Abstract
The cell cycle inhibitor p21 plays an important role in monocytic cell differentiation, during which it translocates from the nucleus to cytoplasm. This process involves the negative regulation of the p21 nuclear localization signal (NLS). Here, we sought to determine the relationship between the cytoplasmic translocation of p21 and another molecule, Brap2, a cytoplasmic protein which binds the NLS of BRCA1 and was recently reported to inactivate KSR in the Ras-activating signal pathway under the name of IMP. We report that p21 and Brap2 directly interact, both in vitro and in vivo, in a manner requiring the NLS of p21 and the C-terminal portion of Brap2. When it is cotransfected with Brap2, p21 is expressed in the cytoplasm. Monocytic differentiation of the promyelomonocytic cell lines U937 and HL60 is associated with the upregulation of Brap2 expression concomitantly with the upregulation and cytoplasmic relocalization of p21. Our results underscore the role played by Brap2 in the process of cytoplasmic translocation of p21 during monocyte differentiation.
The hormone 1,25-dihydroxyvitamin D3 (VD3) can induce differentiation of hematopoietic cell lines such as HL60 and U937 along a macrophage-monocyte pathway. In a search for VD3 target genes, the cell cycle inhibitor p21 and the homeobox gene product HoxA10 were identified as direct transcriptional targets of the VD3 receptor (15, 19). HoxA10 can directly bind to the p21 promoter, together with its trimeric partners PBX1 and MEIS1, and activate p21 transcription (5). It has been shown that VD3-induced monocytic differentiation is associated with the initial nuclear expression and subsequent cytoplasmic translocation of p21 (3). Furthermore, we have demonstrated that peripheral blood monocytes express p21 in the cytoplasm, which appears important for their survival and for specific function. Cytoplasmic p21 expression protects monocytes by preventing the induction of the activated mitogen-activated protein kinase pathway by reactive oxygen species. This protection is accomplished in part by binding to and inhibiting ASK1, which otherwise triggers cell death.
Several tumor suppressor genes, including BRCA1, encode nuclear proteins, the functions of which are critically dependent on their correct nuclear localization. BRCA1 is normally located in the nucleus and plays important roles in DNA damage monitoring and repair (20). The mechanisms regulating the nuclear localization of BRCA1 are prerequisite to its tumor suppressor activity, and their dysregulation may lead to cellular transformation. In contrast to normal breast epithelial cells, where BRCA1 is found in the nucleus, in many advanced breast cancer cells BRCA1 is mislocated to the cytoplasmic compartment (6). In an attempt to identify the underlying mechanism for BRCA1 mislocation, Li et al. searched for proteins that interacted with the nuclear localization signal (NLS) of BRCA1 and identified Brap2 (BRCA1-associated protein 2), which is predominantly localized to the cytoplasm (13). Subsequent studies, however, failed to show any direct link between Brap2 and the intracellular localization of BRCA1. Nonetheless, Brap2 is a unique cytoplasmic protein whose properties include the ability to bind the NLS motif and, as was recently reported, to inactivate KSR, a scaffold or adaptor protein that couples activated Raf to its substrate MEK (16).
During the investigation of nuclear p21 translocation to the cytoplasm, we found that Brap2 binds p21 and, moreover, that this binding is required for the cytoplasmic localization of p21.
MATERIALS AND METHODS
Plasmids.
Green fluorescent protein (GFP)-fused p21 expression vector was constructed by ligating PCR fragments of p21 with pEGFP-C2 vector (Clontech). Corresponding p21 fragments are as follows: nuclear export signal (NES)-NLS (amino acids [aa] 71 to 164), NES-dNLS (deletion of the NLS) (aa 71 to 140), dNES-NLS (aa 79 to 164), dNES-dNLS (aa 79 to 140), C-terminal NLS (aa 111 to 164), and C-terminal dNLS (aa 111 to 140). GFP-fused Brap2 expression vector was constructed by ligating full-length Brap2 amplified by PCR using differentiated U937 cDNA as a template. Myc-tagged Brap2 was constructed using pCMV-Tag1 vector (Stratagene). pCMV-Brap2 vector was also constructed using pcDNA3.1 (Invitrogen). For in vitro translation, Myc-tagged Brap2 was cloned into pBluescript-KS vector (Stratagene). C-terminal Myc-tagged full-length p21 and flag-tagged dNLS p21 (aa 1 to 140) were also constructed using PCR fragments as inserts. The glutathione S-transferase (GST) fusion system was used to generate fusion proteins. Several expression plasmids were constructed using the pGEX-5X vector (Pharmacia): GST-p21C (aa 87 to 164), GST-p21C dNLS (aa 87 to 140), and GST-p21 (aa 1 to 164). Expression of the fusion protein was induced by the addition of isopropyl-β-d-thiogalactopyranoside to a final concentration of 0.4 mM in an exponentially growing bacterial culture at 30°C.
Cell culture and transfection.
HeLa and HEK293 cells were cultured in minimum essential medium (Sigma) with 10% fetal bovine serum in 5% CO2 humidified atmosphere at 37°C. WEHI3B D+, HL60, and U937 cells were cultured in RPMI 1640 (GIBCO/BRL) with 10% fetal bovine serum in 5% CO2 humidified atmosphere at 37°C. HeLa and HEK293 cells were transfected by using Effectene (QIAGEN). WEHI3B D+ and U937 cells were transfected by electroporation.
In vitro differentiation of WEHI3B D+, HL60, and U937 cells.
Twenty-four hours after electroporation of WEHI3B D+ cells, medium was replaced with 1 mg of G418-containing medium/ml. Cells were seeded into 96-well plates. GFP signals were checked by fluorescence microscopy, and GFP-positive wells were maintained and expanded. Several wells with GFP-positive cells were selected, and cells were transferred to glass-bottomed culture dishes, to which tetradecanoyl phorbol acetate (TPA) and VD3 were then added at final concentrations of 50 ng/ml and 50 nM, respectively. Cells were then examined by confocal microscopy.
HL60 and U937 cells were induced to differentiate in the presence of 50 nM VD3. U937/CB6-p21 cells were induced to differentiate after the addition of zinc to the culture medium, as reported previously (2).
Confocal microscopy.
Nuclei were visualized by the addition of Hoechst 33258. Fluorescence images were recorded by a confocal laser-scanning microscope (LSM-GB200; Olympus).
Brap2 mRNA expression analysis.
Brap2 mRNA expression was analyzed by reverse transcriptase (RT)-PCR using the following primers: Brap2 5′ primer, 5′-ATGAGTGTGTCACTGGTTGTTATCC-3′; Brap2 3′ primer, 5′-TCAGGGATGTCTGTTGCTCTGA-3′.
In vitro binding assay.
Myc-tagged Brap2 was translated in vitro from the pBluescript-KS vector using a TNT-coupled transcription-translation system (Promega) in the presence of [35S]methionine (Dai-ichi Pharmaceuticals Inc.). Glutathione-Sepharose beads containing about 20 μg of GST or GST fusion proteins were preincubated with Tris-buffered saline buffer (25 mM Tris-HCl [pH 8.0], 120 mM NaCl, 10% bovine serum albumin) also containing protease inhibitors for 30 min at room temperature with rotation. The beads were then incubated with translated in vitro products in standard lysis buffer (100 mM NaCl, 50 mM Tris [pH 7.4], 5 mM EDTA, 0.5% Triton X-100, and protease inhibitors) for 1 h at room temperature with rotation. Complexes were washed extensively with standard lysis buffer, boiled in protein loading buffer, separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and detected with image analyzer Bas 2000 (Fuji-Xerox).
Antibody preparation.
The cDNA sequence of Brap2 corresponding to aa 423 to 570, amplified by PCR from RNA derived from differentiated U937 cells, was cloned in frame with a six-His tag sequence in the expression vector pTrc-His-B (Invitrogen). After transformation with the resulting pTrc-His-Brap2-C plasmid and 6 h of induction with 100 mM isopropyl-β-d-thiogalactopyranoside), bacteria were lysed in 6 M urea, sonicated, and centrifuged, and the recombinant protein was purified from the supernatant by Ni-nitrilotriacetic acid agarose (Amersham-Pharmacia) and 100 to 500 mM imidazole elution. Sera from rabbits immunized with the recombinant protein were affinity purified.
Western blotting and immunoprecipitation.
Cell lysates were prepared in a lysis buffer (150 mM NaCl, 1% Nonidet P-40, 0.1% SDS, 1% sodium deoxycholate, 5 mM EDTA, 10 mM Tris [pH 7.4]) containing protease inhibitors. Proteins were separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes (Millipore). Western blotting was performed with the following primary antibodies: anti-GFP polyclonal antibody (PAb) (Living Colors; Clontech), anti-p21 monoclonal antibody (MAb) (C24420; Transduction Laboratories), anti-Myc PAb (MBL), and anti-Flag MAb (M2; Sigma). Binding of the primary antibody was detected by using a commercial ECL kit (Amersham-Pharmacia). Immune complexes were immunoprecipitated from clarified cell lysates with mouse immunoglobulin G (IgG) M280 magnetic beads (Dynal) preincubated with antibody to p21. The beads were washed extensively with washing buffer (150 mM NaCl, 10 mM Tris [pH 7.4], 0.5% Triton X-100, 5 mM EDTA), boiled in loading buffer, separated by SDS-PAGE, and subjected to Western blot analysis.
RNA interference.
Double-stranded RNAs were produced by in vitro transcription using the Silencer siRNA construction kit (Ambion) according to the manufacturer's instructions. We prepared siRNA targeting human Brap2; the target sequence was 5′-AACCAATATATGGTGCTGATA-3′. The control siRNA sequence was 5′-AACCAATGGTATATGCTGATA-3′. U937 cells were transfected with 400 pmol of double-stranded siRNA using GenomOne HVJ envelope vector kit (Ishihara Sangyo Kaisha) according to the manufacturer's instructions.
Fluorescence-activated cell sorter analysis.
Aliquots of cells were stained with fluorescein isothiocyanate-conjugated anti-CD14 (Nichirei). For the cell survival assay, aliquots of cells were stained with rhodamine 123 and merocyanine 540 (Molecular Probes). Cells that showed a low level of staining with rhodamine 123 and a high level of staining with merocyanine 540 were counted for dead cells. The analysis was performed with a FACSCalibur cytometer (Becton Dickinson).
RESULTS
Localization of p21 is determined by NLS.
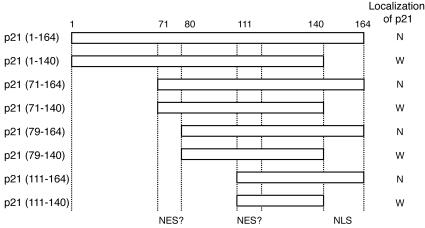
p21 contains several functional regions, including cyclin-binding domain 1 (aa 21 to 26), cyclin-binding domain 2 (aa 153 to 159), Cdk-binding domain (aa 49 to 71), PCNA-binding domain (aa 141 to 160), and NLS (aa 140 to 159). We also found that p21 carries NES-like sequences (aa 71 to 79 and 111 to 120). We constructed several GFP-p21 deletion mutant-fusion vectors as depicted in Fig. 1. The introduction of these expression vectors into HeLa cells revealed that p21 localized exclusively to the nucleus in the presence of NLS. In the absence of NLS, GFP signals were detected in all cell compartments.
FIG. 1.
Subcellular localization of p21 is determined by its nuclear localization signal. Predicted nuclear export-like signals are located at aa 71 to 79 and 111 to 119. NLS is located at aa 141 to 164. Fragments from p21 depicted were C terminally fused with GFP in pEGFP plasmids. These vectors were introduced into HeLa cells, and the subcellular localization of GFP signals was determined. N, nucleus; W, whole-cell compartment.
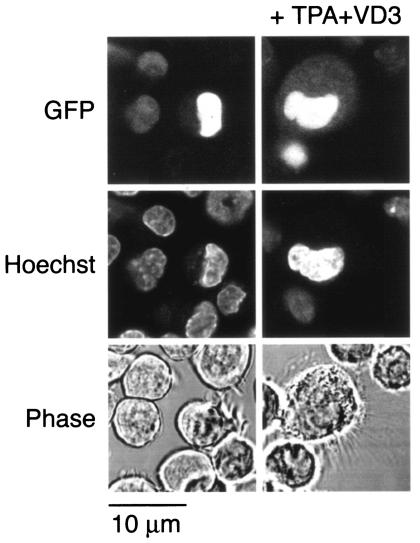
Because changes in the subcellular localization of p21 were observed in a monocyte differentiation system, we took advantage of the murine promonocytic cell line WEHI3B D+, which has been demonstrated to differentiate to monocytic cells associated with characteristic morphological change in the presence of TPA plus VD3 (4). These cells were treated with TPA plus VD3 to determine the effect of differentiation on the behavior of the GFP-p21 fusion proteins. To this end, the GFP-p21 fusion vectors depicted in Fig. 1 were electroporated into WEHI3B D+ cells, and after incubation with TPA, the subcellular localization of GFP signals was analyzed by confocal microscopy. In the presence of NLS, GFP signals were seen exclusively in the nucleus before treatment with TPA plus VD3. However, after treatment with TPA plus VD3, GFP signals appeared in the cytoplasm associated with the morphological change of WEHI3B D+ (Fig. 2, top panel). These results suggest that the negative regulation of NLS is involved in the determination of the subcellular location of p21 during monocytic differentiation. In the absence of NLS, GFP signals were detected in both the nucleus and the cytoplasm. TPA plus VD3 induced no significant difference in GFP signals in cells transfected with the constructs without NLS.
FIG. 2.
Cytoplasmic expression of p21 depends on NLS in monocytic differentiation. GFP signals in cells transfected with GFP-tagged p21-NLS constructs are shown. WEHI3B D+ cells were transfected with a construct containing GFP fused with p21 (aa 80 to 164). GFP-positive cells were selected and cultured with 50 ng of TPA/ml and 50 nM VD3 and then examined by confocal microscopy. Photos in the left and right columns show transfected cells before and after culture with TPA plus VD3, respectively. Monocytic differentiation induction resulted in the cytoplasmic expression of GFP signal in cells transfected with p21-NLS. TPA plus VD3 induced no difference of GFP signal in cells transfected with the p21 construct without NLS. Top, middle, and bottom panels show GFP signals, nuclear staining with Hoechst 33258, and a phase-contrast view of the corresponding fields, respectively.
Brap2 binds to NLS of p21 and functions as a cytoplasmic retention protein.
We could not detect any mobility shift of p21 protein in SDS-PAGE and subsequent Western blot analyses before or after monocytic differentiation of U937 cells. This result renders modifications of p21, such as phosphorylation, unlikely.
Brap2 is a unique cytoplasmic protein isolated as the NLS-interacting protein of BRCA1 (13) (data not shown). Brap2 has been characterized as binding to several NLSs, including bipartite types. We therefore assessed whether Brap2 interacts with p21 protein by using an in vitro GST pull-down assay. An in vitro-translated full-length Brap2 carrying a c-Myc epitope was incubated with GST alone, GST-p21-dNLS-C (aa 87 to 140), GST-p21-C (aa 87 to 164), or GST-p21 (aa 2 to 164). As shown in Fig. 3A, Brap2 binds to GST-p21-C and GST-p21 but not to GST alone or GST-p21-dNLS-C. These results suggest that Brap2 directly binds p21 through its NLS motif. To further confirm this interaction in vivo, cotransfection immunoprecipitation experiments were done using HEK293 cells. GFP-Brap2 vectors were cotransfected with C-terminally Myc-tagged p21 (aa 1 to 164) or Flag-tagged dNLS-p21 (aa 1 to 140). As shown in Fig. 3B and C, Brap2 binds full-length p21 but not dNLS-p21. HEK293 cells express a C-terminal truncated form of p21 (17), which no longer interacts with Brap2 (Fig. 3C and data not shown). These results support our finding that Brap2 binds to p21 through NLS.
FIG. 3.
Brap2 interacts with p21 in vitro and in vivo. (A) Brap2 was translated in vitro in the presence of [35S]methionine and was incubated with glutathione agarose coated with GST alone (lane 2), GST-p21 (aa 87 to 140) (lane 3), GST-p21 (aa 87 to 164) (lane 4), or GST-p21 (aa 2 to 164) (lane 5). Boiled beads were electrophoresed. The input product is shown in lane 1. The top band (about 60 kDa) represents Brap2. Asterisks indicate degradation products of Brap2. (B and C) HEK293 cells were transiently cotransfected with pEGFP-Brap2 and pCMV-p21-Myc vectors (lanes 2, 4, 6, and 8), cotransfected with pEGFP and pCMV-p21-Myc (lanes 1 and 3), or cotransfected with pEGFP-Brap2 and pCMV-dNLS-p21 (lanes 5 and 7). Cell lysate from each sample was immunoprecipitated (IP) with anti-p21 MAb. WB, Western blotted with. (B) p21 interacts with GFP-tagged Brap2 but not GFP alone. The expression of GFP-fused Brap2 in whole lysates (lanes 1 and 2) and p21 immune complexes (lanes 3 and 4) is demonstrated by anti-GFP PAb (upper panel). The expression of C-terminally Myc-fused p21 in whole lysates (lanes 1 and 2) and p21 immune complexes (lanes 3 and 4) is demonstrated by anti-Myc PAb (lower panel). The heavy and light chains of the precipitating antibody (IgH and IgL, respectively) are indicated. (C) Brap2 interacts with NLS of p21. The expression of GFP-fused Brap2 in whole lysates (lanes 5 and 6) and p21 immune complexes (lanes 7 and 8) is demonstrated by anti-GFP PAb (upper panel). The expression of p21 and dNLS-p21 in whole lysate (lanes 5 and 6) and p21 immune complexes (lanes 7 and 8) is demonstrated by anti-p21 MAb (lower panel). The heavy and light chains of the precipitating antibody (IgH and IgL) are indicated.
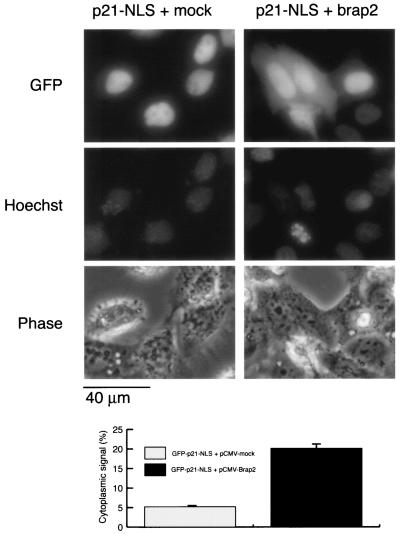
We then tested the hypothesis that p21 NLS function could be blocked by interaction with Brap2. GFP signals were found exclusively in the nucleus when the GFP-p21-NLS (aa 80 to 164) expression vector was transfected with a mock vector, pCMV-Tag1, into HeLa cells. In contrast, a GFP signal was detected in cytoplasm when the GFP-p21-NLS vector was cotransfected with the Brap2 expression vector, pCMV-Tag1/Brap2 (Fig. 4). Total GFP-positive cells made up about 20% and 10% of the cells in cotransfectants with mock vector and Brap2, respectively. About 20% of GFP-positive cells showed cytoplasmic GFP signals in the cotransfectants with pCMV-Tag1/Brap2, while only about 5% showed weak cytoplasmic GFP signals in the cotransfectants with mock vector.
FIG. 4.
Brap2 retains p21 in cytoplasm. HeLa cells were cotransfected with GFP-p21-NLS vector with mock or Brap2 vector. GFP signals were detected by fluorescence microscopy. Photographs in the left columns show cells transfected with mock vector, and those in the right columns show cells transfected with Brap2. Top panels show GFP signals, middle panels show DNA stained with Hoechst 33258, and bottom panels show a phase-contrast view of the corresponding fields. The bar graph shows the mean percentage of cells with GFP signals in the cytoplasm ± standard deviation.
C-terminal site of Brap2 binds to NLS of p21.
Next we mapped the p21 binding site on Brap2. The latter possesses C2H2 zinc fingers in the middle and leucine heptad repeats in the C terminus. Three truncation mutants of Brap2, designated Brap2-N (aa 2 to 187) and Brap2-M (aa 168 to 420), which include zinc fingers, and Brap2-C (aa 406 to 600), which includes leucine heptad repeats, were expressed as GFP-tagged fusions. Subcellular localization of GFP signals with Brap2-N and Brap2-M were detected in the whole-cell compartment, but Brap2-C was detected exclusively in cytoplasm (data not shown). These vectors together with Myc-tagged p21 were transfected into HEK293 cells, and their associations were analyzed with a coimmunoprecipitation assay. Only the C-terminal domain of Brap2 was able to bind to p21 (Fig. 5).
FIG. 5.
p21 interaction domain on Brap2. pEGFP vectors containing each of the deletion mutant constructs of Brap2, including Brap2 (F; aa 2 to 600), Brap2-N (N; aa 2 to 187), Brap2-M (M; aa 168 to 420), and Brap2-C (C; aa 406 to 600) were transiently cotransfected with pCMV-p21-Myc into HEK293 cells. Cell lysates from each sample were immunoprecipitated (IP) by anti-Myc PAb. The left panel shows the expression of each of the GFP-fused Brap2 deletion mutants demonstrated by anti-GFP PAb (upper panel) and the expression of p21-Myc by anti-Myc PAb (lower panel) in whole lysate. The right panel shows the interaction of Myc-tagged p21 and the deletion mutant of Brap2 in cells. The immune complex of Myc-tagged p21 was analyzed by Western blotting (WB) with anti-GFP PAb.
Upregulation of Brap2 expression during monocytic differentiation.
When we examined the expression of Brap2 in U937 and HL60 cells, we found a modest increase (about 1.5- and 2-fold, respectively) of mRNA levels following VD3 treatment for 3 days. At this stage, 58% of U937 cells and 62% of HL60 cells were CD14 positive. Under the same conditions, p21 mRNA levels significantly increased (2, 10, 22) (Fig. 6).
FIG. 6.
Expression of Brap2 mRNA is increased during monocytic differentiation of U937 cells. Brap2 expression was analyzed by RT-PCR before and after monocytic differentiation induction of U937 cells (top gel). RNA was extracted from U937 or HL60 cells cultured with or without VD3 for 3 days. p21 expression was induced by VD3 in U937 and HL60 cells, as demonstrated by RT-PCR (middle gel). Actin mRNA was amplified by RT-PCR and shown as an internal control (bottom gel). The bar graph shows the increase (n-fold) of VD3-induced Brap2 transcript standardized to samples without VD3 treatment. The percentage of cells that were CD14 positive is shown below the bars.
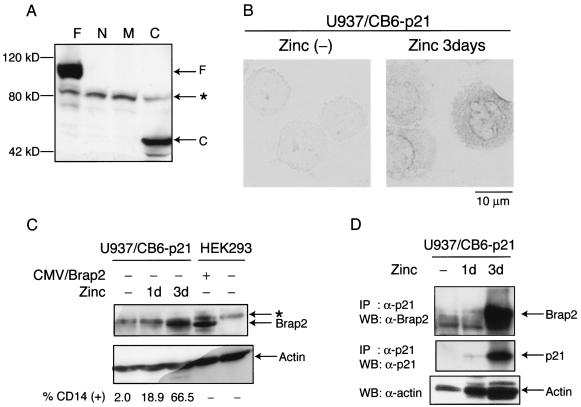
To examine the expression of Brap2 at the protein level, we raised an antibody against the C-terminal region (aa 423 to 570) of human Brap2, which recognizes full-length Brap2 and Brap2-C but not Brap2-N or Brap2-M (Fig. 7A). To assess differentiation-associated expression of Brap2, U937/CB6-p21 and U937/CB6-mock cells were treated with zinc. In this system, the addition of zinc leads to p21 expression, and zinc treatment for 3 days induces monocyte differentiation only in U937/CB6-p21 cells (2). As shown by immunocytochemical staining, zinc treatment induced Brap2 expression only in U937/CB6-p21 but not U937/CB6 cells with mock protein (U937/CB6-mock) (Fig. 7B; data not shown for U937/CB6-mock). U937/CB6-p21 cells treated with zinc for 3 days showed significant induction of Brap2 expression and differentiation (Fig. 7C). Furthermore, induced Brap2 was coimmunoprecipitated with induced p21 (Fig. 7D).
FIG. 7.
Expression of Brap2 protein is increased during monocytic differentiation of U937 cells. (A) Anti-Brap2 antibody was generated against the C-terminal portion (aa 423 to 570) of Brap2. This antibody recognizes F or C fragments of the Brap2 fusion protein. Aliquots of the lysates used in the procedures described in the legend to Fig. 5 were subjected to Western blotting with anti-Brap2 antibody. The asterisk indicates a nonspecific band. (B) Immunohistological expression of Brap2 in zinc-treated U937/CB6-p21 cells. (C) Brap2 expression is increased during monocyte differentiation. Cell lysates from U937/CB6-p21 cells treated with 120 μM ZnSO4 for 1 day or 3 days were extracted. As a positive control, HEK293 cells were transfected with pCMV/Brap2. Cell lysates were subjected to Western blotting with anti-Brap2 antibody. The percentage of cells that were CD14 positive is shown at the bottom. (D) Brap2 was coimmunoprecipitated with p21 in differentiating U937/CB6-p21 cells. Zinc-treated U937/CB6-p21 cells were lysed and immunoprecipitated (IP) with anti-p21 antibody. Immune complexes were electrophoresed and subjected to Western blotting (WB) with anti-Brap2 or anti-p21 antibody (top and middle panels). The loading control was subjected to Western blotting with antiactin antibody (bottom panel).
siRNA against Brap2 does not inhibit differentiation but inhibits apoptosis resistance.
To address the significance of Brap2 expression for cytoplasmic p21 localization, we set up small interfering RNAs (siRNAs) targeting Brap2 (9). Brap2-targeting siRNA but not control siRNA significantly reduced GFP-Brap2 expression without any reduction of control GFP expression (unpublished data). U937/CB6-p21 cells were transfected with an siRNA targeting Brap2 or control siRNA, and then p21 expression was induced by zinc. After 3 days of incubation with Zn, we analyzed monocytic differentiation by examining CD14 expression, p21 subcellular localization, and cell viability after hydrogen peroxide treatment. Cells treated with any siRNAs showed no significant change in CD14 expression. Brap2-targeting siRNA-treated cells showed considerable reduction of cytoplasmic p21 expression compared to control siRNA-treated cells concomitantly with decreased apoptosis resistance to hydrogen peroxide (Fig. 8).
FIG. 8.
Inhibition of Brap2 by siRNA does not inhibit p21 expression but reduces cytoprotective activity of cytoplasmic p21. (A) Percentage of cytoplasmic p21-expressing cells in U937/CB6-p21 cells treated with zinc for 3 days in the presence of siRNA. U937/CB6-p21 cells were transfected with control or Brap2-targeting siRNA and then treated with 120 μM ZnSO4 for 3 days. The results from one of three experiments are shown. (B) Brap2 expression was reduced in the presence of siRNA. Cell lysates from U937/CB6-p21 cells treated with zinc for 3 days in the presence of siRNA were extracted. Cell lysates were immunoprecipitated (IP) with anti-Brap2 antibody and subjected to Western blotting (WB) with anti-Brap2. Whole-cell lysates were blotted with anti-p21 antibody, demonstrating significant expression induction of p21 in both transfectants. The percentage of cells that were CD14 positive is shown under the blot. The asterisk indicates a nonspecific band. (C) Inhibition of Brap2 expression by siRNA reduces resistance against hydrogen peroxide-induced apoptosis of differentiated U937/CB6-p21 cells. U937/CB6-p21 cells were transfected with either control or Brap2-targeting siRNA and were cultured with Zn for 3 days. Cells were treated with 300 μM hydrogen peroxide for 16 h. Dead cells were scored by fluorescence-activated cell sorting analysis. The results from one of three experiments are shown.
DISCUSSION
The NLS and NES system is one of the major mechanisms for active transport of molecules into and/or out of the nucleus. This system is regulated by shuttling carriers, such as importin and exportin complexes in cooperation with the Ran-GTP and GTP hydrolysis system (11). Subcellular localization of proteins is determined not only by identifying the presence of NLS or NES sequences but also by altering the accessibility of these regions, for example, as a result of cell signaling. NF-κB NLS is masked by interaction with IκB, which is degraded by IκB kinase activated by proinflammatory cytokines. NF-AT resides in the cytoplasm in unstimulated T lymphocytes. NF-AT NLS in resting T cells is dephosphorylated in response to stimulation, and this causes its nuclear translocation (8). Similar phosphorylation- or dephosphorylation-dependent modification of NLS and NES are involved in modifications of cyclin B and cyclin D during cell cycle progression (1, 24). Thus, the subcellular localization of NLS- and/or NES-bearing proteins is controlled by the accessibility of their NLS and/or NES to the transport machinery.
During monocyte differentiation, the cell cycle inhibitory protein p21 translocates from the nucleus to the cytoplasm (3). Interestingly, whereas in the nucleus p21 functions as a cell cycle brake by binding to multicyclin/Cdk complexes and PCNA-DNA polymerase δ subunit (21), in the cytoplasm p21 can promote the assembly of cyclin D/Cdk4 complexes and their nuclear translocation (12). This function is reportedly essential for cyclin D/Cdk4 activation (7). It was previously reported that in monocytes, p21 is expressed in the cytoplasm, where it acts as an inhibitor of apoptosis (3). Another novel function of cytoplasmic p21 associated with increased neurite outgrowth in developing neurons has been recently reported (23). Thus, cytoplasmic p21 exerts biological effects distinct from those of nuclear p21, and it is important to determine the regulatory mechanisms responsible for its cytoplasmic or nuclear expression.
Because p21 is essentially a nuclear protein, it is translated in the cytoplasm and then translocates into the nucleus by means of the NLS. This is not the case in differentiated monocytes where p21 is expressed in cytoplasm. There are at least two possible mechanisms for cytoplasmic expression of p21, one being cytoplasmic retention of p21, i.e., inhibition of its nuclear translocation, and the other being nuclear export of p21. We failed to show involvement of p21 nuclear export in monocytic differentiation because of the lack of appropriate experimental designs. Therefore, the involvement of monocyte-specific nuclear export of p21 cannot be ruled out completely. In the present study, however, we demonstrated that the inhibition of p21 nuclear transport by Brap2 is a possible mechanism for cytoplasmic p21 expression.
Brap2 was shown to be a unique cytoplasmic protein with the ability to bind to both simple and bipartite NLS. Furthermore, Brap2 reportedly possesses a higher NLS binding affinity than importin α (13). This characteristic supports the hypothesis that Brap2 can function as a cytoplasmic retention protein for NLS-bearing molecules by interfering with their interaction with importin α. To our knowledge, p21 is the first partner of Brap2 demonstrated to physically interact with it, leading to the retention of p21 in cytoplasm. It should also be noted that Brap2 expression is upregulated in a differentiation-associated manner in U937 and HL60 cells, thus indicating a coordinated Brap2 expression is upregulated in a differentiation-associated manner in U937 and HL60 cells, thus indicating a coordinated expression of p21 and Brap2 during monocyte differentiation. Furthermore, treatment with Brap2-targeting siRNA reduced cytoplasmic p21 expression concomitantly with reduction of apoptosis resistance.
In coexpression studies of GFP-p21-NLS in HeLa cells, the transfection efficiency with Brap2 appeared significantly lower than that of mock vector. This lower efficiency is probably because the overexpression of Brap2 is toxic to cells; this toxicity might arise from the fact that Brap2 could target nuclear proteins, whose physiological function is impaired when they are retained in the cytoplasm, thus inhibiting cell survival and/or cell growth. Alternatively, as has been reported recently, Brap2 (IMP) inactivates KSR, a scaffold or adaptor protein that couples activated Raf to its substrate MEK (16), and thus the overexpression of Brap2 may inhibit cell proliferation through inactivating Ras activation signals.
Recently, a mechanism in breast cancer cells for cytoplasmic p21 expression involving the Akt system has been reported (25). Akt phosphorylates at Thr-145 in the NLS of p21 and inhibits nuclear translocation. Breast cancer cells overexpressing HER-2/neu, which activates Akt, could escape from p21-induced cell cycle arrest and acquire apoptosis resistance. It is hypothesized that this represents one of the mechanisms for clonal growth of cancer cells, though it is still controversial in the light of the findings of others (14, 18). Monocyte differentiation is associated with cell cycle arrest, which, in contrast to cell growth systems, is usually examined. In the system using VD3, we did not detect any phosphorylated active form of Akt (data not shown), though some monocyte differentiation signals such as those induced by TPA could activate Akt (data not shown). These findings make it unlikely that Akt signaling is involved in our system.
Binding to apoptosis signal-regulating kinase 1 (ASK1) exerts at least the cytoprotective ability of p21. ASK1 binds p21 at aa 1 to 140 (3), and Brap2 binds at aa 140 to 164. Thus, ASK1 and Brap2 bind p21 at close but distinct domains. As ASK1 is a cytoplasmic protein, one might speculate that it acts as a cytoplasmic retention protein for p21. However, this seems unlikely for the following reasons: first, the level of ASK1 expression does not change during monocyte differentiation (data not shown), and second, at an early stage of monocyte differentiation (3), p21 expression was detected in the nucleus even in the presence of ASK1. Thus, we hypothesize that during monocyte differentiation, which depends on p21 expression (2), p21 and Brap2 expression are concomitantly induced and the expressed Brap2 protects the NLS of p21, allowing p21 to remain in the cytoplasm and subsequently bind to ASK1.
There may be many other Brap2-like proteins, localizing to cytoplasm and interacting with NLS motifs. Thus, cytoplasmic retention proteins may comprise a unique functional family. They function as inhibitors and/or sequestering factors for nuclear proteins but mediate novel biological functions of the nuclear proteins in the cytoplasm.
Acknowledgments
This work was supported by a Grant-in-Aid for Cancer Research from the Ministry of Health and Welfare, Japan, as part of a comprehensive 10-year strategy for cancer control; by a Grant-in-Aid from the Ministry of Education, Science and Culture, Japan; by a grant from the Japan Leukemia Research Fund; and by the Italian Association for Cancer Research (AIRC) and Telethon grants E.337 and E764.
REFERENCES
- 1.Alt, J. R., J. L. Cleveland, M. Hannink, and J. A. Diehl. 2000. Phosphorylation-dependent regulation of cyclin D1 nuclear export and cyclin D1-dependent cellular transformation. Genes Dev. 14:3102-3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asada, M., T. Yamada, K. Fukumuro, and S. Mizutani. 1998. p21Cip1/WAF1 is important for differentiation and survival of U937 cells. Leukemia 12:1944-1950. [DOI] [PubMed] [Google Scholar]
- 3.Asada, M., T. Yamada, H. Ichijo, D. Delia, K. Miyazono, K. Fukumuro, and S. Mizutani. 1999. Apoptosis inhibitory activity of cytoplasmic p21(Cip1/WAF1) in monocytic differentiation. EMBO J. 18:1223-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bettens, F., E. Schlick, W. Farrar, and F. Ruscetti. 1986. 1,25-Dihydroxycholecalciferol-induced differentiation of myelomonocytic leukemic cells unresponsive to colony stimulating factors and phorbol esters. J. Cell. Physiol. 129:295-302. [DOI] [PubMed] [Google Scholar]
- 5.Bromleigh, V. C., and L. P. Freedman. 2000. p21 is a transcriptional target of HOXA10 in differentiating myelomonocytic cells. Genes Dev. 14:2581-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, Y., C.-F. Chen, D. J. Riley, D. C. Allred, P.-L. Chen, D. Von Hoff, C. K. Osborne, and W.-H. Lee. 1995. Aberrant subcellular localization of BRCA1 in breast cancer. Science 270:789-791. [DOI] [PubMed] [Google Scholar]
- 7.Cheng, M., P. Oliver, J. A. Diehl, M. Fero, M. F. Roussel, J. M. Roberts, and C. J. Sherr. 1999. The p21 Cip1 and p27 Kip1 CDK ‘inhibitors’ are essential activators of cyclin D-dependent kinases in murine fibroblasts. EMBO J. 18:1571-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cyert, M. S. 2001. Regulation of nuclear localization during signaling. J. Biol. Chem. 276:20805-20808. [DOI] [PubMed] [Google Scholar]
- 9.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 10.Jiang, H., J. Lin, Z.-Z. Su, F. R. Collart, E. Huberman, and P. B. Fisher. 1994. Induction of differentiation in human promyelocytic HL-60 leukemia cells activates p21, WAF1/CIP1, expression in the absence of p53. Oncogene 9:3397-3406. [PubMed] [Google Scholar]
- 11.Kuersten, S., M. Ohno, and I. W. Mattaj. 2001. Nucleocytoplasmic transport: Ran, beta and beyond. Trends Cell Biol. 11:497-503. [DOI] [PubMed] [Google Scholar]
- 12.LaBaer, J., M. D. Garrett, L. F. Stevenson, J. M. Slingerland, C. Sandhu, H. S. Chou, A. Fattaey, and E. Harlow. 1997. New functional activities for the p21 family of CDK inhibitors. Genes Dev. 11:847-862. [DOI] [PubMed] [Google Scholar]
- 13.Li, S., C. Y. Ku, A. A. Farmer, Y. S. Cong, C. F. Chen, and W. H. Lee. 1998. Identification of a novel cytoplasmic protein that specifically binds to nuclear localization signal motifs. J. Biol. Chem. 273:6183-6189. [DOI] [PubMed] [Google Scholar]
- 14.Li, Y., D. Dowbenko, and L. A. Lasky. 2002. AKT/PKB phosphorylation of p21Cip/WAF1 enhances protein stability of p21Cip/WAF1 and promotes cell survival. J. Biol. Chem. 277:11352-11361. [DOI] [PubMed] [Google Scholar]
- 15.Liu, M., M. H. Lee, M. Bommakanti, and L. P. Freedman. 1996. Transcriptional activation of the Cdk inhibitor p21 by vitamin D3 leads to the differentiation of the myelomonocytic cell line U937. Genes Dev. 10:142-153. [DOI] [PubMed] [Google Scholar]
- 16.Matheny, S. A., C. Chen, R. L. Kortum, G. L. Razidlo, R. E. Lewis, and M. A. White. 2004. Ras regulates assembly of mitogenic signalling complexes through the effector protein IMP. Nature 427:256-260. [DOI] [PubMed] [Google Scholar]
- 17.Poon, R. Y. C., and T. Hunter. 1998. Expression of a novel form of p21Cip1/Waf1 in UV-irradiated and transformed cells. Oncogene 16:1333-1343. [DOI] [PubMed] [Google Scholar]
- 18.Rössig, L., A. S. Jadidi, C. Urbich, C. Badorff, A. M. Zeiher, and S. Dimmeler. 2001. Akt-dependent phosphorylation of p21Cip1 regulates PCNA binding and proliferation of endothelial cells. Mol. Cell. Biol. 21:5644-5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rots, N. Y., M. Liu, E. C. Anderson, and L. P. Freedman. 1998. A differential screen for ligand-regulated genes: identification of HoxA10 as a target of vitamin D3 induction in myeloid leukemic cells. Mol. Cell. Biol. 18:1911-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scully, R., and D. M. Livingston. 2000. In search of the tumour-suppressor functions of BRCA1 and BRCA2. Nature 408:429-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sherr, C. J., and J. M. Roberts. 1995. Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 9:1149-1163. [DOI] [PubMed] [Google Scholar]
- 22.Steinman, R. A., B. Hoffman, A. Iro, C. Guillouf, D. A. Liebermann, and M. E. El-Houseini. 1994. Induction of p21(WAF1/CIP1) during differentiation. Oncogene 9:3389-3396. [PubMed] [Google Scholar]
- 23.Tanaka, H., T. Yamashita, M. Asada, S. Mizutani, H. Yoshikawa, and M. Tohyama. 2002. Cytoplasmic p21Cip1/WAF1 regulates neurite remodeling by inhibiting Rho-kinase activity. J. Cell Biol. 158:321-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toyoshima-Morimoto, F., E. Taniguchi, N. Shinya, A. Iwamatsu, and E. Nishida. 2001. Polo-like kinase 1 phosphorylates cyclin B1 and targets it to the nucleus during prophase. Nature 410:215-220. [DOI] [PubMed] [Google Scholar]
- 25.Zhou, B. P., Y. Liao, W. Xia, B. Spohn, M. H. Lee, and M. C. Hung. 2001. Cytoplasmic localization of p21Cip1/WAF1 by Akt-induced phosphorylation in HER-2/neu-overexpressing cells. Nat. Cell Biol. 3:245-252. [DOI] [PubMed] [Google Scholar]