild-type and snf2Δ backgrounds, with an MNase cleavage pattern for both strains similar to the active, wild-type form at the normal SUC2 location (Fig. 3). In particular, in both strains MNase cleavage occurs over the TATA and the region between the TATA and the UAS. These results suggest that Swi/Snf is not required to maintain SUC2 in an active chromatin structure when it is in the rDNA; however, this active structure is not sufficient to allow expression in the presence of wild-type Swi/Snf (see Discussion).
FIG. 3.
MNase analysis of SUC2 chromatin structure. Strains were grown in YPD medium to 107 cells/ml and then shifted to derepressing conditions as described in Materials and Methods. Spheroplasts were isolated and then incubated with increasing amounts of MNase as described in Materials and Methods. DNA was purified, digested with HinfI, and subjected to indirect end-labeling analysis using a probe that anneals to +140 to +296 (+1 = ATG) in the coding sequence of SUC2. The SUC2 genomic region is diagramed on the left. The positions of two prominent sites that differ in levels of MNase sensitivity for SUC2 in its wild-type location are marked with arrows. N denotes the naked DNA controls (lanes 1 and 26). The strains used were FY78 (SNF2; lanes 2 to 7), FY328 (snf2Δ; lanes 8 to 13), FY2313 (SNF2 rDNA::SUC2; lanes 14 to 19), and FY2316 (snf2Δ rDNA::SUC2; lanes 20 to 25).
Swi/Snf is a general repressor in rDNA.
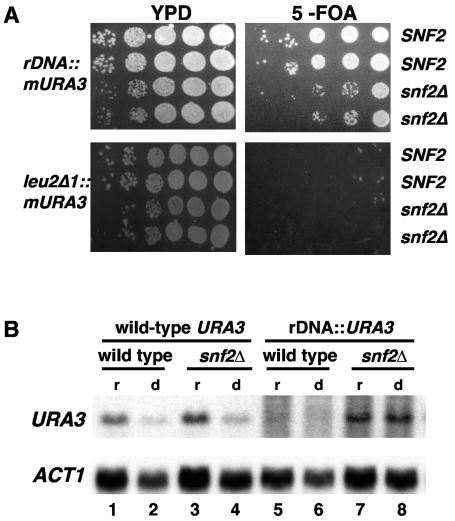
Since Swi/Snf silences the transcription of SUC2 in the rDNA, we asked whether Swi/Snf has a general role in rDNA silencing. To do this, we examined the expression of two forms of URA3, a gene not normally regulated by Swi/Snf. First, we used a previously established rDNA-silencing assay that measures expression of a modified URA3 gene, mURA3, by spot tests on solid media (60). In this case, URA3 expression is under control of a minimal TRP1 promoter (60). We found that in SNF2 strains, as expected, expression of the mURA3 gene in the rDNA is reduced relative to its expression when integrated at leu2Δ1, reflecting transcriptional silencing in rDNA (Fig. 4A). In contrast, in snf2Δ strains, expression of mURA3 in the rDNA is significantly greater than in the SNF2 strains. In addition, we measured mRNA levels for the wild-type URA3 gene present on the cassette that contains SUC2 (Fig. 4B). In this construct, the URA3 promoter is 1.9 kb from the SUC2 promoter (Fig. 1) and therefore is unlikely to be regulated the same as SUC2. When URA3 is in its natural location, there is no significant difference in the URA3 mRNA levels between SNF2 and snf2Δ strains. In contrast, when URA3 is located in the rDNA, it is strongly silenced in SNF2 strains and has a significantly increased mRNA level in snf2Δ mutants. These experiments provide strong evidence that Swi/Snf silences URA3 specifically when it is located in the rDNA, suggesting that Swi/Snf is generally required for rDNA silencing of RNA polymerase II-transcribed genes.
FIG. 4.
Swi/Snf silences mURA3 and URA3 in the rDNA. (A) Swi/Snf silences expression of mURA3 in the rDNA. Tenfold serial dilutions of stationary-phase cultures of SNF2 (L1081 and L1084) or snf2Δ (L1079) strains containing the mURA3-LEU2 cassette at the rDNA and SNF2 (L1082 and L1083) or snf2Δ (L1085 and L1086) containing the mURA3-LEU2 cassette at leu2Δ1 were spotted onto 5-FOA and YPD media to monitor expression of mURA3. Loss of silencing is indicated by less growth on 5-FOA. (B) Swi/Snf represses the transcription of URA3 in the rDNA. URA3 mRNA levels were measured in four strains by Northern hybridization analysis. For URA3 at its natural location, FY78 (wild type; lanes 1 and 2) and FY328 (snf2Δ; lanes 3 and 4) were used. For URA3 in the rDNA, FY2313 (wild type; lanes 5 and 6) and FY2316 (snf2Δ; lanes 7 and 8) were used. Strains were grown under conditions both repressing (YPD) and depressing (shifted to 0.05% glucose) for SUC2 transcription (see Materials and Methods). In low glucose, URA3 at its natural location is transcribed at a low level whereas URA3 in the rDNA is not significantly affected. ACT1 served as a loading control.
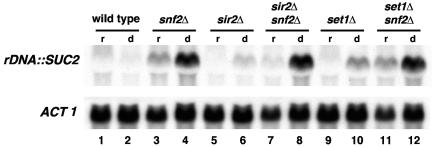
Analysis of the effect of snf2Δ, sir2Δ, and set1Δ mutations on rDNA silencing of SUC2.
Several other factors have been previously shown to be required for rDNA silencing (45, 57). The factor most extensively characterized and that is known to function directly in rDNA silencing is Sir2, a histone deacetylase (45, 57). To determine whether the Swi/Snf complex affects rDNA silencing indirectly by affecting other genes known to be required for rDNA silencing, we compared rDNA silencing of SUC2 between snf2Δ and two other previously characterized silencing mutants, sir2Δ and set1Δ (7, 8, 60). We found that while each mutation caused increased SUC2 mRNA levels, the snf2Δ mutation caused a significantly greater increase (Fig. 5; compare lane 4 to lanes 6 and 10). In snf2Δ sir2Δ and snf2Δ set1Δ double mutants, the SUC2 mRNA levels are similar to those of the snf2Δ single mutant (Fig. 5; compare lane 4 to lanes 8 and 12). The greater defect in the snf2Δ mutant strongly suggests that at least a component of the control of silencing by Swi/Snf is independent of Sir2 and Set1.
FIG. 5.
Analysis of the effects of sir2Δ and set1Δ mutations on rDNA silencing of SUC2. Northern analysis of SUC2 when integrated in the rDNA. Strains were grown in YPD (repressing conditions; lanes r) and then shifted to YEP plus 0.05% glucose (derepressing conditions; lanes d) for 2 h and 45 min. ACT1 serves as a loading control. The strains analyzed are as follows: SNF2 (FY2312), snf2Δ (FY2315), sir2Δ (L1075), sir2Δ snf2Δ (L1076), set1Δ (FY2318), and set1Δ snf2Δ (FY2319).
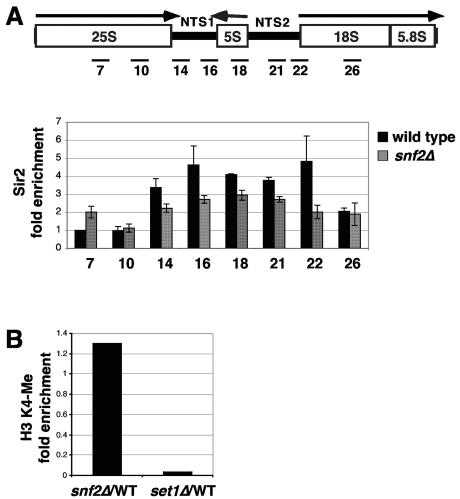
We also used ChIP to test whether loss of Swi/Snf affects Sir2- or Set1-mediated silencing. First, we found that Sir2 is still associated with the rDNA repeat in snf2Δ mutants, although the distribution of Sir2 along the rDNA repeat in snf2Δ mutants is modestly different from that seen with wild-type strains (Fig. 6A) (28). This small change seems unlikely to account for the loss of silencing of RNA polymerase II-transcribed genes in the rDNA in snf2Δ mutants. This is particularly true for SUC2, as it is integrated in a position in the rDNA repeat that normally has very low levels of Sir2 (Fig. 6A) (28). We also examined the levels of histone H3 K4 methylation in wild-type and snf2Δ strains and found that they are the same (Fig. 6B). These results suggest that Swi/Snf controls rDNA silencing independently of Sir2 and Set1.
FIG. 6.
Analysis of Sir2 and histone H3 methylation in snf2Δ mutants. (A) ChIP analysis of Sir2 in the rDNA. ChIP was performed across the rDNA repeat in both wild-type (FY2313) and snf2Δ (FY2316) strains. The positions of the regions analyzed by PCR in the ChIP experiments are shown at the top. The comparison between the wild-type and snf2Δ strains, normalized to an untranscribed region of the genome, is shown below (36). The PCR products correspond to those previously used (28). (B) ChIP analysis of histone H3 K4 methylation in wild-type and snf2Δ strains. ChIP was performed in three strains: the wild type (FY2313), snf2Δ (FY2316), and set1Δ (FY2318). Comparisons of snf2Δ and set1Δ to the wild type are shown.
rDNA recombination is reduced in snf2Δ strains.
Previous studies have shown that most mutations that impair rDNA silencing elevate the rate of mitotic recombination at the rDNA. This relationship has been demonstrated for mutations in SIR2, TOP1, UBC2, and ZDS2 (7, 21, 56). The correlation is not perfect, however, as mutations in SET1 impair rDNA silencing and yet have no effect on rDNA recombination (8), and mutations in FOB1 also impair rDNA silencing and decrease rDNA recombination (15, 28, 35). To determine whether snf2Δ causes an effect on rDNA recombination, we compared rDNA mitotic recombination levels in wild-type, snf2Δ, and sir2Δ strains (see Materials and Methods). Surprisingly, in a snf2Δ mutant there is dramatic reduction in the rate of rDNA mitotic recombination, approximately 50-fold below that of the wild type (Table 2). In a sir2Δ mutant, as expected, the rate was elevated compared to that of the wild type. To test the epistatic relationship between snf2Δ and sir2Δ with respect to rDNA recombination, we also tested snf2Δ sir2Δ double mutants. Our results (Table 2) show that the snf2Δ sir2Δ double mutant still has a recombination rate below that of the wild type although greater than that of the snf2Δ single mutant. These results are consistent with the conclusion that the role of the Swi/Snf complex in rDNA silencing is distinct from that of Sir2 and other rDNA silencing factors previously identified.
TABLE 2.
Deletion of SNF2 reduces mitotic recombination in the rDNA
| Relevant genotypea | No. of Ura auxotrophs/total no. of cells analyzed | No. of URA3 markers lost/generationb | Mutant loss rate/ wild-type loss rate |
|---|---|---|---|
| Wild type | 144/4132 (0.035%) | 2.9 × 10−4 | |
| sir2Δ | 2288/5261 (0.43%) | 3.6 × 10−3 | 12.4 |
| snf2Δ | 4/6412 (6.2 × 10−4) | 5.1 × 10−6 | 0.02 |
| snf2Δ sir2Δ | 21/3099 (6.8 × 10−3) | 5.6 × 10−5 | 0.19 |
The strains used for these experiments were FY2313, wild type; L1075, sir2Δ; FY2316, snf2Δ; and L1076 and L1077, snf2Δ sir2Δ.
The rate of mitotic recombination was determined by measuring the rate of loss of the URA3 marker (number of Ura− auxotrophs/total number of cells analyzed) after 120 generations of growth in nonselective medium (as described in Materials and Methods).
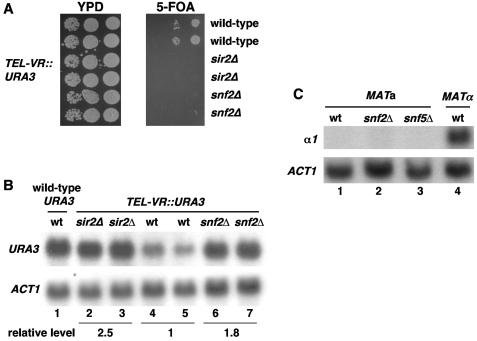
Snf2 is also required for silencing at telomeres but not at HM loci.
We also tested whether Swi/Snf is required for silencing at the two other known silenced regions in S. cerevisiae, telomeres and HM loci. To detect whether Swi/Snf has a role in telomeric silencing, we first performed spot tests using strains that have URA3 near the right telomere of chromosome V. The results (Fig. 7A) show that a snf2Δ mutation causes increased expression of the URA3 reporter compared to a wild-type background. To determine whether the increased URA3 expression is caused at the transcriptional level, we performed Northern hybridization analysis. Our results show that the level of URA3 mRNA is modestly but significantly increased in the snf2Δ mutants compared to the wild-type strain results (Fig. 7B). Thus, Swi/Snf is required for telomeric silencing. To determine whether Swi/Snf has a role in the silencing of the HM loci, we performed Northern hybridization analysis to assay for α1 mRNA expressed from HMLα in a MATa strain. Our results (Fig. 7C) show that there is no detectable α1 mRNA in the swi/snf mutants tested, suggesting that the Swi/Snf complex is not required for silencing of the HM loci.
FIG. 7.
SNF2 is required for telomeric silencing. (A) Telomeric silencing phenotypes determined using a telomeric URA3 reporter gene. Tenfold serial dilutions of stationary-phase cultures of wild-type (L1087 and L1088), sir2Δ (L1091 and L1092), or snf2Δ (L1089 and L1090) strains containing the URA3 at the right telomere of chromosome V were spotted onto 5-FOA and YPD medium to monitor expression of the URA3. Loss of silencing is indicated by reduced growth on 5-FOA. (B) Swi/Snf represses URA3 transcription at the telomere. Strains with URA3 at its normal genomic location (lane 1) or integrated at the right telomere of chromosome V (lanes 2 to 7) were grown in SC medium supplemented with 100 mg of uracil/liter. mRNA levels of URA3 and the loading control, ACT1, were measured by Northern analysis. The strains used in the experiment were as follows: lane 1, FY78; lane 2, L1091; lane 3, L1092; lane 4, L1087; lane 5, L1088; lane 6, L1089; and lane 7, L1090. The quantitation represents the relative level of URA3 mRNA normalized to the level of ACT1. The numbers represent the averages for the pairs of strains shown. wt, wild type. (C) Swi/Snf has no detectable effect on HMLα silencing. Three MATa strains in lanes 1 to 3 (wild type [wt], FY78; snf2Δ, FY328; snf5Δ, FY1658) and a MATα strain, FY1856, were grown in YPD to 2 × 107 cells/ml. α1 and ACT1 mRNA levels were measured by Northern analysis.
DISCUSSION
Our results have demonstrated that the Swi/Snf complex, previously shown to be required for the normal activation and repression of many genes in S. cerevisiae, also regulates transcriptional silencing in the rDNA and at telomeres. Our results provide strong evidence that the Swi/Snf-dependent mechanism acts independently of the histone-modifying enzymes Sir2 and Set1. First, snf2Δ causes a significantly greater defect in the rDNA silencing of SUC2 than either sir2Δ or set1Δ. Second, in snf2Δ mutants, Sir2 is still associated with the rDNA and Set1-dependent histone methylation levels are normal. Third, in contrast to sir2Δ, snf2Δ does not alter nucleolar structure nor does it affect the association of Net1 with the nucleolus (data not shown). Finally, a snf2Δ mutation dramatically reduces rDNA recombination, a phenotype distinct from the increased levels in sir2Δ mutants and the unaffected levels in set1Δ mutants. These findings support the existence of a Swi/Snf-dependent mechanism for rDNA transcriptional silencing that acts independently of Sir2 or Set1.
The role of Swi/Snf in SUC2 chromatin structure when SUC2 is in its normal genomic location differs from that seen when SUC2 is in the rDNA. At the normal SUC2 genomic location, an active MNase cleavage pattern is dependent upon Swi/Snf, while in the rDNA, an active pattern is independent of Swi/Snf. Therefore, SUC2 chromatin structure and its Swi/Snf dependence can be determined by genomic location. Furthermore, since SUC2 chromatin structure is in the active conformation in either the presence or absence of Swi/Snf, the role of Swi/Snf in rDNA silencing must occur at a level other than that assayed by MNase sensitivity. Finally, the Swi/Snf independence of SUC2 chromatin structure when SUC2 is in the rDNA suggests that this active conformation may be dependent upon a different chromatin remodeling complex.
Our results have demonstrated that the absence of the Swi/Snf complex causes a drastic reduction in rDNA mitotic recombination. While mutations in FOB1 and HRM2-HRM4 also reduce rDNA mitotic recombination (42), those effects are not as severe as those caused by a mutation in SNF2. However, in similarity to snf2Δ, fob1Δ also impairs rDNA silencing (28). Our finding that snf2Δ is largely epistatic to sir2Δ with respect to recombination suggests that snf2Δ causes a change in rDNA chromatin that makes it inaccessible to recombination enzymes, even in the absence of Sir2 activity. This finding, combined with our MNase results, hints that the control of rDNA chromatin structure by Swi/Snf might occur at a higher-order level, a role that has been previously suggested for Swi/Snf (27).
An important question regarding the function of Swi/Snf in rDNA silencing is whether its role is direct or indirect. One obvious direct role is for Swi/Snf to directly control chromatin structure of nucleolar DNA. However, by ChIP experiments, neither Snf2 nor Snf5 were detectably associated with the rDNA (data not shown). In the most extensive experiments, Snf2 association was assayed across the entire rDNA repeat, at the SUC2 promoter, and at the URA3 promoter. In addition, by immunolocalization experiments, Snf2 was nuclear, in consistency with earlier findings (29, 38); however, Snf2 also appeared nucleolar in only a low percentage of cells (data not shown). While these negative results do not rule out the possibility that Swi/Snf functions directly in rDNA silencing, they leave open the possibility of a less direct role. For example, Swi/Snf might regulate a gene required for rDNA silencing. Regardless of the specific mechanism by which Swi/Snf controls rDNA silencing, our results have shown that it plays a prominent role in rDNA silencing that is independent of previously identified factors.
Acknowledgments
We thank Mary Bryk, Julie Huang, and Jessica Pamment for helpful comments on the manuscript. We also thank Julie Huang and Danesh Moazed for advice on the Sir2 ChIP experiments, Robert Schiestl for advice on nonhomologous integration, and Lorraine Pillus for helpful discussions.
This work was supported by National Institutes of Health grant GM32967.
REFERENCES
- 1.Aparicio, O. M., B. L. Billington, and D. E. Gottschling. 1991. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell 66:1279-1287. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1998. Current protocols in molecular biology. Greene/Wiley-Interscience, New York, N.Y.
- 3.Becker, P. B., and W. Horz. 2002. ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 71:247-273. [DOI] [PubMed] [Google Scholar]
- 4.Biggar, S. R., and G. R. Crabtree. 1999. Continuous and widespread roles for the Swi-Snf complex in transcription. EMBO J. 18:2254-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 6.Briggs, S. D., M. Bryk, B. D. Strahl, W. L. Cheung, J. K. Davie, S. Y. Dent, F. Winston, and C. D. Allis. 2001. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 15:3286-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bryk, M., M. Banerjee, M. Murphy, K. E. Knudsen, D. J. Garfinkel, and M. J. Curcio. 1997. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev. 11:255-269. [DOI] [PubMed] [Google Scholar]
- 8.Bryk, M., S. D. Briggs, B. D. Strahl, M. J. Curcio, C. D. Allis, and F. Winston. 2002. Evidence that Set1, a factor required for methylation of histone H3, regulates rDNA silencing in S. cerevisiae by a Sir2-independent mechanism. Curr. Biol. 12:165-170. [DOI] [PubMed] [Google Scholar]
- 9.Burns, L. G., and C. L. Peterson. 1997. Protein complexes for remodeling chromatin. Biochim. Biophys. Acta 1350:159-168. [DOI] [PubMed] [Google Scholar]
- 10.Cairns, B. R., N. L. Henry, and R. D. Kornberg. 1996. TFG/TAF30/ANC1, a component of the yeast SWI/SNF complex that is similar to the leukemogenic proteins ENL and AF-9. Mol. Cell. Biol. 16:3308-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carlson, M., and D. Botstein. 1982. Two differentially regulated mRNAs with different 5′ ends encode secreted with intracellular forms of yeast invertase. Cell 28:145-154. [DOI] [PubMed] [Google Scholar]
- 12.Cote, J., C. L. Peterson, and J. L. Workman. 1998. Perturbation of nucleosome core structure by the SWI/SNF complex persists after its detachment, enhancing subsequent transcription factor binding. Proc. Natl. Acad. Sci. USA 95:4947-4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cote, J., J. Quinn, J. L. Workman, and C. L. Peterson. 1994. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science 265:53-60. [DOI] [PubMed] [Google Scholar]
- 14.Dammann, R., R. Lucchini, T. Koller, and J. M. Sogo. 1995. Transcription in the yeast rRNA gene locus: distribution of the active gene copies and chromatin structure of their flanking regulatory sequences. Mol. Cell. Biol. 15:5294-5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Defossez, P. A., R. Prusty, M. Kaeberlein, S. J. Lin, P. Ferrigno, P. A. Silver, R. L. Keil, and L. Guarente. 1999. Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol. Cell 3:447-455. [DOI] [PubMed] [Google Scholar]
- 16.Dimova, D., Z. Nackerdien, S. Furgeson, S. Eguchi, and M. A. Osley. 1999. A role for transcriptional repressors in targeting the yeast Swi/Snf complex. Mol. Cell 4:75-83. [DOI] [PubMed] [Google Scholar]
- 17.Dudley, A. M., C. Rougeulle, and F. Winston. 1999. The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes Dev. 13:2940-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fritze, C. E., K. Verschueren, R. Strich, and R. Easton Esposito. 1997. Direct evidence for SIR2 modulation of chromatin structure in yeast rDNA. EMBO J. 16:6495-6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gavin, I. M., and R. T. Simpson. 1997. Interplay of yeast global transcriptional regulators Ssn6p-Tup1p and Swi-Snf and their effect on chromatin structure. EMBO J. 16:6263-6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geng, F., Y. Cao, and B. C. Laurent. 2001. Essential roles of Snf5p in Snf-Swi chromatin remodeling in vivo. Mol. Cell. Biol. 21:4311-4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gottlieb, S., and R. E. Esposito. 1989. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell 56:771-776. [DOI] [PubMed] [Google Scholar]
- 22.Harlow, E., and D. Lane. 1999. Using antibodies, a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Hassan, A. H., K. E. Neely, M. Vignali, J. C. Reese, and J. L. Workman. 2001. Promoter targeting of chromatin-modifying complexes. Front. Biosci. 6:D1054-D1064. [DOI] [PubMed] [Google Scholar]
- 24.Hirschhorn, J. N., S. A. Brown, C. D. Clark, and F. Winston. 1992. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev. 6:2288-2298. [DOI] [PubMed] [Google Scholar]
- 25.Holstege, F. C., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717-728. [DOI] [PubMed] [Google Scholar]
- 26.Hoppe, G. J., J. C. Tanny, A. D. Rudner, S. A. Gerber, S. Danaie, S. P. Gygi, and D. Moazed. 2002. Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol. Cell. Biol. 22:4167-4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horn, P. J., K. A. Crowley, L. M. Carruthers, J. C. Hansen, and C. L. Peterson. 2002. The SIN domain of the histone octamer is essential for intramolecular folding of nucleosomal arrays. Nat. Struct. Biol. 9:167-171. [DOI] [PubMed] [Google Scholar]
- 28.Huang, J., and D. Moazed. 2003. Association of the RENT complex with nontranscribed and coding regions of rDNA and a regional requirement for the replication fork block protein Fob1 in rDNA silencing. Genes Dev. 17:2162-2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huh, W. K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson, J. S. Weissman, and E. K. O'Shea. 2003. Global analysis of protein localization in budding yeast. Nature 425:686-691. [DOI] [PubMed] [Google Scholar]
- 30.Imai, S., C. M. Armstrong, M. Kaeberlein, and L. Guarente. 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403:795-800. [DOI] [PubMed] [Google Scholar]
- 31.Kent, N. A., L. E. Bird, and J. Mellor. 1993. Chromatin analysis in yeast using NP-40 permeabilised sphaeroplasts. Nucleic Acids Res. 21:4653-4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kent, N. A., and J. Mellor. 1995. Chromatin structure snap-shots: rapid nuclease digestion of chromatin in yeast. Nucleic Acids Res. 23:3786-3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khavari, P. A., C. L. Peterson, J. W. Tamkun, D. B. Mendel, and G. R. Crabtree. 1993. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature 366:170-174. [DOI] [PubMed] [Google Scholar]
- 34.Kingston, R. E., and G. J. Narlikar. 1999. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 13:2339-2352. [DOI] [PubMed] [Google Scholar]
- 35.Kobayashi, T., and T. Horiuchi. 1996. A yeast gene product, Fob1 protein, required for both replication fork blocking and recombinational hotspot activities. Genes Cells 1:465-474. [DOI] [PubMed] [Google Scholar]
- 36.Komarnitsky, P., E. J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 14:2452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kormanec, J., I. Schaaff-Gerstenschlager, F. K. Zimmermann, D. Perecko, and H. Kuntzel. 1991. Nuclear migration in Saccharomyces cerevisiae is controlled by the highly repetitive 313 kDa NUM1 protein. Mol. Gen. Genet. 230:277-287. [DOI] [PubMed] [Google Scholar]
- 38.Kumar, A., S. Agarwal, J. A. Heyman, S. Matson, M. Heidtman, S. Piccirillo, L. Umansky, A. Drawid, R. Jansen, Y. Liu, K. H. Cheung, P. Miller, M. Gerstein, G. S. Roeder, and M. Snyder. 2002. Subcellular localization of the yeast proteome. Genes Dev. 16:707-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuras, L., and K. Struhl. 1999. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature 399:609-613. [DOI] [PubMed] [Google Scholar]
- 40.Landry, J., A. Sutton, S. T. Tafrov, R. C. Heller, J. Stebbins, L. Pillus, and R. Sternglanz. 2000. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc. Natl. Acad. Sci. USA 97:5807-5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Larschan, E., and F. Winston. 2001. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 15:1946-1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin, Y. H., and R. L. Keil. 1991. Mutations affecting RNA polymerase I-stimulated exchange and rDNA recombination in yeast. Genetics 127:31-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Manivasakam, P., and R. H. Schiestl. 1998. Nonhomologous end joining during restriction enzyme-mediated DNA integration in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:1736-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martens, J. A., and F. Winston. 2003. Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr. Opin. Genet. Dev. 13:136-142. [DOI] [PubMed] [Google Scholar]
- 45.Moazed, D. 2001. Common themes in mechanisms of gene silencing. Mol. Cell 8:489-498. [DOI] [PubMed] [Google Scholar]
- 46.Natarajan, K., B. M. Jackson, H. Zhou, F. Winston, and A. G. Hinnebusch. 1999. Transcriptional activation by Gcn4p involves independent interactions with the SWI/SNF complex and the SRB/mediator. Mol. Cell 4:657-664. [DOI] [PubMed] [Google Scholar]
- 47.Neely, K. E., A. H. Hassan, C. E. Brown, L. Howe, and J. L. Workman. 2002. Transcription activator interactions with multiple SWI/SNF subunits. Mol. Cell. Biol. 22:1615-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neely, K. E., A. H. Hassan, A. E. Wallberg, D. J. Steger, B. R. Cairns, A. P. Wright, and J. L. Workman. 1999. Activation domain-mediated targeting of the SWI/SNF complex to promoters stimulates transcription from nucleosome arrays. Mol. Cell 4:649-655. [DOI] [PubMed] [Google Scholar]
- 49.Petes, T. D., and D. Botstein. 1977. Simple Mendelian inheritance of the reiterated ribosomal DNA of yeast. Proc. Natl. Acad. Sci. USA 74:5091-5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pollard, K. J., and C. L. Peterson. 1997. Role for ADA/GCN5 products in antagonizing chromatin-mediated transcriptional repression. Mol. Cell. Biol. 17:6212-6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prochasson, P., K. E. Neely, A. H. Hassan, B. Li, and J. L. Workman. 2003. Targeting activity is required for SWI/SNF function in vivo and is accomplished through two partially redundant activator-interaction domains. Mol. Cell 12:983-990. [DOI] [PubMed] [Google Scholar]
- 52.Quinn, J., A. M. Fyrberg, R. W. Ganster, M. C. Schmidt, and C. L. Peterson. 1996. DNA-binding properties of the yeast SWI/SNF complex. Nature 379:844-847. [DOI] [PubMed] [Google Scholar]
- 53.Richmond, E., and C. L. Peterson. 1996. Functional analysis of the DNA-stimulated ATPase domain of yeast SWI2/SNF2. Nucleic Acids Res. 24:3685-3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roberts, S. M., and F. Winston. 1997. Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics 147:451-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 56.Roy, N., and K. W. Runge. 2000. Two paralogs involved in transcriptional silencing that antagonistically control yeast life span. Curr. Biol. 10:111-114. [DOI] [PubMed] [Google Scholar]
- 57.Rusche, L. N., A. L. Kirchmaier, and J. Rine. 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72:481-516. [DOI] [PubMed] [Google Scholar]
- 58.Schiestl, R. H., and T. D. Petes. 1991. Integration of DNA fragments by illegitimate recombination in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 88:7585-7589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smith, J. S., and J. D. Boeke. 1997. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 11:241-254. [DOI] [PubMed] [Google Scholar]
- 61.Smith, J. S., C. B. Brachmann, I. Celic, M. A. Kenna, S. Muhammad, V. J. Starai, J. L. Avalos, J. C. Escalante-Semerena, C. Grubmeyer, C. Wolberger, and J. D. Boeke. 2000. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc. Natl. Acad. Sci. USA 97:6658-6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith, J. S., E. Caputo, and J. D. Boeke. 1999. A genetic screen for ribosomal DNA silencing defects identifies multiple DNA replication and chromatin-modulating factors. Mol. Cell. Biol. 19:3184-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Straight, A. F., W. Shou, G. J. Dowd, C. W. Turck, R. J. Deshaies, A. D. Johnson, and D. Moazed. 1999. Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell 97:245-256. [DOI] [PubMed] [Google Scholar]
- 64.Sudarsanam, P., Y. Cao, L. Wu, B. C. Laurent, and F. Winston. 1999. The nucleosome remodeling complex, Snf/Swi, is required for the maintenance of transcription in vivo and is partially redundant with the histone acetyltransferase, Gcn5. EMBO J. 18:3101-3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sudarsanam, P., V. R. Iyer, P. O. Brown, and F. Winston. 2000. Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97:3364-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sun, Z. W., and M. Hampsey. 1999. A general requirement for the Sin3-Rpd3 histone deacetylase complex in regulating silencing in Saccharomyces cerevisiae. Genetics 152:921-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Swanson, R. N., C. Conesa, O. Lefebvre, C. Carles, A. Ruet, E. Quemeneur, J. Gagnon, and A. Sentenac. 1991. Isolation of TFC1, a gene encoding one of two DNA-binding subunits of yeast transcription factor tau (TFIIIC). Proc. Natl. Acad. Sci. USA 88:4887-4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Treitel, M. A., S. Kuchin, and M. Carlson. 1998. Snf1 protein kinase regulates phosphorylation of the Mig1 repressor in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:6273-6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Winston, F., and M. Carlson. 1992. Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet. 8:387-391. [DOI] [PubMed] [Google Scholar]
- 70.Winston, F., C. Dollard, and S. L. Ricupero-Hovasse. 1995. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11:53-55. [DOI] [PubMed] [Google Scholar]
- 71.Wu, L., and F. Winston. 1997. Evidence that Snf-Swi controls chromatin structure over both the TATA and UAS regions of the SUC2 promoter in Saccharomyces cerevisiae. Nucleic Acids Res. 25:4230-4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yudkovsky, N., C. Logie, S. Hahn, and C. L. Peterson. 1999. Recruitment of the SWI/SNF chromatin remodeling complex by transcriptional activators. Genes Dev. 13:2369-2374. [DOI] [PMC free article] [PubMed] [Google Scholar]