Abstract
Three kinases, Pak1, Tos3, and Elm1, activate Snf1 protein kinase in Saccharomyces cerevisiae. This cascade is conserved in mammals, where LKB1 activates AMP-activated protein kinase. We address the specificity of the activating kinases for the three forms of Snf1 protein kinase containing the β-subunit isoforms Gal83, Sip1, and Sip2. Pak1 is the most important kinase for activating Snf1-Gal83 in response to glucose limitation, but Elm1 also has a significant role; moreover, both Pak1 and Elm1 affect Snf1-Sip2. These findings exclude the possibility of a one-to-one correspondence between the activating kinases and the Snf1 complexes. We further identify a second, unexpected role for Pak1 in regulating Snf1-Gal83: the catalytic activity of Pak1 is required for the nuclear enrichment of Snf1-Gal83 in response to carbon stress. The nuclear enrichment of Snf1 fused to green fluorescent protein (GFP) depends on both Gal83 and Pak1 and is abolished by a mutation of the activation loop threonine; in contrast, the nuclear enrichment of Gal83-GFP occurs in a snf1Δ mutant and depends on Pak1 only when Snf1 is present. Snf1-Gal83 is the only form of the kinase that localizes to the nucleus. These findings, that Pak1 both activates Snf1-Gal83 and controls its nuclear localization, implicate Pak1 in regulating nuclear Snf1 protein kinase activity.
The Snf1/AMP-activated protein kinase (AMPK) family is important for responses to metabolic stress (for reviews, see references 10 and 16). In mammals, AMPK is activated by increases in AMP during metabolic stress, by the hormones leptin and adiponectin (24, 44), and by drugs used in the treatment of type 2 diabetes (46). AMPK coordinates energy homeostasis, regulating lipid and glucose metabolism. In the yeast Saccharomyces cerevisiae, Snf1 kinase is also activated by stresses, notably glucose limitation (14, 23, 41, 43). Snf1 regulates the transcription of many genes and the activity of metabolic enzymes in response to carbon stress, and Snf1 is required for the utilization of alternate carbon sources (2, 8). Snf1 also affects meiosis and sporulation, aging (1), haploid invasive growth (4), and diploid pseudohyphal growth (18).
Snf1 is regulated by three upstream kinases, Pak1, Tos3, and Elm1, which phosphorylate the activation loop threonine of the catalytic subunit (Thr210) and activate Snf1 protein kinase (13, 25, 33). Any one of these three upstream kinases suffices for Snf1 function in vivo. A mammalian ortholog, the LKB1 tumor suppressor kinase, similarly phosphorylates and activates AMPK (11, 13, 42). Snf1 catalytic activity is also regulated by at least two other mechanisms. Protein phosphatase 1 (Reg1-Glc7) controls the phosphorylation and activity of Snf1 kinase (22, 23, 29), and the Std1 (Msn3) protein, which interacts with glucose sensors (31), plays a modest role in regulating Snf1 catalytic activity (19).
Snf1 protein kinase, like AMPK, is heterotrimeric, and the kinase comprises the Snf1 catalytic subunit, the Snf4 activating subunit, and one of three β-subunit isoforms, Gal83, Sip1, or Sip2. The different forms of Snf1 kinase are here designated, according to their β subunit, as Snf1-Gal83, Snf1-Sip1, and Snf1-Sip2. The three β subunits exhibit considerable functional redundancy, as all three must be mutated to confer a Snf− phenotype, but they also play distinct roles (30, 45). Gal83 mediates the interaction of Snf1 kinase with Sip4, a transcriptional activator of gluconeogenic genes (30, 36), and with the transcriptional apparatus (38); Sip2 has been implicated in aging (1, 20); and each of the three β subunits has a distinct role in haploid invasive growth (39). A major function of the β subunits is to regulate the subcellular localization of the kinase (38), thereby governing access to different substrates. All three β subunits are cytoplasmic when cells are grown in abundant glucose, but when cells are shifted to low glucose or a nonfermentable carbon source, Snf1-Gal83 becomes enriched in the nucleus, Snf1-Sip1 relocalizes around the vacuole, and Snf1-Sip2 remains cytoplasmic. The cyclic AMP-dependent protein kinase (protein kinase A) regulates the localization of Snf1-Sip1 (12) but does not affect Snf1-Gal83 (38). The existence of three functionally distinct β-subunit isoforms with different, and differently regulated, subcellular localizations contributes to the functional versatility of Snf1 kinase in regulating multiple cellular processes and responses to stress.
Why do yeast cells have three upstream activating kinases for Snf1? Pak1, Tos3, and Elm1 appear to exhibit considerable functional redundancy, as all three cognate genes must be deleted to confer a Snf− mutant phenotype (13, 33). However, this finding does not exclude the possibility that the kinases indeed have distinct functions. The existence of three different forms of Snf1 protein kinase raised the possibility that each upstream kinase exhibits specificity for Snf1 protein that is associated with a particular β subunit.
We have examined the functional correspondence between the three upstream kinases and the three forms of Snf1 protein kinase. We show that Pak1 is the most critical kinase for activation of Snf1-Gal83 but that Elm1 also has a significant role; moreover, we show that Pak1 also affects Snf1-Sip2, as does Elm1. These findings rule out the possibility of a one-to-one correspondence between upstream kinases and forms of Snf1 protein kinase with respect to activation. We further identify a second, unexpected role for Pak1 in regulating Snf1-Gal83: phosphorylation of Snf1 by Pak1 is required for the relocalization of Snf1-Gal83 to the nucleus in response to carbon stress. Thus, Pak1 not only activates Snf1-Gal83 but also controls its localization and thereby regulates nuclear Snf1 protein kinase activity.
MATERIALS AND METHODS
Strains and genetic methods.
S. cerevisiae strains used in this work are listed in Table 1. Unless otherwise indicated, strains carrying tos3Δ, pak1Δ, and elm1Δ mutations were constructed by genetic crossing, and segregants were genotyped by PCR with genomic DNA as the template and with primers specific for the mutant loci. To construct MCY4999, primers flanking the stop codon of PAK1 were used to amplify the GFP-KanMX6 sequence from pFA6a-GFPS65T-KanMX6 (21) by PCR, and the resulting DNA fragment was used to transform strain W303-1A; the gene fusion is functional for the nuclear localization of Gal83. Rich medium (yeast extract-peptone [YEP]) and selective synthetic complete (SC) media (27) contained 2% concentrations of the carbon sources indicated below except in the case of glycerol-ethanol, which was 2% glycerol plus 3% ethanol.
TABLE 1.
S. cerevisiae strains used in this studya
| Strain | Genotype | Source or reference |
|---|---|---|
| W303-1A | MATaade2 can1 his3 leu2 trp1 ura3 | 34 |
| W303-1B | MATα ade2 can1 his3 leu2 trp1 ura3 | 34 |
| CTY10-5d | MATaade2-101 his3-Δ200 leu2-Δ1 trp1-Δ901 gal4 gal80 URA3::lexAop-lacZ | Gift of R. Sternglanz |
| MCY4024 | CTY10-5d gal83Δ::TRP1 | 38 |
| MCY4093 | MATα gal83Δ::TRP1 ade2 can1 his3 leu2 trp1 ura3 | H. Wiatrowski |
| MCY4094 | MATasip1Δ::kanMX6 sip2Δ::kanMX4 ade2 can1 his3 leu2 trp1 ura3 | H. Wiatrowski |
| MCY4095 | MATasip1Δ::kanMX6 gal83Δ::TRP1 ade2 can1 his3 leu2 trp1 ura3 | H. Wiatrowski |
| MCY4096 | MATasip2Δ::kanMX4 gal83Δ::TRP1 ade2 can1 his3 leu2 trp1 ura3 | H. Wiatrowski |
| MCY4099 | MATα gal83Δ::TRP1 sip1Δ::kanMX6 sip2Δ::kanMX4 ade2 can1 his3 leu2 trp1 ura3 | 40 |
| MCY4908 | W303-1A snf1Δ10 | 12 |
| MCY4950 | MATapak1Δ::kanMX4 sip2Δ::kanMX4 gal83Δ::TRP1 ade2 can1 his3 leu2 trp1 ura3 | This study |
| MCY4951 | MATapak1Δ::kanMX4 sip1Δ::kanMX6 sip2Δ::kanMX4 ade2 can1 his3 leu2 trp1 ura3 | This study |
| MCY4952 | MATapak1Δ::kanMX4 sip1Δ::kanMX6 gal83Δ::TRP1 ade2 can1 his3 leu2 trp1 ura3 | This study |
| MCY4958 | MATaelm1Δ::URA3 sip1Δ::kanMX6 sip2Δ::kanMX4 ade2 can1 his3 leu2 trp1 ura3 | This study |
| MCY4967 | MATα pak1Δ::kanMX4 elm1Δ::URA3 sip2Δ::kanMX4 gal83Δ::TRP1 ade2 can1 his3 leu2 trp1 ura3 | This study |
| MCY4969 | MATatos3Δ::kanMX4 sip2Δ::kanMX4 gal83Δ::TRP1 ade2 can1 his3 leu2 trp1 ura3 | This study |
| MCY4975 | MATα tos3Δ::kanMX4 sip1Δ::kanMX6 gal83Δ::TRP1 ade2 can1 his3 leu2 trp1 ura3 | This study |
| MCY4981 | MATapak1Δ::kanMX4 gal83Δ::TRP1 ade2 can1 his3 leu2 trp1 ura3 | This study |
| MCY4999 | W303-1A PAK1-GFP::kanMX6 | This study |
| MCY5018 | MATaelm1Δ::URA3 sip2Δ::kanMX4 gal83Δ::TRP1 ade2 can1 his3 leu2 trp1 ura3 | This study |
| MCY5025 | MATα pak1Δ::kanMX4 snf1Δ10 ade2 can1 his3 leu2 trp1 ura3 | This study |
| MCY5114 | W303-1A tos3Δ::kanMX4 | 13 |
| MCY5115 | W303-1B pak1Δ::kanMX4 | 13 |
| MCY5122 | MATatos3Δ::kanMX4 elm1Δ::URA3 ade2 can1 his3 leu2 trp1 ura3 | This study |
| MCY5123 | MATapak1Δ::kanMX4 elm1Δ::URA3 tos3Δ::kanMX4 ade2 can1 his3 leu2 trp1 ura3 | This study |
| MCY5125 | W303-1A elm1Δ::kanMX4 | This study |
| MCY5134 | CTY10-5d pak1Δ::kanMX4 | This study |
| MCY5137 | MATα tos3Δ::kanMX4 sip1Δ::kanMX6 sip2Δ::kanMX4 ade2 can1 his3 leu2 trp1 ura3 | This study |
| MCY5139 | MATaelm1Δ::URA3 sip1Δ::kanMX6 gal83Δ::TRP1 ade2 can1 his3 leu2 trp1 ura3 | This study |
| MCY5140 | MATα elm1Δ::URA3 sip1Δ::kanMX6 gal83Δ::TRP1 ade2 can1 his3 leu2 trp1 ura3 | This study |
All strains have the W303 genetic background except MCY2649, MCY2693, and MCY2916, which have the S288C background, and CTY10-5d. Alleles are ade2-1, can1-100, his3-11,15, leu2-3,112, trp1-1, and ura3-1, except where otherwise noted.
Plasmids.
The carbon source-responsive element (CSRE)-lacZ reporter was pOV22 (37). LexA-Snf1G53R was expressed from pRJ216 (17). Green fluorescent protein (GFP)-tagged Gal83 and Snf1 were expressed from their native promoters on centromeric plasmids pRT12, pRT13, or pOV72 and pOV84, respectively (12, 38). pKH43, expressing Snf1T210A-GFP, was constructed by recombination in yeast (MCY4908) between the MluI-AflII fragment of pOV84 and an overlapping PCR fragment containing the T210A mutation, derived from pRJ217 (13, 19); the mutation was confirmed by sequencing. pKH44 was constructed similarly by using the SacII-MluI fragment of pOV84. pSK119A, a mutant derivative of pSK119 (35), and pRJ217 expressed hemagglutinin (HA)- and LexA-tagged Snf1T210A, respectively, from the ADH1 promoter. pRH108, expressing LexA-tagged Tos3 from the ADH1 promoter, is a derivative of pLexA(1-202)+PL (28).
Plasmid pRH104, expressing triple-HA-tagged Pak1 from the ADH1 promoter, was constructed by cloning a PCR product containing the PAK1 open reading frame flanked by MfeI sites into pWS93 (32) at the EcoRI site. pRH119 is a derivative of pRH104 with a mutation changing Asp277 to Ala and was constructed essentially as described previously (15), with the mutagenic primer Pak1D277A-R1 (5′-AGGCTTAATAGCTCGGTGAATGATTCC-3′) and primer Pak1D277A-F1 (5′-CTAGAGTATTGTTCTCGAGGC-3′) to produce a mega-primer, which was then used in combination with primer Pak1D277A-R2 (5′-CGGTAAGTTTTGGTTGCGTGAATTCATTCGGCC-3′) to amplify the coding sequence. pRH119 expresses kinase-dead HA-Pak1D277A, which contains the same mutation that has been characterized previously (25). pRH131, a derivative of pRH104 with a mutation changing Asp295 to Ala, was constructed by using the QuikChange XL site-directed mutagenesis kit (Stratagene) with the mutagenic primer Pak1D295A-1 (5′-GGCACTGTTAAGATTTCCGCTTTTGGTGTTTCTTTAGC-3′) and its complement. Both mutations were confirmed by sequencing; the plasmids expressed proteins of the expected size and did not provide Pak1 function in pak1Δ tos3Δ elm1Δ mutant cells.
Assay of Snf1 kinase activity by phosphorylation of the SAMS peptide.
Cells (100 ml) were grown in YEP-2% glucose to an optical density at 600 nm (OD600) of 1, collected by centrifugation or filtration as noted below, incubated in YEP-0.05% glucose for 30 min, and then collected by filtration. Extracts were prepared from two independent cultures, and assays were performed essentially as described previously (13, 43). Briefly, cells were broken by vortexing them with glass beads in 0.6 ml of buffer A (50 mM Tris-HCl [pH 7.5], 50 mM NaF, 5 mM sodium pyrophosphate, 1 mM EDTA, 1 mM dithiothreitol, 0.1 mM phenylmethylsulfonyl fluoride, 0.5% Triton X-100, 10% [vol/vol] glycerol). After centrifugation (20,000 × g for 10 min), the extract was applied to a 1-ml column of DEAE-Sepharose (Amersham Biosciences) and washed with 10 ml of buffer A. Snf1 activity was eluted from the column with buffer A containing 0.2 M NaCl. All steps of the purification were performed at 4°C. Pooled peak fractions (2 ml) were assayed for phosphorylation of the SAMS peptide (HMRSAMSGLHLVKRR) (5) in the presence of [γ-32P]ATP as described previously (13). Assays were performed at least in triplicate, using different protein concentrations to confirm linearity. Kinase activity is expressed as nanomoles of phosphate incorporated into the peptide per minute per milligram of protein (5). The assayed fractions were subjected to immunoblot analysis, which was carried out as described previously (36) with anti-Snf1 antibody (3).
β-Galactosidase assay.
Transformants were grown to mid-log phase in selective SC medium-2% glucose and were shifted to SC medium-0.05% glucose for 3 or 5.5 h. β-Galactosidase activity was assayed in permeabilized cells and is expressed in Miller units.
Microscopy.
Cultures were grown to mid-log phase in selective SC medium-2% glucose. Nuclei were stained for 5 min by the addition of 4′,6′-diamidino-2-phenylindole (DAPI; 0.8 μg/ml). Cells from 1 ml of culture were harvested by brief centrifugation and resuspended in residual medium (∼20 μl), and an aliquot was placed on a microscope slide. For shift experiments, cells were grown in selective SC medium-2% glucose, collected by centrifugation, and resuspended in 1 to 5 ml of medium containing a different carbon source. Cells were incubated for 30 min, stained with DAPI, and collected for examination as described above. Cells were viewed using a Nikon Eclipse E800 fluorescence microscope. Images were taken with an Orca100 (Hamamatsu) camera by using Open Lab (Improvision) software, and they were processed with Adobe Photoshop 5.5. Images of GFP fluorescence were taken with exposure times of 0.8 s.
RESULTS
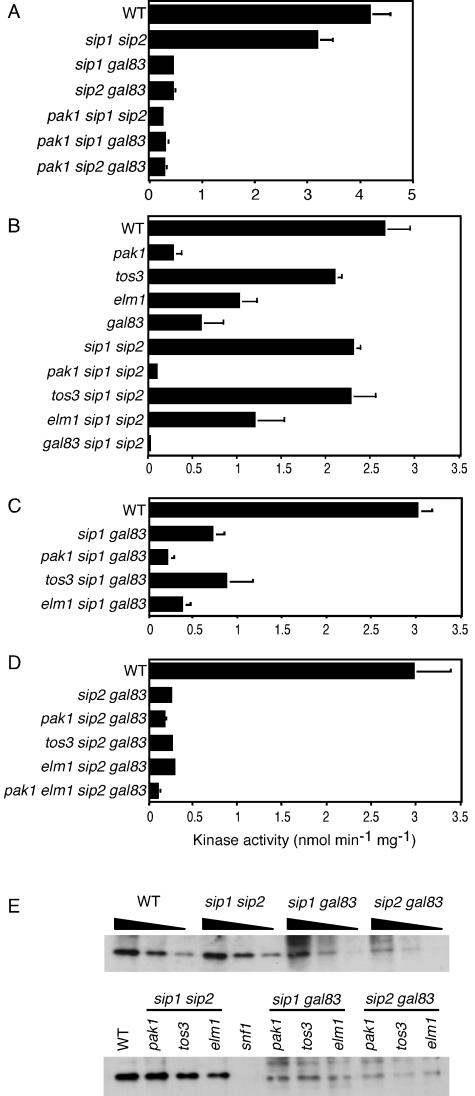
Pak1 is the major kinase required for the activity of Snf1-Gal83 kinase in vitro. Gal83 is the most abundant β subunit in glucose-grown cells (9, 38). Assays of mutants expressing only one β subunit showed that Snf1-Gal83 kinase is, correspondingly, responsible for the majority of Snf1 kinase activity (Fig. 1A). Cells were grown in glucose, collected by centrifugation, and shifted to 0.05% glucose for 30 min; both centrifugation and carbon deprivation are stresses that activate Snf1 (41, 43). The kinase was partially purified and catalytic activity was assayed by phosphorylation of the SAMS synthetic peptide substrate (5). In the sip1Δ sip2Δ mutant, which expresses only Snf1-Gal83, the level of activity was 75% of wild-type levels. Conversely, in sip2Δ gal83Δ and sip1Δ gal83Δ strains, which express only Snf1-Sip1 and Snf1-Sip2, respectively, activity was reduced to 10% of that of the wild type. Part of this decrease can be attributed to the somewhat lower levels of Snf1 protein in the absence of the major β subunit, Gal83 (Fig. 1E).
FIG. 1.
Assay of Snf1 kinase activity. Cells of the indicated genotype were grown in YEP-2% glucose, collected by centrifugation (A) or by filtration (B to D), and shifted to YEP-0.05% glucose for 30 min. Cells were then collected by filtration, extracts were prepared, and Snf1 kinase was partially purified. Snf1 catalytic activity was assayed by phosphorylation of the SAMS peptide substrate (5). In each case, extracts were prepared from two independent cultures, and values are averages of results of four to six assays from the two experiments. (A) Low values were as follows: 0.45 ± 0.04 for the sip1 gal83 mutant, 0.31 ± 0.06 for the pak1 sip1 gal83 mutant, 0.45 ± 0.06 for the sip2 gal83 mutant, 0.30 ± 0.05 for the pak1 sip2 gal83 mutant, and 0.26 ± 0.02 for the pak1 sip1 sip2 mutant. Kinase activity is expressed as nanomoles of phosphate incorporated into the peptide per minute per milligram of protein. (E) Proteins (2 μg) from the fractions assayed inpanels B to D were subjected to immunoblot analysis with anti-Snf1 antibody. The upper panel shows serial twofold dilutions. WT, wild type.
Although deletion of all three upstream kinase genes is necessary to abolish Snf1 kinase activity, mutation of PAK1 causes the greatest decrease (13) (see Fig. 1B). To assess the role of Pak1 in the activation of Snf1-Gal83, we assayed the pak1Δ sip1Δ sip2Δ mutant for in vitro kinase activity (Fig. 1A). In the triple mutant, activity was reduced 12-fold relative to that of the sip1Δ sip2Δ mutant, indicating that Pak1 has a major role in the activation of Snf1-Gal83. The pak1Δ mutation also appeared to cause some reduction of Snf1-Sip1 and Snf1-Sip2 activities, as judged by a comparison of the corresponding double and triple mutants (Fig. 1A; see the legend for values). The absence of Pak1, or of the other upstream kinases, did not affect levels of Snf1 protein (Fig. 1E), in accord with previous results (13).
Snf1-Gal83 kinase is not exclusively activated by Pak1.
To determine whether Snf1-Gal83 kinase is exclusively activated by Pak1, we assayed Snf1-Gal83 activity in mutants lacking each of the three upstream kinases. Cells were grown in 2% glucose, collected by filtration, and shifted to 0.05% glucose for 30 min (Fig. 1B). The pak1Δ sip1Δ sip2Δ mutant again showed much lower activity than the sip1Δ sip2Δ double mutant, and the elm1Δ sip1Δ sip2Δ mutant showed a twofold reduction in activity. The activity level of the tos3Δ sip1Δ sip2Δ mutant was not significantly different from that of the sip1Δ sip2Δ mutant, although it remains possible that Tos3 has a minor effect. We conclude that Pak1 is the most important of the three upstream kinases for activating Snf1-Gal83 but that Elm1 also has a role.
We also compared the three upstream kinases with respect to their activation of Snf1-Sip2 and Snf1-Sip1. In this assay, when cells were collected by filtration, Snf1-Sip2 activity was reduced more than threefold by the pak1Δ mutation and twofold by elm1Δ (Fig. 1C). The activity of Snf1-Sip1 was very low in this assay, so the apparent effects of Pak1 and Elm1 are not compelling (Fig. 1D).
Pak1 affects Snf1-Gal83 function in vivo.
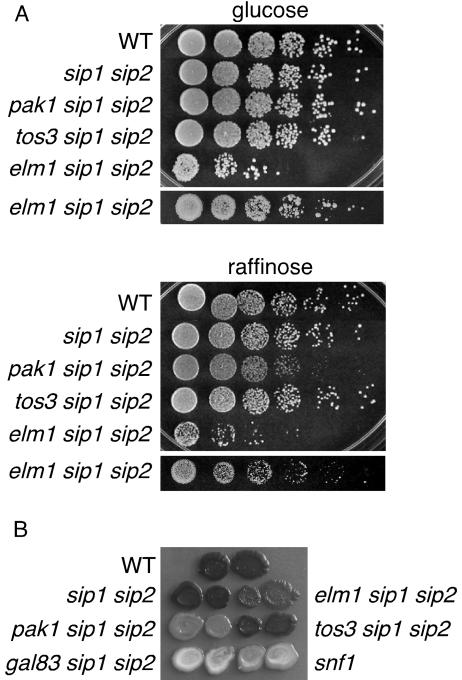
The in vitro assays suggest that Pak1 has a major physiological role in activating Snf1-Gal83 kinase. To assess the role of Pak1 in vivo, we compared sip1Δ sip2Δ and pak1Δ sip1Δ sip2Δ cells with respect to several phenotypes. First, we tested growth on different carbon sources. The absence of Pak1 diminished growth on raffinose (Fig. 2A) but did not affect growth on glycerol-ethanol (data not shown). We noted no effect of a tos3 deletion and a small effect of an elm1 deletion, as judged by comparisons to the glucose control plate (elm1Δ strains are clumpy and spot differently from the others). The absence of Pak1 did not affect growth on raffinose when all β subunits were present (references 13 and 25 and data not shown).
FIG. 2.
Growth phenotypes and glycogen accumulation. Cells of the indicated genotype were grown overnight in YEP-2% glucose. (A) Cells were adjusted to an OD600 of 0.5 and spotted with serial dilutions (one 10-fold dilution followed by successive 5-fold dilutions) on YEP-2% glucose and YEP-2% raffinose in the presence of the respiratory inhibitor antimycin (1 μg/ml). Because elm1 cells are clumpy, a second dilution series was also spotted; cells were adjusted to an OD600 of 2.5 and then spotted with successive fivefold dilutions (lower panels). Plates were incubated at 30°C and photographed on successive days. Photographs show growth after 2 days on glucose and 4 days on raffinose. (B) Cells were spotted in duplicate on YEP-2% glucose, incubated for 3 days, and exposed to iodine vapor to stain the glycogen. WT, wild type.
We next assayed glycogen accumulation, which requires Snf1 (7). Cells grown on solid medium containing 2% glucose were exposed to iodine vapor; cells containing glycogen stain dark (Fig. 2B). A comparison of sip1Δ sip2Δ and pak1Δ sip1Δ sip2Δ strains showed that Pak1 is required for Snf1-Gal83 function in glycogen accumulation. A very modest effect for Elm1 was also apparent upon direct examination of the plate.
Activation of the CSRE of gluconeogenic genes, which binds the activators Cat8 and Sip4 (26, 37), depends on Snf1-Gal83 (40). Cells carrying a CSRE-lacZ reporter were induced by a shift from 2 to 0.05% glucose for 5.5 h. β-Galactosidase activity was undetectable prior to the shift. After induction, activity was modestly lower in pak1Δ sip1Δ sip2Δ cells than in sip1Δ sip2Δ cells (4 ± 1 and 13 ± 1 U, respectively; values are averages for seven transformants). Activity was <1 U in sip1Δ sip2Δ gal83Δ cells and was slightly lower in wild-type cells (10 U) than in sip1 sip2 cells, where all the kinase is in the Snf1-Gal83 form that activates the CSRE.
Finally, we used an assay in which LexA-tagged Snf1G53R, a catalytically hyperactive mutant kinase, activates transcription of a lacZ reporter with LexA binding sites in its promoter (17). This activation requires Gal83 (38) and most likely reflects the interaction of the kinase with the transcriptional apparatus (17). We expressed LexA-Snf1G53R in strain CTY10-5d, which carries the lacZ reporter, and in pak1Δ and gal83Δ derivatives of CTY10-5d. Transformants were grown in 2% glucose, shifted to 0.05% glucose for 3 h, and assayed for β-galactosidase activity. Activation of the reporter was reduced 35-fold in pak1Δ mutant cells relative to that in the wild type (2.6 ± 0.1 and 92 ± 3 U, respectively; values are averages for three transformants). Activation was virtually abolished in the gal83Δ mutant (0.4 ± 0.1 U); for all strains, activity was negligible in glucose-grown cells (pak1Δ mutant, 0.3 U; wild type, 1.0 U; gal83Δ mutant, 0.1 U). Thus, Pak1 was important for the function of Snf1G53R-Gal83 in this assay.
Pak1 affects Snf1-Sip2 function in vivo.
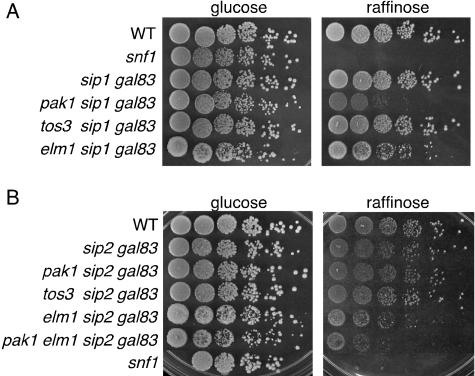
Assays of Snf1 catalytic activity in vitro suggested that Pak1 contributes to the activation of Snf1-Sip2 and, possibly, Snf1-Sip1 (Fig. 1). To confirm that Pak1 affects other forms of the kinase besides Snf1-Gal83, we assayed growth phenotypes. In cells expressing only Snf1-Sip2, the pak1Δ mutation impaired growth on raffinose (Fig. 3A; compare sip1Δ gal83Δ and pak1Δ sip1Δ gal83Δ cells). The defect is more severe than for pak1Δ sip1Δ sip2Δ cells, suggesting that low levels of Snf1-Sip2 activity are less effective in providing functions required for growth on raffinose than low levels of Snf1-Gal83 activity. In cells expressing Snf1-Sip1, no effect of pak1Δ was evident (Fig. 3B), but this negative result does not exclude the possibility of effects on other phenotypes not assayed here. These findings, together with the results of the kinase assays, indicate that Pak1 is not dedicated to the activation of Snf1-Gal83 but rather also regulates the activity of at least one other form of Snf1 protein kinase.
FIG. 3.
Growth phenotypes. Cells of the indicated genotype expressing only Snf1-Sip2 (A) or Snf1-Sip1 (B) were grown overnight in YEP-2% glucose. Cells were adjusted to an OD600 of 0.25, except for strains carrying elm1, which were adjusted to an OD600 of 2.5, and were spotted with serial fivefold dilutions on YEP-2% glucose and YEP-2% raffinose-antimycin (1 μg/ml). Plates were incubated at 30°C. Photographs show growth after 2 days on glucose and 4 days on raffinose. WT, wild type.
We also examined the effects of the other two upstream kinases. In cells expressing only Snf1-Sip2 or Snf1-Sip1, the elm1Δ mutation also appeared to impair growth on raffinose, whereas a deletion of tos3 had no effect (Fig. 3). These results are consistent with a role for Elm1 in activating these kinase forms.
Upstream kinases are required for nuclear enrichment of Gal83 in response to carbon stress.
The β subunits regulate the subcellular localization of Snf1 protein kinase in response to the carbon source, and localization presumably affects the access of the kinase to particular sets of substrates. All β subunits are cytoplasmic during growth in 2% glucose, but upon a shift to glucose-limiting conditions, Gal83 rapidly becomes enriched in the nucleus, Sip1 relocalizes around the vacuole, and Sip2 remains cytoplasmic (38). Protein kinase A negatively regulates the localization of Sip1 to the vacuolar membrane (12) but does not affect the localization of Gal83 (38). The evidence has suggested glucose-6-phosphate as a candidate signal for the nuclear exclusion of Gal83 (38), but the pathway controlling the localization of Gal83 has not been identified.
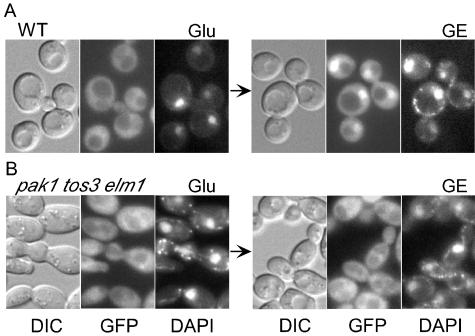
To assess the role of the upstream kinases, we expressed functional, GFP-tagged Gal83 from its native promoter on a centromeric plasmid in wild-type and pak1Δ tos3Δ elm1Δ cells. During growth in glucose, Gal83-GFP was cytoplasmic and excluded from nuclei in both wild-type and mutant cells (Fig. 4A and B). Upon a shift to glycerol-ethanol, Gal83-GFP became enriched in the nuclei of wild-type cells. Figure 4A shows cells photographed 30 min after the shift, but enrichment was evident within 5 min (data not shown). In contrast, Gal83-GFP remained excluded from the nuclei of triple mutant cells at 30 min (Fig. 4B). Thus, the triple mutant exhibited a striking defect in the nuclear enrichment of Gal83-GFP.
FIG. 4.
Nuclear localization of Gal83-GFP in response to carbon stress depends on upstream kinases. Cells of the indicated genotype expressed Gal83-GFP. Strains used were W303-1A (wild type [WT]) (A) and MCY5123 (B). Cells were grown to mid-log phase in SC medium-2% glucose (Glu) (left panels) and then were shifted to SC medium-2% glycerol-3% ethanol (GE) for 30 min (right panels); the arrows indicate the shifts. Differential interference contrast (DIC), GFP fluorescence, and DAPI staining are shown.
The nuclear localization of Gal83 is defective in pak1Δ mutants.
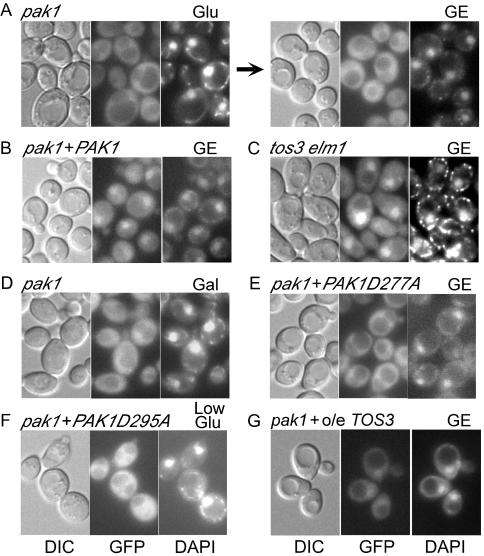
To identify the upstream kinase(s) responsible for regulating the localization of Gal83, we examined Gal83-GFP in each of the single mutants. Nuclear enrichment was observed in tos3Δ and elm1Δ mutants after a shift to glycerol-ethanol (data not shown), but the pak1Δ mutant was clearly defective (Fig. 5A), as were the tos3Δ pak1Δ and elm1Δ pak1Δ mutants (data not shown). Expression of HA-tagged Pak1 from a plasmid complemented the pak1Δ mutant defect, thereby confirming that the mutation is responsible for the phenotype (Fig. 5B). Conversely, nuclear localization occurred normally in the tos3Δ elm1Δ double mutant, indicating that Pak1 is sufficient for localization (Fig. 5C). To determine whether Pak1 is required for the nuclear enrichment of Gal83 under different conditions, we shifted glucose-grown cells to low (0.05%) glucose for 30 min or to glycerol-ethanol or galactose for extended growth. In all cases, the pak1Δ mutant failed to exhibit a nuclear enrichment of Gal83-GFP (Fig. 5D and data not shown).
FIG. 5.
Pak1 is required for the nuclear localization of Gal83-GFP. Cells of the indicated genotype expressed Gal83-GFP. Strains used were MCY5115 (A and D), MCY5115 expressing HA-Pak1 from pRH104 (B), MCY5122 (C), MCY5115 expressing HA-Pak1D277A from pRH119 (E), MCY5115 expressing HA-Pak1D295A from pRH131 (F), and MCY5115 overexpressing (o/e) LexA-tagged Tos3 from the ADH1 promoter on the multicopy plasmid pRH108 (G). (A) Cells were grown to mid-log phase in SC medium-2% glucose (Glu) (left panel) and then were shifted to SC medium-2% glycerol-3% ethanol (GE) for 30 min (right panel); the arrow indicates a shift. (B, C, and E to G) Cells were grown as described for panel A and were shifted to GE or low (0.05%) glucose (Low Glu) for 30 min. Panels show cells after the shift; Gal83-GFP was excluded from the nuclei of glucose-grown cells as described above (not shown). (D) Glucose-grown cells were shifted to 2% galactose (Gal) for 72 h, with repeated dilutions to maintain the cells in exponential growth; similar results were observed after a shift to GE for 50 h (not shown). DIC, GFP fluorescence, and DAPI staining are shown.
To determine whether the catalytic activity of Pak1 is required for nuclear enrichment, we introduced mutations into the HA-Pak1 expression plasmid to replace Asp277 with Ala, which inactivates Pak1 with respect to the phosphorylation of Snf1 on Thr210 (25), and to change the highly conserved Asp295 to Ala. The expression of the wild-type HA-Pak1 served as a control (Fig. 5B). Gal83-GFP did not become enriched in the nuclei of pak1Δ mutant cells expressing the kinase-dead HA-Pak1D277A or HA-Pak1D295A upon shifts to glycerol-ethanol or low glucose (Fig. 5E and F). Thus, the catalytic activity of Pak1 is required for the nuclear enrichment of Gal83.
Evidence for a minor role for Tos3 in the localization of Gal83.
Although Tos3 and Elm1 do not have primary roles in the nuclear enrichment of Gal83, evidence suggests that in the absence of Pak1 they play minor roles under some conditions. In the pak1Δ mutant, Gal83-GFP was clearly excluded from the nucleus during growth in glycerol-ethanol or galactose, whereas after a shift from 2% glucose to 0.05% glucose or glycerol-ethanol, nuclear exclusion was less easily apparent (Fig. 5A). In contrast, exclusion was clearly evident in the pak1Δ tos3Δ elm1Δ mutant after a shift (Fig. 4B). Moreover, overexpression of kinase-dead HA-Pak1 enhanced nuclear exclusion in the pak1Δ mutant (Fig. 5E and F). These observations suggested that during acute carbon stress, Tos3 and/or Elm1 partially compensates for the absence of Pak1 and promotes the presence of a very low level of Gal83 in the nucleus. To test this possibility, we overexpressed Tos3 (LexA tagged) from the ADH1 promoter in a pak1Δ mutant. We detected weak nuclear enrichment in some cells after a shift to glycerol-ethanol, indicating that overexpression of Tos3 partially suppressed the pak1Δ defect (Fig. 5G). A similar experiment with Elm1 gave negative, and hence inconclusive, results. Together, these data suggest a minor degree of overlap of the functions of Tos3 and Pak1 with respect to localization, which is evident in the absence of Pak1.
Pak1 is not localized in the nucleus in response to carbon stress.
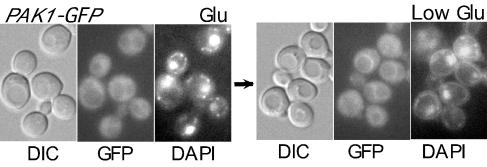
One possible explanation for the profound effects of Pak1 on nuclear localization is that Pak1 itself is enriched in the nucleus and is responsible for the colocalization of Snf1-Gal83. To test this idea, we constructed a gene fusion expressing Pak1-GFP from the genomic locus. Glucose-grown cells showed low-level cytosolic fluorescence (Fig. 6). After a shift to limiting glucose, many cells exhibited enhanced fluorescence around the vacuolar membrane, similar to that of Sip1-GFP under these conditions (12); however, we detected no nuclear enrichment of Pak1-GFP (Fig. 6).
FIG. 6.
Localization of Pak1-GFP. Cells of strain MCY4999, expressing Pak1-GFP from the genomic locus, were grown to mid-log phase in SC medium-2% glucose (Glu) and were shifted to 0.05% glucose (Low Glu) for 30 min. DIC, GFP fluorescence, and DAPI staining are shown.
The nuclear localization of Gal83 does not depend on Snf1.
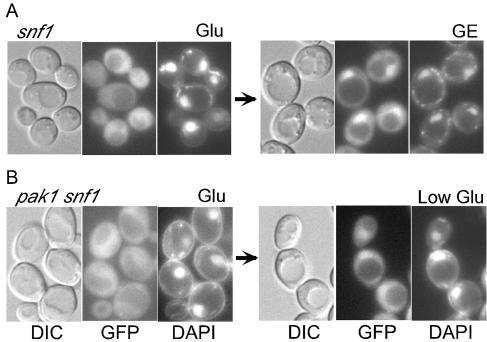
A simple model is that Pak1 causes the nuclear localization of Gal83, which is present in Snf1-Gal83 kinase complexes, by activating Snf1. However, previous studies of snf1Δ mutant cells of the S288C background showed that nuclear enrichment of Gal83-GFP does not require Snf1 (38). Because differences in genetic backgrounds can have profound effects, we reexamined this issue with a snf1Δ mutant of the W303 background used in this study. Gal83-GFP was excluded from nuclei in glucose-grown snf1Δ cells and became enriched in nuclei after a shift to glycerol-ethanol (Fig. 7A). The fraction of cells showing strong enrichment was somewhat lower than for the wild type, which may simply reflect the general unhealthiness of snf1Δ cells or may reflect a contributing role of Snf1. Nonetheless, Snf1 is clearly not required, as cells exhibited regulated nuclear enrichment of Gal83 in the absence of Snf1.
FIG. 7.
Nuclear localization of Gal83-GFP in snf1Δ and pak1Δ snf1Δ mutant cells. Cells expressed Gal83-GFP. Strains used were MCY4908 (A) and MCY5025 (B). Cells were grown to mid-log phase in SC medium-2% glucose (Glu) and then were shifted to SC medium-0.05% glucose (Low Glu) for 30 min. DIC, GFP fluorescence, and DAPI staining are shown.
In the absence of Snf1, the nuclear localization of Gal83 does not depend on Pak1.
The findings described above suggested that Pak1 regulates Gal83 directly; alternatively, it remains possible that in the absence of Snf1, the nuclear localization of Gal83 does not depend on Pak1. We indeed found that in pak1Δ snf1Δ double mutant cells, Gal83-GFP became enriched in nuclei upon a shift to low glucose (Fig. 7B). Enrichment was not as pronounced as in the wild type but was not noticeably different from that observed in the snf1Δ mutant. Thus, in the absence of Snf1, the nuclear localization of Gal83 is regulated by a mechanism that is independent of Pak1.
Both Pak1 and Gal83 are required for the nuclear enrichment of Snf1.
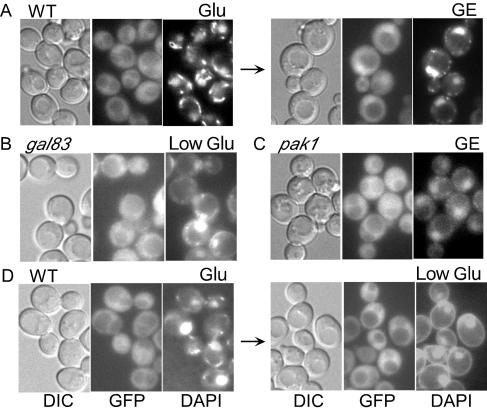
To examine the localization of Snf1, we used functional, GFP-tagged Snf1 expressed from its native promoter on a centromeric plasmid (38). In the S288C background, Snf1-GFP was enriched in the nucleus after a shift to glycerol or during growth in glycerol-ethanol, and enrichment was strongly reduced in gal83Δ mutant cells (38). In the W303 background, the nuclear enrichment of Snf1-GFP was similarly evident in wild-type cells after a shift to a poor carbon source (Fig. 8A); enrichment was less striking than for Gal83-GFP because some Snf1 is complexed with Sip1 and Sip2, which do not relocalize to the nucleus (38). No nuclear enrichment occurred in gal83Δ cells after a shift to low glucose or glycerol-ethanol (Fig. 8B and data not shown). We also observed no nuclear enrichment of Snf1-GFP in pak1Δ, pak1Δ sip1Δ sip2Δ, or pak1Δ gal83Δ cells (Fig. 8C and data not shown). Thus, both Pak1 and Gal83 are required for the nuclear enrichment of Snf1.
FIG. 8.
Localization of Snf1-GFP and Snf1T210A-GFP. Cells expressed Snf1-GFP (A to C) and Snf1T210A-GFP (D). Strains used were W303-1A (A and D), MCY4093 (B), and MCY5115 (C). Cells were grown in SC medium-2% glucose (Glu) and were shifted to 2% glycerol-3% ethanol (GE) or 0.05% glucose (Low Glu) for 30 min. Arrows indicate the shifts. (B and C) Cells after the shift. Results were the same for both shift conditions (data not shown). DIC, GFP fluorescence, and DAPI staining are shown. WT, wild type.
Mutation of Thr210 of Snf1 abolishes the nuclear localization of Snf1-Gal83.
A parsimonious model is that Pak1 controls the localization of Snf1-Gal83 through phosphorylation of the activation loop threonine (Thr210) of Snf1. This model predicts that the nuclear enrichment of Gal83 would be impaired in the presence of Snf1T210A, which has Thr210 replaced with Ala (6). To maximize the association of Gal83 with Snf1T210A, we expressed Snf1T210A from the ADH1 promoter in snf1Δ cells; the native level of Gal83 has been reported to be higher than that of Snf1 (9). Expression of Snf1T210A (HA or LexA tagged) prevented the nuclear enrichment of Gal83-GFP, whereas expression of HA-Snf1 had no effect (data not shown). As a more direct test of the model, we expressed Snf1T210A-GFP from its native promoter in both wild-type and snf1Δ cells. Snf1T210A-GFP remained excluded from the cytosol and nucleus after a shift from high to low glucose (Fig. 8D and data not shown). Hence, the nuclear enrichment of Snf1 requires the phosphorylation of Thr210.
Phosphorylation of Thr210 causes conformational changes and activates Snf1. To assess the requirement for Snf1 catalytic activity, we expressed kinase-dead Snf1K84R-GFP, in which Arg replaces the Lys of the ATP-binding site (3). Snf1K84R-GFP did not become enriched in the nucleus after a shift to low glucose (data not shown). In previous experiments, enrichment of overexpressed Gal83-GFP in snf1-K84R cells probably reflected Gal83-GFP that was not complexed with Snf1K84R (38). Together, these findings strongly suggest that the nuclear enrichment of Snf1-Gal83 requires the phosphorylation and activation of Snf1 by Pak1.
DISCUSSION
Yeast cells contain three forms of Snf1 protein kinase, with different β subunits, and three activating kinases for Snf1. We here exclude the model that there is a one-to-one correspondence between the different forms and the upstream kinases. We show that Pak1 is the most important of the three upstream kinases for activating Snf1-Gal83 in response to glucose limitation. Snf1-Gal83 is the form that is responsible for most of the Snf1 activity in glucose-grown cells, so these findings implicate Pak1 as a critical activating kinase for the adaptation of cells to glucose limitation. We further identify a second role for Pak1: Pak1 is both necessary and sufficient for the nuclear accumulation of Snf1-Gal83 in response to glucose limitation. Snf1-Gal83 is the only form of the kinase that localizes to the nucleus; hence, Pak1 regulates nuclear Snf1 protein kinase activity.
We first examined the relationship between the three activating kinases and the different forms of Snf1 protein kinase. Biochemical and genetic analyses demonstrated a close functional association between Pak1 and Snf1-Gal83 but also showed that the activating kinases have overlapping functions with respect to the different forms of Snf1 kinase. Snf1-Gal83 is not activated exclusively by Pak1 in response to glucose limitation, as Elm1 also has a role. Conversely, Pak1 is not dedicated to the activation of Snf1-Gal83 but rather also affects the function of Snf1-Sip2, and possibly Snf1-Sip1, as is also the case for Elm1. Several assays confirmed that Pak1 regulates Snf1-Gal83 in vivo but also revealed various degrees of dependence of Snf1-Gal83's function on Pak1. The extent of dependence most likely reflects the properties of particular Snf1-Gal83 target proteins. For example, the sequence of the recognition site affects the threshold of Snf1-Gal83 activity required for phosphorylation, and the location of a target in the cytosol or nucleus (and whether it shuttles back and forth) affects access to Snf1-Gal83 in a pak1Δ mutant.
The activating kinase Pak1 is both necessary and sufficient for regulation of the localization of Snf1-Gal83 in response to carbon stress, as no significant nuclear enrichment was detected in its absence and no significant defect was detected in the tos3Δ elm1Δ mutant. Evidence suggests that the other kinases have minor roles. Nuclear enrichment of Snf1 requires the catalytic activity of Pak1 and is abolished by a mutation of the activation loop Thr210 of Snf1, indicating that the phosphorylation of Snf1 is critical. Mutation of the ATP-binding site also inhibited enrichment, indicating that Snf1 catalytic activity is required. These findings imply that Pak1 controls the nuclear localization of Snf1 through its phosphorylation and activation of Snf1. The nuclear enrichment of Snf1 also depends on Gal83. In contrast, the nuclear enrichment of Gal83 does not depend on Snf1; moreover, the nuclear enrichment of Gal83 depends on Pak1 only when Snf1 is present, and it is inhibited by the expression of Snf1T210A. These findings suggest that inactive Snf1 anchors Gal83 in the cytosol. Further studies will be required to determine whether the nucleocytoplasmic distribution of Snf1-Gal83 is regulated at the level of nuclear import, export, or retention. Proteins involved in these processes are candidates for targets of Snf1 activity. In addition, Gal83 is a possible candidate because Snf1 phosphorylates Gal83 in vitro (45), and phosphorylation of Snf1-Gal83 may permit nuclear accumulation. Pak1 itself does not become enriched in the nucleus in response to carbon stress, so the dependence of nuclear enrichment on phosphorylation may serve to ensure that Snf1-Gal83 that has not yet been activated does not accumulate in the nucleus.
These findings further show that a Pak1-independent mechanism also regulates the localization of Snf1-Gal83. Gal83 becomes enriched in the nucleus in response to carbon stress in the pak1Δ snf1Δ mutant, indicating that the localization of Gal83 is regulated by a mechanism, as yet unidentified, that is independent of Pak1. However, the possibility of an auxiliary role for Pak1 in regulating Gal83 in the absence of Snf1 is not excluded. Because the nuclear localization of Snf1 depends on Gal83, these data imply that the Pak1-independent mechanism regulates the localization of Snf1-Gal83. Previous studies of localization of Gal83 were carried out in cells containing Snf1 (38), and it is now clear that further investigation of the Pak1-independent mechanism must be carried out with cells lacking Snf1.
Although the three activating kinases exhibit substantial functional overlap, evidence presented here particularly implicates Pak1 in regulating the phosphorylation of targets of Snf1 that reside in the nucleus. Snf1-Gal83 is the only form of the kinase that localizes to the nucleus. Pak1 not only activates Snf1-Gal83 but also controls its nuclear localization and thereby regulates nuclear Snf1 protein kinase activity.
Acknowledgments
We thank Heather Wiatrowski for strains and Kenia de los Santos for technical assistance.
This work was supported by Public Health Service grant GM34095 from the National Institutes of Health to M.C.
REFERENCES
- 1.Ashrafi, K., S. S. Lin, J. K. Manchester, and J. I. Gordon. 2000. Sip2p and its partner Snf1p kinase affect aging in S. cerevisiae. Genes Dev. 14:1872-1885. [PMC free article] [PubMed] [Google Scholar]
- 2.Carlson, M. 1999. Glucose repression in yeast. Curr. Opin. Microbiol. 2:202-207. [DOI] [PubMed] [Google Scholar]
- 3.Celenza, J. L., and M. Carlson. 1986. A yeast gene that is essential for release from glucose repression encodes a protein kinase. Science 233:1175-1180. [DOI] [PubMed] [Google Scholar]
- 4.Cullen, P. J., and G. F. Sprague, Jr. 2000. Glucose depletion causes haploid invasive growth in yeast. Proc. Natl. Acad. Sci. USA 97:13619-13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davies, S. P., D. Carling, and D. G. Hardie. 1989. Tissue distribution of the AMP-activated protein kinase, and lack of activation by cyclic-AMP-dependent protein kinase, studied using a specific and sensitive peptide assay. Eur. J. Biochem. 186:123-128. [DOI] [PubMed] [Google Scholar]
- 6.Estruch, F., M. A. Treitel, X. Yang, and M. Carlson. 1992. N-terminal mutations modulate yeast SNF1 protein kinase function. Genetics 132:639-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.François, J., and J. L. Parrou. 2001. Reserve carbohydrates metabolism in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 25:124-145. [DOI] [PubMed] [Google Scholar]
- 8.Gancedo, J. M. 1998. Yeast carbon catabolite repression. Microbiol. Mol. Biol. Rev. 62:334-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghaemmaghami, S., W. K. Huh, K. Bower, R. W. Howson, A. Belle, N. Dephoure, E. K. O'Shea, and J. S. Weissman. 2003. Global analysis of protein expression in yeast. Nature 425:737-741. [DOI] [PubMed] [Google Scholar]
- 10.Hardie, D. G., D. Carling, and M. Carlson. 1998. The AMP-activated/SNF1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Annu. Rev. Biochem. 67:821-855. [DOI] [PubMed] [Google Scholar]
- 11.Hawley, S. A., J. Boudeau, J. L. Reid, K. J. Mustard, L. Udd, T. P. Mäkelä, D. R. Alessi, and D. G. Hardie. 2003. Complexes between the LKB1 tumor suppressor, STRADα/β and MO25α/β are upstream kinases in the AMP-activated protein kinase cascade. J. Biol. 2:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hedbacker, K., R. Townley, and M. Carlson. 2004. Cyclic AMP-dependent protein kinase regulates the subcellular localization of Snf1-Sip1 protein kinase. Mol. Cell. Biol. 24:1836-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hong, S.-P., F. C. Leiper, A. Woods, D. Carling, and M. Carlson. 2003. Activation of yeast Snf1 and mammalian AMP-activated protein kinase by upstream kinases. Proc. Natl. Acad. Sci. USA 100:8839-8843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang, R., and M. Carlson. 1996. Glucose regulates protein interactions within the yeast SNF1 protein kinase complex. Genes Dev. 10:3105-3115. [DOI] [PubMed] [Google Scholar]
- 15.Ke, S. H., and E. L. Madison. 1997. Rapid and efficient site-directed mutagenesis by single-tube ‘megaprimer’ PCR method. Nucleic Acids Res. 25:3371-3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kemp, B. E., D. Stapleton, D. J. Campbell, Z. P. Chen, S. Murthy, M. Walter, A. Gupta, J. J. Adams, F. Katsis, B. van Denderen, I. G. Jennings, T. Iseli, B. J. Michell, and L. A. Witters. 2003. AMP-activated protein kinase, super metabolic regulator. Biochem. Soc. Trans. 31:162-168. [DOI] [PubMed] [Google Scholar]
- 17.Kuchin, S., I. Treich, and M. Carlson. 2000. A regulatory shortcut between the Snf1 protein kinase and RNA polymerase II holoenzyme. Proc. Natl. Acad. Sci. USA 97:7916-7920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuchin, S., V. K. Vyas, and M. Carlson. 2002. Snf1 protein kinase and the repressors Nrg1 and Nrg2 regulate FLO11, haploid invasive growth, and diploid pseudohyphal differentiation. Mol. Cell. Biol. 22:3994-4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuchin, S., V. K. Vyas, E. Kanter, S.-P. Hong, and M. Carlson. 2003. Std1p (Msn3p) positively regulates the Snf1 kinase in Saccharomyces cerevisiae. Genetics 163:507-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin, S. S., J. K. Manchester, and J. I. Gordon. 2001. Enhanced gluconeogenesis and increased energy storage as hallmarks of aging in Saccharomyces cerevisiae. J. Biol. Chem. 276:36000-36007. [DOI] [PubMed] [Google Scholar]
- 21.Longtine, M. S., A. McKenzie, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 22.Ludin, K., R. Jiang, and M. Carlson. 1998. Glucose-regulated interaction of a regulatory subunit of protein phosphatase 1 with the Snf1 protein kinase in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 95:6245-6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCartney, R. R., and M. C. Schmidt. 2001. Regulation of Snf1 kinase. Activation requires phosphorylation of threonine 210 by an upstream kinase as well as a distinct step mediated by the Snf4 subunit. J. Biol. Chem. 276:36460-36466. [DOI] [PubMed] [Google Scholar]
- 24.Minokoshi, Y., Y. B. Kim, O. D. Peroni, L. G. Fryer, C. Muller, D. Carling, and B. B. Kahn. 2002. Leptin stimulates fatty-acid oxidation by activating AMP-activated protein kinase. Nature 415:339-343. [DOI] [PubMed] [Google Scholar]
- 25.Nath, N., R. R. McCartney, and M. C. Schmidt. 2003. Yeast Pak1 kinase associates with and activates Snf1. Mol. Cell. Biol. 23:3909-3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rahner, A., M. Hiesinger, and H.-J. Schüller. 1999. Deregulation of gluconeogenic structural genes by variants of the transcriptional activator Cat8p of the yeast Saccharomyces cerevisiae. Mol. Microbiol. 34:146-156. [DOI] [PubMed] [Google Scholar]
- 27.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Ruden, D. M., J. Ma, Y. Li, K. Wood, and M. Ptashne. 1991. Generating yeast transcriptional activators containing no yeast protein sequences. Nature 350:250-252. [DOI] [PubMed] [Google Scholar]
- 29.Sanz, P., G. R. Alms, T. A. J. Haystead, and M. Carlson. 2000. Regulatory interactions between the Reg1-Glc7 protein phosphatase and the Snf1 protein kinase. Mol. Cell. Biol. 20:1321-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmidt, M. C., and R. R. McCartney. 2000. β-Subunits of Snf1 kinase are required for kinase function and substrate definition. EMBO J. 19:4936-4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmidt, M. C., R. R. McCartney, X. Zhang, T. S. Tillman, H. Solimeo, S. Wölfl, C. Almonte, and S. C. Watkins. 1999. Std1 and Mth1 proteins interact with glucose sensors to control glucose-regulated gene expression in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:4561-4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song, W., and M. Carlson. 1998. Srb/mediator proteins interact functionally and physically with transcriptional repressor Sfl1. EMBO J. 17:5757-5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sutherland, C. M., S. A. Hawley, R. R. McCartney, A. Leech, M. J. Stark, M. C. Schmidt, and D. G. Hardie. 2003. Elm1p is one of three upstream kinases for the Saccharomyces cerevisiae SNF1 complex. Curr. Biol. 13:1299-1305. [DOI] [PubMed] [Google Scholar]
- 34.Thomas, B. J., and R. Rothstein. 1989. Elevated recombination rates in transcriptionally active DNA. Cell 56:619-630. [DOI] [PubMed] [Google Scholar]
- 35.Treitel, M. A., S. Kuchin, and M. Carlson. 1998. Snf1 protein kinase regulates phosphorylation of the Mig1 repressor in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:6273-6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vincent, O., and M. Carlson. 1999. Gal83 mediates the interaction of the Snf1 kinase complex with the transcription activator Sip4. EMBO J. 18:6672-6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vincent, O., and M. Carlson. 1998. Sip4, a Snf1 kinase-dependent transcriptional activator, binds to the carbon source-responsive element of gluconeogenic genes. EMBO J. 17:7002-7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vincent, O., R. Townley, S. Kuchin, and M. Carlson. 2001. Subcellular localization of the Snf1 kinase is regulated by specific β subunits and a novel glucose signaling mechanism. Genes Dev. 15:1104-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vyas, V. K., S. Kuchin, C. D. Berkey, and M. Carlson. 2003. Snf1 kinases with different β-subunit isoforms play distinct roles in regulating haploid invasive growth. Mol. Cell. Biol. 23:1341-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wiatrowski, H. A., B. J. W. van Denderen, C. D. Berkey, B. E. Kemp, D. Stapleton, and M. Carlson. 2004. Mutations in the Gal83 glycogen-binding domain activate the Snf1/Gal83 kinase pathway by a glycogen-independent mechanism. Mol. Cell. Biol. 24:352-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilson, W. A., S. A. Hawley, and D. G. Hardie. 1996. Glucose repression/derepression in budding yeast: SNF1 protein kinase is activated by phosphorylation under derepressing conditions, and this correlates with a high AMP:ATP ratio. Curr. Biol. 6:1426-1434. [DOI] [PubMed] [Google Scholar]
- 42.Woods, A., S. R. Johnstone, K. Dickerson, F. C. Leiper, L. G. Fryer, D. Neumann, U. Schlattner, T. Wallimann, M. Carlson, and D. Carling. 2003. LKB1 is the upstream kinase in the AMP-activated protein kinase cascade. Curr. Biol. 13:2004-2008. [DOI] [PubMed] [Google Scholar]
- 43.Woods, A., M. R. Munday, J. Scott, X. Yang, M. Carlson, and D. Carling. 1994. Yeast SNF1 is functionally related to mammalian AMP-activated protein kinase and regulates acetyl-CoA carboxylase in vivo. J. Biol. Chem. 269:19509-19516. [PubMed] [Google Scholar]
- 44.Yamauchi, T., J. Kamon, Y. Minokoshi, Y. Ito, H. Waki, S. Uchida, S. Yamashita, M. Noda, S. Kita, K. Ueki, K. Eto, Y. Akanuma, P. Froguel, F. Foufelle, P. Ferre, D. Carling, S. Kimura, R. Nagai, B. B. Kahn, and T. Kadowaki. 2002. Adiponectin stimulates glucose utilization and fatty-acid oxidation by activating AMP-activated protein kinase. Nat. Med. 8:1288-1295. [DOI] [PubMed] [Google Scholar]
- 45.Yang, X., R. Jiang, and M. Carlson. 1994. A family of proteins containing a conserved domain that mediates interaction with the yeast SNF1 protein kinase complex. EMBO J. 13:5878-5886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou, G., R. Myers, Y. Li, Y. Chen, X. Shen, J. Fenyk-Melody, M. Wu, J. Ventre, T. Doebber, N. Fujii, N. Musi, M. F. Hirshman, L. J. Goodyear, and D. E. Moller. 2001. Role of AMP-activated protein kinase in mechanism of metformin action. J. Clin. Investig. 108:1167-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]