Abstract
The recruitment of TATA box-binding protein (TBP) to promoters is one of the rate-limiting steps during transcription initiation. However, the global importance of TBP recruitment in determining the absolute and changing levels of transcription across the genome is not known. We used a genomic approach to explore the relationship between TBP recruitment to promoters and global gene expression profiles in Saccharomyces cerevisiae. Our data indicate that first, RNA polymerase III promoters are the most prominent binding targets of TBP in vivo. Second, the steady-state transcript levels of genes throughout the genome are proportional to the occupancy of their promoters by TBP, and changes in the expression levels of these genes are closely correlated with changes in TBP recruitment to their promoters. Third, a consensus TATA element does not appear to be a major determinant of either TBP binding or gene expression throughout the genome. Our results indicate that the recruitment of TBP to promoters in vivo is of universal importance in determining gene expression levels in yeast, regardless of the nature of the core promoter or the type of activator or repressor that may mediate changes in transcription. The primary data reported here are available at http://www.iyerlab.org/tbp.
Transcription is precisely regulated by the concerted action of general transcription factors, sequence-specific activators or repressors, and accessory proteins that modulate chromatin accessibility. Understanding the relationship between the binding of an individual transcription factor to its target promoters and the expression of all its target genes is important for understanding the mechanisms of transcriptional regulation that operate on a genome-wide scale.
TATA box-binding protein (TBP) is a general transcription factor protein that binds the TATA element located approximately 35 to 50 nucleotides upstream of the transcription start site in Saccharomyces cerevisiae. TBP is highly conserved with more than 80% identity in the 180-amino-acid C-terminal domain across eukaryotic species (11) and is a universal transcription factor that is central to all three RNA polymerase (Pol) systems (6, 41). TBP is associated with a variety of complexes, such as SL1 (5), TFIID, TFIIIB, and SAGA in yeast (28). SL1 functions at RNA Pol I promoters, TFIID is specific for promoters of mRNA genes transcribed by RNA Pol II, and the TFIIIB complex targets RNA Pol III genes. In humans, TBP is also known to be a member of other complexes, such as TFTC, SNAPC, and PCAF (28); however, its role in these complexes is unclear.
Experiments in S. cerevisiae have shown that the TBP occupancy of several RNA Pol II promoters is correlated with transcriptional activity, and the binding of TBP is stimulated by activators and general transcription factors (21, 23). However, the relationship between TBP binding and transcription has not been examined in vivo on a global scale under physiological conditions where hundreds of promoters are known to be activated or repressed. TBP binds not only to canonical TATA elements that are primarily found in Pol II promoters but also to TATA-less Pol II promoters (29) as well as Pol I and Pol III promoters that lack TATA elements (6, 32, 39). Since TBP is a general transcription factor required for all three of the nuclear RNA polymerases, it is expected to be required for the transcription of every gene. However, the relationship between the occupancy of each chromosomal promoter by TBP and the steady-state expression level of the corresponding gene is not known. Moreover, it is not clear what biases, if any, exist in the binding distribution of TBP across the genome, particularly with respect to the type of RNA Pol that transcribes each promoter.
Knowing the dynamic relationship between TBP recruitment to promoters and transcription across the genome has important implications for the mechanism of transcriptional activation and repression, given that the thousands of genes in the genome are likely to have a wide range of core promoter architecture and local chromatin structure and be regulated by a variety of qualitatively different activator or repressor proteins under different physiological conditions. Here we use a combination of chromatin immunoprecipitation (ChIP), genome-wide promoter microarrays, and expression profiling methods (27) in S. cerevisiae to map the chromosomal binding distribution of TBP and determine the global role of TBP recruitment to promoters and corresponding genome-wide gene expression profiles in vivo.
MATERIALS AND METHODS
Yeast strain and culture conditions.
The S. cerevisiae strain used in all experiments was a derivative of W303-1A and contains an influenza virus hemagglutinin epitope (HA)-tagged TBP gene (21) (gift from K. Struhl). TBP expressed in this strain contains three copies of the HA epitope inserted after codon 3 of the TBP open reading frame (ORF). In most cases, cells were grown to mid-log phase (optical density at 600 nm of 0.4 to 0.6) in synthetic complete medium lacking uracil, and half of the culture was used for formaldehyde cross-linking and ChIP, while the other half was used for mRNA isolation for expression profiling. For heat shock treatment, cells grown continuously at 25°C were collected by centrifugation, resuspended in an equal volume of prewarmed 39°C medium, and returned to 39°C for growth. Samples were collected or cross-linked after 10-, 30-, and 60-min time periods. For methyl methanesulfonate (MMS) treatment to induce DNA damage, cells were grown at 30°C to mid-log phase. MMS (0.02%) (Sigma) was added to the cultures, and cells were collected or cross-linked after a 30-min incubation. For stationary phase, cells were grown to an optical density of >5.0 and cells were collected or cross-linked after there was no further increase in culture density.
Yeast DNA microarrays and hybridization.
Microarrays that include nearly every ORF and intergenic element from the yeast genome were manufactured as described previously (16, 18). Microarrays were scanned with a GenePix 4000B scanner (Axon Instruments). Fluorescence intensities were quantified using GenePix Pro software (version 4.0), and data were uploaded to a relational database for further analysis (20). Data were filtered to exclude spots with obvious defects or a signal intensity below an empirically determined threshold. PCR amplification and fluorescence labeling of immunoprecipitated DNA and labeling of cDNA was performed as described previously (17). The reference hybridization probe used in the experiments shown in Fig. 1, 2, 3, and 4 was a common pool of wild-type yeast genomic DNA that had been sonicated. Amplification and labeling of the reference were performed by the same protocols used for the ChIP samples.
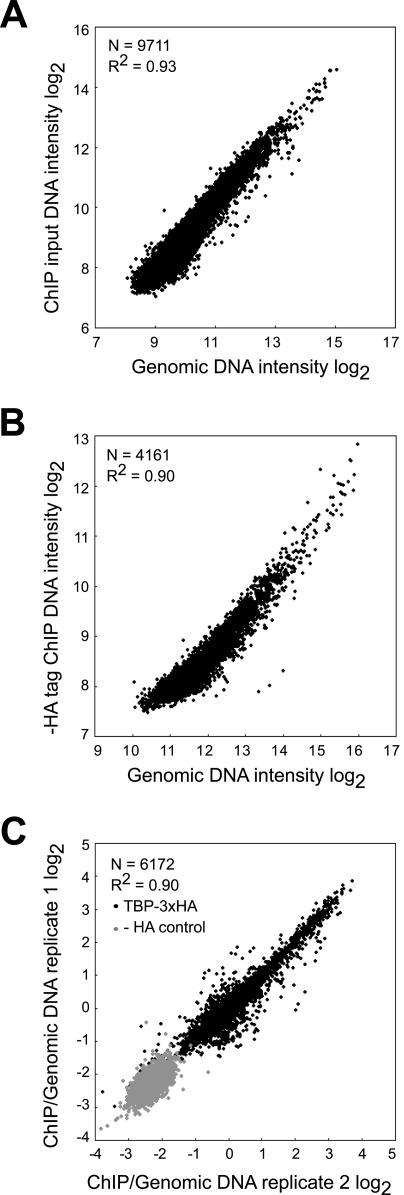
FIG. 1.
ChIP-chip hybridization controls. (A) Sheared genomic DNA labeled with Cy3 and ChIP input DNA labeled with Cy5 were hybridized to yeast whole-genome microarrays containing ORF and intergenic sequences. The median intensities of 9,711 spots from both channels are plotted on a log2 scale. (B) Sheared genomic DNA was labeled with Cy3, and DNA recovered after subjecting an untagged control strain to ChIP was labeled with Cy5 and cohybridized to whole-genome arrays. The median intensities of 4,161 spots on the scatter plot show a strong correlation between the two samples (R2 = 0.90).The small number of spots in this experiment is due to the fact that ChIP using an anti-HA antibody from a strain lacking HA yielded very low amounts of DNA and therefore low signal intensity at many spots that did not pass basic quality thresholds. (C) Independent replicates of TBP ChIP-chip experiments were performed. For each replicate, TBP ChIP DNA was labeled with Cy5, and genomic DNA was labeled with Cy3 as a reference. Cy5/Cy3 ratios (log2) of 6,172 spots are plotted and show a strong correlation (R2 = 0.90), suggesting that TBP ChIP experiments are reproducible when genomic DNA is used as a reference (HA-labeled TBP [TBP-3xHA]). We similarly tested the untagged strain as a control (− HA control). All of these experiments shown in panels A, B, and C were performed at least twice, and similar results were obtained.
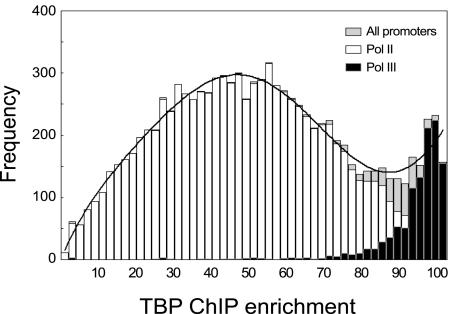
FIG. 2.
Genome-wide binding distribution of TBP. The data shown are from 49 independently grown cultures each of which was separately analyzed in ChIP-chip experiments. TBP ChIP samples were labeled with Cy5 and hybridized to whole-genome arrays together with reference genomic DNA labeled with Cy3. TBP ChIP enrichment was calculated as the median percentile rank of Cy5/Cy3 ratios across all experiments (described in Materials and Methods) and plotted as a histogram. The bimodal distribution of TBP ChIP enrichment (black continuous line) shows strong enrichment of specific fragments, indicating consistent binding of TBP to these loci in all growth conditions. The white bars represent TBP binding to Pol II promoters. The black bars represent loci adjacent to Pol III genes and are the most highly enriched across all ChIP experiments. The complete data set underlying this and other figures is available (http://www.iyerlab.org/tbp).
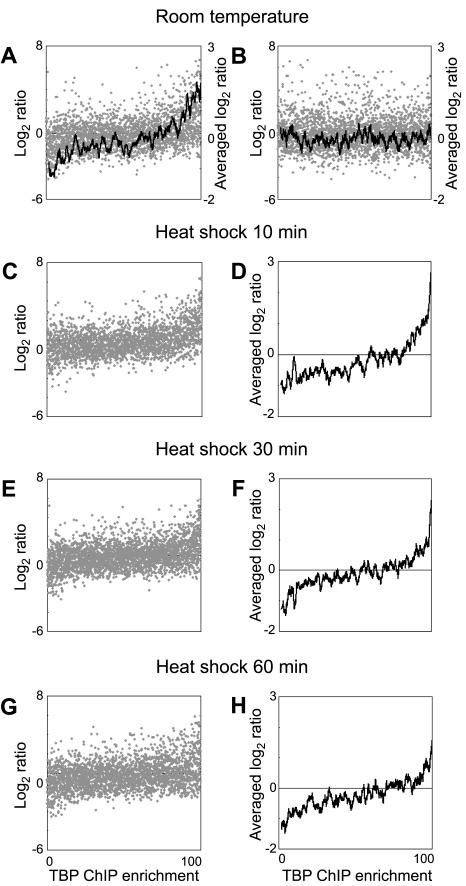
FIG. 3.
TBP recruitment to promoters is proportional to steady-state levels of Pol II gene transcripts throughout the genome. (A) In healthy, exponentially growing cells, TBP recruitment to Pol II promoters was measured in ChIP-chip experiments, and the corresponding mRNA transcript levels were measured by cDNA hybridization to yeast whole-genome microarrays. Genomic DNA was used as a reference. Each gray spot represents the steady-state transcript level of one gene (log2 cDNA/genomic DNA ratio) and TBP occupancy at its promoter (rank of the ChIP/genomic DNA ratio). A moving-window average analysis (window size of 50; step size of 1) (black line) applied to the transcript levels shows the global trend of this relationship. (B) The ChIP enrichment values were scrambled relative to the steady-state transcript level values for each gene, and these scrambled data were plotted as in panel A. The black line shows the moving-window average applied to the transcript levels as described above. (C to H) Similar relationships between TBP recruitment and steady-state transcript levels are evident when the cells were heat shocked for 10 min (C and D), 30 min (E and F), or 60 min (G and H). Relationships between TBP recruitment and steady-state transcript levels for each Pol II gene across the genome are plotted individually on a scatter plot (C, E, and G) or using a moving-window average analysis as described above (D, F, and H).
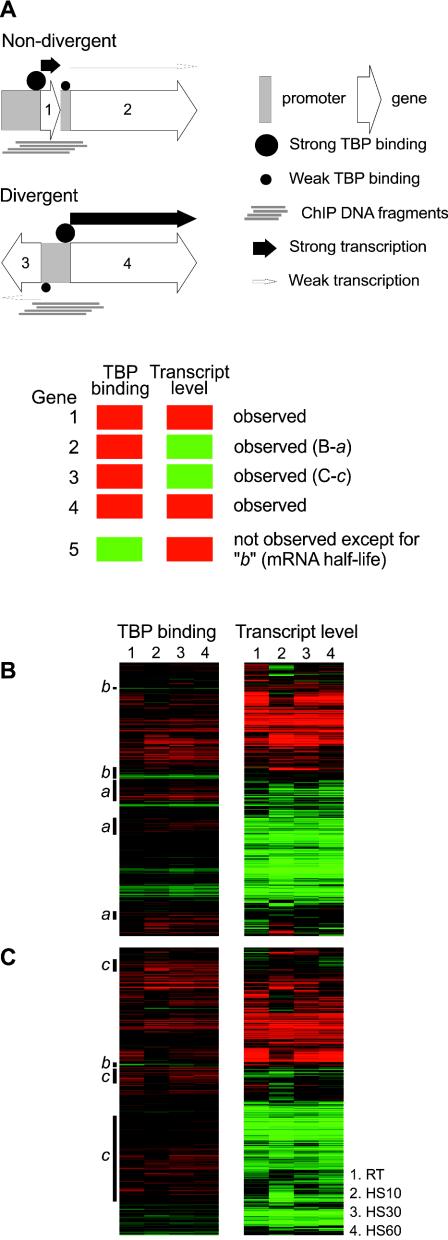
FIG. 4.
Confounding effect of compact genome organization. (A) The compact arrangement of genes in the yeast genome can give rise to an apparent discrepancy between TBP binding and transcrip-tional activity. Strong binding of TBP to the core promoter of gene 1 or gene 4 generates ChIP DNA fragments that can physically overlap and hybridize to the core promoter of gene 2 and gene 3. Even if the promoters of genes 2 and 3 actually have low levels of TBP binding and are transcribed at low levels, the overlapping ChIP DNA hybridization makes it appear as if genes 2 and 3 have strong TBP binding but low transcript levels. (B and C) Relationship between TBP occupancy at promoters and steady-state transcript levels visualized on a gene-by-gene basis by plotting microarray ratios on a red-green color scale. A total of 1,570 genes across the genome with available data points from each of the four mRNA expression microarrays as well as the four ChIP microarrays are shown here. Each column within the two subpanels represents one of four different growth conditions, room temperature (RT) and heat shock for 10, 30, and 60 min (HS10, HS30, and HS60, respectively). (B) Genes that do not share their upstream intergenic region with other genes. (C) Genes that share their upstream intergenic region with another divergently transcribed gene. The labeled black bars to the left of the panels in panels B and C indicate the expected or explainable subsets of genes showing the greatest disparity between transcript levels and TBP recruitment. Class a and c genes are indicated in the figure; these designations were made after manual inspection of their chromosomal loci. Class b genes are genes with higher than average mRNA half-lives.
Cross-linking and immunoprecipitation.
Cross-linking and immunoprecipitation were performed as described previously (17). We used antihemagglutinin antibodies (Santa Cruz) at a 1:100 dilution for ChIP.
mRNA preparation.
Cells were collected by centrifugation at 3,000 × g for 5 min at room temperature. Each 50-ml cell pellet was resuspended in 8 ml of lysis buffer (10 mM Tris-Cl [pH 4.0], 10 mM EDTA, 0.5% sodium dodecyl sulfate) and stored at −80°C until RNA preparation. Total RNA was prepared by acid phenol lysis as previously described (38). Poly(A)+ mRNA was purified using an Oligotex kit (QIAGEN).
Determination of genomic enrichment and gene-promoter associations.
For Fig. 2, we defined the ChIP enrichment score E for each genomic locus as follows: E = median R(Cy5/Cy3)1, R(Cy5/Cy3)2,… .R(Cy5/Cy3)n, where R(Cy5/Cy3)i is the percentile rank of the Cy5-to-Cy3 ratio of the microarray element corresponding to that locus among all genomic loci in the ith independent ChIP experiment. E is expressed as a percentage with values between 0 and 100 and is thus a measure of consistent in vivo association of TBP with a given genomic locus in 49 independent experiments. For combining data from replicate experiments, we averaged the percentile rank and took that to be the enrichment value (see Fig. 3, 4, and 5). Each experiment in these figures represents the average of two to four independent replicate experiments (cultures, ChIP, and microarray hybridizations). Since more than one locus spotted on the microarrays could be the promoter for a given gene, we assigned the locus that was immediately upstream of each ORF as the promoter of that gene. This pairing of each gene with a single promoter was used for all analyses.
FIG. 5.
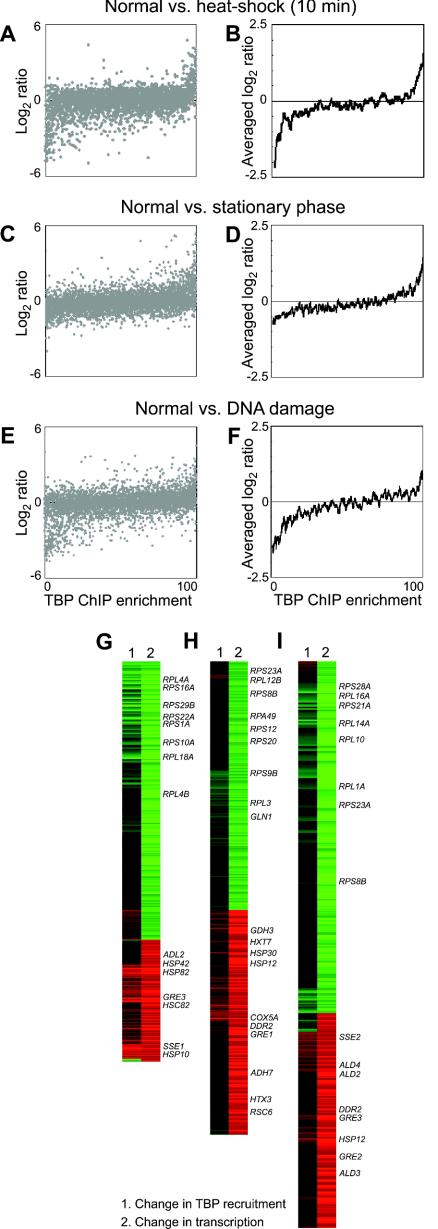
Relationship between changes in TBP recruitment and changes in gene expression after stress perturbations. Changes in TBP recruitment in stressed cells relative to normally growing cells were determined by hybridizing Cy3-labeled ChIP DNA derived from thenormally grown cells together with Cy5-labeled ChIP sample from stressed cells onto whole-genome microarrays. Corresponding cDNA samples prepared from aliquots of the same cultures harvested just prior to formaldehyde cross-linking were analyzed in parallel to determine the global changes in gene expression. The changes in gene expression (log2 Cy5/Cy3 ratio) of each gene and changes in TBP binding (percentile rank of Cy5/Cy3 ratio) at its promoter are plotted individually (A, C, and E) or with a moving-window average (window size of 50; step size of 1) applied to the change in gene expression levels (B, D, and F). A gene-by-gene visualization of the correlation between changes in gene expression and changes in TBP recruitment during heat shock (G), stationary phase (H), and DNA damage (I) is shown on a red-green color scale as in Fig. 4. These panels show only the subset of genes from panels A to F that had significant expression changes (more than twofold). Representative gene names are indicated to the right of the panels.
RESULTS
TBP binds predominantly to RNA Pol III promoters.
We first determined the genome-wide binding distribution of TBP under steady-state conditions to identify any bias in terms of the promoters or genomic loci that it binds to. In all the experiments described here, we used cells bearing a HA-tagged derivative of TBP as the only functional copy of TBP (21). We performed a large number of independent ChIP-chip experiments from independently grown cultures in a range of different growth conditions. ChIP DNA from each culture was amplified separately and fluorescently labeled with Cy5. Each independent sample was combined separately with sheared genomic reference DNA that had been amplified and fluorescently labeled with Cy3 and then cohybridized to whole-genome yeast microarrays that contained nearly every ORF as well as every intergenic sequence. We calculated the consistent enrichment value for each genomic locus across all the independent ChIP-chip experiments (see Materials and Methods).
To ensure that the sheared genomic DNA was an appropriate microarray reference, we explicitly compared it to two other kinds of reference samples in control hybridizations. Sheared genomic DNA was virtually identical to ChIP input DNA (Fig. 1A) and to ChIP-recovered DNA from a control strain lacking HA (Fig. 1B). Independent ChIP-chip experiments using sheared genomic DNA as a reference were also highly reproducible (Fig. 1C). Thus, sheared genomic DNA is an appropriate reference sample but has an advantage over the other kinds of reference samples in that the same genomic DNA reference can be used in many independent experiments, allowing quantitative cross comparisons to be made.
Figure 2 shows a bimodal distribution of enrichment in 49 biologically independent TBP ChIP-chip experiments, suggesting that there are specific genomic loci that TBP consistently and strongly associated with. Most of these loci are Pol III promoters, such as tRNAs, 5S rRNA, and U6 snRNA, or loci adjacent to Pol III promoters (Fig. 2). We considered 1 kb on either side of the Pol III transcripts to be adjacent loci, because TBP-associated fragments enriched after random shearing and ChIP may overlap and hybridize not only to the DNA segment containing the actual binding site but also to neighboring loci. In the top 6% of highly enriched loci, 598 of 619 segments (96.6%) were Pol III promoters or adjacent loci. Of the remaining 21 segments, 13 were next to transposon (Ty) long terminal repeats (Ty LTRs), 2 represented snoRNAs, and 6 loci represented other Pol II targets. RNA Pol III genes in S. cerevisiae consist of 299 tRNAs and at least 9 other genes, such as snR6, RPR1, SCR1, and six 5S rRNA genes. Of all the tRNA genes represented on the arrays, 92% (252 of 274 retained after data filtering) were in the top 6% of most highly enriched loci. Furthermore, eight of the nine non-tRNA Pol III targets were highly enriched. Most of the remaining tRNA genes which were not in the top 6% also had relatively high ChIP enrichment values (>88).
The strong and consistent enrichment of genomic loci adjacent to nearly every RNA Pol III promoter under all the different growth conditions we tested suggests that TBP has a very strong preference for binding to all of the RNA Pol III promoters in vivo. This pronounced bias in the chromosomal binding distribution of TBP is likely to be related to the high rate of transcription from RNA Pol III promoters (4). Similar conclusions were reached in other recent studies that have specifically profiled the genome-wide binding distribution of the Pol III transcription machinery (12, 33).
Steady-state transcript levels of coding genes throughout the genome are correlated with TBP binding.
The vast majority of promoters in the genome occur upstream of protein-coding genes and are transcribed by RNA Pol II. Precise regulation of the level of transcription from these promoters is critical for normal cellular functioning. We determined the relationship between the binding of TBP to promoters and the corresponding steady-state level of transcription. The binding distribution of TBP to promoters across the genome was measured as described earlier. To determine the relationship of TBP binding to steady-state mRNA levels, we simultaneously analyzed mRNAs from aliquots of the same culture harvested just prior to formaldehyde cross-linking, in conjunction with a genomic DNA reference probe. mRNA was reverse transcribed into cDNA, labeled with Cy5 dye, and hybridized to whole-genome microarrays together with amplified, Cy3-labeled, sheared genomic DNA. Thus, the Cy5/Cy3 ratio of each ORF spot reflects the steady-state level of the transcript corresponding to that gene. Since almost all Pol III promoters had a high occupancy rate by TBP under all the conditions we tested (Fig. 2), we excluded data from loci adjacent to Pol III target genes in this analysis.
We first examined the relationship between TBP-binding and transcript levels in exponentially growing yeast cells at room temperature in synthetic complete medium lacking uracil. We found that there is a marked tendency for genes that are expressed at high steady-state transcript levels to have high levels of TBP occupancy at their promoters (Fig. 3A). Conversely, genes with low steady-state transcript levels show correspondingly low levels of TBP binding at their promoters. The apparent noise and the outliers in this plot of transcript levels against ChIP enrichment arise largely from identifiable sources and are discussed in greater detail below. The correlation between TBP-binding and transcript levels was evident when we applied a moving-window average analysis to the set of data that linked fluorescence ratios from the ChIP experiments with the ratios from the cDNA hybridizations (Fig. 3A). As a control, the same analysis applied to the same data set, which had been scrambled to break the link between the ratios for a gene and its promoter, did not reveal any correlation between transcript levels and TBP enrichment (Fig. 3B). Under three other conditions, in cells that were subjected to heat shock for 10, 30, or 60 min, during which transcript levels for several hundred genes are known to be altered, we observed similar patterns of correlation between the TBP occupancy of promoters and the corresponding steady-state level of Pol II gene transcripts (Fig. 3C to H). The data sets in Fig. 3 together include about 61% of all genes in the genome. Importantly, the only genes that have been excluded are those failing to meet basic data quality criteria or those that are adjacent to Pol III promoters. This data set is thus an unbiased representation of the entire genome and reflects the global relationship of TBP binding and transcriptional activity.
Compact genome organization and mRNA half-life effects contribute to apparent noise.
Nearly one half of all yeast genes have promoters within an upstream intergenic region that is shared with another divergently transcribed gene. In our analysis, the TBP binding data from such a common intergenic region are assigned to both potential downstream genes. Moreover, the compact nature of the yeast genome can mean that ChIP enrichment of one locus because of TBP binding to a site within it can also appear as the enrichment of an unrelated promoter region a short distance away because of overlaps with randomly fragmented ChIP DNA (Fig. 4A).
To examine the possibility that this inherent ambiguity in linking the TBP binding data from many intergenic regions with downstream transcript levels could complicate our analysis of the relationship between TBP binding and expression levels shown in Fig. 3 and contribute to the apparent noise, we analyzed the relationship in distinct subsets of genes. First, we divided all genes into two sets, one containing only genes that do not share a promoter with other genes (Fig. 4B) and the other containing all the genes in divergently transcribed gene pairs (Fig. 4C). Hierarchical clustering was then used to group the rows of data so that loci where TBP binding and expression levels were not correlated could be easily identified.
The three prominent classes of genes for which TBP binding and expression levels are apparently not well correlated are indicated in Fig. 4B and C. Class a genes showed strong TBP binding but low transcript levels. Most of the intergenic loci in class a were either near Pol III genes but just outside our distance threshold of 1 kb or were near Ty LTRs or other strongly transcribed genes with an intervening short gene (Fig. 4A). Class b genes exhibited the reverse pattern of low TBP binding but high expression levels. Genes in this category had a higher than average mRNA half-life (36 min compared to 27 min overall based on the data of Wang et al. [40]; P = 0.014). Class c genes also exhibited strong TBP binding and low transcript levels, and these loci are not near Pol III promoters or Ty LTRs. However, this large group of apparently discordant loci appears only among the divergently transcribed genes (Fig. 4C). Class c thus represents the expected class of genes where the promoter of only one member of a divergently transcribed gene pair is strongly bound and regulated by TBP, but the same binding data (high Cy5/Cy3 ratio) from the ChIP-chip experiment gets assigned to the other member of the pair as well (Fig. 4A). Strikingly, with the exception of subset b above, there was no large class of genes and promoters with the reverse type of discrepancy (weak TBP binding coupled with strong transcription), suggesting that when one or both of a pair of divergent genes are strongly transcribed, recruitment of TBP to the promoter in the shared intergenic region is always necessary.
No other features of the genes, such as their length, biological function, presence of a TATA box, presence of other promoter motifs, or the type of activator that regulated them, were correlated with the subsets of genes described above. Since these subsets of genes with apparent discordance between TBP binding and expression levels are all included in the plots shown in Fig. 3, they contribute to the apparent noise in the data. Outside of these predictable subsets of genes, there are a few exceptions where TBP binding is not correlated with expression, but in the overwhelming majority of cases, strong expression levels correspond to strong TBP recruitment, and conversely, low transcript levels correspond to low TBP recruitment. Therefore, our results indicate that the extent of binding of TBP to Pol II promoters is generally proportional to the levels of transcription throughout the genome, suggesting that the recruitment of TBP is the main determinant of the strength of a promoter.
Global changes in mRNA levels are correlated with changes in TBP recruitment to promoters.
The yeast genome gets transcriptionally reprogrammed in response to a variety of environmental and growth signals. One example is the heat shock and other stress responses which result in the rapid induction or repression of hundreds of genes across the genome (9). We examined the relationship between changes in gene expression and changes in TBP recruitment to promoters under three different perturbations. ChIP DNA from unstressed exponentially growing cells was labeled with Cy3 and hybridized to microarrays together with Cy5-labeled ChIP DNA from cells that were either heat shocked for 10 min, treated with the DNA-damaging agent MMS, or grown to stationary phase. We measured transcriptional induction and repression in the same samples by hybridizing Cy3-labeled cDNA from the unstressed cells and Cy5-labeled cDNA from the corresponding stressed cells together onto whole-genome microarrays. The change in TBP ChIP enrichment at each locus was plotted along the x axis, and the change in the mRNA expression level of the corresponding gene was plotted along the y axis in a scatter plot (Fig. 5A, C, and E). We also analyzed the same data sets using a moving-window average analysis to better represent the overall trend of this relationship (Fig. 5B, D, and F). The gene-by-gene visualization of the data indicates that the correlation between change in TBP binding and transcriptional activation or repression is a robust phenomenon observed throughout the genome and is not an artifact of the moving-window average analysis (Fig. 5G, H, and I).
As shown in Fig. 5, strong changes in gene expression at the level of mRNA are generally well correlated with changes in TBP recruitment to their promoters. Heat shock causes rapid changes in gene expression levels, for instance, it strongly induces the stress response genes, such as SSE1, HSP10, and HSP82, while repressing ribosomal protein gene expression (9). As early as 10 min after heat shock, TBP recruitment to promoters is well correlated with the strong changes observed in gene expression (Fig. 5A, B, and G). This result is consistent with a model where the induction or repression of hundreds of genes across the genome in response to heat shock occurs as a consequence of alterations in TBP recruitment to their promoters. A similar relationship between changes in TBP recruitment and strong changes in gene expression was observed during the global response to stationary phase (Fig. 5C, D, and H) or MMS treatment (Fig. 5E, F, and I).
We examined several cases where there was an apparent lack of correlation between changes in TBP recruitment and changes in gene expression. In most cases, this apparent discrepancy could be attributed to genes with marked differences in regulation located in close proximity on the genome or to the binding of TBP to distinct core promoters within the same intergenic region (as depicted in Fig. 4A). Another factor contributing to the apparent noise is the fact that across the few growth conditions we examined, there were still several hundred genes whose expression levels did not change appreciably with the perturbation. We therefore expect there to be some fluctuation about the average for spots with relative mRNA expression and ChIP ratios close to 1, leading to some scatter around the central values. Notably, we did not observe any large classes of genes showing strong changes in gene expression levels but without a corresponding change in TBP recruitment to their promoters. Taken together, our results indicate that changes in TBP binding across the genome are well correlated with similar changes in mRNA expression levels in response to a number of physiological transitions and strongly support the idea that TBP is a universal rate-limiting determinant of gene activation and repression.
The TATA element is a minor determinant of TBP binding strength and Pol II transcriptional activity.
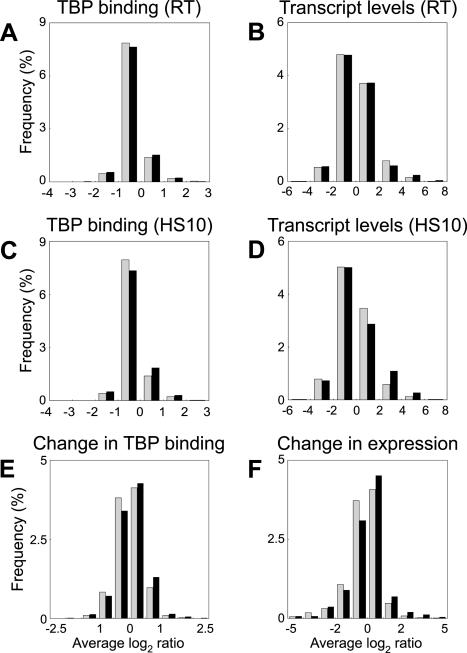
Although TBP is known to be required for the initiation of transcription from every RNA Pol II promoter, the quality of the core promoter can vary. Many promoters contain a consensus TATA element, but many others have a weak or no TATA element (25, 29). We examined the global relationship between the quality of the core promoter and the strength or responsiveness of the promoter. First, we partitioned all genes into two classes based on whether they contained or lacked a canonical TATA element (1) within 200 bp upstream of the start codon. We then examined whether genes in each class showed the same behavior with respect to TBP binding or transcriptional activity. In exponentially growing cells at room temperature, there was no observable difference in the tendency of TATA-containing and TATA-less genes to show extreme values for either TBP binding or steady-state transcript levels (Fig. 6). When yeast cells were heat shocked for 10 min, it appeared that TATA-containing genes were slightly more likely to show high steady-state transcript levels (Fig. 6D) and TBP binding (Fig. 6C). If we considered the change in expression or a change in TBP binding when cells were heat shocked (relative to normal cells), there was a small increase in the tendency of TATA-containing genes to be better bound by TBP (Fig. 6E) and to be transcriptionally induced (Fig. 6F). If we relaxed the definition of a TATA box to TATA(A/T)A, we did not observe even this modest difference in the behavior of TATA-containing versus TATA-less genes (data not shown).
FIG. 6.
Role of a canonical TATA element in determining the strength of TBP binding to promoters or Pol II gene expression. The distribution of values for TBP binding to its promoter or gene expression (log2 Cy5/Cy3 ratios) is shown. All genes were scored for the presence of a canonical TATA element within 200 bp upstream of the start codon. Data for genes containing the canonical TATA element in their promoters (black bars) and the genes lacking this TATA element (shaded bars) are plotted separately. The graphs show the relationship between the presence of a TATA element and TBP binding in exponentially growing yeast cells at room temperature (RT) (A), steady-state transcript levels in exponentially growing yeast cells at room temperature (B), TBP binding 10 min after heat shock (HS10) (C), steady-state transcript levels 10 min after heat shock (D), changes in TBP recruitment during a 10-min heat shock (E), and changes in gene expression levels during the 10-min heat shock (F).
Since the differences in the behavior of TATA-containing genes appeared to be restricted to induced genes, we examined this bias across all three conditions for which we had genome-wide data. Approximately 17% of all yeast genes have the canonical TATA element in their promoters. However, among the genes that were induced in the three different growth conditions, heat shock for 10 min, stationary phase, and DNA damage by MMS, 27, 27, and 32%, respectively, had the canonical TATA element, while the proportion of TATA-containing promoters among the genes that were repressed in these conditions did not vary from the overall value. Thus, although TBP binding is itself strongly correlated with both the transcript level and change in expression level, the quality of the TATA element of core promoters has only a modest effect in determining the extent of TBP binding or RNA Pol II transcriptional activity and may be more relevant in gene induction rather than repression.
DISCUSSION
We have combined genome-wide binding distribution assays and expression profiling methods to study the genomic role of TBP in vivo. The promoters of tRNA genes comprise the overwhelming majority of RNA Pol III promoters in the cell, and their strong and consistent occupancy by TBP likely reflects the high rate of transcription of tRNA genes. S. cerevisiae RNA Pol III has been shown to have high transcription efficiency in vitro due to facilitated recycling of the Pol on TFIIIB-DNA complexes that are stable through multiple rounds of initiation (4, 8). Strong binding of the promoter by TBP could maintain the stability of TFIIIB complexes and may explain the high transcription efficiency of yeast RNA Pol III. The 275 nuclear tRNA genes encoded in the genome comprise 39 distinct anticodon specificities which suffice to recognize all 61 codons (26). Multiple tRNA promoters from each of the specific tRNA classes have high TBP occupancy rates, regardless of their codon usage bias in protein-coding sequences in the genome. Thus, there appears to be no differential transcription of different tRNA genes, and our data support the notion that the various cellular levels of different isoacceptor tRNAs reflect their respective copy numbers in the genome (19). Interestingly, we observed a moderate decrease in the TBP occupancy of tRNA promoters after heat shock or DNA damage but a slight increase during stationary phase. Ribosomal protein gene transcription and protein synthesis are dramatically reduced during all three conditions. The reason for this apparent discrepancy between the transcription of tRNA genes and ribosomal protein genes is not clear, but it may reflect the fact that tRNA promoters are optimized for high levels of transcription and lack significant upstream control elements for specific regulation. Analysis by pattern-finding programs such as MEME or MDscan did not reveal any overrepresented motifs within or upstream of the Pol III genes that are strongly bound by TBP other than the previously established internal A and B or C boxes.
The genome-wide correlation between TBP occupancy of a promoter and transcriptional activity is striking in light of the potential diversity of mechanisms for regulating transcript levels and achieving activation or repression. Several transcription factors, such as Hsf1, Skn7, Msn2/Msn4, Crt1, Ssn6, Tup1, Cat8, Sip4, Mot1, Abf1, and Rap1, are each involved in the activation or repression of dozens of target genes during the growth conditions we have examined here (7, 13, 14, 22, 24, 30, 31, 34, 35, 37). One model for the mechanism for transcriptional activation in vivo is that different kinds of activator or repressor proteins affect different steps of the initiation process, such as TBP recruitment or a step after the TBP binding step, such as recruitment of the RNA Pol II holoenzyme complex, formation of the open complex, or transcription elongation, to maintain constitutive levels of transcription or achieve transcriptional activation or repression. According to this model, it is possible that some activators alter other steps but do not alter the recruitment of TBP to the promoter. If this were indeed the case, we would expect to see distinct classes of promoters for which TBP occupancy is not correlated with transcriptional activity. However, with the exception of known and predictable classes of genes with this apparent discrepancy arising due to compact genome organization (Fig. 4), we do not generally see such behavior. The results of our genome-wide binding study by itself do not distinguish whether TBP binding to the core promoter is a cause or effect of transcriptional activity. However, TBP recruitment to the core promoter is known to be an important prerequisite for transcriptional initiation. Therefore, our genome-wide data are consistent with a model where TBP recruitment is the dominant mechanism of achieving transcriptional control in vivo, regardless of the kind of activator or promoter that is being utilized. Our data do not exclude the possibility that transcription factors alter other steps of the initiation process in addition to altering TBP recruitment. Also, we do not distinguish between distinct mechanisms of affecting TBP recruitment, such as through direct protein-protein interactions of an activation domain with TBP or its associated factors (3, 10) or by recruiting other chromatin-modifying factors and altering local chromatin structure and indirectly affecting TBP occupancy of the promoter (15).
Given that mRNA stability is an important mode of regulating gene expression (2, 36), it is striking that steady-state RNA levels in the cell are predominantly and globally correlated to TBP binding, which may be considered an indicator of the rate of initiation. The steady-state level of a transcript is expected to depend on both the rate of initiation and the half-life of the message. However, our data indicate that with the exception of a small subset of messages with high mRNA half-lives, the steady-state levels of most transcripts are more strongly correlated with the rate of initiation. This could reflect the predilection of the cell to exercise control over gene expression at the earliest step, transcription initiation, rather than to transcribe messages and differentially regulate mRNA stability.
Our results indicate that yeast promoters lacking a canonical TATA element are capable of having a high TBP occupancy rate and initiating transcripts at levels comparable to those of promoters with canonical TATA boxes, although TATA-containing genes are somewhat more likely to show higher levels of TBP binding and induction (Fig. 6). Moreover, the TBP occupancy of the TATA-less promoters is equally capable of being modulated in response to physiological signals, suggesting that even at TATA-less promoters, activation and repression can be achieved through the modulation of TBP recruitment to the promoter. The slightly increased likelihood that we have observed for TATA-containing promoters to have a strong TBP occupancy rate or be highly expressed during heat shock is consistent with observations from independent recent studies (1). However, it is unclear whether this is a difference that truly affects only transcription induction as opposed to repression. We observed that TATA-less genes repressed by stress (e.g., ribosomal protein-coding genes) show reduction of TBP binding similar to TATA-containing genes when they are turned off; when cells return to favorable growth conditions, these TATA-less genes must conversely show increased TBP binding as they are turned on again. It is possible that TBP binds at these so-called TATA-less promoters to non-consensus elements. TBP-associated factors can promote the binding of TBP to TATA-less promoters, and it is possible that they have a greater relevance at TATA-less promoters (25).
Although the importance of TBP recruitment in the activation process has been studied previously for a small number of genes, our data and analysis represent the first detailed look at its global role in determining steady-state transcript levels and modulation of gene expression at the level of transcription. Our results are consistent with a model where transcriptional activation and repression in yeast involve TBP recruitment to the core promoter as the predominant, if not exclusive, mechanism for modulating transcript levels. This step of the initiation process appears to be of global importance across the genome, without regard to the nature of the activator, the quality of the core promoter, or the physiological state of the cell. Given the strong conservation of TBP and the rest of the general transcription machinery, it will be interesting to examine whether this situation prevails in other organisms as well.
Acknowledgments
We thank J. Davies and other members of the Iyer lab for all aspects of microarray production.
This work was supported in part by grants from the National Institutes of Health (AA13518 and CA95548) and from the Texas Higher Education Coordinating Board.
REFERENCES
- 1.Basehoar, A. D., S. J. Zanton, and B. F. Pugh. 2004. Identification and distinct regulation of yeast TATA box-containing genes. Cell 116:699-709. [DOI] [PubMed] [Google Scholar]
- 2.Beelman, C. A., and R. Parker. 1995. Degradation of mRNA in eukaryotes. Cell 81:179-183. [DOI] [PubMed] [Google Scholar]
- 3.Chen, X., G. Farmer, H. Zhu, R. Prywes, and C. Prives. 1993. Cooperative DNA binding of p53 with TFIID (TBP): a possible mechanism for transcriptional activation. Genes Dev. 7:1837-1849. [DOI] [PubMed] [Google Scholar]
- 4.Cloutier, T. E., M. D. Librizzi, A. K. Mollah, M. Brenowitz, and I. M. Willis. 2001. Kinetic trapping of DNA by transcription factor IIIB. Proc. Natl. Acad. Sci. USA 98:9581-9586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Comai, L., N. Tanese, and R. Tjian. 1992. The TATA-binding protein and associated factors are integral components of the RNA polymerase I transcription factor, SL1. Cell 68:965-976. [DOI] [PubMed] [Google Scholar]
- 6.Cormack, B. P., and K. Struhl. 1992. The TATA-binding protein is required for transcription by all three nuclear RNA polymerases in yeast cells. Cell 69:685-696. [DOI] [PubMed] [Google Scholar]
- 7.Dasgupta, A., R. P. Darst, K. J. Martin, C. A. Afshari, and D. T. Auble. 2002. Mot1 activates and represses transcription by direct, ATPase-dependent mechanisms. Proc. Natl. Acad. Sci. USA 99:2666-2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dieci, G., S. Giuliodori, M. Catellani, R. Percudani, and S. Ottonello. 2002. Intragenic promoter adaptation and facilitated RNA polymerase III recycling in the transcription of SCR1, the 7SL RNA gene of Saccharomyces cerevisiae. J. Biol. Chem. 277:6903-6914. [DOI] [PubMed] [Google Scholar]
- 9.Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen, G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodrich, J. A., T. Hoey, C. J. Thut, A. Admon, and R. Tjian. 1993. Drosophila TAFII40 interacts with both a VP16 activation domain and the basal transcription factor TFIIB. Cell 75:519-530. [DOI] [PubMed] [Google Scholar]
- 11.Greenblatt, J. 1991. Roles of TFIID in transcriptional initiation by RNA polymerase II. Cell 66:1067-1070. [DOI] [PubMed] [Google Scholar]
- 12.Harismendy, O., C. G. Gendrel, P. Soularue, X. Gidrol, A. Sentenac, M. Werner, and O. Lefebvre. 2003. Genome-wide location of yeast RNA polymerase III transcription machinery. EMBO J. 22:4738-4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haurie, V., M. Perrot, T. Mini, P. Jeno, F. Sagliocco, and H. Boucherie. 2001. The transcriptional activator Cat8p provides a major contribution to the reprogramming of carbon metabolism during the diauxic shift in Saccharomyces cerevisiae. J. Biol. Chem. 276:76-85. [DOI] [PubMed] [Google Scholar]
- 14.Huang, M., Z. Zhou, and S. J. Elledge. 1998. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell 94:595-605. [DOI] [PubMed] [Google Scholar]
- 15.Imbalzano, A. N., H. Kwon, M. R. Green, and R. E. Kingston. 1994. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature 370:481-485. [DOI] [PubMed] [Google Scholar]
- 16.Iyer, V. R. 2003. Isolation and amplification of array material from yeast, p. 30-34. In D. Bowtell and J. Sambrook (ed.), DNA microarrays: a molecular cloning manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 17.Iyer, V. R. 2003. Microarray-based detection of DNA protein interactions: chromatin immunoprecipitation on microarrays, p. 453-463. In D. Bowtell and J. Sambrook (ed.), DNA microarrays: a molecular cloning manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 18.Iyer, V. R., C. E. Horak, C. S. Scafe, D. Botstein, M. Snyder, and P. O. Brown. 2001. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature 409:533-538. [DOI] [PubMed] [Google Scholar]
- 19.Kanaya, S., Y. Yamada, Y. Kudo, and T. Ikemura. 1999. Studies of codon usage and tRNA genes of 18 unicellular organisms and quantification of Bacillus subtilis tRNAs: gene expression level and species-specific diversity of codon usage based on multivariate analysis. Gene 238:143-155. [DOI] [PubMed] [Google Scholar]
- 20.Killion, P. J., G. Sherlock, and V. R. Iyer. 20 August 2003, posting date. The Longhorn Array Database (LAD): an open-source, MIAME compliant implementation of the Stanford Microarray Database (SMD). BMC Bioinformatics 4:32. [Online.] http://www.biomedcentral.com/1471-2105/4/32. [DOI] [PMC free article] [PubMed]
- 21.Kuras, L., and K. Struhl. 1999. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature 399:609-613. [DOI] [PubMed] [Google Scholar]
- 22.Li, B., and J. C. Reese. 2000. Derepression of DNA damage-regulated genes requires yeast TAFIIs. EMBO J. 19:4091-4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, X. Y., A. Virbasius, X. Zhu, and M. R. Green. 1999. Enhancement of TBP binding by activators and general transcription factors. Nature 399:605-609. [DOI] [PubMed] [Google Scholar]
- 24.Lieb, J. D., X. Liu, D. Botstein, and P. O. Brown. 2001. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat. Genet. 28:327-334. [DOI] [PubMed] [Google Scholar]
- 25.Martinez, E., C. M. Chiang, H. Ge, and R. G. Roeder. 1994. TATA-binding protein-associated factor(s) in TFIID function through the initiator to direct basal transcription from a TATA-less class II promoter. EMBO J. 13:3115-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Percudani, R., A. Pavesi, and S. Ottonello. 1997. Transfer RNA gene redundancy and translational selection in Saccharomyces cerevisiae. J. Mol. Biol. 268:322-330. [DOI] [PubMed] [Google Scholar]
- 27.Pollack, J. R., and V. R. Iyer. 2002. Characterizing the physical genome. Nat. Genet. 32(Suppl.):515-521. [DOI] [PubMed] [Google Scholar]
- 28.Pugh, B. F. 2000. Control of gene expression through regulation of the TATA-binding protein. Gene 255:1-14. [DOI] [PubMed] [Google Scholar]
- 29.Pugh, B. F., and R. Tjian. 1991. Transcription from a TATA-less promoter requires a multisubunit TFIID complex. Genes Dev. 5:1935-1945. [DOI] [PubMed] [Google Scholar]
- 30.Raitt, D. C., A. L. Johnson, A. M. Erkine, K. Makino, B. Morgan, D. S. Gross, and L. H. Johnston. 2000. The Skn7 response regulator of Saccharomyces cerevisiae interacts with Hsf1 in vivo and is required for the induction of heat shock genes by oxidative stress. Mol. Biol. Cell 11:2335-2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reid, J. L., V. R. Iyer, P. O. Brown, and K. Struhl. 2000. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol. Cell 6:1297-1307. [DOI] [PubMed] [Google Scholar]
- 32.Rigby, P. W. 1993. Three in one and one in three: it all depends on TBP. Cell 72:7-10. [DOI] [PubMed] [Google Scholar]
- 33.Roberts, D. N., A. J. Stewart, J. T. Huff, and B. R. Cairns. 2003. The RNA polymerase III transcriptome revealed by genome-wide localization and activity-occupancy relationships. Proc. Natl. Acad. Sci. USA 100:14695-14700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roth, S., and H. J. Schuller. 2001. Cat8 and Sip4 mediate regulated transcriptional activation of the yeast malate dehydrogenase gene MDH2 by three carbon source-responsive promoter elements. Yeast 18:151-162. [DOI] [PubMed] [Google Scholar]
- 35.Schmitt, A. P., and K. McEntee. 1996. Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 93:5777-5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sheth, U., and R. Parker. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300:805-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sorger, P. K. 1990. Yeast heat shock factor contains separable transient and sustained response transcriptional activators. Cell 62:793-805. [DOI] [PubMed] [Google Scholar]
- 38.Spellman, P. T., G. Sherlock, M. Q. Zhang, V. R. Iyer, K. Anders, M. B. Eisen, P. O. Brown, D. Botstein, and B. Futcher. 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9:3273-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Struhl, K. 1994. Duality of TBP, the universal transcription factor. Science 263:1103-1104. [DOI] [PubMed] [Google Scholar]
- 40.Wang, Y., C. L. Liu, J. D. Storey, R. J. Tibshirani, D. Herschlag, and P. O. Brown. 2002. Precision and functional specificity in mRNA decay. Proc. Natl. Acad. Sci. USA 99:5860-5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.White, R. J., P. W. Rigby, and S. P. Jackson. 1992. The TATA-binding protein is a general transcription factor for RNA polymerase III. J. Cell Sci. Suppl. 16:1-7. [DOI] [PubMed] [Google Scholar]