Abstract
Serum response factor (SRF) regulates genes involved in cell proliferation, migration, cytoskeletal organization, and myogenesis. Myocardin and myocardin-related transcription factors (MRTFs) act as powerful transcriptional coactivators of SRF in mammalian cells. We describe an MRTF from Drosophila, called DMRTF, which shares high homology with the functional domains of mammalian myocardin and MRTFs. DMRTF forms a ternary complex with and stimulates the activity of Drosophila SRF, which has been implicated in branching of the tracheal (respiratory) system and formation of wing interveins. A loss-of-function mutation introduced into the DMRTF locus by homologous recombination results in abnormalities in tracheal branching similar to those in embryos lacking SRF. Misexpression in wing imaginal discs of a dominant negative DMRTF mutant also causes a diminution of wing interveins, whereas overexpression of DMRTF results in excess intervein tissue, abnormalities reminiscent of SRF loss- and gain-of-function phenotypes, respectively. Overexpression of these DMRTF mutants in mesoderm and in the tracheal system also perturbs mesoderm cell migration and tracheal branching, respectively. We conclude that the interaction of MRTFs with SRF represents an ancient protein partnership involved in cytoplasmic outgrowth and cell migration during development.
Serum response factor (SRF) is an ancient and evolutionarily conserved member of the MADS (MCM1, Agamous, Deficiens, SRF) box family of transcription factors that regulates a wide range of genes involved in cell growth, migration, cytoskeletal organization, and myogenesis (reviewed in ref. 1). The ability of SRF to regulate such diverse sets of genes depends on its association with cell-type-specific and signal-responsive transcription factors.
SRF is required for expression of cardiac and smooth muscle genes during development of the vertebrate cardiovascular system (2–7). The SRF homologue in Drosophila (DSRF) is also required for development of the tracheal system (8, 9), a branching tubular network that functions as a respiratory system by mediating gaseous exchange in the embryo (reviewed in ref. 10). In the DSRF mutants pruned and blistered, tracheal branching is disrupted due to failure of terminal tracheal cells to extend cytoplasmic projections toward target tissues (8, 9). SRF also plays a key role in wing development in Drosophila by promoting the formation of intervein tissue and suppressing wing vein formation (11, 12).
Myocardin (13) and myocardin-related transcription factors (MRTFs) (14) are SAP (SAF-A/B, Acinus, PIAS) domain transcription factors that interact with SRF and potently enhance the expression of SRF-dependent genes (13–17). Myocardin is expressed specifically in the developing cardiovascular system in vertebrate embryos (13), whereas MRTFs are expressed in a wide range of cell types (14). Targeted deletion of the mouse myocardin gene blocks expression of SRF target genes in vascular smooth muscle cells with consequent embryonic lethality (18). MRTF-A (14), also called megakaryoblastic leukemia (MAL)/MKL-1/BSAC (19–22), has also been shown to transduce the stimulatory effects of Rho signaling and actin polymerization to SRF and its target genes (21, 23, 24).
A complete understanding of the role of myocardin and MRTFs in the mouse is complicated by potential functional redundancy among these factors. Therefore, to further explore the functions of the myocardin family in vivo, we searched for and identified an MRTF gene in Drosophila. Here we describe the only MRTF in that organism, referred to as DMRTF, which, like its mammalian counterparts, interacts with SRF and stimulates SRF-dependent transcription. Consistent with its potential role as a coactivator for SRF, loss of DMRTF function results in abnormalities in tracheal development similar to those that result from DSRF loss-of-function mutations. Through overexpression of hypo- and hypermorphic DMRTF alleles in the developing wing and mesoderm, we also implicate DMRTF in specification of wing intervein tissue and migration of mesodermal cells. These findings suggest that the association of SRF with MRTFs represents an evolutionarily conserved regulatory mechanism involved in the development of tissues that undergo extensive cytoplasmic extension or migration.
Materials and Methods
Isolation of the DMRTF Gene and Homologous Recombination. The DMRTF gene was identified by sequence similarity by using mouse myocardin sequences to blast the Drosophila protein database. Ends-out homologous recombination at the DMRTF locus was performed as described (25). Briefly, two arms (≈4kb each) obtained from genomic PCR were subcloned into the pw25 vector to create the construct DMRTFko-pw25 for generating transformants. Transgenic flies bearing this construct on chromosome 2 were crossed to yw; 70FLP, 70I-SceI, Sco/CyO flies, and the progeny (F1) were heat-shocked at 37°C for 1 h at days 3, 4, and 5 after egg laying. The DMRTFko-pw25/70FLP, 70I-SceI, Sco virgins of the F1 progeny were crossed to w1118 males. Approximately 0.04% of the F2 progeny appeared with red eyes (9 of ≈24,300 flies). Two independent lines showed translocation of the w+ marker from chromosome 2 to 3, where DMRTF is located. Targeted homologous recombination was verified by genomic PCR and sequencing. Primer information is available on request.
Drosophila Strains. Overexpression of transgenes was accomplished by using the Gal4-UAS system (26). The following fly lines were used: twi-Gal4; 24B-Gal4 (conferring panmesodermal expression) (27); Daughterless (da)-Gal4 (conferring early ubiquitous expression); btl-Gal4 (conferring tracheal expression; a gift from M. Affolter, Universität Basel, Basel); and ms1096-Gal4 (conferring imaginal disk expression). bs14 (a null mutant of DSRF) was from Bloomington Stock Center. Oregon-R was used as the wild-type reference strain. The UAS-myocardin, UAS-DMRTF, UAS-DMRTFΔN, UAS-DMRTFΔC, and UAS-DMRTFΔNΔC constructs were generated by cloning the corresponding cDNAs into pUAST followed by standard transformation.
Immunohistochemistry and Microscopy. Embryos from different lines were collected and stained with various antibodies as described (28). The following primary antibodies were used: mouse IgG anti-DSRF (1:500, Geneka Biotechnology, New York); rabbit anti-Dmef2, 1:1,000 (29); mouse IgM 2A12 (labels tracheal lumen; Developmental Studies Hybridoma Bank, University of Iowa, Iowa City). Cy2, Cy3, Cy5, or biotin-conjugated secondary antibodies (The Jackson Laboratory) were used to recognize the primary antibodies. Images were obtained with a Zeiss LSM510-meta confocal microscope or a Leica (Deerfield, IL) DMRXE compound microscope.
Transfection and Coimmunoprecipitation Assays. Cell transfection, luciferase assays, and coimmunoprecipitation experiments were performed as described (15). Unless otherwise indicated, 100 ng of reporter plasmid and 100 ng of each activator plasmid were used. The SM22-luciferase and atrial natriuretic factor (ANF)-luciferase reporters have been described (13). Luciferase activities are expressed as mean ± standard deviation from three experiments.
Gel Mobility Shift. DSRF and DMRTF were translated in vitro with a TNT T7-coupled reticulocyte lysate system (Promega). Gel mobility-shift assays were performed by using radiolabeled probes for the distal CArG box from the SM22 promoter as described (30).
RNA Interference. Double-strand RNA interference and immunostaining of injected embryos were performed according to the online protocol available at www.biochem.northwestern.edu/carthew/manual/RNAi_Protocol.html. Primer sequences for generating the template for double-strand RNA synthesis are available on request.
Results
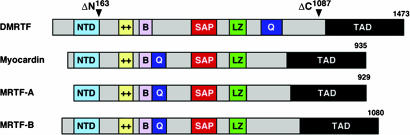
Structural Conservation of Mammalian and Drosophila MRTFs. By analysis of publicly available Drosophila genome and cDNA sequence, we identified a single Drosophila gene encoding a potential ortholog of mammalian myocardin and MRTFs, which we refer to as DMRTF. By RT-PCR of embryonic mRNA, we obtained a complete ORF of DMRTF with the potential to encode a protein of 1,473 aa (Fig. 1). DMRTF shares homology with the N-terminal domain (NTD, 62% identity), basic region (36% identity), and SAP domain (68% identity) of myocardin and MRTFs. Like the mammalian proteins, DMRTF also contains a leucine zipper domain between the SAP domain and the C-terminal transcription activation domain, as well as a glutamine (Q)-rich domain located closer to the C terminus than in the mammalian proteins. There is no recognizable homology between DMRTF and the mammalian proteins outside the above domains. DMRTF shares comparable homology to mouse myocardin, MRTF-A and MRTF-B, making it unclear whether it represents one of these factors in particular. The sequence alignment of DMRTF, myocardin, MRTF-A, and MRTF-B is provided as Fig. 7, which is published as supporting information on the PNAS web site.
Fig. 1.
Structural similarities between Drosophila and mammalian MRTFs. NTD contains three RPEL regions. ++, basic region; B, B-box-like region; LZ, leucine zipper; Q, glutamine-rich region; TAD, transcription activation domain. The positions of deletion mutations used to generate DMRTF mutant proteins are shown at the top.
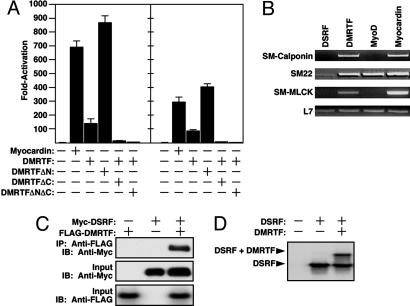
DMRTF Is a Cofactor of SRF. To determine whether DMRTF was able to stimulate SRF activity like mammalian myocardin and MRTFs, we performed transfection assays in COS cells with luciferase reporters linked to the SM22 and ANF promoters, both of which contain a pair of SRF-binding sites (3, 31). As shown in Fig. 2A, DMRTF stimulated the activity of these promoters by ≈100-fold. Mutation of the SRF-binding sites in these promoters abolished responsiveness to DMRTF (data not shown).
Fig. 2.
DMRTF is a cofactor for SRF. (A) COS cells were transiently transfected with expression vectors encoding mouse myocardin, DMRTF, or DMRTF mutants and luciferase reporters linked to the SM22 (Left) or ANF (Right) promoters as indicated, and luciferase activity was determined. (B) 10T1/2 cells were transiently transfected with expression vectors and transcripts for the indicated genes were detected by RT-PCR. DMRTF and myocardin induce the expression of smooth muscle markers (SM-calponin, SM22, and SM-MLCK), whereas MyoD activates expression only of SM22, which is expressed in smooth and skeletal muscle. L7 transcripts were detected as a loading control. (C) Drosophila S2 cells were transiently transfected with expression vectors encoding Myc-tagged DSRF and FLAG-tagged DMRTF, and immunoprecipitation (IP) and immunoblotting (IB) were performed as indicated. (D) Gel mobility-shift assays were performed with a radiolabeled SRF-binding site as probe and DSRF and DMRTF proteins translated in vitro. Only the region of the gel containing the DNA-protein complexes is shown. DMRTF forms a ternary complex with DSRF.
The NTD of MAL/MRTF-A has been shown to sequester the protein in the cytoplasm via its association with G-actin, and its deletion enhances transcriptional activity of MAL (23). Similarly, deletion of the NTD of DMRTF (mutant DMRTFΔN) resulted in its localization to the nucleus (data not shown) and an increase in transcriptional activity (Fig. 2 A). Deletion of the transcription activation domain from the C terminus of DMRTF (mutant DMRTFΔC) abolished transcriptional activity and created a dominant negative mutant (Fig. 2 A). This mutant, like wild-type DMRTF, was present in the cytoplasm and nucleus of transfected COS cells (data not shown). A mutant lacking the N and C termini (mutant DMRTFΔNΔC) was localized to the nucleus (data not shown) and also acted in a dominant negative manner (Fig. 2 A).
In mouse embryonic stem cells homozygous for a null mutation in the SRF gene (32), neither DMRTF nor myocardin was able to stimulate expression of SM22-luciferase, whereas addition of either mammalian or Drosophila SRF restored transcriptional activity to DMRTF and myocardin (data not shown). Like myocardin and MRTF-A (15), DMRTF was also able to activate expression of endogenous smooth muscle genes (SM-calponin, SM22, and SM-MLCK) in 10T1/2 fibroblasts assayed by RT-PCR (Fig. 2B). In immunoprecipitation assays, DMRTF associated with DSRF, and the two proteins formed a ternary complex on an SRF-binding site in gel mobility-shift assays (Fig. 2 C and D). Thus, DMRTF, like the mammalian members of the myocardin family, acts as a potent SRF coactivator.
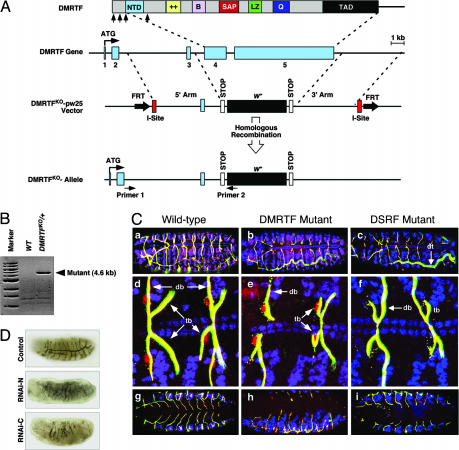
Inactivation of the DMRTF Gene by Homologous Recombination. DMRTF is expressed ubiquitously through embryogenesis, as shown by both in situ hybridization and antibody staining (data not shown). The DMRTF gene maps to region 62F2-3 on chromosome 3 and contains five protein-coding exons (Fig. 3A). There were no available P element insertions in the region of the DMRTF gene, so we designed a targeting vector to introduce a loss-of-function mutation in the gene by ends-out homologous recombination (Fig. 3A and ref. 25). The DMRTFko-pw25 construct contained two genomic arms that flank exons 4 and 5 of the gene, which encode all of the identified domains of DMRTF from amino acid 73 to 1473. Homologous recombination deleted this region of the gene, resulting in a null mutation. Sequencing of the mutant allele showed that the mini-white gene replaced the coding region (exons 4 and 5) of DMRTF between the two arms in the DMRTFko-pw25 construct. We refer to this mutant allele as DMRTFko. The targeted mutation was confirmed by PCR of genomic DNA from heterozygous mutant flies (Fig. 3B) and DNA sequencing (data not shown).
Fig. 3.
Inactivation of the DMRTF gene by homologous recombination. Elimination of DMRTF expression causes defects in tracheal branching. (A) The structures of the DMRTF gene, targeting vector, and targeted locus are shown. Positions of intron/exon junctions in the coding region are shown by vertical arrows beneath the protein. The five coding exons of the DMRTF gene are numbered. Positions of primers used for PCR in B are shown at the bottom. (B) PCR of genomic DNA from wild-type (WT) and DMRTFko/+ flies. A product of the predicted size is generated only with genomic DNA from mutant flies, because primer 1 sequence is located outside the 5′ arm of the targeting vector, and primer 2 sequence is within the w+ gene. (C) Stage 16 embryos were stained with mouse IgM anti-2A12 antibody (recognized by FITC-streptavidin for biotin-goat-anti-mouse IgM, green) to reveal the tracheal system, anti-DSRF (recognized by Cy3-goat-anti-mouseIgG, red) to reveal terminal tracheal cells, and anti-Dmef2 (Cy5-goat-anti-rabbit, blue) to show the heart and muscles. Tracheal branches appear yellow because Cy3-goat-anti-mouse IgG also recognizes mouse-IgM-anti-2A12. (a, d, and g) Wild-type. (b, e, and h) DMRTF mutant. (c, f, and i) DSRF mutant. a–f are dorsal views and g–i are ventral views of stage 16 embryos. d–f show high-magnification views of dorsal branches (db) and terminal branches (tb) of the tracheal system. DMRTF mutants (b, e, and h) often show truncated terminal branches on both the dorsal and ventral sides of the embryos, similar to the phenotype of DSRF mutants (c, f and i). The dorsal vessel and somatic muscles, which are unaffected in both mutants, are purple in the wild-type and DMRTF mutants but blue in the DSRF mutants, because both DSRF and Dmef2 are expressed in these tissues. (D) Stage 16 embryos were stained with anti-2A12 to reveal the tracheal system. Embryos were injected with buffer as control or dsRNA corresponding to coding sequences for the N- and C-terminal regions of DMRTF, as indicated. Both interfering RNAs perturbed tracheal branching.
DMRTF Loss-of-Function Phenotypes. Using a balancer chromosome with actin-GFP, we were able to identify homozygous DMRTF mutant embryos and larvae. Flies heterozygous for the DMRTFko allele showed no phenotype, whereas the homozygous DMRTFko allele was lethal at the first instar larval stage.
To identify possible defects of the homozygous mutant, we stained the DMRTFko embryos using antibodies against Dmef2 (cardiac and somatic muscle cells), Even-skipped (subset of pericardial cells, dorsal muscles, and central nervous system), Tinman (early cardiac and visceral mesoderm), DSRF (terminal tracheal cells), and 2A12 (tracheal lumen). We found no significant defects in the mesoderm or central nervous system or in the overall DSRF expression pattern in homozygous DMRTFko embryos (data not shown). However, homozygous DMRTF mutant embryos displayed defects in the tracheal system at late stage 16 that included a tortuous dorsal trunk (≈80% penetrance) and truncated terminal branches from the dorsal and ganglionic branches (≈60% penetrance) (Fig. 3C b, e, and h). The dorsal anastomosis, which connects the contralateral hemisegment of the tracheal system, was also frequently missing in DMRTFko mutants (≈50% penetrance) (Fig. 3Cb, e, and h). The phenotype of truncated terminal branches was also observed in the bs14 DSRF mutant (Fig. 3C c, f, and i) with a higher level of penetrance (≈95% penetrance) (9), suggesting that the lack of zygotic DMRTF affects DSRF function in the developing tracheal system.
To further explore the functions of DMRTF, we reduced the expression of maternal and zygotic DMRTF by double-strand RNA interference, using interfering RNAs representing sequences coding for the N- and C-terminal regions of the DMRTF protein. More than 1,000 embryos were injected with buffer alone (Fig. 3D, control), allowed to develop to stage 16, and stained with 2A12 to ensure that injection and immunostaining procedures did not cause tracheal defects. Approximately 400 embryos each were then injected with double-stranded RNA derived from coding sequences for the N and C termini (RNAi-N and RNAi-C, respectively) of DMRTF. Both interfering RNAs resulted in a severe tracheal phenotype characterized by lack of fusion and migration of tracheal branches (Fig. 3D). The phenotype resulting from RNAi was much more severe than the DMRTFko phenotype, which may be due to elimination of both maternal and zygotic DMRTF transcripts that sustain MRTF function during early embryogenesis.
Ectopic Expression of DMRTF Affects Cell Migration in the Mesoderm. We also examined DMRTF functions by overexpressing various forms of the protein in different tissues using the Gal4/UAS system (26). These forms included wild-type DMRTF, the dominant negative DMRTF mutant in which the C-terminal transcription activation domain was deleted (DMRTFΔC), the hyperactive form in which the inhibitory NTD was deleted (DMRTFΔN), and the mutant containing only the SRF-binding domain (DMRTFΔNΔC). Overexpression of wild-type DMRTF in the mesoderm using twi-Gal4; 24B-Gal4 drivers did not evoke a phenotype (data not known), whereas DMRTFΔC interfered with migration of Dmef2-expressing cells, such that only a small fraction of these cells migrated dorsally, and the majority remained in the ventral lateral region of the embryo at stage 14 (Fig. 4C). Only a few cardiac cells reached the dorsal midline, and most dorsal muscles were not formed at stage 16 in the presence of this mutant protein (Fig. 4D). Interestingly, when the hyperactive form of DMRTF was overexpressed in the mesoderm, the precisely aligned row of cardiac cells and segmentally distributed muscle pattern was disrupted (Fig. 4E). Large groups of muscle cells were clustered and located more dorsally than usual, leaving obvious gaps in the ventral lateral region (Fig. 4E). Although all of the cardiac cells migrated to the dorsal midline at stage 16, they failed to align and form a heart tube (Fig. 4F). These phenotypes are likely caused by inappropriate (or accelerated) migration of mesodermal cells. Overexpression of mouse myocardin in the mesoderm caused a phenotype similar to that of the activated form of DMRTF (Fig. 4 G and H), suggesting mouse myocardin can function similarly to the active form of DMRTF in Drosophila.
Fig. 4.
Abnormalities in mesoderm migration from DMRTF misexpression. (A) Wild-type. DMRTF mutant proteins or mouse myocardin were misexpressed in the developing mesoderm by using the twi; 24B-Gal4 drivers. Embryos were stained for Dmef2 (red). (C and D) Misexpression of DMRTFΔC resulted in a reduction in ventrodorsal migration and a consequent lack of cardiac cells along the dorsal midline of stage 16 embryos. Misexpression of the active mutant DMRTFΔN or myocardin caused excessive migration, leaving consequent gaps in the mesodermal layer of stage 14 embryos and misaligned cardiac cells at stage 16 (E–H). Arrows point to abnormalities in the distribution of mesodermal cells. A, C, E and G are lateral views of Stage 14 embryos. B, D, F, and H are dorsal views of stage 16 embryos.
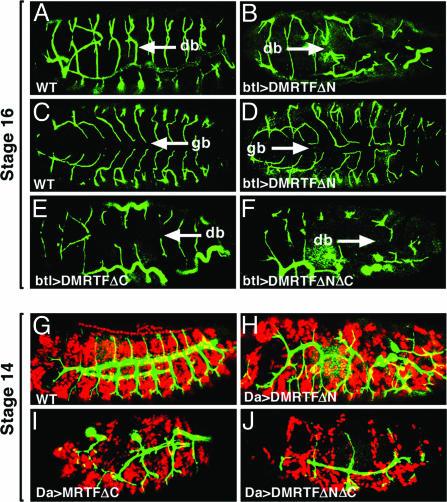
Ectopic Expression of DMRTF Affects Tracheal Branch Migration and Fusion. We also misexpressed the various DMRTF mutants in the tracheal system using the btl-Gal4 driver. Overexpression of wild-type DMRTF did not cause an obvious phenotype (data not shown). In contrast, the normal pattern of the tracheal system was disrupted by overexpression of the hyperactive form of DMRTF (DMRTFΔN). Abnormally long dorsal branches were often seen on the dorsal side of these embryos, whereas the extension of the ganglionic branches appeared undirectional and enhanced (Fig. 5 C and D). Overexpression of the dominant negative forms of DMRTF (DMRTFΔC and DMRTFΔNΔC) in the tracheal system caused the opposite phenotype, in which both the dorsal and the ganglionic branches were often truncated or missing (Fig. 5 E and F, data not shown). These findings suggest that activated DMRTF is necessary for cytoplasmic outgrowth of the tracheal branches, but the direction of extension requires additional cues.
Fig. 5.
Tracheal defects resulting from DMRTF misexpression. (A) Wild-type. (B, D–F, and H–J) DMRTF mutant proteins were misexpressed in the developing tracheal system by using the btl-Gal4 driver (B and D–F) or throughout the embryo using the Da-Gal4 driver (H–J). Embryos were stained for 2A12 (green) and Dmef2 (red) expression. (B and D) Misexpression of the active mutant DMRTFΔN perturbed the normal pattern of the tracheal system and caused excessive branching in both dorsal branches (B) and ventral ganglionic branches (D). (E and F) Misexpression of either dominant negative form of DMRTF (DMRTFΔC or DMRTFΔNΔC) inhibited tracheal branching or migration. (G–J) When overexpressed ubiquitously in the embryo, these DMRTF mutants caused defects in both mesoderm migration and tracheal branching. A, B, E, and F are dorsal–lateral views, and C and D are ventral views of stage 16 embryos. G–J are lateral views of stage 14 embryos. db, dorsal tracheal branches; gb, ganglionic tracheal branches.
We also overexpressed mutants of DMRTF using a Da-Gal4 driver, which is expressed early and ubiquitously in the embryo. The hyperactive mutant DMRTFΔN caused a disruption in patterning of the tracheal system associated with excessive and unpatterned branches (Fig. 5H). In contrast, the DMRTFΔC and DMRTFΔNΔC mutants inhibited branching (Fig. 5 I and J). These mutants also disrupted somatic muscle patterning.
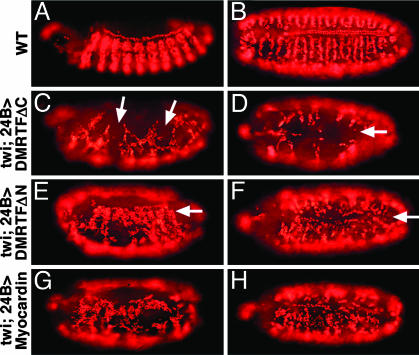
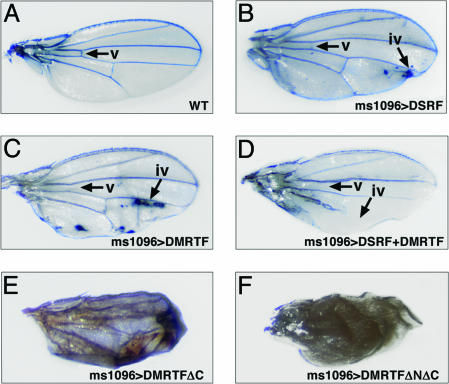
DMRTF and DSRF Genetically Interact During Wing Development. DSRF has been shown to act in a dosage-sensitive manner within the developing wing to suppress formation of wing veins and to promote the development of intervein tissue (11, 12). Overexpression of both the wild-type and active forms of DMRTF in the wing imaginal discs using ms1096-Gal4 caused a reduction of veins and overproliferation of intervein tissues in the adult wing, similar to the phenotype resulting from DSRF overexpression (Fig. 6 A–C and data not shown). This phenotype was more dramatic when DSRF and DMRTF were overexpressed together in the wing discs (Fig. 6D), consistent with the interdependence of these factors. In contrast, misexpression of the dominant negative forms of DMRTF (DMRTFΔC or DMRTFΔNΔC) dramatically reduced the intervein tissue and often resulted in a failure of the two layers of the wing to fuse completely (Fig. 6 E and F). This phenotype is highly similar to that of bs12 (a hypomorphic DSRF allele) (11), suggesting that the dominant negative form of DMRTF inhibits DSRF function during wing development.
Fig. 6.
Wing defects resulting from DMRTF misexpression. (A) Wild-type. DMRTF mutant proteins or DSRF were misexpressed in the wing imaginal discs by using the ms1096-Gal4 driver. (B and C) Misexpression of DSRF or DMRTF resulted in reduced vein (B) and excess intervein (C). (D) This phenotype was enhanced by coexpression of DSRF and DMRTF. (E and F) Misexpression of the inactive mutant proteins DMRTFΔC or DMRTFΔNΔC resulted in a loss of intervein and adhesion between the two cell layers of the wing. iv, intervein tissue; v, vein.
Discussion
The results of this study show that the Drosophila genome encodes a single member of the myocardin family, DMRTF, which acts as a transcriptional coactivator for SRF. Based on loss- and gain-of-function phenotypes, it appears that DMRTF is required for proper patterning of the tracheal system and for the formation of wing intervein tissue, which also depend on DSRF. It is likely that the strong maternal contribution of DMRTF mRNA and protein to the embryo diminishes potential phenotypes resulting from a targeted mutation of the DMRTF locus until the larval stage. The phenotypes from RNAi and misexpression of dominant negative mutant proteins strongly implicate DMRTF in the control of SRF activity during developmental processes involving cytoplasmic outgrowth and cell migration.
Involvement of DMRTF in Tracheal Development. The Drosophila tracheal system is comprised of a network of interconnected epithelial tubes that undergo sequential sprouting (10). Terminal branches of the tracheal network are formed from individual cells that express high levels of DSRF and extend long cytoplasmic processes toward target tissues. Signaling from fibroblast growth factor (FGF) to SRF has been shown to regulate cytoplasmic outgrowth of terminal cells during tracheal branching (9, 10). The similarities between the tracheal branching phenotypes of DMRTF and DSRF mutant embryos suggest that DMRTF and DSRF may function together during terminal tracheal branching. The small GTPase Rac, which regulates cell adhesion and actin-based cytoskeletal motility, has also been shown to act in a signaling pathway that interconnects FGF signaling with SRF during tracheal branching (33). The DMRTF RNAi tracheal phenotype is similar to that of embryos lacking Rac1 and -2 (33), suggesting DMRTF may act in the Rac signaling pathway during tracheal development. Given the role of the mammalian DMRTF ortholog, MRTF-A/MAL, in transduction of growth signals and changes in actin treadmilling to SRF (21, 23, 24), these findings point to a similar role for DMRTF during cytoplasmic branching of the tracheal system in Drosophila.
Involvement of DMRTF in Wing Development. The wings of Drosophila are derived from sheets of epithelial cells that become subdivided into vein and intervein cells, giving rise to a stereotypical pattern of veins that provide structural support to the wings. Intervein cells are required for adherence of the two surfaces of the wing. As in the developing tracheal system, signaling by FGF to SRF has been shown to be required for vein/intervein formation (11, 12); the absence of either of these factors results in a deficiency of intervein cells.
Consistent with the proposed role of DMRTF as a coactivator of SRF, DMRTF mutant embryos or embryos expressing dominant negative DMRTF displayed a loss of intervein cells. Conversely, misexpression of hypermorphic DMRTF alleles resulted in excess intervein cells and the concomitant loss of vein cells. The latter phenotype was also observed in response to misexpression of DSRF and DMRTF together.
Involvement of DMRTF in Cell Migration. Mesodermal cells form in the ventral region of the Drosophila embryo and migrate in a dorsolateral direction to give rise to a uniform monolayer. This process is essential for the regional specification of different mesodermal derivatives, such as cardiac, somatic, and visceral muscles. Like the dependence of tracheal branching on FGF signaling, the mesoderm-specific FGF receptor, heartless, was shown to be required for mesoderm migration (34). Recently, a guanyl nucleotide exchange factor, pebble, was found to be required for mesoderm migration (35, 36), likely through modification of a small GTPase such as Rho and Rac. Interestingly, the activity of DMRTF, like that of its mammalian orthologue MRTF-A/MAL, is also stimulated by Rho-actin signaling in Drosophila cultured cell assays (Z.H. and E.N.O., unpublished work). Because misexpression of dominant negative and hyperactive forms of DMRTF results in opposite migratory phenotypes, DMRTF may act downstream of heartless and pebble in mesoderm migration.
Evolutionary Conservation of the SRF–MRTF Partnership. The target genes of DMRTF responsible for the diverse DMRTF gain- and loss-of-function phenotypes remain to be determined. We imagine that many of these target genes are directly regulated by SRF and, indeed, we have identified several such genes with multiple SRF-binding sites in their control regions (Z.H. and E.N.O., unpublished work). However, it is also possible that transcription factors in addition to SRF serve as cofactors for DMRTF.
SRF integrates diverse signals for cell growth, migration, cytoskeletal organization, and myogenesis via its association with transcriptional cofactors. The mammalian MRTF MAL has been shown to associate with G-actin in the cytoplasm and to translocate to the nucleus in response to growth factor signaling and actin treadmilling (23). It is intriguing that embryonic stem cells lacking SRF display defects in spreading, adhesion, and migration, which correlate with abnormalities in actin stress fibers (37). Thus, the striking parallels between the roles of SRF and myocardin family members in mammalian cells and Drosophila suggest that these factors comprise an ancient and evolutionarily conserved system for coupling changes in cell shape and extracellular signaling with cell migration during development.
Acknowledgments
We thank the following individuals for reagents: M. Affolter (University of Tübingen, Tübingen, Germany), A. Nordheim (Universitär Basel, Basel), K. Golic (University of Utah, Salt Lake City), and B. Paterson (National Institutes of Health, Bethesda). We thank R. Cripps and D. McKearin for critical comments on the manuscript, A. Tizenor for graphics, and J. Page for editorial assistance. Z.H. was supported by a grant from The American Heart Association, and E.N.O. was supported by grants from the National Institutes of Health and the D. W. Reynolds Center for Clinical Cardiovascular Research.
Abbreviations: SRF, serum response factor; DSRF, Drosophila SRF; MRTF, myocardin-related transcription factor; SAP, SAF-A/B, Acinus, PIAS; MAL, megakaryoblastic leukemia; DMRTF, Drosophila MRTF; NTD, N-terminal domain; FGF, fibroblast growth factor.
Data deposition: The DMRTF cDNA and protein sequences reported in this paper have been deposited in the GenBank database (accession no. AY682915).
Note Added in Proof. After completion of this work, Somogyi and Rorth (38) reported that DSRF and DMRTF, which they refer to as MAL-D, are required for migration of border cells during Drosophila oogenesis. They also show that MAL-D accumulates in the nucleus in response to cell stretching, and that MAL-D/SRE activity is required for formation of the cytoskeleton in migrating cells. Their findings are consistent with and complementary to those of the present study.
References
- 1.Miano, J. M. (2003) J. Mol. Cell Cardiol. 35, 577–593. [DOI] [PubMed] [Google Scholar]
- 2.Wang, D., Passier, R., Liu, Z.-P., Shin, S. H., Wang, Z., Li, S., Sutherland, L. B., Small, E., Krieg, P. A. & Olson, E. N. (2002) Cold Spring Harb. Symp. Quant. Biol. 67, 97–105. [DOI] [PubMed] [Google Scholar]
- 3.Li, L., Liu, Z., Mercer, B., Overbeek, P. & Olson, E. N. (1997) Dev. Biol. 187, 311–321. [DOI] [PubMed] [Google Scholar]
- 4.Kim, S., Ip, H. S., Lu, M. M., Clendenin, C. & Parmacek, M. S. (1997) Mol. Cell. Biol. 17, 2266–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mack, C. P. & Owens, G. K. (1999) Circ. Res. 84, 852–861. [DOI] [PubMed] [Google Scholar]
- 6.Lilly, B., Olson, E. N. & Beckerle, M. C. (2001) Dev. Biol. 240, 531–547. [DOI] [PubMed] [Google Scholar]
- 7.Landerholm, T. E., Dong, X. R., Lu, J., Belaguli, N. S., Schwartz, R. J. & Majesky, M. W. (1999) Development (Cambridge, U.K.) 126, 2053–2062. [DOI] [PubMed] [Google Scholar]
- 8.Affolter, M., Montagne, J., Walldorf, U., Groppe, J., Kloter, U., LaRosa, M. & Gehring, W. J. (1994) Development (Cambridge, U.K.) 120, 743–753. [DOI] [PubMed] [Google Scholar]
- 9.Guillemin, K., Groppe, J., Ducker, K., Treisman, R., Hafen, E., Affolter, M. & Krasnow, M. A. (1996) Development (Cambridge, U.K.) 122, 1353–1362. [DOI] [PubMed] [Google Scholar]
- 10.Ghabrial, A., Luschnig, S., Metzstein, M. M. & Krasnow, M. A. (2003) Annu. Rev. Cell Dev. Biol. 19, 623–647. [DOI] [PubMed] [Google Scholar]
- 11.Fristrom, D., Gotwals, P., Eaton, S., Kornberg, T. B., Sturtevant, M., Bier, E. & Fristrom, J. W. (1994) Development (Cambridge, U.K.) 120, 2661–2671. [DOI] [PubMed] [Google Scholar]
- 12.Montagne, J., Groppe, J., Guillemin, K., Krasnow, M. A., Gehring, W. J. & Affolter, M. (1996) Development (Cambridge, U.K.) 122, 2589–2597. [DOI] [PubMed] [Google Scholar]
- 13.Wang, D., Chang, P. S., Wang, Z., Sutherland, L., Richardson, J. A., Small, E., Krieg, P. A. & Olson, E. N. (2001) Cell 105, 851–862. [DOI] [PubMed] [Google Scholar]
- 14.Wang, D. Z., Li, S., Hockemeyer, D., Sutherland, L., Wang, Z., Schratt, G., Richardson, J. A., Nordheim, A. & Olson, E. N. (2002) Proc. Natl. Acad. Sci. USA 99, 14855–14860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang, Z., Wang, D. Z., Pipes, G. C. & Olson, E. N. (2003) Proc. Natl. Acad. Sci. USA 100, 7129–7134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoshida, T., Sinha, S., Dandre, F., Wamhoff, B. R., Hoofnagle, M. H., Kremer, B. E., Wang, D. Z., Olson, E. N. & Owens, G. K. (2003) Circ. Res. 92, 856–864. [DOI] [PubMed] [Google Scholar]
- 17.Du, K. L., Ip, H. S., Li, J., Chen, M., Dandre, F., Yu, W., Lu, M. M., Owens, G. K. & Parmacek, M. S. (2003) Mol. Cell. Biol. 23, 2425–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, S., Wang, D.-Z., Wang, Z., Richardson, J. & Olson, E. N. (2003) Proc. Natl. Acad. Sci. USA 100, 9366–9370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma, Z., Morris, S. W., Valentine, V., Li, M., Herbrick, J. A., Cui, X., Bouman, D., Li, Y., Mehta, P. K., Nizetic, D., et al. (2001) Nat. Genet. 28, 220–221. [DOI] [PubMed] [Google Scholar]
- 20.Mercher, T., Coniat, M. B., Monni, R., Mauchauffe, M., Khac, F. N., Gressin, L., Mugneret, F., Leblanc, T., Dastugue, N., Berger, R., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 5776–5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cen, B., Selvaraj, A., Burgess, R. C., Hitzler, J. K., Ma, Z., Morris, S. W. & Prywes, R. (2003) Mol. Cell. Biol. 23, 6597–6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sasazuki, T., Sawada, T., Sakon, S., Kitamura, T., Kishi, T., Okazaki, T., Katano, M., Tanaka, M., Watanabe, M., Yagita, H., Okumura, K. & Nakano, H. (2002) J. Biol. Chem. 277, 28853–60. [DOI] [PubMed] [Google Scholar]
- 23.Miralles, F., Posern, G., Zaromytidou, A. I. & Treisman, R. (2003) Cell 113, 329–342. [DOI] [PubMed] [Google Scholar]
- 24.Du, K. L., Chen, M., Li, J., Lepore, J. J., Mericko, P. & Parmacek, M. S. (2004) J. Biol. Chem. 279, 17578–17586. [DOI] [PubMed] [Google Scholar]
- 25.Gong, W. J. & Golic, K. G. (2003) Proc. Natl. Acad. Sci. USA 100, 2556–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brand, A. H. & Perrimon, N. (1993) Development (Cambridge, U.K.) 118, 401–415. [DOI] [PubMed] [Google Scholar]
- 27.Greig, S. & Akam, M. (1993) Nature 362, 630–632. [DOI] [PubMed] [Google Scholar]
- 28.Han, Z., Fujioka, M., Su, M., Liu, M., Jaynes, J. B. & Bodmer, R. (2002) Dev. Biol. 252, 225–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lilly, B., Zhao, B., Ranganayakulu, G., Paterson, B. M., Schulz, R. A. & Olson, E. N. (1995) Science 267, 688–693. [DOI] [PubMed] [Google Scholar]
- 30.Wang, Z., Wang, D. Z., Hockemeyer, D., McAnally, J., Nordheim, A. & Olson, E. N. (2004) Nature 428, 185–189. [DOI] [PubMed] [Google Scholar]
- 31.Sprenkle, A. B., Murray, S. F. & Glembotski, C. C. (1995) Circ. Res. 77, 1060–1069. [DOI] [PubMed] [Google Scholar]
- 32.Arsenian, S., Weinhold, B., Oelgeschlager, M., Ruther, U. & Nordheim, A. (1998) EMBO J. 17, 6289–6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chihara, T., Kato, K., Taniguchi, M., Ng, J. & Hayashi, S. (2003) Development (Cambridge, U.K.) 130, 1419–1428. [DOI] [PubMed] [Google Scholar]
- 34.Gisselbrecht, S., Skeath, J. B., Doe, C. Q. & Michelson, A. M. (1996) Genes Dev. 10, 3003–3017. [DOI] [PubMed] [Google Scholar]
- 35.Schumacher, S., Gryzik, T., Tannebaum, S. & Muller, H.-A. J. (2004) Development (Cambridge, U.K.) 131, 2631–2640. [DOI] [PubMed] [Google Scholar]
- 36.Smallhorn, M., Murray, M. J. & Saint, R. (2004) Development (Cambridge, U.K.) 131, 2641–2651. [DOI] [PubMed] [Google Scholar]
- 37.Schratt, G., Philippar, U., Berger, J., Schwarz, H., Heidenreich, O. & Nordheim, A. (2002) J. Cell Biol. 156, 737–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Somogyi, K. & Rorth, P. (2004) Dev. Cell 7, 85–93. [DOI] [PubMed] [Google Scholar]