Abstract
Human embryonic stem (hES) cells are defined by their extensive self-renewal capacity and their potential to differentiate into any cell type of the human body. The challenge in using hES cells for developmental biology and regenerative medicine has been to direct the wide differentiation potential toward the derivation of a specific cell fate. Within the nervous system, hES cells have been shown to differentiate in vitro into neural progenitor cells, neurons, and astrocytes. However, to our knowledge, the selective derivation of any given neuron subtype has not yet been demonstrated. Here, we describe conditions to direct hES cells into neurons of midbrain dopaminergic identity. Neuroectodermal differentiation was triggered on stromal feeder cells followed by regional specification by means of the sequential application of defined patterning molecules that direct in vivo midbrain development. Progression toward a midbrain dopamine (DA) neuron fate was monitored by the sequential expression of key transcription factors, including Pax2, Pax5, and engrailed-1 (En1), measurements of DA release, the presence of tetrodotoxin-sensitive action potentials, and the electron-microscopic visualization of tyrosinehydroxylase-positive synaptic terminals. High-yield DA neuron derivation was confirmed from three independent hES and two monkey embryonic stem cell lines. The availability of unlimited numbers of midbrain DA neurons is a first step toward exploring the potential of hES cells in preclinical models of Parkinson's disease. This experimental system also provides a powerful tool to probe the molecular mechanisms that control the development and function of human midbrain DA neurons.
The isolation of human embryonic stem (hES) cells (1) has stimulated research aimed at the selective generation of specific cell types for regenerative medicine. Although protocols have been developed for the directed differentiation of mouse embryonic stem (ES) cells into therapeutically relevant cell types, such as dopamine (DA) neurons (2, 3), motor neurons (4), and oligodendrocytes (5), the efficient generation of these cell types from hES cells has not yet been reported (6). Earlier studies demonstrating efficient neural differentiation from hES cells (7, 8) have yielded largely γ-aminobutyric acid (GABA)ergic and glutamatergic neurons with a maximum of 3% DA neurons reported (9). A very recent study (10) reported up to 20% tyrosine hydroxylase (TH)-positive cells from hES cells but did not confirm midbrain DA neuron identity. A bias toward the generation of GABAergic and glutamatergic neurons is also observed in primary rodent and human neural precursor cells isolated from the CNS after expansion in the presence of epidermal growth factor and fibroblast growth factor (FGF). Similar to the work with primary neural precursors, current hES differentiation protocols require expansion of ES-derived neural precursors in FGF2. We have recently shown that extended FGF2 expansion of mouse ES-derived neural precursors selects for forebrain fates, including GABAergic differentiation (11). The generation of ventral midbrain, hindbrain, and spinal cord-type neurons requires the ventralizing signal sonic hedgehog (SHH), in conjunction with factors that define anterior–posterior patterning, such as FGF8, FGF4, and retinoic acid (2, 4, 11, 12).
Here, we demonstrate that pathways important for in vivo midbrain development can be systematically applied to direct hES cell differentiation into DA neurons in vitro. Neural differentiation was induced by means of a modified stromal feeder-based differentiation system (11). Such stromal cells, derived from the bone marrow (13) or the aorta-gonad-mesonephros region, have been used to maintain hematopoietic stem cells in culture. The same stromal cells promote neural differentiation in mouse and monkey ES cells (3, 11, 14). The molecular nature of the neural-inducing, stromal-derived-inducing activity (3) remains unknown. Here we report that coculture of hES cells on MS5 stroma yields highly efficient differentiation into neuroepithelial structures, termed neural rosettes. Cells in these structures express markers compatible with a neural plate identity and show extensive self-renewal capacity. Ventral midbrain/hindbrain fates are induced upon replating of rosettes and exposure to FGF8 and SHH followed by terminal differentiation into midbrain DA neurons. The availability of unlimited numbers of DA neurons that express the full complement of midbrain DA neuron markers and exhibit in vitro functionality provides the basis for assessing the therapeutic potential of hES cells in preclinical models of Parkinson's disease (15). This in vitro differentiation assay also offers a unique tool for mechanistic studies on human midbrain DA neuron development.
Materials and Methods
Culture of Undifferentiated Primate ES Cells. hES cell lines H1 (WA-01, XY, passages 40–65), H9 (WA-09, XX, passages 25–35), and HES-3 (ES-03, XX, passages 50–65); rhesus monkey line R366 (XY, passages 15–35); and the cynomolgus parthenogenetic line Cyno1 (XX, passages 15–42) were cultured on mitotically inactivated mouse embryonic fibroblasts (MEF, Specialty Media, Lavellette, NJ). Undifferentiated hES and monkey ES cells were maintained under growth conditions and passaging techniques described in refs. 8 and 16–18.
Neural Induction. MS5 and S2 stromal cells were maintained in α-MEM medium containing 10% FBS and 2 mM l-glutamine (11). For some studies, transgenic MS5 cells were used that stably overexpress Wnt1 after transfection (Fugene-6) of a Wnt1 expression construct followed by G418 selection. Neural differentiation of hES cells was induced by means of coculture on MS5, MS5-Wnt, or S2 stroma at comparable efficiencies. hES cells were plated at 5–20 × 103 cells on a confluent layer of irradiated (50 Gy) stromal cells in 6-cm cell culture plates in serum replacement medium containing DMEM, 15% knockout serum replacement (Invitrogen), 2 mM l-glutamine and 10 μM β-mercaptoethanol. After 16 days in serum replacement medium, cultures were switched to N2 medium modified according to ref. 19. Medium was changed every 2–3 days, and growth factors were added in various combinations and at various time points as described: 200 ng/ml SHH, 100 ng/ml FGF8, 20 ng/ml brain-derived neurotrophic factor (BDNF), 10–20 ng/ml glial cell line-derived neurotrophic factor, 1 ng/ml transforming growth factor type β3 (R & D Systems), 0.5–1.0 mM dibutyryl cAMP, and 0.2 mM ascorbic acid (AA) (Sigma–Aldrich). Rosettes structures were harvested mechanically from feeders at day 28 of differentiation and gently replated on 15 μg/ml polyornithine/1 μg/ml laminin-coated culture dishes in N2 medium supplemented with SHH, FGF8, AA, and BDNF (passage 1). After 7–9 days (≈80% confluency), cells were mechanically passaged after exposure to Ca2/Mg2-free Hanks' balanced salt solution for 1 h at room temperature and spun at 200 × g for 5 min. Cells were resuspended in N2 medium, replated again onto polyornithine/laminin-coated culture dishes (50–100 × 103 cells per cm2) in the presence of SHH, FGF8, AA, and BDNF (passage 2). After an additional 7–9 days of culture, cells were differentiated in the absence of SHH and FGF8 but in the presence of BDNF, glial cell line-derived neurotrophic factor, transforming growth factor type β3, dibutyryl cAMP, and AA.
Immunocytochemistry. Cells were fixed in 4% paraformaldehyde/0.15% picric acid and stained with the following primary antibodies. Rabbit polyclonal antibodies included TH and vesicular monoamine transporter 2 (VMAT2, Pel-Freez Biologicals); nestin 130 (R. McKay, National Institutes of Health, Bethesda); glial fibrillary acidic protein and DA (Chemicon); GABA and serotonin (Sigma); Pax2, Pax6, and β-tubulin III (Covance); Pax5 (clone A2; M. Busslinger, Institute of Molecular Pathology, Vienna); aromatic l-amino acid decarboxylase (AADC, Protos Biotech, New York); synaptic vesicle 2 (SV2, Developmental Studies Hybridoma Bank, University of Iowa, Iowa City); and synapsin (Calbiochem). Mouse monoclonal antibodies (IgG) included Oct4 (Santa Cruz Biotechnology), TH (Sigma), class III β-tubulin (Tuj1, Covance, Princeton), En1, and Lmx (Developmental Studies Hybridoma Bank). Mouse monoclonal antibodies (IgM) included Ki67 (Sigma) and O4 (Chemicon). Appropriate Alexa488- and Alexa555-labeled secondary antibodies (Molecular Probes) and 4′,6-diamidino-2-phenylindole counterstain were used for visualization.
RT-PCR, Electrophysiology, and Electron Microscopy. Total RNA was extracted by using the RNeasy kit and DNase I treatment (Qiagen, Valencia, CA). Total RNA (2 μg each) was reverse transcribed (SuperScript, Invitrogen). PCR conditions were optimized and linear amplification range was determined for each primer by varying annealing temperature and cycle number. PCR products were identified by size, and identity was confirmed by DNA sequencing. MS5 cells were negative for all primers used in this study except for 18S ribosomal RNA. Under coculture conditions (days 0–28), hES cell progeny was mechanically separated from feeders to avoid cross contamination of the RNA pool by feeder cells. Primer sequences, cycle numbers, and annealing temperatures are provided upon request. For quantitation, gels were imaged by using a 12-bit charge-coupled device camera (Alpha Innotech, San Leandro, CA). Data are presented as means of normalized expression levels (18S ratios) obtained from three independent experiments and scaled such that the maximum level of expression during the observed time period was arbitrarily set at 1. Semiquantitative RT-PCR data were from H1 cells. Similar data were derived from H9 and HES-3 cells. Electrophysiological analyses were carried out as described recently for characterizing mouse ES-derived neurons (11). Freeze substitution-postembedding-ImmunoGold labeling for TH was performed according to ref. 20 (for details, see Supporting Materials and Methods, which is published as supporting information on the PNAS web site).
HPLC Analysis. Reversed-phase HPLC for the detection of DA in the supernatant was performed as described in ref. 21. Samples were collected at day 50 of differentiation, stabilized in orthophosphoric acid and metabisulfite, and extracted by aluminum adsorption (Chromosystems, Munich). Separation of the injected samples (ESA Autosampler 540) was achieved by isocratic elution in MD-TM mobile phase (ESA, Bedford, MA) at 0.5 ml/min. The oxidative potential of the analytical cell (ESA Mod. 5011, Coulochem II) was set at 350 mV. Results were validated by coelution with catecholamine standards under various buffer conditions and detector settings.
Cell Counts and Statistical Analyses. Data for percentage of TH-positive neurons were derived from a total of 20 independent experiments for the H1 line, from 10 independent experiments for H9, and from 2 independent experiments for HES-3. No significant variation in TH yield was observed between low- or high-passage cells for each hES line tested (within the range of passages indicated above). Cells were selected for quantification in a uniform random fashion (fractionator). Each field was scored first for 4′,6-diamidino-2-phenylindole-positive nuclei, followed by Tuj1 and, subsequently, TH colocalization. For each experiment, an average of 2,000 cells were scored. The total number of cells analyzed in this study exceeded 50,000 cells. Percentages of TH-positive neurons were compared with ANOVA and Newman–Keuls post hoc analysis (statistica 5.5, StatSoft, Tulsa, OK). Data are presented as mean ± SEM.
Results
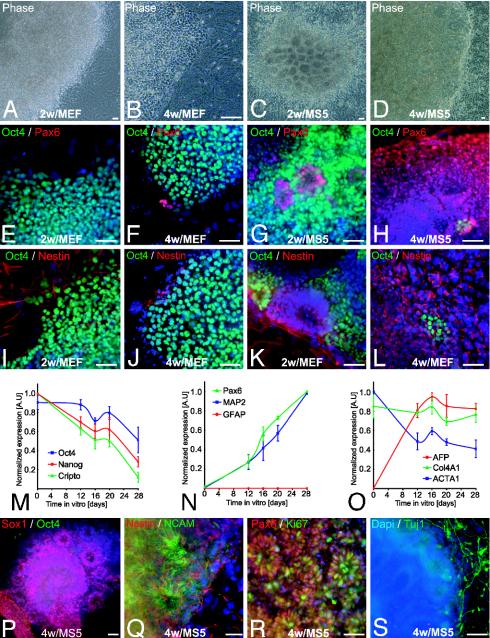
Stromal Feeder-Induced Neural Differentiation of hES Cells. We have recently demonstrated that neural induction by means of coculture on stromal feeder cells is highly efficient and reproducible across a wide range of mouse ES and nuclear transfer ES cell lines of various genetic origins (11). Nonhuman primate ES cells induced on the stromal line PA6 yielded differentiation into a variety of neuronal cell types (22) as well as pigmented epithelial cells (14). Here, we report hES cell differentiation on MS5 stroma. hES lines H1 (WA-01) or H9 (WA-09) were plated at low density under serum-free conditions on MS5 (13) or MEF control stroma (Fig. 1). hES cells plated on MEF in serum replacement medium displayed immature cell morphologies (Fig. 1 A and B), continued to express the ES cell marker Oct4 (POU5F1), and were devoid of neural markers. (Fig. 1 E, F, I, and J). In contrast, hES cells plated on MS5 or MS5-Wnt formed neuroepithelial structures that were arranged in rosettes and expressed the neural markers Pax6, nestin, neural cell adhesion molecule (NCAM), and Sox1 (Fig. 1 C, D, G, H, K, L, P, and Q). At two weeks of differentiation, rosettes were still surrounded by groups of Oct4-positive cells (Fig. 1 G and K). After an additional 2 weeks of differentiation in N2 medium, >90% of the hES cells on MS5 or MS5-Wnt expressed neural markers and formed large clusters (1–8 mm in diameter) composed of hundreds of individual rosettes. However, rare clusters of persisting Oct4-positive cells could still be detected among rosettes (Fig. 1 H and L).
Fig. 1.
Stromal feeder-induced neural differentiation of hES cells. (A–D) Representative phase contrast images of hES cells (line H1) cocultured for 2 or 4 weeks on MEF (A and B) or MS5 (C and D). (E–H) Pax6 and Oct4 expression in hES cells differentiated on MEF or MS5. (I–L) Oct4 and nestin expression in hES cells on MEF or MS5. (M–O) Semiquantitative RT-PCR analysis for genes characteristic of undifferentiated ES cells (M); neural lineage (N); and non-neural, endodermal, and mesodermal differentiation (O). Data are presented as normalized values (see Materials and Methods) and derived from three independent experiments, line H1, days 0–28. GFAP, glial fibrillary acidic protein; AFP, α-fetoprotein; Col4A1, collagen type IV α; ACTA1, α1-actin. (P–S) Characterization of hES-derived rosettes on MS5. Expression of Sox1 (P) and nestin and NCAM (Q) confirmed the neural identity of the rosettes. (R) Cell proliferation was assessed by Ki67 labeling. (S) Tuj1-positive cells were observed surrounding rosettes. Dapi, 4′,6-diamidino-2-phenylindole. (Scale bars, 50 μm.)
Semiquantitative RT-PCR analysis confirmed a time-dependent decrease in the expression of ES cell markers, such as Oct4, Cripto (TDGF1), and Nanog, upon stromal feeder-mediated differentiation (Fig. 1M). A delayed decrease in Oct4 expression was observed consistent with recent work in mouse ES cells, suggesting that transiently sustained Oct4 levels are required for neural induction (23). The decrease in ES cell markers was associated with increased expression of neural markers, including Pax6 and the neuronal marker MAP2 (Fig. 1N). No glial fibrillary acidic protein was detected, suggesting absence of astrocytic differentiation. As expected, control genes related to endodermal and mesodermal differentiation did not show consistent increases or decreases in gene expression during stromal feeder-mediated differentiation of hES cells (Fig. 1O).
At 4 weeks of differentiation, neural rosettes expressing Sox1 (Fig. 1P), NCAM (Fig. 1Q), and Pax6 were composed of proliferating Ki67-positive precursors (Fig. 1R). Cells within the rosettes were devoid of neuronal markers, such as Tuj1 or MAP2. However, cells emanating from the borders of individual rosettes were often positive for Tuj1 and exhibited neuronal morphologies (Fig. 1S). Comparable results were obtained with stromal feeder cells derived from the aorta-gonad-mesonephros region, such as the line S2 (11). The neural inducing effects were confirmed in multiple hES and nonhuman primate ES cell lines, including the hES cell lines H1, H9, and HES-3, and the monkey lines R366 and Cyno1.
Patterning and Differentiation of Neural Precursors from ES-Derived Rosettes. Previous work identified SHH and FGF8 as crucial factors in the specification of midbrain DA neurons (24). These studies in explant culture and subsequent work with mouse ES cells (2, 11, 22) demonstrated that the effect of SHH and FGF8 on dopaminergic differentiation is limited to distinct developmental windows that correspond to the establishment of the isthmic organizer and events controlling dorsoventral patterning during neural tube closure. Based on the work in mouse ES cells, we predicted that in hES cells SHH and FGF8 might act between days 12 and 20 of differentiation to specify human midbrain DA neuron fate. This time period corresponds to the onset of neurulation until the time of neural tube closure in human development, taking into account the presumptive age of hES cells at the time of isolation (5–7 days) (25).
Exposure to SHH and FGF8 from day 12 to day 20 of differentiation, followed by differentiation in the presence of AA and BDNF, resulted in a 3-fold increase in TH-positive cells (see Fig. 2H, P0), TH being the rate-limiting enzyme in the synthesis of DA. However, no expression of other midbrain-related markers, such as Pax2, Pax5, En1, or AADC was observed by immunocytochemistry, suggesting that early exposure to SHH and FGF8 is not sufficient to induce a full midbrain DA neuron phenotype. Interestingly, neural rosettes persisted in culture for several weeks, even in the absence of any exogenous mitogens or feeder cells, and remained devoid of cells expressing distinct region-specific transcription factors. This result suggested that neural rosettes may retain the capacity to respond to patterning cues beyond the predicted temporal window.
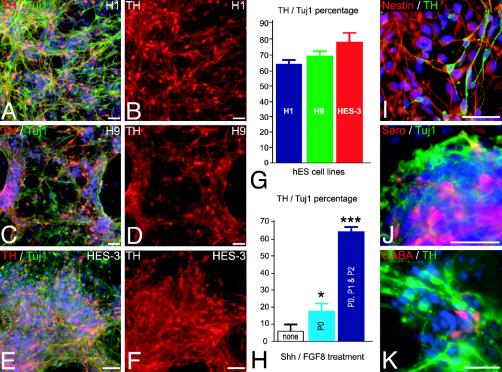
Fig. 2.
High-yield derivation of TH-positive neurons from hES-derived rosettes. (A–F) Representative images of TH/Tuj1-positive (red, TH; green, Tuj1) neurons derived from the hES cell lines H1 (A and B), H9 (C and D), and HES-3 (E and F). (G) Percentage of hES-derived TH neurons for each of the hES cell lines upon differentiation at passage 2. (H) Percentage of TH neurons upon differentiation of rosettes obtained in the absence of SHH and FGF8 (none, white bar), in the presence of SHH and FGF8 during passage 0 (P0, light blue bar), or in the presence of SHH and FGF8 during passages 0, 1, and 2 (P0, P1 and P2, dark blue bar). *, P < 0.05; ***, P < 0.001 compared to none. (I) Most cells negative for TH (green) expressed the neural precursor marker nestin (red). Other neuronal subtypes present were serotonergic (Sero, red) (J) and GABAergic neurons (red) (K). (Scale bars, 50 μm.)
To test whether rosettes respond to midbrain patterning cues, clusters of rosettes were mechanically isolated at day 28 (passage 0), replated on polyornithine/laminin precoated dishes (passage 1), and cultured for 7–10 days in N2 medium containing SHH and FGF8. After the next passage (passage 2) cells were grown in SHH and FGF8 for an additional week before differentiation in N2 medium supplemented with glial cell line-derived neurotrophic factor, dibutyryl cAMP, and transforming growth factor type β3 [previously found to enhance En1 expression in the developing rodent midbrain (26)]. After differentiation of passage 2 cells (50 days in culture) 30–50% of the total cells derived from H1, H9, or HES-3 (Fig. 2 A–F) lines were Tuj1-positive neurons. All Tuj1-postitive cells were negative for Ki67. Virtually all cells negative for TH or Tuj1 expressed the intermediate filament nestin, suggesting neural precursor identity (Fig. 2I). Among the Tuj1-positive neuronal population 64–79% of the cells expressed TH (H1, 64 ± 2%; H9, 70 ± 2%; HES-3, 79 ± 7%) (Fig. 2G). Similar data were obtained with the two monkey ES cell lines R366 and Cyno1 (data not shown). The percentage of TH expressing neurons was highly dependent on exposure to SHH and FGF8 during passages 1 and 2 (Fig 2H). In addition to TH-positive neurons, ≈5% serotonin-positive (Fig. 2J) and 1–2% GABA-positive neurons (Fig. 2K) were detected. No coexpression of GABA and TH was observed, compatible with midbrain dopaminergic differentiation. GABA and TH coexpression is observed in olfactory DA neurons (27) and transiently in striatal GABAergic neurons (28). No cholinergic or glutamatergic neurons were detected at that stage. The neurotransmitter phenotype of the other Tuj1-positive neurons (29%) remains unknown. No glial fibrillary acidic protein-positive cells were observed up to passage 2. However, astrocytic differentiation could be readily induced in long-term cultures (>70 days) exposed to either serum or ciliary neurotrophic factor (Fig. 6 A and B, which is published as supporting information on the PNAS web site). A few O4-positive oligodendrocytes displaying characteristic star-like morphologies could also be detected in long-term serum-free cultures (Fig. 6C), confirming that neural rosettes can yield progeny in all three CNS lineages.
Gene expression analysis from day 28 (passage 0) to day 50 (passage 2) of differentiation showed a complete loss of ES markers, including Oct4 (Fig. 3A). Pax6 expression reached a plateau at the rosette stage, whereas MAP2 expression continued to increase up to passage 2 (Fig. 3A). This finding suggests that neural induction is largely completed by day 28, whereas neurogenesis continues at least up to passage 2. Induction of markers characteristic of midbrain DA neuron development was readily detected during this period (Fig. 3B).
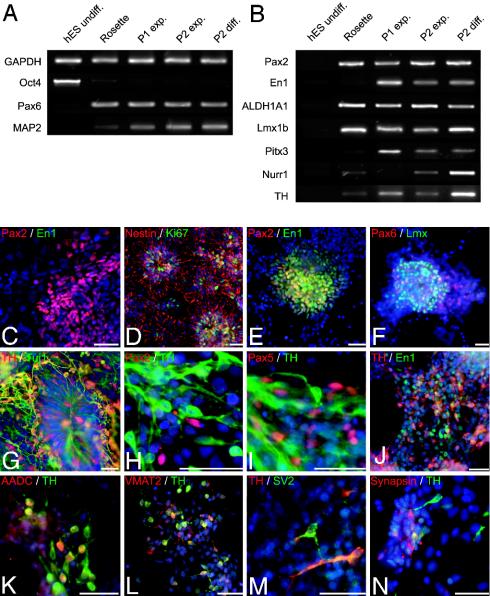
Fig. 3.
Phenotypic characterization of midbrain dopaminergic differentiation. (A and B) RT-PCR analyses of undifferentiated hES cells, passage 0 cells (day 28, Rosette), passage 1 cells during cell expansion (day 35, P1 exp.), passage 2 cells during cell expansion (day 42, P2 exp.), and passage 2 cells during cell differentiation (day 50, P2 diff.). Shown are ES-cell, neural, and neuronal markers (A) and markers of midbrain DA neuron development (B). ALDH1A1, aldehyde dehydrogenase 1; Nurr1, nuclear orphan receptor 1. (C) Pax2-positive cells (red) at passage 1 did not coexpress En1 (green). (D and E) Cells at passage 2 expressed Ki67 (green) and nestin (red) (D) and coexpressed Pax2 (red) and En1 (green) (E). (F) Pax6 (red) and Lmx (green) expression in passage 2 expansion cells. (G–N) Differentiated cells at passage 2: TH/Tuj1-positive (red, TH; green, Tuj1) neurons with radially oriented processes emanating from a rosette (G); Pax2- or Pax5-positive (red) immature precursors next to TH-positive (green) neurons (H and I); En1-p (green) cells coexpressing TH (red) (J); TH-positive (red) cells coexpressing AADC (green) (K) or VMAT2 (L); TH-positive neurons expressing the synaptic markers SV2 (green) (M) or synapsin (red) (N). Similar results were obtained with three independent hES lines. C–E show H9-derived cells; G–I and M show H1-derived cells; and F, J–L, and N show HES-3-derived cells. (Scale bars, 50 μm.)
Immunocytochemical data of passage 1 and passage 2 cells revealed that, unlike at day 28 of differentiation, hES-derived cells started to express key markers of midbrain DA neuron development. A developmental progression was observed with Pax2 appearing first in passage 1 cells in the absence of En1 expression (Fig. 3C). Cells at passages 1 and 2 continued to exhibit neural precursor characteristics expressing Ki67 and nestin (Fig. 3D). At passage 2, cells coexpressing Pax2 and En1 (Fig. 3E) and cells expressing Lmx (Fig. 3F) were observed. The total number of En1-positive cells was highly dependent on the developmental stage. The percentage of En1 cells in the total population varied from 0% (passage 0 and early passage 1) up to 30% (passage 2). At day 50, the few remaining rosettes were surrounded by TH-positive cells often radially oriented toward the center of the rosette (Fig. 3G). Coexpression studies showed that Pax2- and Pax5-expressing cells were negative for TH (Fig. 3 H and I). In contrast, cells positive for En1/TH were readily detected (Fig. 3J). Our data suggest that, similar to mouse in vivo development, human midbrain DA neurons in vitro are derived from proliferating progenitors that sequentially express Pax2 and Pax5 and, upon exiting the cell cycle, become positive for En1 and eventually TH. Single-cell lineage analysis will be required to fully confirm this finding. Additional markers expressed in TH-positive cells were AADC (Fig. 3K), VMAT2 (Fig. 3L), and synaptic markers, such as SV2 (Fig. 3M) and synapsin (Fig. 3N). Although 100% of the TH-positive cells coexpressed VMAT2, only 50% of the TH-positive cells coexpressed AADC (Fig. 3K), and 74% were immunoreactive for DA (data not shown). At day 50 of differentiation, 15% of TH-positive cells expressed SV2, confirming the immature state of synaptic development in these cells. No colocalization of TH and DA-β hydroxylase was observed at any stage of differentiation, confirming the absence of noradrenergic and adrenergic neurons (data not shown).
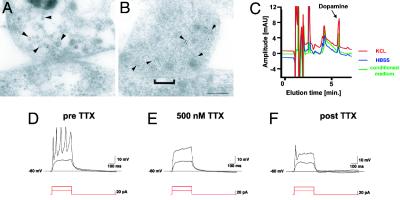
The generation of synapses is a definite marker of neuronal identity. Electron microscopic analysis at day 50 showed immature synaptic contacts in ≈10–20% of all cells. Mature synaptic contacts with many distinct postsynaptic densities were detected in 2–4% of the cells. TH-ImmunoGold-labeled particles were associated with neurotransmitter-containing vesicles and clustered near the cell membrane (Fig. 4 A and B). The generation and maturation of synapses in the majority of hES-derived DA neurons required several weeks of additional in vitro differentiation (data not shown). Reversed-phase HPLC analysis demonstrated both basal and KCl-inducible release of DA from hES-derived neurons (Fig. 4C). Cells never exposed to SHH/FGF8 (Fig. 2H, compare to none) did not yield detectable levels of DA by HPLC. Electrophysiological properties of ES-derived neurons were assessed in single-cell recordings. Passage 2 cells were grown on glass coverslips in the presence of SHH and FGF8 and differentiated for an additional 12–20 days. Recordings were obtained from a total of 81 cells. Depolarization-induced action potentials were detected in 65% (53/81) of the cells (Fig. 4D). Fast, repetitive firing was observed in 11% of cells. In all cells tested, the action potentials were blocked by 500 nM tetrodotoxin (TTX) (n = 11; Fig. 4 D–F). These data demonstrate the in vitro derivation of functional midbrain DA neurons from hES cells.
Fig. 4.
Ultrastructural and functional characterization of hES-derived DA neurons. (A and B) Electron micrographs of TH ImmunoGold-labeling (5 nm) at synaptic contacts formed between neurons derived from hES cells. Gold particles are associated with small vesicles presumably containing neurotransmitters located at the presynaptic terminal (arrowheads). (A and B) Two examples of synapses are shown: A shows recently formed immature contact with a faint membrane density and few synaptic vesicles, and B shows a more differentiated synaptic contact with a distinct postsynaptic density (bracket) and a larger number of vesicles clustered in the proximity of the cell membrane. (Scale bar, 125 nm.) (C) Representative HPLC chromatogram showing high levels of DA in the medium of rosette-derived neurons (green line: medium conditioned for 24 h, 751 ± 239 pg/ml). Relatively low levels of basal DA release were detected (blue line: exposure to buffer for 15 min, 147 ± 78 pg/ml) in contrast to very high levels of DA after 15 min of KCL-evoked depolarization (red line: 1,283 ± 421 pg/ml). Data were derived from three independent experiments. Rosette-derived neurons were patched in the whole-cell configuration in current clamp mode. (D) Action potentials were evoked by depolarizing currents at a threshold of –40 mV. (E and F) Action potentials were tetrodotoxin (TTX)-sensitive.
Discussion
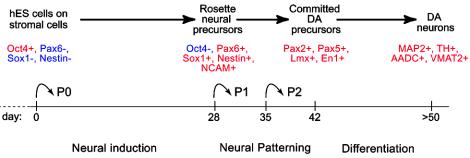
This study demonstrates the efficient derivation of midbrain DA neurons from hES cells by means of the neural-inducing properties of stromal feeder cells and the sequential application of patterning and differentiation molecules. Up to 79% of all of the neurons express TH, the rate-limiting enzyme in the synthesis of DA. In addition to TH expression, cells in these cultures express key markers associated with normal midbrain DA neuron (Fig. 5). Unlike neural differentiation of mouse ES, hES cell differentiation on MS5 is marked by the formation of large (up to 8 mm in diameter) neuroepithelial structures composed of hundreds of individual rosettes that can be isolated, propagated, and induced to undergo midbrain dopaminergic differentiation. The dopaminergic phenotype is confirmed by various morphological and functional in vitro assays, including electron microscopic visualization of TH-positive ImmunoGold-labeled synaptic terminals, measurement of KCl-evoked DA release, and the presence of depolarization-induced and tetrodotoxin (TTX)-sensitive action potentials.
Fig. 5.
Developmental progression, in vitro neural induction, neural patterning, and differentiation were monitored by the expression of specific lineage markers in hES-derived progeny as indicated. Neural induction was achieved under serum-free conditions on stromal feeder cells. At passages 1 and 2, hES-derived rosettes were plated on a polyornithine/laminin substrate in the absence of feeder cells. Neural patterning occurred in the presence of SHH/FGF8 (days 28–42). Neural differentiation required withdrawal of SHH/FGF8 and exposure to various factors that increase dopaminergic differentiation and survival (see text for details).
Stromal feeder-mediated neural induction was first described for mouse ES cells and has been proposed to specifically induce neurons of midbrain DA neuron phenotype (3). Later studies in mouse and nonhuman primate ES cells demonstrated that the neural-inducing effects are separate from the effects on patterning and that differentiation conditions can be adapted for a wide range of neuronal and glial subtypes (11, 14, 22). Here, we show that neural-inducing effects for hES cells can be provided by bone marrow and aorta-gonad-mesonephros-derived stromal feeder cells. In contrast to previous work on the differentiation of cynomolgus monkey ES cells on PA6 (14), we did not observe significant differentiation of hES cells into retinal pigment epithelial cells. However, the hES line HES-3 and the rhesus monkey ES cell line R366 yielded some (<5%) pigmented epithelial cells if maintained on MS5 beyond day 28 of differentiation (data not shown).
Based on their distinctive morphology, it has been suggested that hES derived rosettes might mimic the developing neural tube (8). Dorsoventral patterning characterized by the formation of distinct transcription factor expression domains occurs during neurulation and is largely completed at the time of neural tube closure (29). Expression of Sox1, Pax6, nestin, and NCAM in the absence of specific dorsoventral markers within rosettes suggests that these cells might correspond to a neural plate rather than a neural tube stage. This interpretation is compatible with the robust response of these cells to the patterning effects of SHH and FGF8.
Our study did not show a differentiation bias toward glutamatergic or GABAergic neurons as observed previously with hES-derived precursors maintained under neurosphere-like conditions (7, 8). Although the reasons for these differences remain to be elucidated, we speculate that the absence of FGF2-mediated proliferation might be an important component (11). Our results suggest that rosettes maintained in the absence of FGF2 remain amenable to both anterior–posterior and dorsoventral patterning and may serve as a universal source for neuronal subtype specific differentiation. Contamination of the dopaminergic population with serotonergic neurons is not surprising given the close developmental relationship between these two ventral neuron subtypes (24) and confirms the validity of the developmental model proposed here. Serotonergic differentiation might be further enhanced by using developmentally relevant factors, such as FGF4.
The sequence of transcription factor expression involved in midbrain DA neuron development in vitro is similar to the expression patterns in vivo with Pax2, aldehyde dehydrogenase 1, and Lmx1b expression preceding gradual increases in the expression of markers characteristic of early postmitotic neurons, such as En1, nuclear orphan receptor 1, Pitx3, and, finally, TH. Although robust En1 expression was observed during DA neuron differentiation, not all TH-positive neurons remained En1-positive, in contrast to rodent midbrain development (30). It will be essential to test whether loss of En1 reflects a feature of normal human midbrain development or an idiosyncrasy of our in vitro conditions. Not surprisingly, DA transporter immunoreactivity was absent in our TH-positive cell population. This finding is probably due to the relatively immature state of the TH-positive cell population, the serum-free culture conditions, and the absence of a striatal target, known to be essential for DA transporter expression in vivo (31). Exposure of hES-derived neural precursors to SHH/FGF8 dramatically increased the percentage of TH-positive neurons and subsequently the percentage of mature human DA neurons. The high DA neuron yield is illustrated by the fact that for every single undifferentiated hES cell plated at the start of the protocol, >100 TH-positive neurons can be harvested at day 50 of differentiation. Starting with a single 6-cm culture dish, the number of TH-positive cells generated by day 50 amounts to the total number of DA neurons present in the adult human substantia nigra (≈1 × 106 cells). The time frame for midbrain DA neuron differentiation (7–8 weeks in vitro) may appear long. However, midbrain DA neurons are born in vivo at 6.5–8.5 weeks after conception (32), a time frame comparable to our in vitro data.
One important application of the high-yield midbrain DA neuron derivation protocol reported here will be transplantation into preclinical animal models of Parkinson's disease. The conditions developed were highly reproducible for three independent hES and two monkey ES cell lines. A full complement of midbrain DA neurons markers both at the RNA and protein level and robust in vitro functionality also suggest great in vivo potential. However, cell survival and long-term maintenance of phenotype will be essential parameters to test in vivo. The therapeutic use of human cells exposed to mouse feeders raises safety concerns, and clinical consideration might require the use of a human feeder-based or feeder-free neural induction system. MS5 cells could be eliminated by fluorescence-activated cell sorter analysis for endoglin or ScaI, although we typically cannot detect any MS5 cells in passage 2 cultures. Positive selection strategies could also be developed to further increase the safety and purity of the dopaminergic neurons pool for transplantation. The demonstration that neural rosettes respond to developmental patterning cues suggests that our protocol may be easily adapted for the generation of other relevant neuronal subtypes, such as motor neurons, cerebellar neurons, or forebrain GABA neurons. The controlled in vitro differentiation and the availability of genetic tools for loss and gain of function in hES cells will provide a unique environment for the systematic studies of human brain development.
Supplementary Material
Acknowledgments
We thank H. Varmus (Sloan–Kettering Institute) for the Wnt1 cDNA expression construct, R. McKay (National Institutes of Health, Bethesda) for nestin antibodies, M. Busslinger (Institute of Molecular Pathology, Vienna) for Pax5 antibodies, R. Lovell-Badge (National Institute for Medical Research, London) for Sox1 antibodies, and K. Wiesel and M. Moore (Sloan–Kettering Institute) for MS5 and S2 stromal cells. We also thank V. Pratomo, T. Bajwa, and P. Song for excellent technical assistance and M. Tomishima for critical review of the manuscript. This work was supported by Michael J. Fox Foundation for Parkinson's Research Grant 01.2002.07, the Parkinson's Disease Foundation, and by National Institutes of Health Grants K08 GM68018-1 (to N.T.), NS40300, and NS044819.
Abbreviations: ES, embryonic stem; hES, human ES; DA, dopamine; TH, tyrosine hydroxylase; GABA, γ-aminobutyric acid; FGF, fibroblast growth factor; MEF, mouse embryonic fibroblasts; AA, ascorbic acid; SHH, sonic hedgehog; BDNF, brain-derived neurotrophic factor; AADC, aromatic l-amino acid decarboxylase; SV2, synaptic vesicle 2; Tuj1, class III β-tubulin; NCAM, neural cell adhesion molecule; En1, engrailed-1.
References
- 1.Thomson, J. A., Itskovitz-Eldor, J., Shapiro, S. S., Waknitz, M. A., Swiergiel, J. J., Marshall, V. S. & Jones, J. M. (1998) Science 282, 1145–1147. [DOI] [PubMed] [Google Scholar]
- 2.Lee, S.-H., Lumelsky, N., Studer, L., Auerbach, J. M. & McKay, R. D. G. (2000) Nat. Biotechnol. 18, 675–679. [DOI] [PubMed] [Google Scholar]
- 3.Kawasaki, H., Mizuseki, K., Nishikawa, S., Kaneko, S., Kuwana, Y., Nakanishi, S., Nishikawa, S. I. & Sasai, Y. (2000) Neuron 28, 31–40. [DOI] [PubMed] [Google Scholar]
- 4.Wichterle, H., Lieberam, I., Porter, J. A. & Jessell, T. M. (2002) Cell 110, 385–397. [DOI] [PubMed] [Google Scholar]
- 5.Brustle, O., Jones, K. N., Learish, R. D., Karram, K., Choudhary, K., Wiestler, O. D., Duncan, I. D. & McKay, R. G. (1999) Science 285, 754–756. [DOI] [PubMed] [Google Scholar]
- 6.Studer, L. (2001) Nat. Biotechnol. 19, 1117–1118. [DOI] [PubMed] [Google Scholar]
- 7.Reubinoff, B. E., Itsykson, P., Turetsky, T., Pera, M. F., Reinhartz, E., Itzik, A. & Ben Hur, T. (2001) Nat. Biotechnol. 19, 1134–1140. [DOI] [PubMed] [Google Scholar]
- 8.Zhang, S. C., Wernig, M., Duncan, I. D., Brustle, O. & Thomson, J. A. (2001) Nat. Biotechnol. 19, 1129–1133. [DOI] [PubMed] [Google Scholar]
- 9.Carpenter, M. K., Inokuma, M. S., Denham, J., Mujtaba, T., Chiu, C. P. & Rao, M. S. (2001) Exp. Neurol. 172, 383–397. [DOI] [PubMed] [Google Scholar]
- 10.Park, S., Lee, K. S., Lee, Y. J., Shin, H. A., Cho, H. Y., Wang, K. C., Kim, Y. S., Lee, H. T., Chung, K. S., Kim, E. Y., et al. (2004) Neurosci. Lett. 359, 99–103. [DOI] [PubMed] [Google Scholar]
- 11.Barberi, T., Klivenyi, P., Calingasan, N. Y., Lee, H., Kawamata, H., Loonam, K., Perrier, A. L., Bruses, J., Rubio, M. E., Topf, N., et al. (2003) Nat. Biotechnol. 21, 1200–1207. [DOI] [PubMed] [Google Scholar]
- 12.Kim, J. H., Auerbach, J. M., Rodriguez-Gomez, J. A., Velasco, I., Gavin, D., Lumelsky, N., Lee, S. H., Nguyen, J., Sanchez-Pernaute, R., Bankiewicz, K., et al. (2002) Nature 418, 50–56. [DOI] [PubMed] [Google Scholar]
- 13.Itoh, K., Tezuka, H., Sakoda, H., Konno, M., Nagata, K., Uchiyama, T., Uchino, H. & Mori, K. J. (1989) Exp. Hematol. 17, 145–153. [PubMed] [Google Scholar]
- 14.Kawasaki, H., Suemori, H., Mizuseki, K., Watanabe, K., Urano, F., Ichinose, H., Haruta, M., Takahashi, M., Yoshikawa, K., Nishikawa, S. I., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 1580–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perrier, A. L. & Studer, L. (2003) Semin. Cell Dev. Biol. 14, 181–189. [DOI] [PubMed] [Google Scholar]
- 16.Reubinoff, B. E., Pera, M. F., Fong, C. Y., Trounson, A. & Bongso, A. (2000) Nat. Biotechnol. 18, 399–404. [DOI] [PubMed] [Google Scholar]
- 17.Thomson, J. A., Kalishman, J., Golos, T. G., Durning, M., Harris, C. P., Becker, R. A. & Hearn, J. P. (1995) Proc. Natl. Acad. Sci. USA 92, 7844–7848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cibelli, J. B., Grant, K. A., Chapman, K. B., Cunniff, K., Worst, T., Green, H. L., Walker, S. J., Gutin, P. H., Vilner, L., Tabar, V., et al. (2002) Science 295, 819. [DOI] [PubMed] [Google Scholar]
- 19.Johe, K. K., Hazel, T. G., Müller, T., Dugich-Djordjevic, M. M. & McKay, R. D. G. (1996) Genes Dev. 10, 3129–3140. [DOI] [PubMed] [Google Scholar]
- 20.Rubio, M. E. & Soto, F. (2001) J. Neurosci. 21, 641–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Studer, L., Psylla, M., Buhler, B., Evtouchenko, L., Vouga, C. M., Leenders, K. L., Seiler, R. W. & Spenger, C. (1996) Brain Res. Bull. 41, 143–150. [DOI] [PubMed] [Google Scholar]
- 22.Mizuseki, K., Sakamoto, T., Watanabe, K., Muguruma, K., Ikeya, M., Nishiyama, A., Arakawa, A., Suemori, H., Nakatsuji, N., Kawasaki, H., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 5828–5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shimozaki, K., Nakashima, K., Niwa, H. & Taga, T. (2003) Development (Cambridge, U.K.) 130, 2505–2512. [DOI] [PubMed] [Google Scholar]
- 24.Ye, W. L., Shimamura, K., Rubenstein, J. R., Hynes, M. A. & Rosenthal, A. (1998) Cell 93, 755–766. [DOI] [PubMed] [Google Scholar]
- 25.O'Rahilly, R. & Müller, F. (1987) Developmental Stages in Human Embryos (Carnegie Inst. Washington, Washington, DC)
- 26.Farkas, L. M., Dunker, N., Roussa, E., Unsicker, K. & Krieglstein, K. (2003) J. Neurosci. 23, 5178–5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gall, C. M., Hendry, S. H., Seroogy, K. B., Jones, E. G. & Haycock, J. W. (1987) J. Comp. Neurol. 266, 307–318. [DOI] [PubMed] [Google Scholar]
- 28.Max, S. R., Bossio, A. & Iacovitti, L. (1996) Dev. Brain Res. 91, 140–142. [DOI] [PubMed] [Google Scholar]
- 29.Briscoe, J. & Ericson, J. (2001) Curr. Opin. Neurobiol. 11, 43–49. [DOI] [PubMed] [Google Scholar]
- 30.Simon, H. H., Saueressig, H., Wurst, W., Goulding, M. D. & O'Leary, D. D. M. (2001) J. Neurosci. 21, 3126–3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perrone-Capano, C., Tino, A., Amadoro, G., Pernas-Alonso, R. & Di Porzio, U. (1996) Brain Res. Mol. Brain Res. 39, 160–166. [DOI] [PubMed] [Google Scholar]
- 32.Freeman, T. B., Spence, M. S., Boss, B. D., Spector, D. H., Strecker, R. E., Olanow, G. W. & Kordower, J. H. (1991) Exp. Neurol. 113, 344–353. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.