Abstract
Pyrrolysine, the 22nd cotranslationally inserted amino acid, was found in the Methanosarcina barkeri monomethylamine methyltransferase protein in a position that is encoded by an in-frame UAG stop codon in the mRNA. M. barkeri encodes a special amber suppressor tRNA (tRNAPyl) that presumably recognizes this UAG codon. It was reported that Lys-tRNAPyl can be formed by the aminoacyl-tRNA synthetase-like M. barkeri protein PylS [Srinivasan, G., James, C. M. & Krzycki, J. A. (2002) Science 296, 1459–1462], whereas a later article showed that Lys-tRNAPyl is synthesized by the combined action of LysRS1 and LysRS2, the two different M. barkeri lysyl-tRNA synthetases. Pyrrolysyl-tRNAPyl formation was presumed to result from subsequent modification of lysine attached to tRNAPyl. To investigate whether pyrrolysine can be directly attached to tRNAPyl we chemically synthesized pyrrolysine. We show that PylS is a specialized aminoacyl-tRNA synthetase for charging pyrrolysine to tRNAPyl; lysine and tRNALys are not substrates of the enzyme. In view of the properties of PylS we propose to name this enzyme pyrrolysyl-tRNA synthetase. In contrast, the LysRS1:LysRS2 complex does not recognize pyrrolysine and charges tRNAPyl with lysine. These in vitro data suggest that Methanosarcina cells have two pathways for acylating the suppressor tRNAPyl. This would ensure efficient translation of the in-frame UAG codon in case of pyrrolysine deficiency and safeguard the biosynthesis of the proteins whose genes contain this special codon.
The Methanosarcinaceae are methanogenic archaea that are distinguished by their ability to reduce a wide variety of compounds other than carbon dioxide to methane. The first described species, Methanosarcina barkeri, can produce methane from carbon dioxide, acetate, methanol, methylated thiols, or methylated amines (1). Methanogenesis from methylamines (mono-, di-, or trimethylamine) requires substrate-specific methyltransferases: monomethylamine methyltransferase (mtmB), dimethylamine methyltransferase (mtbB), or trimethylamine methyltransferase (mttB) (1). The genes encoding these enzymes are unique in that they possess an in-frame amber stop codon that Methanosarcina cells need to read through (suppress) to produce a full-length protein (2). This phenomenon was first observed for the M. barkeri MS mtmB gene and mRNA; the corresponding protein contains a lysine residue at the position specified by the in-frame UAG (3). However, the crystal structure of the monomethylamine methyltransferase protein revealed a modified lysine, pyrrolysine, in this position (4). At the same time it was shown that a special amber suppressor tRNA (tRNAPyl encoded by pylT) existed in the organism presumably for the purpose of reading the special UAG codon (5). The same study reported that the M. barkeri pylS gene encoded the enzyme PylS, later named lysine-tRNAPyl ligase (EC 6.1.1.25), that charged the pylT suppressor tRNA specifically with lysine (5). In addition, a different way of attaching lysine to pylT suppressor tRNA was later reported (6); lysyl-tRNAPyl could be formed by the combined action of the two classes of LysRS enzymes present in the Methanosarcinaceae, LysRS1 and LysRS2. Even though these results showed different ways to charge tRNAPyl with lysine, both studies supported the principle of an indirect, tRNA-dependent amino acid modification pathway of pyrrolysine synthesis, similar to that of selenocysteine formation (7). However, the possible occurrence of a direct pathway, and of pyrrolysine being a normal metabolite in Methanosarcinaceae (8), could not be dismissed without testing the direct attachment of pyrrolysine to tRNAPyl.
Here we show that PylS is an aminoacyl-tRNA synthetase-like enzyme specific for pyrrolysine (but not lysine) and tRNAPyl (but not tRNALys). However, the LysRS1:LysRS2 complex does not recognize pyrrolysine and charges tRNAPyl only with lysine (6). These in vitro data support the view that Methanosarcina cells have two strategies for charging the suppressor tRNAPyl: to ensure efficient read-through of the in-frame UAG codon and to safeguard the biosynthesis of full-length methylamine methyltransferase protein.
Materials and Methods
General. Oligonucleotide synthesis and DNA sequencing was performed by the Keck Foundation Biotechnology Resource Laboratory at Yale University. Uniformly labeled sodium [32P]pyrophosphate [1–60 Ci/mmol (1 Ci = 37 GBq)] and [γ-32P]ATP (6,000 Ci/mmol) were from Amersham Biosciences. Lysine was from Sigma.
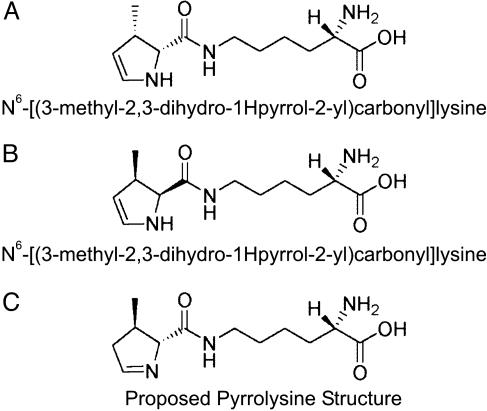
Pyrrolysine Synthesis. N6-[(3-methyl-2,3-dihydro-1H-pyrrol-2-yl)-carbonyl]lysine (Fig. 1) was chemically synthesized(A.B., unpublished results). The product was purified by HPLC and freed from any contaminating lysine (as shown by MS). The product was characterized by NMR. The spectra are as follows: 1H NMR (400 MHz, CD3OD) δ 6.21 (dd, J = 3.2, 7.9 Hz, 1H), 5.04 (dd, J = 2.0, 7.8 Hz, 1H), 3.92–3.88 (m, 2H), 3.72 (d, J = 14.1 Hz, 1H), 3.65–3.54 (m, 2H), 3.49–3.37 (m, 2H), 3.27–3.17 (m, 1H), 2.76–2.68 (m, 1H), 1.93–1.82 (m, 2H), 1.64–1.57 (m, 2H), 1.44–1.39 (m, 2H), 1.22 (d, J = 6.9 Hz, 3H); 13C NMR (100 MHz, CD3OD) δ 172.76, 166.62, 130.14, 112.72, 79.87, 58.35, 57.07, 54.04, 47.34, 32.52, 31.66, 29.18, 28.58, 28.05, 23.74, 23.37, 18.92, 18.77, and 18.24 ppm.
Fig. 1.
Structures of pyrrolysine derivatives. (A and B) Diastereomers of the chemically synthesized N6-[(3-methyl-2,3-dihydro-1H-pyrrol-2-yl)carbonyl]-lysine. (C) Proposed structure of pyrrolysine. The chiral arrangement in the pyrrole ring was patterned after the crystallographic image in figure 2C of ref. 5.
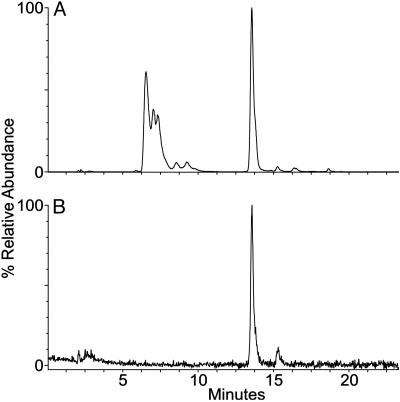
Preliminary data suggested an alkali-facilitated conversion of the enamine (Fig. 1 A and B) into the imine form (Fig. 1C), by a treatment with 40 mM NaOH for 30 min at 37°C and subsequent neutralization. For the experiments in this paper, this sample was called “pyrrolysine.” Electrospray ionization (ESI) MS (see below) indicated the presence of several pyrrolysine isomers: Mr 255.2 calc. for C12H21N3O3; obs. m/z 256.2 (MH+) (Fig. 2A), m/z 278.3 (MNa+), m/z 294 (MK+).
Fig. 2.
Reconstructed ion chromatograms for m/z 256 (MH+ for pyrrolysine) from LC/ESI MS analysis of synthetic pyrrolysine (A) and the amino acid in pryrrolysyl-tRNAPyl generated by PylS in the presence of the synthetic pyrrolysine preparation (B).
Purification of Mature M. barkeri MS tRNAPyl. M. barkeri MS cells were extracted, and unfractionated tRNA was purified as described in ref. 6. The tRNAPyl species was isolated by using a sequence-specific biotinylated oligonucleotide as reported in ref. 6. Preliminary sequence analysis showed the tRNA to be >50% pure.
In Vitro Transcription of tRNAPyl Gene. The M. barkeri Fusaro pylT was cloned and transcribed as reported in ref. 6.
Overexpression and Purification of M. barkeri Fusaro PylS. The M. barkeri Fusaro pylS gene previously cloned (6) into the pET15b vector (Novagen) was transformed into Epicurian coli BL21-CodonPlus (DE3)-RIL (Stratagene). The transformant was grown at 37°C in Luria broth supplemented with ampicillin (100 μg/ml) and chloramphenicol (34 μg/ml) to a cell density of A600 = 1.4. The expression of the recombinant protein was then induced by isopropyl-β-d-thio-galactopyranoside (final concentration of 1 mM) for 3 h at 37°C. The cells were harvested by centrifugation and washed with PBS; this and subsequent steps were performed at 4°C. The cell paste was resuspended into lysis buffer [100 mM Hepes, pH 7.2/500 mM NaCl/5 mM 2-mercaptoethanol/protease inhibitor (Hoffmann–La Roche)] and sonicated for 10 cycles of 30 s. The cell extract was obtained by ultracentrifugation at 40,000 × g for 1 h and subsequently loaded onto a Ni-NTA agarose column (Qiagen, Valencia, CA), according to the standard procedure. The desired protein was eluted with lysis buffer containing 750 mM imidazole. The fractions containing the protein were pooled, and the elution buffer was exchanged on a PD-10 column (Amersham Biosciences) for 100 mM Hepes (pH 7.2), 10 mM MgCl2, 30 mM KCl, 5 mM DTT, and 40% glycerol. The PylS protein preparation was stored at –20°C. The enzyme was >95% pure as judged by Coomassie staining of an SDS/PAGE gel.
To ascertain that the PylS preparation was not contaminated with Escherichia coli LysRS, aminoacylation of unfractionated E. coli tRNA was carried out in the presence of saturating amounts of lysine. Because the PylS preparation was unable to lysylate the tRNA (data not shown), we concluded that it was free of any contaminating LysRS.
Overexpression and Purification of LysRS Proteins. The M. barkeri lysK (encoding LysRS1) and lysS (encoding LysRS2) genes cloned into the pET15b vector (Novagen) and transformed into Epicurian coli BL21-CodonPlus (DE3)-RIL (Stratagene) were overexpressed and purified as described in ref. 9.
ATP-32PPi Exchange with Pyrrolysine and Lysine. The reactions were performed at 37°C, as described in ref. 9, in 50 mM Hepes-NaOH (pH 7.2), 10 mM MgCl2, 50 mM potassium chloride, 5 mM DTT, 2 mM KF, 2 mM ATP, 2 mM 32PPi (2 cpm/pmol), 1 μM of the enzymes, and 500 μM pyrrolysine or l-lysine in a final volume of 0.1 ml. [32P]ATP formation was followed by specific absorption on acid-washed Norit [0.2 ml of a 1% suspension (wt/vol) of Norit in a solution of 0.4 M sodium pyrophosphate solution containing 15% (vol/vol) perchloric acid], filtration on Whatman GF/C filters, and washing with 15 ml of water and 5 ml of ethanol.
Aminoacylation Assays. These were performed for 90 min at 37°C in 100 mM Hepes (pH 7.2), 30 mM KCl, 15 mM MgCl2, 5 mM ATP, 5 mM DTT, 500 μM lysine or 500 μM pyrrolysine, 2 μM tRNAPyl transcript, and 1.6 μM PylRS, LysRS1, or LysRS2. All reactions were started by addition of the amino acid.
Analysis of Amino Acid Charged to tRNAPyl. tRNAPyl was acylated with pyrrolysine. The reaction mixture (1 ml) contained 3 μM tRNAPyl transcript, 500 μM pyrrolysine, and 3.2 μM M. barkeri Fusaro PylS in 100 mM Hepes (pH 7.2), 30 mM KCl, 15 mM MgCl2, 5 mM ATP, and 5 mM DTT at 37°C for 90 min. The reaction was stopped by the addition of 1 vol of 0.3 M NaOAc (pH 5.0)/10 mM EDTA. After phenol extraction and ethanol precipitation, the sample was deacylated at 37°C for 1 h in water. The amino acid was separated from the tRNA on a Microcon YM-10 (Millipore). The filtrate was recovered and dried in speed vacuum, and the content was analyzed by MS.
Acid Urea Gel Electrophoresis of Aminoacyl-tRNA and Northern Hybridization. This method (10) allows the separation of charged and uncharged tRNAs based on the electrophoretic mobility difference seen between the two species. Hybridization to a sequence-specific probe permits the determination of the identity of the aminoacylated tRNA.
tRNAPyl transcripts were used in lysylation or pyrrolysylation reactions. The reactions were stopped with 1 vol of 0.3 M sodium acetate (pH 4.5)/10 mM EDTA. After phenol extraction and ethanol precipitation, the aminoacyl-tRNAs were dissolved in 2× loading buffer [7 M urea/0.3 M sodium acetate (pH 4.5)/10 mM EDTA/0.1% bromophenol blue/0.1% xylene cyanol] and were loaded (0.4 μg of transcript and 20 μg of unfractionated tRNA) on a 6.5% polyacrylamide gel (50 × 20 cm, 0.4 mm thick) containing 7 M urea and 0.1 M sodium acetate (pH 5.0). The gel wasrunat4°C, 500 V, in 0.1 M sodium acetate (pH 5.0) for 18 h. Detection of the tRNAs was performed by Northern blotting. For this purpose the portion of the gel containing the tRNAs was electroblotted onto a Hybond N+ membrane (Amersham Biosciences) by using a Hoefer electroblot apparatus (Amersham Biosciences) at 10 V for 15 min and then at 30 V for 2 h with 10 mM Tris-acetate (pH 8.0)/5 mM sodium acetate/0.5 mM Na-EDTA as the transfer buffer. The membranes were cross-linked for 2 min in a Stratalinker UV Crosslinker 2400 (Stratagene). The tRNAs were detected by hybridization with a 5′ [32P]-labeled oligodeoxyribonucleotide probe. The probes were complementary to nucleotides 1–17 of tRNAPyl.
Nuclease Digestion of tRNAPyl. Mature tRNAPyl (1 μg) was hydrolyzed to nucleosides by using nuclease P1, phosphodiesterase I, and Antarctic phosphatase (New England Biolabs) as described in ref. 11. Ten picomoles of tRNAPyl in 10 mM Tris/1 mM EDTA (pH 7) was totally digested to Gp-terminated oligonucleotides by using 1,000 units of RNase T1 (Ambion, Austin, TX) in a total volume of 6 μl; the solution was incubated for 30 min at 37°C. Each of the tRNA digests was analyzed directly by MS, without any cleanup.
Liquid Chromatography (LC)/ESI MS. Amino acid analysis. Pyrrolysine solutions and the native tRNAPyl nucleoside hydrolysate were analyzed on an Hewlett–Packard 1090 liquid chromatograph coupled directly to a Micromass (Beverly, MA) Quattro II mass spectrometer. Analytes were separated on a Develosil C30 column by using a multilinear gradient essentially as described in ref. 12, except that the flow rate was 0.3 ml/min and the concentration of buffer A was 5 mM. The ion source and nebulizer temperatures were 140°C and 275°C, respectively. Nucleosides were identified as described in ref. 12. The pyrrolysine samples were characterized by generating time vs. abundance profiles (a “reconstructed ion chromatogram,” RIC) for m/z 147 (MH+ of lysine) and for m/z 256 (MH+ of pyrrolysine). The relative amounts of each of the pyrrolysine isomers were determined as the integrated area fraction of the RIC of each selected peak relative to the sum of all of the responses.
Analysis of modified nucleosides. Oligonucleotides from purified tRNAPyl were analyzed by using a Waters CapLC liquid chromatograph coupled without solvent stream splitting to a Micromass Q-Tof 2 mass spectrometer. Both instruments were controlled by using a Micromass MassLynx V.3.4 data system. Separations were carried out on a Luna C18 (2) column (150 × 0.5 mm; Phenomenex, Belmont, CA) at a flow rate of 17 μl/min by using a hexaf luoroisopropanol/tetraethylammonium/methanol gradient as described in ref. 13, except that the initial concentration of hexafluordisopropanol/tetraethylammonium buffer was 0.4 M. Ion source and nebulizer temperatures were 80°C and 135°C, respectively; negative ions were detected. The eluate was conducted directly into the mass spectrometer. Data were collected over a mass range of m/z 300–1,900.
Results
Chemical Synthesis of Pyrrolysine. The crystal structure of M. barkeri monomethylamine methyltransferase suggested a structure for pyrrolysine as a potentially labile (4R,5R)-4-substituted-pyrroline-5-carboxylate with a possible methyl, hydroxyl, or amino substitution (4). Because chemical synthesis of the 4-hydroxyl compound was unsuccessful in our hands, we decided to synthesize the 4-methyl derivative (Fig. 1). The synthetic product turned out to be N6-[(3-methyl-2,3-dihydro-1H-pyrrol-2-yl)carbonyl]lysine (A.B., unpublished work; see Materials and Methods and Fig. 1 A and B), the enamine isomer of pyrrolysine. To facilitate conversion to the imine form of pyrrolysine, we treated our compound with alkali (see Materials and Methods) and analyzed the mixture by LC/ESI MS (Fig. 2 A). It needs to be emphasized that our “pyrrolysine” sample (used in the assays below) consists of several isomers of our putative pyrrolysine but is free from lysine contamination (data not shown). The question of what residue resides in the 4 position of natural pyrrolysine is unresolved (4).
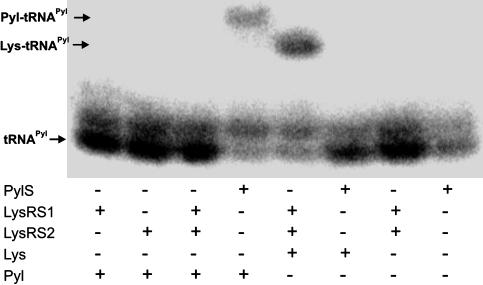
PylS Forms Pyrrolysyl-tRNAPyl. Because our pyrrolysine sample was nonradioactive, we decided to analyze the M. barkeri PylS-catalyzed acylation reactions by acidic urea gel electrophoresis, followed by Northern hybridization with tRNAPyl-specific probes (10). The results show that PylS acylates pyrrolysine but not lysine on to tRNAPyl (compare lanes 4 and 6 in Fig. 3). However, the LysRS1:LysRS2 combination (Fig. 3, lane 5), which forms lysyl-tRNAPyl as reported in ref. 6, is unable to use pyrrolysine (Fig. 3, lane 3). These results further support our finding (6) that LysRS1 and LysRS2 form a specific complex that charges tRNAPyl with lysine. Individually, LysRS1 or LysRS2 do not charge tRNAPyl with either lysine or pyrrolysine. The nucleotide modifications present in mature tRNAPyl are not essential for PylS recognition, as the data in Fig. 3 were obtained with the unmodified tRNAPyl transcript. Periodate treatment of the tRNAPyl transcript prevented subsequent acylation with pyrrolysine (data not shown), indicating that successful acylation takes place at the tRNA's 3′ end.
Fig. 3.
Northern blot analysis of the M. barkeri Fusaro tRNAPyl transcript charged with lysine (Lys) or pyrrolysine (Pyl) by the M. barkeri Fusaro PylS, LysRS1, and LysRS2 enzymes. The resulting tRNA products were loaded onto an acid gel. After transfer to a nitrocellulose membrane, the samples were hybridized with a tRNAPyl-specific oligonucleotide probe. The positions of tRNAPyl, pyrrolysyl-tRNAPyl, and lysyl-tRNAPyl are indicated. The slower-moving band (above tRNAPyl) is an extended transcription product.
Given that our pyrrolysine preparation contained several isomers, we wanted to check whether PylS would discriminate among them. Therefore, a sample of pyrrolysyl-tRNAPyl was deacylated, and the resulting pyrrolysine fraction was analyzed by MS. It was satisfying to see (Fig. 2B) that PylS specifically recognized only one of the isomers.
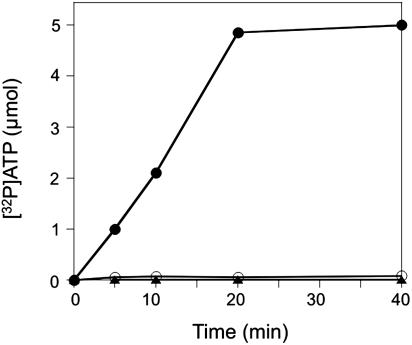
PylS Activates Pyrrolysine. To investigate the amino acid specificity of PylS, we measured amino acid activation, the first step of the aminoacylation reaction, by the [32P]ATP-PPi exchange reaction (14). PylS efficiently activated pyrrolysine but was inactive with lysine (Fig. 4). Unfractionated M. barkeri tRNA did not affect the activation reaction; thus, PylS resembles the majority of aminoacyl-tRNA synthetases in this respect. Furthermore, only ATP supported amino acid activation; UTP, CTP, and GTP had no effect. A steady-state enzyme kinetic analysis allowed the determination of apparent Km (1.1 mM) and kcat (6.6/min) values for pyrrolysine (the concentration of the active pyrrolysine isomer in the pyrrolysine preparation is ≈50% as determined from the LC MS separation in Fig. 2 A). These values are much lower than those for lysine by E. coli LysRS2 (15). However, once the structure of the active pyrrolysine isomer is known (see below) and the compound is available in pure form, precise kinetic parameters will be determined.
Fig. 4.
Pyrrolysyl-AMP synthesis by PylS as measured in the ATP-PPi exchange reaction. The experimental conditions are described in Materials and Methods. The reaction was conducted in the presence of either 500 μM pyrrolysine (•) or 500 μM lysine (○). Background level was determined in the absence of amino acid (▴).
Structure of tRNAPyl. The sequence of tRNAPyl suggests a structure somewhat different from other tRNA molecules that might aid in the formation of the ternary complex with LysRS1 and LysRS2 (discussed in detail in ref. 6). In this RNA molecule, the junction between the acceptor stem and D stem is shorter by one nucleotide, the link between the D stem and anticodon stem is longer by one nucleotide, and the D loop is considerably shorter than that found in canonical lysine tRNAs. Modeling suggests that these changes (compared to normal tRNALys), nevertheless, allow a conventional L-shaped tRNA fold. This secondary structure is reminiscent of bovine mitochondrial tRNASer (16). Because the flexibility and conformation of tRNA sequence is decisively affected by the degree and nature of nucleotide modification (17), we collected some information on the modification state of M. barkeri tRNAPyl. The masses of RNase T1-derived oligonucleotides were determined by LC/ESI MS and compared with those calculated for the unmodified transcript, for detection of modified oligonucleotides from mass shifts (18); pseudouridine is mass-silent and its presence is not revealed by this analysis. Only two modified nucleosides were found, based on the presence of mass increments to unmodified oligos and reconciliation of the modified nucleoside content (data not shown). AACCUGp (with an additional 16 Da) is located in position 5–9 of the tRNA. Thus, it is likely that 4-thiouridine occupies position 8, as it does in most bacterial tRNAs. Furthermore, the oligonucleotide UUAGp (positions 50–53) has an additional 14-Da mass; the nucleotide is tentatively assigned as 1-methyl-pseudouridine, a characteristic modification of archaeal tRNAs in position 50. The lack of nucleotide modifications, especially in the anticodon stem/loop, should allow tRNAPyl to assume a more flexible structure, a property that would suit the needs of this special tRNA for interaction with other cellular components during the process of decoding the special UAG in the mtmB mRNA. The lack of nucleotide modification in mature tRNAPyl may also explain the suitability of the tRNAPyl transcript for acylation.
Discussion
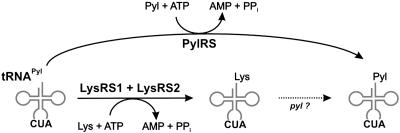
Parallel Pathways of tRNAPyl Acylation Safeguard Efficient Methylamine Methyltransferase Biosynthesis in Vivo. Two distinct pathways exist for the aminoacylation of the suppressor tRNAPyl (Fig. 5). Both pyrrolysyl-tRNAPyl and lysyl-tRNAPyl can be obtained in vitro, raising the question of whether this also takes place in vivo. Given that acid gels can distinguish between these two aminoacyl-tRNA species (Fig. 3), it will be possible to test this in the future. However, the fact that methylamine-dependent growth induces lysK and mtmB (19) and pylS expression (5), and consequently LysRS1 and PylS synthesis, makes it plausible that both lysine and pyrrolysine insertion directed by the special UAG codon of various Methanosarcina genes will serve a practical function in the organism. As long as it is not established (e.g., by mutagenesis of the in-frame UAG codon) that pyrrolysine provides the only functional solution for the enzymes requiring pylT for expression, the existence of parallel pathways is a credible solution to ensure that pyrrolysine limitation will not result in accumulation and, presumably, subsequent degradation of prematurely truncated MtmB protein.
Fig. 5.
Two routes of tRNAPyl aminoacylation. (Upper) PylRS acylates tRNAPyl with Pyl. (Lower) lysyl-tRNAPyl is formed by LysRS1 and LysRS2 acting together (6). Further modification of lysyl-tRNAPyl (stippled arrow) by additional uncharacterized pyl genes has been suggested as a route to pyrrolysyl-tRNAPyl (5).
PylS Is a Pyrrolysyl-tRNA Synthetase. PylS has been reported to form lysyl-tRNAPyl (5) and was therefore named lysine-tRNAPyl ligase (EC 6.1.1.25). The work presented here shows PylS to be an aminoacyl-tRNA synthetase dedicated to acylating pyrrolysine to the pylT suppressor tRNAPyl. Lysine (Fig. 5) and the other M. barkeri tRNALys species (data not shown) are not PylS substrates. Thus, unlike some canonical aminoacyl-tRNA synthetases that recognize amino acid analogs in addition to the unmodified amino acid [e.g., phenylalanyl-tRNA synthetase can charge p-fluorophenylalanine in addition to phenylalanine (20)], PylS is specialized for charging solely a modified amino acid. Pyrrolysine activation is tRNA-independent, as is the activation step of most canonical aminoacyl-tRNA synthetases (14). Sequence analysis of PylS proteins reveals similarities to a class II synthetase-like catalytic domain located in the C-terminal part of the protein. In view of these properties, we propose to name this enzyme pyrrolysyl-tRNA synthetase (PylRS).
Because the chemical structure of the naturally occurring pyrrolysine is unknown, we cannot judge the degree of difference between the biological product and the “pyrrolysine isomer” selected by PylS from our synthetic sample (Fig. 2 A). Thus, it is certainly possible that the enzymatic activity observed in this work may only be a fraction of what the natural substrate would yield. The answer to this question will be necessary for a complete definition of pyrrolysyl-tRNA synthetase.
Acknowledgments
We thank William Whitman for advice, Kevin Sowers for growing Methanosarcina cells, and Jeffrey Sabina, Senyene Hunter, and Shipra Bunjun-Srihari for reviewing this manuscript. This work was supported by grants from the National Institute of General Medical Sciences (to D.S. and J.A.M.) and the Department of Energy (to D.S.). J.L.W. thanks Bristol-Myers Squibb, Amgen, Yamanouchi, Pfizer, and Merck for funding.
Abbreviations: ESI, electrospray ionization; LC, liquid chromatography.
References
- 1.Burke, S. A., Lo, S. L. & Krzycki, J. A. (1998) J. Bacteriol. 180, 3432–3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paul, L., Ferguson, D. J., Jr., & Krzycki, J. A. (2000) J. Bacteriol. 182, 2520–2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.James, C. M., Ferguson, T. K., Leykam, J. F. & Krzycki, J. A. (2001) J. Biol. Chem. 276, 34252–34258. [DOI] [PubMed] [Google Scholar]
- 4.Hao, B., Gong, W., Ferguson, T. K., James, C. M., Krzycki, J. A. & Chan, M. K. (2002) Science 296, 1462–1466. [DOI] [PubMed] [Google Scholar]
- 5.Srinivasan, G., James, C. M. & Krzycki, J. A. (2002) Science 296, 1459–1462. [DOI] [PubMed] [Google Scholar]
- 6.Polycarpo, C., Ambrogelly, A., Ruan, B., Tumbula-Hansen, D., Ataide, S. F., Ishitani, R., Yokoyama, S., Nureki, O., Ibba, M. & Söll, D. (2003) Mol. Cell 12, 287–294. [DOI] [PubMed] [Google Scholar]
- 7.Böck, A., Thanbichler, M., Rother, M. & Resch, A. (2004) in Aminoacyl-tRNA Synthetases, eds. Ibba, M., Francklyn, C. S. & Cusack, S. (Landes Bioscience, Georgetown, TX), pp. 320–327.
- 8.Ibba, M. & Söll, D. (2004) Genes Dev. 18, 731–738. [DOI] [PubMed] [Google Scholar]
- 9.Ambrogelly, A., Korencic, D. & Ibba, M. (2002) J. Bacteriol. 184, 4594–4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Varshney, U., Lee, C. P. & RajBhandary, U. L. (1991) J. Biol. Chem. 266, 24712–24718. [PubMed] [Google Scholar]
- 11.Salazar, J. C., Ambrogelly, A., Crain, P. F., McCloskey, J. A. & Söll, D. (2004) Proc. Natl. Acad. Sci. USA 101, 7536–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pomerantz, S. C. & McCloskey, J. A. (1990) Methods Enzymol. 193, 796–824. [DOI] [PubMed] [Google Scholar]
- 13.Apffel, A., Chakel, J. A., Fischer, S., Lichtenwalter, K. & Hancock, W. S. (1997) Anal. Chem. 69, 1320–1325. [DOI] [PubMed] [Google Scholar]
- 14.Ibba, M. & Söll, D. (2000) Annu. Rev. Biochem. 69, 617–650. [DOI] [PubMed] [Google Scholar]
- 15.Brevet, A., Chen, J., Leveque, F., Blanquet, S. & Plateau, P. (1995) J. Biol. Chem. 270, 14439–14444. [DOI] [PubMed] [Google Scholar]
- 16.Watanabe, Y., Kawai, G., Yokogawa, T., Hayashi, N., Kumazawa, Y., Ueda, T., Nishikawa, K., Hirao, I., Miura, K. & Watanabe, K. (1994) Nucleic Acids Res. 22, 5378–5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Agris, P. F. (2004) Nucleic Acids Res. 32, 223–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kowalak, J. A., Pomerantz, S. C., Crain, P. F. & McCloskey, J. A. (1993) Nucleic Acids Res. 21, 4577–4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Srinivasan, G. (2003) Ph.D. thesis (Ohio State Univ., Columbus).
- 20.Kast, P. & Hennecke, H. (1991) J. Mol. Biol. 222, 99–124. [DOI] [PubMed] [Google Scholar]