Abstract
The current diffusion-retention model for protein trafficking to the inner nuclear membrane (INM) proposes that INM proteins diffuse laterally from the membrane of the endoplasmic reticulum into the INM and are then retained in the INM by binding to nuclear proteins or DNA. Because some data indicate that the sorting of baculovirus envelope proteins to the INM is protein-mediated, we have examined the early stages of INM protein integration and sorting by using photocrosslinking. Both viral and host INM-directed proteins were integrated cotranslationally through the endoplasmic reticulum translocon, and their nonrandom photocrosslinking to two translocon proteins, Sec61α and translocating chain-associated membrane protein (TRAM), revealed that the first transmembrane sequence (TMS) of each viral and host INM-directed protein occupied a very similar location within the translocon. Because few TMSs of non-INM-directed membrane proteins photocrosslink to TRAM, it seems that the INM-directed TMSs occupy different sites within the translocon than do non-INM-directed TMSs. The distinct proximities of translocon components to INM-directed TMSs strongly suggest that such TMSs are recognized and initially sorted within the translocon. Taken together, these data indicate that membrane protein sorting to the INM is an active process involving specific nonnuclear proteins.
Most membrane proteins in eukaryotic cells are cotranslationally integrated into the membrane of the endoplasmic reticulum (ER) at sites termed translocons (1–3). These proteins are then sorted and distributed to cellular locations where they function, typically, by means of vesicular trafficking to the Golgi compartments, other organelles, and the plasma membrane (4, 5). However, in some cases, newly synthesized membrane proteins are directed to the inner nuclear membrane (INM). Because the INM is contiguous with the ER membrane, it is generally presumed that, after leaving the translocon, membrane proteins destined for the INM diffuse through the ER membrane, the outer nuclear membrane, and the nuclear pore membrane to reach the INM (6). Proteins are then retained in the INM by binding to nuclear proteins or DNA that prevent their diffusion back into the ER membrane. This model for protein sorting to the INM is termed the “diffusion–retention” model (6).
The envelope proteins of the baculovirus occlusion-derived virus (ODV) also integrate into the ER and transit to virally induced intranuclear membranes for envelope assembly. A minimum sequence required to direct proteins to the INM was determined by using the viral envelope protein ODV-E66 (E66), and its N-terminal 33 aa were found to be sufficient to traffic fusion proteins to the INM and ODV envelope with an efficiency similar to wild-type protein (7). This sequence has therefore been termed an INM sorting motif (SM) (8). The SM consists of 18 hydrophobic amino acids that form a transmembrane sequence (TMS) with positively charged amino acids that are located four to eight residues from the TMS on the nucleoplasmic or cytoplasmic face of the membrane. Notably, the SM also directs proteins to the INM in the absence of infection (8). Furthermore, a comparison of the viral SM sequence with the sequences of cellular proteins revealed that well-characterized cellular INM proteins have an SM-like sequence, even though the TMSs may be oriented in opposite directions in the bilayer and may be inserted into the bilayer by different mechanisms (8). Hence, fusion proteins containing either viral or cellular SM sequences may constitute reasonable substrates for examining the molecular mechanisms that mediate protein movement into the INM.
Although diffusion–retention could explain ODV envelope protein trafficking, transport to the INM may be regulated by something more complex than passive diffusion and retention. The 33-residue E66 SM sequence was crosslinked to FP25K and BV/ODV-E26 (E26), thereby demonstrating that these two viral proteins are adjacent to the SM while it is still in the ER membrane (8). Moreover, when the gene coding for FP25K was deleted from the viral genome, trafficking of E66 during infection seemed to be blocked at the nuclear envelope: E66 accumulated in punctate regions associated with the ONM and was not detected in the INM or within the virally induced intranuclear membranes (9). Thus, the trafficking of E66 to the INM may involve the active participation of other proteins.
We have here used the viral and cellular SM-like sequences to address three issues that are critical to our understanding of protein sorting to the INM. First, we have used photochemical and chemical crosslinking to detect and identify proteins that are located adjacent to, and presumably interact with, nascent INM proteins at different stages of integration. Second, to determine whether viral and host INM proteins use the same sorting machinery and are sorted in the same way, we have compared the photoadducts obtained with two host and two viral INM proteins. Third, to ascertain at what point, if any, during integration the SM sequence directs INM membrane proteins into a different pathway than that taken by proteins destined for other cellular membranes, we have compared the photocrosslinking of nascent chains directed to either the INM or another membrane. The combined data reveal that viral and host INM proteins interact similarly with translocon proteins, but they interact differently with the translocon than do most other membrane proteins. Thus, the translocon seems to discriminate between INM and other membrane proteins while they are nascent chains, which suggests that sorting is initiated very early during the integration process.
Methods
Materials. Plasmids containing the E66, E66G, and E66SM (this plasmid was termed “SM-cassette” in ref. 8) coding sequences have been described, as has the plasmid coding for Lep1 (10). Microsomes and mRNAs coding for E25, Nur1, and lamin B receptor (LBR) 1 were prepared as described in Supporting Text, which is published as supporting information on the PNAS web site. Nε-(5-azido-2-nitrobenzoyl)-Lys-tRNAamb (εANB-Lys-tRNAamb) and truncated mRNAs were prepared as before (10–12).
Photocrosslinking. In vitro translations (50 μl; 26°C; 40 min) were performed in wheat germ cell-free extract in the presence of 40 nM canine signal recognition particle (SRP), 100 μCi of [35S]Met (1 Ci = 37 GBq), 8 equivalents of canine microsomes, and 72 pmols of εANB-Lys-tRNAamb as indicated (refs. 10 and 13 and Supporting Text). Samples were photolyzed on ice for 15 min by using a 500-W mercury arc lamp as before (13). After photolysis, microsomes were pelleted (4°C, 3 min, 100,000 rpm, Beckman TLA 100 rotor) through a 120-μl sucrose cushion [0.5 M sucrose/20 mM Hepes, pH 7.5/100 mM KOAc/3.6 mM Mg(OAc)2] and then resuspended and immunoprecipitated as before (10). Photoadducts and nascent chains were detected and quantified by using a Bio-Rad FX PhosphorImager.
Results
Experimental Rationale. Any proteins directly involved in the sorting of newly synthesized polypeptides (hereafter termed substrates) into the INM or other membranes will recognize some structural feature(s) of the substrate that directs each protein to a particular location. Because the SM sequence is sufficient to direct a polypeptide to the INM (8), this sequence must contain the structural features that constitute an INM-sorting signal. If sorting is protein-mediated, then recognition of this sequence would require a direct interaction between the SM sequence and a protein involved in sorting. A direct method for detecting interacting proteins is crosslinking because a substrate can react covalently only with proteins that are in close proximity. Furthermore, the use of photoactivatable crosslinking reagents allows one to create fully assembled intermediates at different stages of substrate synthesis and sorting before initiating the crosslinking reaction.
Crosslinking experiments in a complex biochemical system that includes the substrate, ribosomes, ER microsomes, and many associated factors can yield a myriad of crosslinked proteins. However, by selectively positioning photoreactive probes only in the substrate, one can limit the crosslinking targets solely to those proteins adjacent to the substrate. Because the nascent or full-length substrate typically comprises <0.1% of the total protein in an in vitro translation incubation containing microsomes, selective labeling of the substrate can be achieved only by incorporating the photoreactive probes into the substrate as it is being made by the ribosome. To accomplish this result, the translation incubation must contain a modified aminoacyl-tRNA (aa-tRNA) that recognizes a particular mRNA codon, but incorporates an amino acid with a photoreactive probe covalently attached to the side chain. By using this approach, a probe will be incorporated into the substrate wherever its mRNA contains a codon recognized by the aa-tRNA analogue, as shown (1, 14).
Fully assembled integration intermediates with nascent chains of a homogeneous, defined length can be prepared in vitro by translating, in the presence of ER microsomes and SRP, mRNAs that are truncated within the coding region. Ribosomes halt when they reach the end of the mRNA, but the nascent chains do not dissociate from the tRNA and ribosome because the absence of a stop codon prevents normal termination from occurring. The length of the nascent chain in the ribosome–nascent chain complex (RNC) is therefore determined by the length of the truncated mRNA added to the translation. Increasing the length of the truncated mRNAs that code for the substrate in in vitro translations yields intermediates with longer nascent chains that are further along the processing pathway. Thus, the proteinaceous environment surrounding the substrate can be monitored at different stages of integration by varying the length of the mRNA. Moreover, by comparing different lengths of INM and non-INM substrates, the stage at which the processing of an INM substrate diverges from that of a protein directed elsewhere can be determined experimentally. Translations contain [35S]Met to radiolabel the synthesized substrate and εANB-Lys-tRNAamb to incorporate a photoreactive probe into the substrate (11, 12). The εANB-Lys-tRNAamb amber suppressor tRNA recognizes and translates an amber stop codon, so the position of the probe in the substrate is dictated by the location of the single amber stop codon in the mRNA. To identify proteins that may interact with the SM sequence, we positioned an amber stop codon within the nonpolar region of the SM-containing proteins for many experiments in this study. (Note that modification by 5-azido-2-nitrobenzoyl (ANB) eliminates the charge on the lysine side chain.) Moreover, to examine all sides of the putative α-helix formed by a hydrophobic TMS, a single amber codon per mRNA was substituted in place of each of four sequential in-frame codons in each mRNA examined (10). In other experiments, proximity to the unmodified SM sequence was examined by using chemical crosslinking.
We first determined that ribosomes synthesizing E66 are targeted to the ER membrane by SRP and that the integration of E66 into the bilayer is cotranslational. The experimental results that document these conclusions are included in Fig. 5, which is published as supporting information on the PNAS web site.
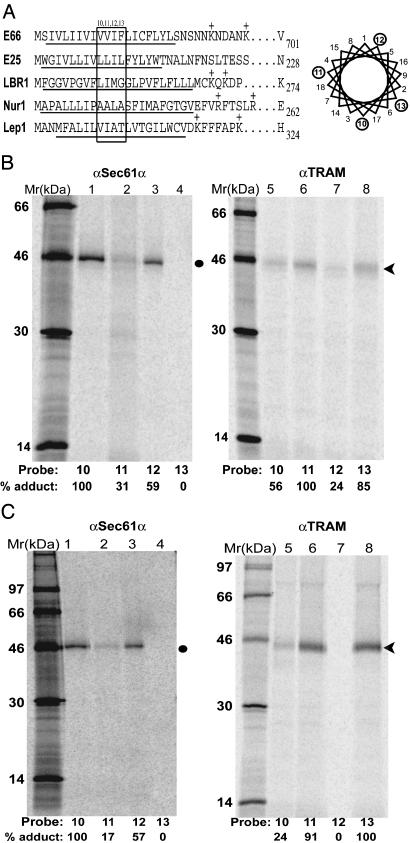
Viral SM Sequence Proximity to Translocon Proteins. A number of studies by different groups have demonstrated that a signal sequence or a signal-anchor (SA) sequence (an uncleaved signal sequence that is sufficiently nonpolar to integrate and form a TMS in the bilayer) is transiently adjacent to translocon proteins after SRP-dependent targeting (1, 3). To determine whether the SM sequence of an INM protein also passes through the translocon, parallel samples of E66 mRNAs containing amber codons at positions 10, 11, 12, or 13 were translated in the presence of SRP, microsomes, and εANB-Lys-tRNAamb, resulting in a 70-residue nascent chain (Fig. 1A). After photolysis, the extent of photocrosslinking of each E66 derivative to both Sec61α and TRAM was determined by immunoprecipitation by using affinity-purified antibodies specific for Sec61α and translocating chain-associated membrane protein (TRAM) (SDS/PAGE analyses of the total samples did not reveal any other major photoadducts, and no photoadducts were observed in the absence of light or ANB; data not shown). As shown in Fig. 1B, E66 reacts covalently with Sec61α primarily by means of probes positioned at residues 10 and 12. Probes at positions 11, 13, and, to a lesser extent, 10 photocrosslink to TRAM. As we have observed and discussed elsewhere (10), the great disparity in the magnitude of photocrosslinking from adjacent positions in the TMS of the SM reveals that it is not oriented randomly within the translocon, nor is the SM TMS free to rotate in or next to the translocon. Instead, the SM TMS seems to be bound in a fixed orientation by one or more translocon proteins. If the SM TMSs were randomly oriented and not bound to the translocon, then one would expect to see a symmetric photocrosslinking pattern in which each probe position reacts more or less equally with a given target protein (note that the probes are at the end of a 12-Å-long flexible lysine side chain, so a 1.5-Å difference in bilayer depth of probes located at adjacent residues in the TMS helix could not explain the dramatic differences in photoadduct formation for adjacent probe locations seen in Fig. 1B). Thus, after SRP-dependent targeting, the TMS of the E66 SM sequence seems to occupy and be bound to a specific site within the translocon.
Fig. 1.
Photocrosslinking of viral INM-directed TMSs to translocon proteins. (A) The N-terminal sequences of E66 and E25 are shown with the TMS underlined, as are the N-terminal sequences of the constructs containing the first TMS of LBR (LBR1), nurim (Nur1), and Lep (Lep1). In each case, an amber codon was substituted for the codon shown at position 10, 11, 12, or 13 (boxed in figure) to position the photoreactive probes at a single nascent chain location in each sample. The probes extend from different sides of the TMS α-helix surface as shown in the helical wheel representation. (B) Integration intermediates containing 70-residue E66-A10, E66-A11, E66-A12, or E66-A13 nascent chains were photolyzed, immunoprecipitated with affinity-purified antibodies to Sec61α (lanes 1–4) or TRAM (lanes 5–8), and analyzed by SDS/PAGE. Photoadducts to Sec61α and TRAM are indicated by the closed circle and arrowhead, respectively. The relative extent of photoadduct formation was quantified by comparing each photoadduct band intensity with that of the most intense photoadduct band in the gel (assigned 100%). (C) Integration intermediates containing 70-residue E25-A10, E25-A11, E25-A12, or E25-A13 nascent chains were photolyzed and analyzed as in B.
To assess the generality of these results, we examined the proximity to the translocon of nascent ODV-E25 (E25), a viral membrane protein that is also sorted to the INM. As above, an amber stop codon was substituted into position 10, 11, 12, or 13 (Fig. 1 A) to yield constructs designated E25-A10, etc. When truncated mRNAs coding for 70 residues of each of these nascent chains were translated in parallel and then photolyzed, the photocrosslinking patterns were very similar to those of nascent E66. Sec61α was in close proximity to residues 10 and 12 in the E25 TMS, whereas residues 11 and 13 were adjacent to TRAM (Fig. 2C). Thus, the locations of the E66 and E25 TMS sequences within the translocon are indistinguishable early in integration. Furthermore, both viral TMSs were clearly adjacent to TRAM: an average of 11% of the nascent chains photocrosslinked to translocon proteins, and approximately half of the photoadducts contained TRAM.
Fig. 2.
Photocrosslinking of mammalian LBR1 and Nur1 TMSs to translocon proteins. (A) Integration intermediates containing 70-residue LBR1-A10, -A11, -A12, and -A13 nascent chains were photolyzed and analyzed as in Fig. 1. (B) Integration intermediates containing 70-residue Nur1-A10, -A11, -A12, or -A13 nascent chains were photolyzed and analyzed as in Fig. 1.
The E66 TMS also photocrosslinks to phospholipids in the translocon, as shown by the presence of photoadducts in these samples that are sensitive to treatment with phospholipase A2 (data not shown). In this respect, the environment of the E66 TMS in the translocon does not differ from that which has been reported for other TMSs (10, 15–17).
Mammalian INM TMS Proximity to Translocon Proteins. The unexpected photocrosslinking of viral TMSs to TRAM raised two questions. Is the close association with TRAM a property of viral proteins, or is this close association a property of INM-directed proteins, but not other membrane proteins? To ascertain which of these two possibilities, if either, is correct, the same approach was used to examine two mammalian proteins that localize in the INM: LBR and nurim. Both LBR and nurim are multispanning membrane proteins, but the first TMS and the flanking charged amino acids of both LBR (18) and nurim (19) are important for directing the protein to the INM. Thus, constructs that retained these features were generated and termed LBR1 and Nur1, respectively. After cotranslational insertion into the membrane, the LBR1 sequence adopts an orientation opposite to that of the first TMS in native LBR (6, 20); however, this difference in TMS orientation has no detectable effect on protein sorting to the INM because both LBR and LBR1 are each directed to the INM (S. T. Williamson, S.C.B., and M.D.S., unpublished results). Derivatives of LBR1 and Nur1 were then prepared by substituting an amber stop codon for a codon at positions 10, 11, 12, or 13 (Fig. 1 A).
When 70-residue nascent chains of the LBR1 constructs were translated, targeted, and photolyzed, SDS/PAGE analyses of the TRAM- and Sec61α-specific immunoprecipitates revealed that the photocrosslinking was asymmetric and the pattern was very similar to that seen with the viral INM TMSs. Sec61α was adjacent to positions 10 and 12 in the LBR TMS, whereas positions 11, 13, and, to a lesser extent, 10 were in close proximity to TRAM (Fig. 2 A). As was seen with the viral INM-directed proteins, 11–12% of the total LBR1 and Nur1 nascent chains synthesized were photocrosslinked to TRAM plus Sec61α. The first LBR TMS and the viral TMSs therefore seem to occupy similar sites in the translocon.
When the Nur1 TMS sequence was examined, the photocrosslinking targets were the same, but the photocrosslinking efficiencies at different probe locations differed significantly from those of other INM TMSs. The extent of photocrosslinking to TRAM from positions 11 and 13 of Nur1 was similar to those observed with LBR1 (Fig. 2 A and B), whereas photocrosslinking to Sec61α was most efficient from position 12 and much less efficient from position 11 (Fig. 2B). Only a trace of photocrosslinking to Sec61α was obtained from position 10, in contrast to the trace photocrosslinking obtained from position 11 in the other three INM TMSs (Figs. 1 B and C and 2 A). The asymmetry of the photocrosslinking to Sec61α and TRAM shows that the first nurim TMS is bound in a fixed orientation within the translocon. Furthermore, the nurim TMS clearly occupies a site in the translocon adjacent to TRAM. But the differences in photocrosslinking yields from different probe positions indicate that the nurim TMS does not interact with the translocon in exactly the same way as the viral TMSs.
Nascent Chain Length Dependence of Photocrosslinking. We have observed that TMSs of non-INM proteins are retained in the translocon even when the length of the nascent chain is far in excess of that required to release the TMS into the lipid bilayer (10, 13). Moreover, the lengthening of the nascent chain did not substantially alter the asymmetric photocrosslinking patterns obtained with different TMSs (10). To assess the retention of INM TMSs in the translocon, we monitored the photocrosslinking of E66SM-A10 as a function of nascent chain length in parallel incubations (E66SM was created by fusing the N-terminal 33 aa of E66 to a lysine-free sequence described in ref. 8). After photolysis, the samples were split for immunoprecipitation with antibodies specific for either Sec61α or TRAM. The extent of E66SM-A10 (Fig. 3) and E66SM-A12 (data not shown) photocrosslinking to Sec61α decrease rapidly as the nascent chain lengthens. At 70 residues, the TMS has just emerged from the ribosome, and lengthening the chain by 5–17 residues results in a substantial decrease in nascent chain photocrosslinking to Sec61α. By the time the nascent chain reaches 100 residues, the E66 TMS has moved away from Sec61α.
Fig. 3.
Nascent chain-length dependence of E66 photocrosslinking to translocon proteins. Photoadducts were detected by immunoprecipitation with antisera specific for either Sec61α (A) or TRAM (B). Nascent chain lengths (amino acids) in the photolyzed E66SM-A10 (A) or -A11 (B) integration intermediates are indicated below the gel. Normally terminated full-length 136-residue E66SM is shown in lanes A6 and B5. Photoadducts are identified as in Fig. 1.
In contrast, the E66 TMS was adjacent to TRAM until later in the process. The extent of E66SM-A11 (Fig. 3) and E66SM-A13 (data not shown) photocrosslinking to TRAM was the same for nascent chain lengths of 75–87, but photoadducts were still visible when the nascent chain reached 120 residues (Fig. 3). Thus, during its passage through the translocon, the E66 SM seems to remain adjacent to TRAM for a longer period than to Sec61α.
SM Sequence Crosslinking to Non-Translocon Proteins. Another approach that can be used to identify proteins that associate with the SM sequence is chemical crosslinking. Because the E66SM construct contains only two lysine codons near the C-terminal end of the SM sequence (Fig. 5A), the crosslinking of the E66SM substrate to another protein by means of bis(sulfosuccinimidyl)suberate, a lysine-specific homobifunctional reagent, would require a Lys residue on the target protein to be in close proximity to a Lys residue in the substrate. Because the sample contains many proteins with surface-exposed lysine amino groups, the detection of a specific covalent complex would be explained most reasonably by the association of the substrate SM sequence with a particular target protein.
By using this approach, we showed that the SM sequence in full-length E66SM was chemically crosslinked to viral proteins FP25K and/or E26 (8). This crosslinking was observed either in nuclei from insect Sf9 cells that had been infected with a recombinant baculovirus expressing E66SM or when E66SM was cotranslationally integrated into ER microsomes from infected cells in vitro (8). It therefore seems that the SM sequence is associated with FP25K and/or E26 after leaving the translocon, a result that strongly suggests that FP25K and E26 are involved in protein sorting and/or trafficking to the INM during baculovirus infection.
It is therefore pertinent to determine at what point in E66SM integration the SM sequence interacts with FP25K and/or E26. Using microsomes from infected Sf9 cells, we observed essentially no crosslinking of E66SM nascent chains to FP25K (Fig. 4A) or E26 (Fig. 4B). Crosslinked species containing the substrate and one of the two putative sorting factors were seen only with full-length E66SM substrate polypeptides (Fig. 4 and Fig. 6, which is published as supporting information on the PNAS web site). Thus, it seems that FP25K and/or E26 associate with the SM sequence after it has been released from the translocon into the lipid bilayer.
Fig. 4.
Nascent chain-length dependence of SM chemical crosslinking to FP25K and E26. (A) Nascent or full-length E66SM proteins were translated in the presence of virus-infected Sf9 microsomes, chemically crosslinked, and immunoprecipitated with antisera specific for either FP25K (A) or E26 (B) as before (8). Nascent chain lengths in integration intermediates are as indicated. Covalent crosslinks between full-length E66SM and FP25K or E26 are identified by the arrowhead adjacent to lane 5 in each gel.
Discussion
Several important conclusions can be drawn from the data reported here. First, the asymmetric photocrosslinking to TRAM and to Sec61α of each TMS examined here reveals that each is bound to a protein(s) within the mammalian translocon (cf. ref. 10). Second, the proteinaceous environment within the translocon is very similar for both cellular and viral INM TMSs. Third, the proteinaceous environment within the translocon differs markedly for INM TMSs and for TMSs directed elsewhere. Fourth, an integrated viral SM sequence is crosslinked to two viral proteins after being released from the translocon in ER microsomes purified from infected insect cells. Fifth, this study confirms the widely held presumption that some INM-directed proteins are targeted to the ER membrane by SRP and integrated cotranslationally at the translocon. Other INM proteins may be inserted posttranslationally as C-tail-anchored proteins (21). Taken together, these results strongly suggest that some membrane protein sorting to the INM is initiated within the translocon early in the integration process, is protein-mediated, and involves substrate recognition by and interaction with a sequence of proteins that function as sorting factors to facilitate the movement of proteins to the INM.
As discussed elsewhere (10), the binding of a substrate TMS to a translocon protein is revealed by the nonrandom and asymmetric photocrosslinking of the TMS to TRAM and/or Sec61α from different photoreactive probe locations that encircle the middle of the cylindrical TMS α-helix. This high-resolution approach for characterizing a TMS's environment within the translocon has so far been applied to 11 TMSs in this (Figs. 1 and 2, and Fig. 7, which is published as supporting information on the PNAS web site) and our previous study (10). Because each of these 11 TMSs was not free to rotate and randomize its orientation relative to TRAM and Sec61α in the translocon, each TMS binds to a translocon protein(s). This interaction presumably plays a direct (but as-yet undefined) mechanistic and/or regulatory role in the integration process.
When the translocon environments of the viral E66 and E25 TMSs were examined, the photocrosslinking of these two TMSs to TRAM and Sec61α was found to be indistinguishable (Fig. 1). In addition, a similar photocrosslinking pattern was obtained when the proximity of the first TMS of mammalian LBR to translocon proteins was investigated (Fig. 2 A). The close similarity of the photocrosslinking results for these three INM proteins seems more than coincidental. Furthermore, the first TMS of mammalian nurim also photocrosslinked to TRAM and Sec61α from the same probe positions as the other three TMSs, although the extent of Sec61α photocrosslinking from positions 10 and 11 of Nur1 was reversed from that of the other three substrate TMSs (Fig. 2B). Most striking, however, was the observation that 4 of 4 INM TMSs were efficiently photocrosslinked to TRAM. Furthermore, these TMSs were in very similar, but not identical, proteinaceous environments or sites within the translocon based on the photocrosslinking-detected proximities of different TMS surfaces to Sec61α and TRAM.
In contrast, of the 10 total non-INM-directed TMSs whose photocrosslinking to Sec 61α and TRAM has been examined by different groups, only the two native TMSs of the vesicular stomatitis virus G (VSVG) protein (10, 13) and the Ii invariant chain (15) were crosslinked to TRAM by means of photoreactive probes in the middle of the TMS. Two nonnative Lep derivatives with one or two charged residues inserted into the TMS were also photocrosslinked to TRAM (17). But the non-INM VSVG TMS was not located in the same site in the translocon as the INM TMSs. In fact, of the 7 non-INM TMSs that have been examined at high resolution by using photoreactive probes on different TMS surfaces, none gave a photocrosslinking pattern similar to the 4 INM TMSs (10)(Figs. 1, 2, and 7). Although the Ii and two charged Lep TMSs have yet to be examined by using the same approach, the clear distinction between the high-resolution photocrosslinking patterns of the 4 INM and 7 non-INM TMSs strongly suggests that the two classes of TMS occupy different sites within the translocon.
It is, of course, possible that INM and non-INM TMS-binding sites within the translocon overlap to some extent. Consistent with the close juxtaposition of potentially different interaction sites within the translocon, both nascent secretory and nascent membrane proteins have previously been photocrosslinked to TRAM, usually from sites that flank the TMS or the nonpolar signal sequence core (e.g., refs. 1 and 22). On the other hand, because of variations in probe length, reactivity, location, and target atom, one must be cautious in extrapolating from a crosslink to a specific structural arrangement between two macromolecules. For example, the second TMS in opsin was chemically crosslinked to TRAM (23), but no crosslinking to TRAM was detected when photoreactive probes were positioned in the middle of the second opsin TMS in a chimeric protein (10). Thus, high-resolution experiments using multiple different probes will be necessary to assess the extent to which the INM and non-INM TMS binding sites in the translocon overlap spatially and dynamically.
Based on photocrosslinking data obtained with charged Lep1 mutants, Heinrich et al. (17) proposed that TRAM interacts with TMSs that were charged or hydrophilic. Yet, of the four INM-directed TMSs found adjacent to TRAM during integration, none contains a charged amino acid (Fig. 1 A). Furthermore, based on the White–Wimley values (ΔGwoct – ΔGwif) for quantifying amino acid movement from the aqueous to the nonpolar phase, the free energies of transfer of the four INM TMSs range from –3.0 kcal/mole for the first nurim TMS to –10.5 kcal/mole for the E66 TMS (a more negative number indicates a more hydrophobic sequence) (8, 24). Thus, even the E66 TMS, which is the most hydrophobic of the 11 TMSs that we have thoroughly examined to date, is positioned adjacent to and photocrosslinks to TRAM during cotranslational integration. It therefore seems that TRAM functions in a different or an additional role than that proposed by Heinrich et al. (17).
We have here compared the photocrosslinking targets of probes that are in different constructs but are positioned the same number of residues from the N terminus. Another approach would be to compare the photocrosslinking targets for probes positioned the same number of amino acids from another reference point, such as the N-terminal residue in the putative TMS. But, because only small variations in photocrosslinking patterns were observed with E66, E25, LBR1, and Nur1 TMSs (Figs. 1 and 2), it seems that the differences in TMS length and location (i.e., the number of residues between the N terminus and the putative start of the TMS) shown in Fig. 1 A do not significantly alter the positioning of the TMS within the translocon. Instead, TMS–translocon interactions are apparently dictated by some other feature of the TMS (see below). This conclusion is supported most strongly, of course, by the fact that the Lep1 TMS does not photocrosslink to TRAM (Fig. 7).
What, then, is the key structural determinant of the SM that is recognized by the translocon? The sample size is still too small to provide a clear answer to this question, but there are some interesting clues in the results presented here. First, as was noted above, we have here examined constructs containing the first TMSs of LBR and nurim because these TMSs are important in terms of sorting to the INM (18, 19) and we wished to facilitate comparisons with the INM-directed viral proteins that contain only a single TMS. Yet, the orientation of the LBR1 TMS in the bilayer is opposite to the bilayer orientation of the same TMS in native LBR. Strikingly, and unexpectedly, both LBR1 and native LBR are sorted and directed to the INM (S. T. Williamson, S.C.B., and M.D.S., unpublished results). Because the LBR TMS is directed to the INM no matter what its orientation, it seems that the structural features recognized by the sorting machinery as a signal for INM-directed TMSs are not dependent upon the orientation of the TMS in the translocon. Second, both E66 and E25 are directed to the INM, even though E25 lacks the cytosolic positive charge that is characteristic of an SM sequence. Because the E66 and E25 TMSs photocrosslink to TRAM and Sec61α identically (Fig. 1), it seems that the positive charge that flanks the TMS is not involved in positioning an INM TMS at a particular site within the translocon. Moreover, both E66 and Lep1 have positive charges flanking the TMS, yet their TMSs occupy different locations in the translocon (Figs. 1B and 7). It therefore seems that the properties of the TMS itself dictate where it moves in the translocon, whereas the positive charge may be important for interactions that occur after leaving the translocon, such as recognition and/or association with FP25K and/or E26 (Fig. 4, and Fig. 8, which is published as supporting information on the PNAS web site). These results, taken together, strongly suggest that the characteristic structural elements that identify a protein as an INM protein and initiate sorting to the INM are located within the TMS itself. Further experiments with multiple INM-directed TMSs and derivatives will be required to determine what TMS features correlate with INM sorting.
The distinctive photocrosslinking pattern observed with INM TMSs indicates that INM and non-INM TMSs occupy distinctly different sites within the translocon, which in turn suggests that INM-directed TMSs are first identified and sorted by components of the translocon. Because INM TMSs are adjacent to both Sec61α and TRAM (Fig. 1 and 2), either or both could be actively involved in recognizing and sorting INM TMSs; future experiments will clarify their involvement. Also, because an INM TMS remains adjacent to TRAM longer than to Sec61α (Fig. 3), it is possible that TRAM may be involved in the hand-off of INM TMSs to the next participant in the putative INM sorting pathway. Upon leaving the translocon, the viral INM-directed proteins interact with other viral proteins that are required for sorting to the INM. Because viruses typically appropriate host mechanisms to achieve their objectives, the identification of both soluble (FP25K) and membrane-associated (E26) viral proteins that are predicted as required for the sorting of viral proteins to the INM strongly suggests that a similar host protein(s) may also be involved in INM sorting. It will require investigating INM substrate transfer from the translocon to other proteins to further clarify the participants and mechanisms that accomplish membrane protein sorting to the INM.
Supplementary Material
Acknowledgments
We thank Yiwei Miao, Lindsey Buhring, Kristen Pfeiffer, and Matthew Powers for excellent technical assistance; Dr. IngMarie Nilsson (Stockholm University, Stockholm) for plasmids and SRP54-specific antibodies; Dr. J. Ellenberg (European Molecular Biology Laboratory, Heidelberg) for the LBR-GFP plasmid; and Dr. Peter McCormick for advice. This article was submitted by S.S. in partial fulfillment of the Ph.D. degree at Texas A&M University. This work was supported by National Institutes of Health Grant R01 GM26494 (to A.E.J.), the Robert A. Welch Foundation (A.E.J.), and Texas Agricultural Experiment Station Project Grant TEXO-08078 (to M.D.S.).
Abbreviations: ER, endoplasmic reticulum; INM, inner nuclear membrane; ODV, occlusion-derived virus; SM, sorting motif; TMS, transmembrane sequence; E66, ODV-E66; E26, ODV-E26; εANB-Lys-tRNAamb,Nε-(5-azido-2-nitrobenzoyl)-Lys-tRNAamb; SRP, signal recognition particle; TRAM, translocating chain-associated membrane protein; LBR, lamin B receptor.
References
- 1.Johnson, A. E. & van Waes, M. A. (1999) Annu. Rev. Cell Dev. Biol. 15, 799–842. [DOI] [PubMed] [Google Scholar]
- 2.Schnell, D. J. & Hebert, D. N. (2003) Cell 112, 491–505. [DOI] [PubMed] [Google Scholar]
- 3.Alder, N. N. & Johnson, A. E. (2004) J. Biol. Chem. 279, 22787–22790. [DOI] [PubMed] [Google Scholar]
- 4.Mellman, I. & Warren, G. (2000) Cell 100, 99–112. [DOI] [PubMed] [Google Scholar]
- 5.Barlowe, C. (2003) Trends Cell Biol. 13, 295–300. [DOI] [PubMed] [Google Scholar]
- 6.Worman, H. J. & Courvalin, J.-C. (2000) J. Membr. Biol. 177, 1–11. [DOI] [PubMed] [Google Scholar]
- 7.Hong, T., Summers, M. D. & Braunagel, S. C. (1997) Proc. Natl. Acad. Sci. USA 94, 4050–4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braunagel, S. C., Williamson, S. T., Saksena, S., Zhong, Z., Russell, W. K., Russell, D. H. & Summers, M. D. (2004) Proc. Natl. Acad. Sci. USA 101, 8372–8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosas-Acosta, G., Braunagel, S. C. & Summers, M. D. (2001) J. Virol. 75, 10829–10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCormick, P. J., Miao, Y., Shao, Y., Lin, J. & Johnson, A. E. (2003) Mol. Cell 12, 329–341. [DOI] [PubMed] [Google Scholar]
- 11.Flanagan, J. J., Chen, J.-C., Miao, Y., Shao, Y., Lin, J., Bock, P. E. & Johnson, A. E. (2003) J. Biol. Chem. 278, 18628–18637. [DOI] [PubMed] [Google Scholar]
- 12.Krieg, U. C., Walter, P. & Johnson, A. E. (1986) Proc. Natl. Acad. Sci. USA 83, 8604–8608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Do, H., Falcone, D., Lin, J., Andrews, D. W. & Johnson, A. E. (1996) Cell 85, 369–378. [DOI] [PubMed] [Google Scholar]
- 14.Johnson, A. E., Chen, J.-C., Flanagan, J. J., Miao, Y., Shao, Y., Lin, J. & Bock, P. E. (2001) Cold Spring Harbor Symp. Quant. Biol. 66, 531–541. [DOI] [PubMed] [Google Scholar]
- 15.Martoglio, B., Hofmann, M. W., Brunner, J. & Dobberstein, B. (1995) Cell 81, 207–214. [DOI] [PubMed] [Google Scholar]
- 16.Mothes, W., Heinrich, S. U., Graf, R., Nilsson, I., von Heijne, G., Brunner, J. & Rapoport, T. A. (1997) Cell 89, 523–533. [DOI] [PubMed] [Google Scholar]
- 17.Heinrich, S. H., Mothes, W., Brunner, J. & Rapoport, T. A. (2000) Cell 102, 233–244. [DOI] [PubMed] [Google Scholar]
- 18.Soullam, B. & Worman, H. (1995) J. Cell Biol. 130, 15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rolls, M. M., Stein, P. A., Taylor, S. S., Ha, E., McKeon, F. & Rapoport, T. A. (1999) J. Cell Biol. 146, 29–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith, S. & Blobel, G. (1993) J. Cell Biol. 120, 631–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wattenberg, B. & Lithgow, T. (2001) Traffic 2, 66–71. [DOI] [PubMed] [Google Scholar]
- 22.Falcone, M., Do, H., Johnson, A. E. & Andrews, D. W. (1999) J. Biol. Chem. 274, 3361–3367. [DOI] [PubMed] [Google Scholar]
- 23.Meacock, S. L., Lecomte, F. J. L., Crawshaw, S. G. & High, S. (2002) Mol. Biol. Cell 13, 4114–4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.White, S. H. & Wimley, W. C. (1999) Annu. Rev. Biophys. Biomol. Struct. 28, 319–365. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.