Abstract
Glycoprotein D (gD) determines which cells can be infected by herpes simplex virus (HSV) by binding to one of the several cell surface receptors that can mediate HSV entry or cell fusion. These receptors include the herpesvirus entry mediator (HVEM), nectin-1, nectin-2, and sites in heparan sulfate generated by specific 3-O-sulfotransferases. The objective of the present study was to identify residues in gD that are critical for physical and functional interactions with nectin-1 and nectin-2. We found that double or triple amino acid substitutions at positions 215, 222, and 223 in gD caused marked reduction in gD binding to nectin-1 and a corresponding inability to function in cell fusion or entry of HSV via nectin-1 or nectin-2. These substitutions either enhanced or did not significantly inhibit functional interactions with HVEM and modified heparan sulfate. These and other results demonstrate that different domains of gD, with some overlap, are critical for functional interactions with each class of entry receptor. Viral entry assays, using gD mutants described here and previously, revealed that nectins are the principal entry receptors for selected human cell lines of neuronal and epithelial origin, whereas HVEM or nectins could be used to mediate entry into a T lymphocyte line. Because T cells and fibroblasts can be infected via HVEM, HSV strains carrying gD mutations that prevent entry via nectins may establish transient infections in humans, but perhaps not latent infections of neurons, and are therefore candidates for development of safe live virus vaccines and vaccine vectors.
The usual manifestations of human disease caused by herpes simplex viruses (HSV) are mucocutaneous lesions, resulting from replication of virus in epithelial cells of skin or mucosa and influx of leukocytes, which may also become infected. These lesions may be mild or severe, but can recur because of the establishment of latent infections in sensory neurons and subsequent reactivation of replicating virus. Rare manifestations of HSV disease include encephalitis, meningitis, and disseminated disease affecting multiple organ systems. If HSV were unable to invade neurons, the most serious consequences of disease would probably not occur and the latent infections leading to lifelong HSV persistence would likely not occur.
Entry receptors for HSV include human and animal members of three classes of cell surface molecules (reviewed in ref. 1). The herpesvirus entry mediator (HVEM or HveA) (2) is a member of the tumor necrosis factor receptor family, is widely expressed, and has a role in the regulation of immune responses (reviewed in ref. 3). Nectin-1 (Prr1 or HveC) (4, 5) and nectin-2 (Prr2 or HveB) (6), members of the Ig superfamily related to the poliovirus receptor, are cell adhesion molecules that can participate in cadherin-based and other cell junctions, including junctions in the nervous system, and are widely expressed in many organs and tissues (reviewed in ref. 7). Certain isoforms of 3-O-sulfotransferases, such as 3-OST-3A, 3-OST-3B, and 3-OST-5, generate sites in heparan sulfate (3-O-S HS) that can serve as HSV entry receptors (8, 9), whereas others generate sites that can bind to antithrombin to enhance anticoagulation (9, 10). This variety of entry receptors may allow HSV to use different receptors for infection of various target cell types.
There are two serotypes of HSV (HSV-1 and HSV-2) that differ somewhat in natural history of disease and in entry receptor preferences. HSV-1 is the usual cause of oral and corneal lesions and adult encephalitis, whereas HSV-2 is more likely to be the cause of genital herpes, meningitis, and neonatal herpes acquired during delivery. Nectin-1 and HVEM are entry receptors for both HSV-1 and HSV-2 strains, whereas nectin-2 is a better receptor for HSV-2 than for HSV-1 (6), except for some HSV-1 strains isolated from cases of encephalitis (11). On the other hand, 3-O-S HS appears to be a better receptor for HSV-1 than for HSV-2 (8).
Four HSV glycoproteins (gB, gD, gH, and gL) are essential for viral entry (reviewed in ref. 12) and are both necessary and sufficient for HSV-induced cell fusion (13). The viral ligand for all of the entry receptors described above is gD (8, 14–16). It is thought that interaction of gD with one of its receptors somehow triggers the fusogenic activity of one of the other essential HSV glycoproteins (17, 18).
Information about structural requirements for the interactions of gD with some of its receptors has come from x-ray crystallography and mutational analysis of gD. X-ray structures were determined for a large portion of the HSV-1 gD ectodomain, either crystallized alone or in complex with the N-terminal two cysteine-rich domains of HVEM (19). Direct physical contact of HVEM with gD was localized to an N-terminal hairpin in gD and involved amino acids 7–15 and 24–32. Interestingly, this N-terminal region was extended and partly disordered when the gD ectodomain was crystallized alone. Certain amino acid substitutions or insertions within the first 43 aa of HSV-1 or HSV-2 gD can abrogate physical and functional interactions with HVEM (2, 14, 20–22), consistent with the x-ray structure of the gD–HVEM complex. Some of these mutations near the N terminus can also prevent functional interactions of HSV-1 gD with 3-O-S HS (22) and/or even confer ability to use nectin-2 as an entry/fusion receptor (6, 20, 22, 23). Deletion of subsets or all amino acids from positions 7–32 in either HSV-1 or HSV-2 gD abrogated cell fusion activity with all known receptors except nectin-1, leading to the conclusion that the major contact site for nectin-1 must be downstream of the HVEM-binding domain (22). This notion is probably true also for nectin-2, because nectin-1 and nectin-2 are closely related and homologous regions of both are critical for functional interactions with gD (24, 25).
To date, no mutations in gD have been described that abrogate functional interactions with nectin-1 without also affecting activities with other entry receptors such as HVEM (21). The present study was designed to identify such mutations, determine whether they also altered activity with nectin-2, and test whether entry/fusion activity with a nectin was required for infection of selected cell types. We found that double or triple amino acid substitutions at positions 215, 222, and 223 reduced or abolished entry/fusion activity with nectin-1 and nectin-2 without preventing activity with HVEM or 3-O-S HS. Viruses containing these mutant forms of gD were unable to infect human cell lines of neuronal or epithelial origin, whereas viruses containing gD mutants active with nectin-1, but not with any of the other receptors, were able to infect the cells. Thus, nectin-1 and perhaps nectin-2 may be the principal receptors for HSV entry into neurons and viruses unable to use nectins for entry could spare neurons from infection.
Materials and Methods
Cell Lines. Cell lines used included Chinese hamster ovary (CHO-K1) cells; CHO-K1 cells stably expressing human HVEM (2), nectin-1 (4) or nectin-2 (6); a mutant baby hamster kidney cell line (BHK-95-19) (26); Vero cells; a Vero cell line carrying the HSV-1 gD gene (VD60) used for the propagation of gD-negative HSV mutants (27); cell lines derived from human neuroblastomas, SH-SY5Y (28) and IMR-5 (29); cell lines derived from carcinomas (C33A and A431); and Jurkat cells. The CHO cell line and derivatives were grown in Ham's F12 medium supplemented with 10% FBS, and the Jurkat cells were grown in RPMI medium 1640 supplemented with 10% FBS. The other cell lines were grown in DMEM supplemented with 10% FBS.
Virus Strains. HSV-1(KOS)tk12 expresses β-galactosidase (β-gal) from an insert in the viral thymidine kinase gene (6). HSV-1(KOS)gD6 expresses β-gal from an insertion that replaces the gD gene (6). HSV-2(333)UL3/4Gal expresses β-gal from an insert between genes UL3 and UL4 that preserved the polyadenylation sites for both genes (S.M., J. Taylor, A. Zago, and P.G.S., unpublished work). This virus was made gD-negative by isolating an additional recombinant virus in which the gD ORF was replaced by the gene for the enhanced GFP. To do this, VD60 cells were transfected with plasmid pSM152-GFP (see next section), by using Lipofectamine reagent (Invitrogen), and then infected with HSV-2(333)UL3/4Gal at five plaque-forming units (pfu) per cell. Virus was isolated from fluorescent plaques and plaque-purified on VD60 cells. Nucleotide sequencing of PCR products revealed that the gD ORF was replaced with the enhanced GFP ORF as planned. The mutant, HSV-2 (333)gD–Gal, had a titer of 3 × 108 pfu/ml on VD60 cells and <103 on Vero cells.
Plasmids. Plasmid pSM152-GFP was generated from plasmid pMY152, which contains the gD gene and portions of flanking genes (gJ and gI) from HSV-2(333) DNA [nucleotides 140487–143001 based on the published sequence of the related HSV-2(HG52) genome, GenBank accession no. NC_001798], cloned between the EcoRI and HindIII sites of pUC19. To generate pSM152-GFP, nucleotides 140978–141830 of the pMY152 insert were replaced with the enhanced GFP ORF from pEGFP-N1 (Clontech) so that its expression would be driven by the gD promoter. To generate HSV-1 mutant forms of gD, pDM20 was first constructed by PCR amplification of the HSV-1(Patton) gD ORF from pCJ3 (30) and cleavage of the PCR product with HindIII (a natural site upstream of the gD ORF) and XhoI (a site added just after the gD stop codon) for cloning between HindIII and XhoI sites in pGEM 7Zf+ (Promega). HSV-1(Patton) gD is identical in sequence to HSV-1(KOS) gD except for a single amino acid substitution in the cleaved signal sequence. Nucleotide sequence analysis revealed that pDM20 had an unintended mutation (change of codon 215 from GAC to GGC), resulting in a D215G amino acid substitution (the first amino acid after the signal peptidase cleavage site is designated position 1). This mutation, in combination with others, proved to be associated with phenotypes of interest. The QuikChange site-directed mutagenesis kit (Stratagene) was used to repair the mutation in codon 215, generating pDM60. Both pDM20 and pDM60 were used as template for QuikChange mutagenesis to introduce other nucleotide substitutions that resulted in amino acid substitutions, and then the HindIII/XhoI fragment containing the altered gD ORF was subcloned into pcDNA3 to generate expression plasmids pDM73 (D215G), pDM61 (Q132L), pDM27 (R222N), pDM29 (F223I), pDM23 (Q132L/D215G), pDM24 (S140N/D215G), pDM28 (R222N/D215G), pDM30 (F223I/D215G), pDM68 (R222N/F223I), and pDM80 (R222N/F223I/D215G). The pcDNA3-based plasmid expressing wild-type HSV-1 gD was pCJ3 (30). To introduce point mutations into the HSV-2 gD ORF, pMY1, which contains the wild-type HSV-2(333) gD ORF in pUC19 (22), was used as template for QuikChange mutagenesis. The DraIII–BspEI fragment containing the mutation(s) of interest was then excised and substituted for the wild-type HSV-2 gD fragment in pAZD2, which is a pCAGGS-based plasmid expressing wild-type HSV-2(333) gD (31), generating expression plasmids pSM03 (D215G), pSM01 (Q132L/D215G), pSM02 (S140N/D215G), pSM04 (R222N/D215G), pSM05 (F223I/D215G), pSM06 (R222N/F223I), and pSM07 (R222N/F223I/D215G). Regions of each HSV-1 gD ORF encoding the ectodomain were fused to the C-terminal 231 codons of the rabbit IgG heavy chain ORF by subcloning into pDM19 as described (22). The plasmids obtained expressed soluble secreted gD:Fc hybrid proteins. To generate similar hybrids for HSV-2 gD, the EcoRI–BamHI fragment from each of the pCAGGS-based plasmids named above was substituted for the equivalent fragment of pMY12, which expresses wild-type HSV-2 gD:Fc (22). The HSV-1 and HSV-2 constructs, respectively, were pMY80 and pMY12 (wild-type gD), pDM74 and pSM13 (D215G), pDM32 and pSM11 (Q132L/D215G), pDM33 and pSM12 (S140N/D215G), pDM38 and pSM14 (R222N/D215G), pDM40 and pSM15 (F223I/D215G), pDM69 and pSM16 (R222N/F223I), and pCJDM85 and pSM17 (R222N/F223I/D215G). Plasmids expressing other mutant forms of HSV-1 and HSV-2 gD were pMY77 and pMY8 (Q27P) and pMY98 and pMY33 (Δ7–32) (22). Nucleotide sequencing of all plasmids confirmed the presence of desired mutations and absence of unintended mutations. All restriction enzymes were purchased from New England Biolabs.
Preparation of gD:Fc Protein. CHO-K1 cells were plated in 100-mm dishes and transfected with one of the plasmids expressing gD:Fc protein, by using Lipofectamine Plus (Invitrogen) according to the manufacturer's recommendations. After 5 h, 5 ml of DMEM/10% FBS was added to each dish for overnight incubation. The next day, the medium was replaced with Opti-MEM and the cells were incubated for an additional 24 h. The culture supernatants containing the secreted gD:Fc proteins were collected, clarified by low-speed centrifugation, and concentrated five times by using Biomax filters (30-kDa cutoff, Millipore). Concentrations of the gD:Fc proteins were determined by ELISA using an anti-rabbit Fc detection system and rabbit IgG for the standard curve.
Cell Fusion Assay. The transfection and assay conditions and plasmids used were as described (31, 32) except that the gD-expressing plasmids were those expressing the wild-type or mutant forms of HSV-1 or HSV-2 gD. CHO-K1 cells or BHK-95-19 cells (effector cells) were transfected with the HSV-1 or HSV-2 set of plasmids expressing the four glycoproteins required for cell fusion (gB, gD or a gD mutant, gH, and gL) and T7 RNA polymerase [0.8 μg of the gB-expressing plasmid and 0.4 μg of each of the other plasmids per well (6-well plate) with Lipofectamine 2000 for CHO cells or Lipofectamine Plus for BHK-95-19 cells]. Target cells included CHO-HVEM, CHO-nectin-1, and CHO-nectin-2 cells that had been transfected with a plasmid carrying the firefly luciferase gene under control of the T7 promoter (pT7ELCLuc). Other target cells included CHO-K1 cells cotransfected with pT7ELCLuc and a plasmid expressing 3-OST-3A and BHK-95-19 cells cotransfected with pT7ELCLuc and pBEC10 expressing HVEM (2), pBG38 expressing nectin-1 (4), or pMW20 expressing nectin-2 (6). At 6 h after transfection, the effector and target cells were trypsinized and replated at a 1:1 ratio in 96-well plates (5 × 104 cells total per well for CHO and 2.5 × 104 cells for BHK-95-19). After an additional 18 h, the cells were lysed, and expression of luciferase was quantified by using the luciferase assay kit (Promega).
Cell ELISA. The binding of antibodies to cells expressing gD and binding of gD:Fc hybrids to cells expressing HSV entry receptors was quantitated by a cell ELISA. For antibody binding, CHO-K1 cells were transfected with one of the gD-expressing plasmids described above and plasmids expressing T7 RNA polymerase and the other viral glycoproteins required for cell fusion. At 6 h after transfection, the cells were replated (without target cells) in 96-well plates. For gD:Fc binding, CHO-HVEM cells, CHO-nectin-1 cells, or CHO-K1 cells transfected with a plasmid expressing 3-OST-3A (8) were plated directly into 96-well plates. After 18 h (or 42 h for the 3-OST-3A-transfected cells), the cells were washed with PBS and then incubated for 30 min with a rabbit polyclonal anti-gD serum, R7, at a 1:10,000 dilution in PBS/3% BSA, or with serial dilutions of the culture supernatants containing the gD:Fc hybrids. Then, the cells were washed, fixed with PBS containing 2% formaldehyde and 0.2% glutaraldehyde, and incubated sequentially with biotinylated anti-rabbit IgG (Sigma), Amdex streptavidin-conjugated horseradish peroxidase (HRP, Amersham Pharmacia), and HRP substrate (BioFx Lab, Owings Mills, MD). Alternatively, the fixed cells were incubated sequentially with an HRP-coupled anti-rabbit Fc antibody and HRP substrate. HRP product was quantified at 380 nm in a Victor Wallac spectrophotometer (PerkinElmer).
Interference Assay. CHO-K1 cells were cotransfected with a plasmid expressing wild-type or mutant gD and pBG38 expressing human nectin-1 or pcDNA-3 as a control (4:1 ratio using 1.5 μg of plasmid DNA total per well with Lipofectamine reagent in Opti-MEM). After 1 day, the cells were replated into 96-well plates. After an additional 12–24 h, the cells were exposed to serial dilutions of HSV-1(KOS)tk12. Six hours later, the cells were washed, permeabilized, and incubated with the β-gal substrate O-nitrophenyl-β-d-galactopyranoside (Sigma) as described (2). The reaction product was quantified at 405 nm as a measure of viral entry.
Complementation of gD-Negative Viruses for Viral Entry. Vero cells were transfected with plasmids expressing wild-type or mutant forms of gD by using Lipofectamine reagent or Lipofectamine 2000. After 24 h, the cells were infected with the homologous complemented gD-negative virus, either HSV-1(KOS)gD6 [20 plaque-forming units (pfu) per cell] or HSV-2(333)gD-Gal (10 pfu per cell). After 2 h, the virus inocula were removed and residual unpenetrated virus was inactivated by exposure of the cells to 0.1 M citrate buffer (pH 3.0) for 1 min. The cells were washed and incubated in growth medium for 24 h. To prepare virus stocks, the cells were then harvested and sonicated and the cell lysates were centrifuged at low speed to remove debris. Various cell types grown in 96-well plates were inoculated with these stocks (70 μl per well) and incubated for 24 h. Viral entry was then assessed by quantifying the expression of β-gal as described above.
Results
Amino Acid Substitutions in HSV-1 and HSV-2 gD. The amino acid positions targeted for substitutions in gD included Q132, S140, R222, and F223, all of which are conserved between HSV-1 and HSV-2. Substitutions Q132L and S140N were found in HSV-1 mutants selected for resistance to neutralization by certain mAbs (33) that can block the binding of HSV-1 gD to both HVEM and nectin-1 (15). Substitutions R222N and F223I were reported to reduce interactions with nectin-1§ and also targeted a region where three amino acid deletions in HSV-1 gD had been shown to reduce entry activity with both HVEM and nectin-1 (34). Phenotypes of interest were observed, as described below, but not with any single amino acid substitutions. Combinations of these substitutions with each other or with an unintended substitution, D215G, resulted in the novel phenotypes described here. The HSV-1 gD mutations analyzed included Q132L, D215G, R222N, F223I, Q132L/D215G, S140N/D215G, R222N/D215G, F223I/D215G, R222N/F223I, and R222N/F223I/D215G. The HSV-2 gD mutations analyzed included D215G and the double and triple mutations just named. Immunoassays showed that all of the HSV-1 and HSV-2 gD mutants were essentially indistinguishable from wild-type HSV-1 or HSV-2 gD in cell surface expression (values obtained by cell ELISA ranged from 80% to 95% of wild-type controls).
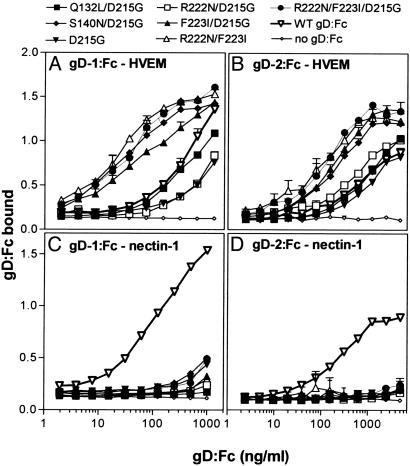
Effects of the gD Mutations on Binding to Entry/Fusion Receptors. Two kinds of experiment were done to determine whether the mutant forms of gD could bind to the various entry receptors. First, CHO cells expressing HVEM, nectin-1 or 3-OST-3A were incubated with soluble forms of gD, hybrids of the gD ectodomains fused to the Fc region of rabbit IgG (gD:Fc hybrids), and binding was quantified by cell ELISA. Fig. 1 shows the results of incubating serial dilutions of the wild-type and mutant forms of gD:Fc with CHO cells that stably express human HVEM or human nectin-1. All of the mutant gD:Fcs were severely impaired for binding to nectin-1 but retained ability to bind to HVEM (Fig. 1). Some of the mutant gD:Fcs (HSV-1 and HSV-2 forms of S140/D215G, F223I/D215G, R222N/F223I, and R222N/F223I/D215G) bound to HVEM more efficiently than did the wild-type gD:Fcs. Two of the HSV-1 mutants (R222N/D215G and D215G) bound to HVEM somewhat less efficiently than wild type. Fig. 2 Upper shows the binding of HSV-1 wild-type and mutant gD:Fcs to CHO-K1 cells expressing 3-OST-3A to generate 3-O-S HS. Binding of wild-type gD:Fc was minimal, barely above the background because of the secondary detection reagents (–gD), but binding of some of the mutant gD:Fcs was significantly elevated, particularly for the R222N/D215G, F223I/D215G, and R222N/F223I/D215G mutants. Binding studies were done with the HSV-2 gD:Fcs and nectin-2 but neither wild-type nor mutant forms of the gD:Fcs exhibited detectable binding as observed for wild type (23, 35).
Fig. 1.
Binding of HSV-1 and HSV-2 wild-type and mutant gD:Fcs to CHO-K1 cells expressing human forms of HVEM and nectin-1. Serial dilutions of concentrated culture supernatants containing known concentrations of the wild-type or mutant forms of HSV-1 gD:Fc (A and C) or HSV-2 gD:Fc (B and D) were incubated with confluent monolayers of CHO-HVEM cells (A and B) or CHO-nectin-1 cells (C and D) in 96-well format. After incubation and washing, the cells were fixed, and then cell-bound gD:Fc was quantified by an Fc detection system. The values presented (HRP reaction product detected at OD380) are means and standard deviations of triplicate determinations and are representative of three independent experiments with similar results.
Fig. 2.
Binding of HSV-1 wild-type and mutant gD:Fcs to CHO cells expressing 3-OST-3A and cell fusion activity of wild-type and mutant HSV-1 gD with target CHO cells expressing 3-OST-3A. (Upper) CHO-K1 cells transfected with a plasmid expressing 3-OST-3A were plated in 96-well dishes and incubated with culture supernatants containing each of the forms of gD:Fc indicated (or no gD:Fc) at 1 μg/ml. Binding of the gD:Fcs to the cells was quantified and the results are presented as described in the legend to Fig. 1. (Lower) CHO-K1 cells were cotransfected with plasmids expressing gB, gD (wild type or mutant or empty vector for the –gD control), gH, gL, and T7 polymerase and were mixed 1:1 with CHO-K1 cells transfected with a plasmid expressing 3-OST-3A and a plasmid carrying the luceriferase gene under control of the T7 promoter or with CHO-K1 cells transfected with just the luciferase plasmid (control CHO cells). The cell mixtures were replated in 96-well dishes at 6 h after transfection. After 18 h of incubation, luciferase activity was quantified as a measure of cell fusion. The bars for the 3-OST-3A-expressing CHO-K1 cells are stacked over the bars for control CHO-K1 cells to show the slightly enhanced fusion of the control cells observed with some mutant forms of HSV-1 gD. The values presented (luciferase activity in arbitrary units) are means and standard deviations of triplicate determinations and are representative of three independent experiments with similar results.
The second type of experiment was to test the ability of membrane-bound forms of wild-type and mutant gD to interact with nectin-1 in an interference assay. When cells are cotransfected with gD and a receptor to which it can bind, the cells can be as resistant to viral entry as if the receptor were not present at all, because of interfering interactions of the cell-associated gD with the receptor (30). CHO-K1 cells were cotransfected with a plasmid expressing human nectin-1 and plasmids expressing wild-type or mutant forms of gD or a control empty vector. After replating in 96-well plates, the cells were inoculated with an HSV-1 recombinant virus that expresses β-gal upon entry, HSV-1(KOS)tk12. Whereas wild-type gD caused severe interference with viral entry (entry was reduced to 10% of that observed for control cells cotransfected with the nectin-1 plasmid and empty vector), mutant R222N/F223I/D215G had no interfering activity. All of the other mutants (D215G, Q132L/D215G, S140N/D215G, R222N/D215G, F223I/D215G, and R222N/F223I) exhibited partial interfering activity, ranging from 40% to 60% of wild-type gD activity.
Effects of the gD Mutations on Cell Fusion Activity with the Various Receptors. Two different cell types were used for cell fusion assays. BHK-95-19 cells were used to test functional activities of the HSV-1 and HSV-2 gD mutants because these cells lack entry/fusion receptors for both serotypes (26). CHO-K1 cells could also be used to test the HSV-1 gD mutants, but not the HSV-2 mutants, because CHO cells lack entry/fusion receptors for HSV-1 but have an endogenous receptor for HSV-2 (31) that gives a very high background level of cell fusion but not of viral entry. CHO-K1 cells had to be used to test functional activities of HSV-1 gD mutants with 3-O-S HS because they express the enzymes required for heparan sulfate modifications that must precede the action of 3-OST-3A (8).
BHK-95-19 cells or CHO-K1 cells were cotransfected with plasmids expressing HSV-1 or HSV-2 gB, gD (mutant or wild type), gH, gL, and T7 polymerase (effector cells) and then detached and replated with a second population of BHK-95–19 (or CHO) cells that had been cotransfected with a plasmid expressing one of the HSV entry receptors and a plasmid carrying the luciferase gene under control of the T7 promoter (target cells). At 18 h after mixing and replating the effector and target cells, the cells were solubilized and substrate was added for the quantification of luciferase activity, as a measure of cell fusion. The results presented in Fig. 3 Middle show that cell fusion activity with nectin-1 as receptor was virtually abolished by the triple mutation, R222N/F223I/D215G, in either HSV-1 or HSV-2 gD. Also, cell fusion activity with nectin-1 was significantly reduced by the double mutations, R222N/D215G, F223I/D215G, and R222N/F223I, especially for HSV-1 gD, whereas activity for the other three mutants (D215G, Q132L/D215G, and S140N/D215G) was >60% that of wild-type gD. These latter mutants supported the fusion of effector cells with nectin-1 target cells at levels higher than would have been predicted from the reduced binding activity shown in Fig. 1, but consistent with the interference results.
Fig. 3.
Cell fusion activities of HSV-1 and HSV-2 gD mutants with target BHK-95-19 cells expressing HVEM, nectin-1, or nectin-2. BHK-95-19 cells were transfected with plasmids expressing the HSV-1 or HSV-2 glycoproteins (gB, gD, gH, and gL) and T7 polymerase (effector cells) or with plasmids expressing one of the entry receptors indicated and the luciferase reporter plasmid (target cells). Controls included effector cells that received empty vector instead of a gD-expressing plasmid and target cells that received empty vector instead of a receptor-expressing plasmid. The cell fusion assay was performed as described in the legend to Fig. 2. The results for each mutant gD (or for the control with no gD) are normalized to the cell fusion activity observed for wild-type gD, set at 100%. Percent of wild-type cell fusion activity = [(luciferase activity for mutant gD in the presence of receptor–luciferase activity for mutant gD in the absence of receptor)/(luciferase activity for wild-type gD in the presence of receptor–luciferase activity for wild-type gD in the absence of receptor)] × 100. The results shown are the means and standard deviations for at least three independent experiments, each done in triplicate.
All of the mutant forms of gD were active for fusion of effector cells with target cells expressing HVEM (Fig. 3 Top), indicating that the mutant proteins were expressed and functional for cell fusion given an appropriate receptor. The levels of cell fusion activity observed ranged from ≈75% to 125% of that associated with wild-type gD, except for HSV-1 mutant F223I/D215G, which was reduced to ≈40% of wild-type activity. Results obtained with the HSV-2 mutants and target cells expressing nectin-2 were essentially the same as those obtained for target cells expressing nectin-1 (Fig. 3 Middle and Bottom). Although the wild-type form of HSV-1 gD was not very active in cell fusion with the receptor generated by 3-OST-3A in CHO-K1 cells (Fig. 2 Lower), three of the mutations (double and triple mutations including the F223I substitution) exhibited significantly enhanced fusion activity with target cells expressing 3-O-S HS, consistent with the gD:Fc binding data (Fig. 2 Upper). Mutation R222N/F223I also enhanced cell fusion mediated by an endogenous receptor, perhaps the same CHO cell receptor that serves, inefficiently, as an entry/fusion receptor for HSV-2 (31).
Use of CHO-HVEM or CHO-nectin-1 cells to assess functional activities of the HSV-1 gD mutants in cell fusion gave results essentially the same as those shown for BHK-95-19 cells in Fig. 3, except that the mutants all exhibited wild-type or enhanced levels of fusion activity with HVEM (data not shown). Three single amino acid substitutions in HSV-1 gD were also tested (Q132L, R222N, and F223I). None of these point mutations had significant effects on cell fusion activity with CHO-HVEM cells (activities observed were 90–130% of that observed for wild-type gD). Cell fusion activity with CHO-nectin-1 cells was greater than wild type for Q132L and R222N (125% and 105%, respectively) and less (75%) for F223I.
Effects of the gD Mutations on Viral Entry via the Various Receptors. Wild-type gD or one of the mutant forms of gD was incorporated into the envelopes of gD-negative mutants of HSV-1 and HSV-2, so that the complemented viruses could be tested for ability to enter cells expressing various receptors. The complemented viruses were obtained by passing the gD-negative mutants once through Vero cells transfected to express either wild-type or mutant gD. Both viral mutants have a lacZ cassette inserted into the viral genome. Viral entry into cells bearing different entry receptors could therefore be compared by quantifying the amount of β-gal expressed at 24 h after inoculation. This process measures the entry of input complemented virus because the mutant viruses generated from the first round of replication would lack gD and would be noninfectious.
Fig. 4 shows that, for both HSV-1 and HSV-2, viruses complemented with the gD mutants having double or triple substitutions in positions D215, R222, and F223 were all significantly impaired for entry into CHO-nectin-1 cells, especially the triple mutant, whereas the other three mutants (D215G, Q132L/D215G, and S140N/D215G) exhibited entry activities equivalent to, or higher than, that observed with wild-type gD. These results parallel the cell fusion results shown in Fig. 3. On the other hand, all of the gD mutants just described were capable of mediating viral entry into cells expressing HVEM, at levels 75–300% of activity associated with wild-type gD. The other two gD mutants shown in Fig. 4 (Q27P and Δ7–32) were previously shown to retain fusion activity with nectin-1 but not with HVEM (22), a result confirmed here for viral entry as well. For each of the replicate experiments summarized in Fig. 4, a single set of fresh complemented virus preparations was used to inoculate both the CHO-HVEM and CHO-nectin-1 cells. Therefore, the failure of some mutants to infect cells via HVEM or nectin-1 was not caused by failure of the mutant gDs to be incorporated into virions, inasmuch as the same complemented virus stock could infect cells via the other receptor.
Fig. 4.
Viral entry activities of the HSV-1 and HSV-2 gD mutants in CHO-HVEM and CHO-nectin-1 cells. HSV-1 and HSV-2 gD-negative mutant viruses were passed through cells expressing each of the wild-type and mutant forms of gD so that the viral envelopes would incorporate the various forms of gD (complementation). Mutants Q27P and Δ7–32 have been described (22). The complemented viruses, which can express β-gal from inserts in the viral genome, were used to inoculate CHO-HVEM and CHO-nectin-1 cells in 96-well dishes. After 24 h, the cells were lysed, and β-gal activity (OD405 for detection of the reaction product) was quantified as a measure of viral entry. The results are normalized to the viral entry activity observed with wild-type gD, set at 100%. Percent of wild-type gD activity = [(absorbance for mutant gD–absorbance in the absence of gD)/(absorbance for wild-type gD–absorbance in the absence of gD)] × 100. The results shown are the means and standard deviations for at least three independent experiments, each done in triplicate.
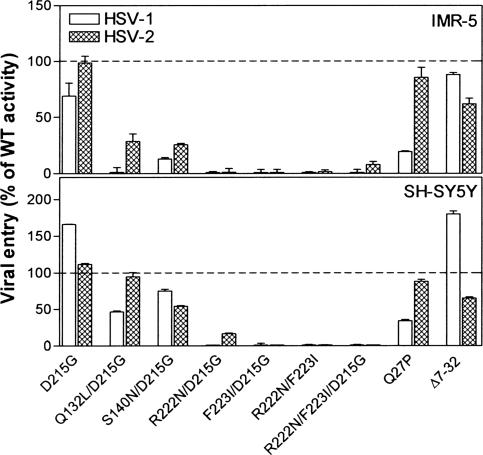
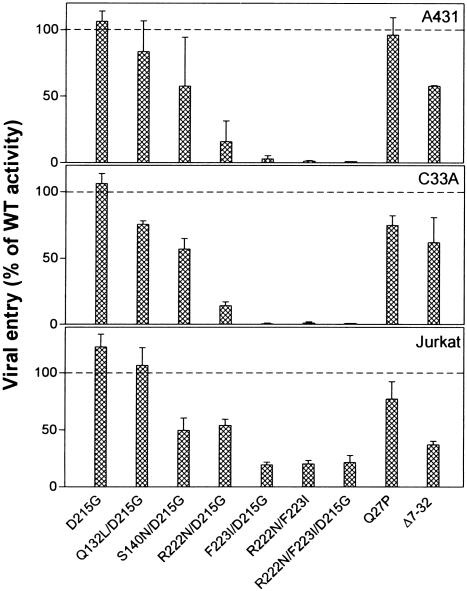
Effects of the gD Mutations on Viral Entry into Various Human Cell Lines. Human cell lines of neuronal, epithelial, and lymphoid origin were selected for entry assays with complemented virus stocks. Two human cell lines derived from neuroblastomas (IMR-5 and SH-SY5Y), the cervical cancer cell line C33A, the vulvar carcinoma cell line A431, and the leukemic Jurkat T cell line were inoculated with the gD-deleted HSV-2 virus preparations complemented with the various HSV-2 gD mutants or wild-type gD. The neuroblastoma cell lines were also inoculated with the analogous complemented HSV-1 virus preparations. Expression of nectin-1 transcripts has been detected in both IMR-5 cells and SH-SY5Y cells and nectin-2 transcripts in SH-SY5Y cells (4, 6). Expression of cell surface nectin-1 has been detected on SH-SY5Y cells and A431 cells and of cell surface HVEM on IMR-5 cells, A431 cells, and Jurkat cells (11, 36).
The results presented in Figs. 5 and 6 demonstrate that viruses complemented with mutants F223I/D215G, R222N/F223I, and R222N/F223I/D215G were severely impaired in ability to infect both the neuroblastoma and epithelial cell lines, despite their ability to infect control CHO-HVEM cells at nearly wild-type efficiency (data not shown) and despite reported expression of HVEM in some of these cells. These mutants were able to infect the Jurkat cells at ≈25% of wild-type gD efficiency. The HSV-2 mutant R222N/D215G exhibited some viral entry activity (≈15% of wild type) with the SH-SY5Y, A431, and C33A cells and ≈50% activity with the Jurkat cells. The other mutants (D215G, Q132L/D215G, and S140N/D215G) exhibited viral entry activities ranging from ≈50% to 100% of wild-type gD levels, except on IMR-5 cells. The Q132L/D215G and S140N/D215G mutants were impaired for entry into IMR-5 cells, but not for entry into cells expressing high levels of nectin-1 (Figs. 4 and 5). It has been noted previously that IMR-5 cells express considerably less nectin-1 than do SH-SY5Y cells, based on mRNA levels (4) and quantitation of receptor levels on cell surfaces (11). The reduced levels of nectin-1 expression on IMR-5 cells may reveal impairment of the Q132L/D215G and S140N/D215G mutants for interactions with nectin-1, an impairment that was evident in the gD:Fc binding assays (Fig. 1).
Fig. 5.
Viral entry activities of the HSV-1 and HSV-2 gD mutants in human neuroblastoma cell lines, IMR-5 and SH-SY5Y. IMR-5 cells and SH-SY5Y were plated in 96-well dishes and inoculated with complemented viruses prepared as described in the legend to Fig. 4. Viral entry was quantified and the results were normalized to viral entry activities obtained with wild-type gD, as described for Fig. 4, for two independent experiments.
Fig. 6.
Viral entry activities of the HSV-2 gD mutants in two human epithelial cell lines, A431 and C33A, and in the Jurkat T cell line. The experiments were done as described in the legends to Figs. 4 and 5. Viral entry was quantified and the results were normalized to viral entry activities obtained with wild-type gD, as described for Fig. 4, for two independent experiments.
HSV-1 and HSV-2 gD-negative viruses complemented with the gD mutants Q27P and Δ7–32 were able to infect all of the human cell lines tested. For reasons we cannot explain, the HSV-1, but not the HSV-2, form of Q27P had reduced ability to mediate viral entry into the neuroblastoma cells. The HSV-2 mutant Δ7–32 had reduced ability to mediate viral entry into the A431 and Jurkat cells. The Q27P mutants retain functional interactions with nectin-1 and nectin-2, whereas the Δ7–32 mutants are functional only with nectin-1 (22). Results obtained with the Jurkat cells indicate that HVEM (used by the double and triple point mutants), nectin-1 (used by Q27P and Δ7–32), and nectin-2 (used by Q27P) all could have a role in HSV-2 entry into these cells.
Discussion
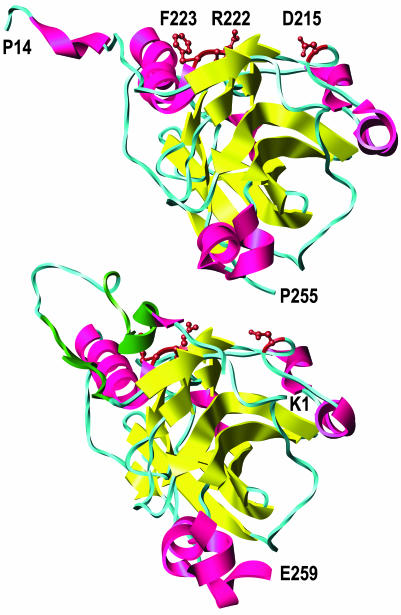
The results presented here, when considered with the x-ray structures of gD (19), identify a potential binding site on gD for the nectins and provide additional evidence that HVEM and the nectins bind to different regions of gD. Fig. 7 presents two models of the gD ectodomain. One model is for gD crystallized alone and the other is for the complex of HVEM with gD. The positions of the amino acid substitutions (R222N/F223I/D215G) that impaired most severely the binding and activity of gD with nectin-1 and nectin-2 are indicated on both models. Note that, in gD crystallized alone, these residues are accessible to solvent and lie together on an exposed surface that could theoretically bind to the nectins. These residues, particularly F223, are exposed only because the N terminus of gD is extended (the first 13 aa are disordered and not shown). In the structure of gD complexed with HVEM, the N terminus assumes a hairpin turn that folds over onto the region of interest, making F223 much less accessible to solvent. These considerations raise the possibility that gD assumes one conformation when bound to HVEM and may have to assume another conformation upon binding to nectin-1 or nectin-2, a possibility previously mentioned by Carfi et al. (19).
Fig. 7.
Locations of the critical amino acid substitutions (D215, R222, and F223, brown) in gD crystallized alone (Upper) and gD crystallized with HVEM (Lower) (HVEM is not shown). β-Strands are yellow, and α-helices are red except for the contact sites with HVEM (amino acids 7–15 and 24–32), which are dark green. The entire ectodomain of gD is ≈316 aa. A truncated form of gD used for crystallization (amino acids 1–285) yielded structures in which only the first 255 (minus 1–13) or 259 aa were visible. The structures shown are based on the coordinates deposited in the Protein Data Bank (37) for entries 1JMA and 1L2G (19). Molecular graphics images were produced by using the ucsf chimera package (38) from the Computer Graphics Laboratory, University of California, San Francisco (supported by National Institutes of Health Grant P41 RR-01081).
Clearly, the principal contact sites on gD for binding to HVEM and nectin-1 are different. The x-ray structure of the HVEM–gD complex revealed that the amino acids of gD directly in contact with HVEM included residues 7–15 and 24–32 in the N-terminal hairpin (19). Amino acid substitutions and deletions that abrogate functional interactions with HVEM lie within this region (2, 14, 20, 22). On the other hand, amino acid substitutions at positions D215, R222, and F223 abrogated physical and functional interactions with nectin-1 and nectin-2 but had little or no effect on such interactions with HVEM.
It should be noted that single amino acid substitutions in the N-terminal domain of HSV-1 gD can confer ability to use nectin-2 as an entry/fusion receptor (6, 20, 22, 23) and that deletion of amino acids 7–32 abrogates the ability of HSV-2 gD to interact functionally with nectin-2 but is without effect on interactions with nectin-1, for either HSV-1 or HSV-2 gD (22). The N-terminal region of HSV-1 or HSV-2 gD somehow influences whether a functional interaction with nectin-2, but not nectin-1, can occur. Perhaps the N terminus provides a secondary contact site for nectin-2 or influences the conformation of the primary contact site in a manner that is critical for nectin-2 binding but irrelevant for nectin-1 binding.
A surprising effect of the amino acid substitutions at positions D215, R222, and F223 (double and triple mutations) was significant enhancement of the ability of HSV-1 gD to bind to 3-O-S HS and to use sites in this modified heparan sulfate as cell fusion receptors. Mutations in the N terminus of HSV-1 gD (L25P, Q27P/R, or Δ7–32) abrogate functional interactions with 3-O-S HS (22). These latter mutations are near a positively charged deep pocket in the gD structure that was occupied by an anion (probably a sulfate ion) and was suggested to be a potential binding site for 3-O-S HS (19).
Entry activities of the complemented HSV-1 and HSV-2 viruses permit the conclusion that nectins are the principal entry receptors for most of the human cell lines tested, namely those of neuronal and epithelial origin. These cells could be infected with viruses complemented by wild-type gD or a mutant form of gD (Δ7–32) capable of using only nectin-1 as an entry receptor although, for HSV-2, entry activities were higher when the Q27P mutant (capable of using both nectin-1 and nectin-2) was used. In contrast, these cell lines failed to be infected by viruses complemented by gD mutants that were unable to infect cells via nectin-1 or nectin-2 but could use HVEM or (in the case of HSV-1) 3-O-S HS as entry receptors. Results obtained with the Jurkat cell line indicate that these cells of T lymphocyte origin can be infected via HVEM, consistent with their reported expression of HVEM (36), or via one or both of the nectins. A potential caveat to these conclusions is the possibility that one or more HSV entry receptors remain undiscovered and that the mutations described here affect usage of these receptors similarly to the nectins.
It is of interest that susceptibilities of some of the cell lines to infection by the complemented HSV-1 or HSV-2 viruses were not necessarily predictable from quantitation of receptor expression on the surfaces of suspended cells (11). Both the SH-SY5Y and A431 cells exhibited high levels of nectin-1 expression on cell surfaces, consistent with their susceptibility to viruses capable of entering cells via nectin-1. On the other hand, nectin-1 was barely, if at all, detectable on the IMR-5 cells. The IMR-5 and A431 cells exhibited significant levels of HVEM expression on cell surfaces. It remains to be explained why these cells could not be infected via HVEM, by the viruses complemented with the double and triple substitution mutants. Evidence was presented that certain human fibroblast types can be infected via either nectins or HVEM (11).
In summary, we have identified a potential contact site on gD for nectin-1 and nectin-2 and demonstrated that structural features of gD critical for interactions with the nectins and HVEM are different. We have generated mutations in HSV-1 and HSV-2 gD that abrogate physical and functional interactions with the nectins, but not with HVEM or 3-O-S HS, and that prevent the infection of certain human cell lines of neuronal and epithelial origin. HSV strains carrying these mutations are under construction and will be useful for characterizing receptor usage in various cell types, exploring the role of various receptors in viral pathogenesis and, potentially, generating nonneurotropic vaccine and vector strains.
Acknowledgments
We thank D. Pleasure (University of Pennsylvania, Philadelphia), B. Herold (Mount Sinai Medical Center, New York), and K. Green, L. Laimins, and R. Longnecker (all of Northwestern University) for various cell lines; R. Eisenberg and G. Cohen (University of Pennsylvania) for the anti-gD antiserum; A. Zago (Northwestern University) for generation of the plasmid used to construct HSV-2(333)UL3/4Gal; J. Taylor (Northwestern University) for construction and isolation of HSV-2(333)UL3/4Gal; N. Susmarski (Northwestern University) for excellent technical assistance with cell culture and virus propagation; and A. Kirschner (Northwestern University) for assistance with the chimera modeling application. This work was supported by U.S. Public Health Service Grants CA-21776, AI-31494, AI-36293, and AI-53774.
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected on April 30, 2002.
Abbreviations: gB, gD, gH, and gL, glycoproteins B, D, H, and L, respectively; HSV, herpes simplex virus; HVEM, herpesvirus entry mediator; 3-OST, 3-O-sulfotransferase; 3-O-S HS, 3-O-sulfated heparan sulfate; CHO, Chinese hamster ovary; BHK, baby hamster kidney; β-gal, β-galactosidase; HRP, horseradish peroxidase.
See accompanying Biography on page 12411.
Footnotes
Bai, Q., Shah, W. A., Cohen, J. B., Eisenberg, R. J., Cohen, G. H. & Glorioso, J. C., 26th International Herpesvirus Workshop, July 28–Aug. 3, 2001, Regensburg, Germany, abstr. 2.10.
References
- 1.Spear, P. G., Eisenberg, R. J. & Cohen, G. H. (2000) Virology 275, 1–8. [DOI] [PubMed] [Google Scholar]
- 2.Montgomery, R. I., Warner, M. S., Lum, B. J. & Spear, P. G. (1996) Cell 87, 427–436. [DOI] [PubMed] [Google Scholar]
- 3.Granger, S. W. & Rickert, S. (2003) Cytokine Growth Factor Rev. 14, 289–296. [DOI] [PubMed] [Google Scholar]
- 4.Geraghty, R. J., Krummenacher, C., Cohen, G. H., Eisenberg, R. J. & Spear, P. G. (1998) Science 280, 1618–1620. [DOI] [PubMed] [Google Scholar]
- 5.Cocchi, F., Menotti, L., Mirandola, P., Lopez, M. & Campadelli-Fiume, G. (1998) J. Virol. 72, 9992–10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Warner, M. S., Geraghty, R. J., Martinez, W. M., Montgomery, R. I., Whitbeck, J. C., Xu, R., Eisenberg, R. J., Cohen, G. H. & Spear, P. G. (1998) Virology 246, 179–189. [DOI] [PubMed] [Google Scholar]
- 7.Takai, Y., Irie, K., Shimizu, K., Sakisaka, T. & Ikeda, W. (2003) Cancer Sci. 94, 655–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shukla, D., Liu, J., Blaiklock, P., Shworak, N. W., Bai, X., Esko, J. D., Cohen, G. H., Eisenberg, R. J., Rosenberg, R. D. & Spear, P. G. (1999) Cell 99, 13–22. [DOI] [PubMed] [Google Scholar]
- 9.Xia, G., Chen, J., Tiwari, V., Ju, W., Li, J.-P., Malmstrom, A., Shukla, D. & Liu, J. (2002) J. Biol. Chem. 277, 37912–37919. [DOI] [PubMed] [Google Scholar]
- 10.Liu, J., Shworak, N. W., Sinay, P., Schwartz, J. J., Zhang, L., Fritze, L. M. S. & Rosenberg, R. (1999) J. Biol. Chem. 274, 5185–5192. [DOI] [PubMed] [Google Scholar]
- 11.Krummenacher, C., Baribaud, F., Ponce de Leon, M., Baribaud, I., Whitbeck, J. C., Xu, R., Cohen, G. H. & Eisenberg, R. J. (2004) Virology 322, 286–299. [DOI] [PubMed] [Google Scholar]
- 12.Spear, P. G. (1993) Semin. Virol. 4, 167–180. [Google Scholar]
- 13.Turner, A., Bruun, B., Minson, T. & Browne, H. (1998) J. Virol. 72, 873–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whitbeck, J. C., Peng, C., Lou, H., Xu, R., Willis, S. H., Ponce de Leon, M., Peng, T., Nicola, A. V., Montgomery, R. I., Warner, M. S., et al. (1997) J. Virol. 71, 6083–6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krummenacher, C., Nicola, A. V., Whitbeck, J. C., Lou, H., Hou, W., Lambris, J. D., Geraghty, R. J., Spear, P. G., Cohen, G. H. & Eisenberg, R. J. (1998) J. Virol. 72, 7064–7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cocchi, F., Lopez, M., Menotti, L., Aoubala, M., Dubreuil, P. & Campadelli-Fiume, G. (1998) Proc. Natl. Acad. Sci. USA 95, 15700–15705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spear, P. G. & Longnecker, R. (2003) J. Virol. 77, 10179–10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cocchi, F., Fusco, D., Menotti, L., Gianni, T., Eisenberg, R. J., Cohen, G. H. & Campadelli-Fiume, G. (2004) Proc. Natl. Acad. Sci. USA 101, 7445–7450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carfi, A., Willis, S. H., Whitbeck, J. C., Krummenacher, C., Cohen, G. H., Eisenberg, R. J. & Wiley, D. C. (2001) Mol. Cell 8, 169–179. [DOI] [PubMed] [Google Scholar]
- 20.Connolly, S. A., Landsburg, D. J., Carfi, A., Wiley, D. C., Cohen, G. H. & Eisenberg, R. J. (2003) J. Virol. 77, 8127–8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jogger, C. R., Montgomery, R. I. & Spear, P. G. (2004) Virology 318, 318–326. [DOI] [PubMed] [Google Scholar]
- 22.Yoon, M., Zago, A., Shukla, D. & Spear, P. G. (2003) J. Virol. 77, 9221–9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lopez, M., Cocchi, F., Menotti, L., Avitabile, E., Dubreuil, P. & Campadelli-Fiume, G. (2000) J. Virol. 74, 1267–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinez, W. M. & Spear, P. G. (2001) J. Virol. 75, 11185–11195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martinez, W. M. & Spear, P. G. (2002) J. Virol. 76, 7255–7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roller, R. J. & Herold, B. C. (1997) J. Virol. 71, 5805–5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ligas, M. W. & Johnson, D. C. (1988) J. Virol. 62, 1486–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ross, R. A., Spengler, B. A. & Biedler, J. L. (1983) J. Natl. Cancer Inst. 71, 741–747. [PubMed] [Google Scholar]
- 29.Gilbert, F. & Balabam-Malenbaum, G. B. (1980) Prog. Cancer Res. Ther. 12, 59–72. [Google Scholar]
- 30.Geraghty, R. J., Jogger, C. R. & Spear, P. G. (2000) Virology 268, 147–158. [DOI] [PubMed] [Google Scholar]
- 31.Zago, A. & Spear, P. G. (2003) J. Virol. 77, 9695–9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pertel, P., Fridberg, A., Parish, M. L. & Spear, P. G. (2001) Virology 279, 313–324. [DOI] [PubMed] [Google Scholar]
- 33.Muggeridge, M. I., Wu, T. T., Johnson, D. C., Glorioso, J. C., Eisenberg, R. J. & Cohen, G. H. (1990) Virology 174, 375–387. [DOI] [PubMed] [Google Scholar]
- 34.Whitbeck, J. C., Muggeridge, M. I., Rux, A. H., Hou, W., Krummenacher, C., Lou, H., van Geelen, A., Eisenberg, R. J. & Cohen, G. H. (1999) J. Virol. 73, 9879–9890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Struyf, F., Martinez, W. M. & Spear, P. G. (2002) J. Virol. 76, 12940–12950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhai, Y., Guo, R., Hsu, T.-L., Yu, G.-L., Ni, J., Kwon, B. S., Jiang, G.-W., Lu, J., Tan, J., Ugustus, M., et al. (1998) J. Clin. Invest. 102, 1142–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. M., Weissig, H., Shindyalov, I. N. & Bourne, P. E. (2000) Nucleic Acids Res. 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang, C. C., Couch, G. S., Pettersen, E. F. & Ferrin, T. E. (1996) chimera, An Extensible Molecular Modeling Application Constructed Using Standard Components (Computer Graphics Lab., Univ. of Calif., San Francisco).