Abstract
A diverse array of mechanisms regulate tissue-specific protein levels. Most research, however, has focused on the role of transcriptional regulation. Here we report systematic differences in synonymous codon usage between genes selectively expressed in six adult human tissues. Furthermore, we show that the codon usage of brain-specific genes has been selectively preserved throughout the evolution of human and mouse from their common ancestor. Our findings suggest that codon-mediated translational control may play an important role in the differentiation and regulation of tissue-specific gene products in humans.
With the advent of mRNA expression arrays, researchers have begun to delineate which genes are selectively expressed in which tissues and, in a fundamental way, distinguish one tissue from another (1, 2). Although such studies help to elucidate expression patterns, the processes underlying differentiation and regulation of tissue-specific proteins remain outstanding problems in developmental and molecular biology. Here, we show that genes selectively expressed in one human tissue can often be discriminated from genes expressed in another tissue purely on the basis of their synonymous codon usage. In particular, we demonstrate that brain-specific genes show a characteristically different codon usage than liver-specific genes; uterus genes differ from testis genes; and ovary genes differ from vulva genes, as well as other pairs of these six tissues.
Codon Bias Across Taxa
Although it came as a surprise to early neutral theorists (3), it is now clear that codon usage is not random: Among synonymous codons, some codons are used preferentially. Moreover, taxa differ in their codon usage. For example, various species of Drosophila each have their own particular codon biases, and their usage differs significantly from Escherichia coli or Saccharomyces cerevisiae (4–6). The dominant theory of codon bias for organisms ranging from E. coli to Drosophila posits that preferred codons correlate with the relative abundances of isoaccepting tRNAs, thereby increasing translational efficiency (7–10).
Synonymous codon choice also affects gene expression in mammals: When nonmammalian genes are to be expressed in mammalian cells, the replacement of mammalian-rare codons with more common synonyms greatly increases gene expression (11–13). Nevertheless, there is little evidence in mammals of selection on synonymous codons for translational efficiency. Instead, mammalian genomes exhibit large-scale variation in GC content [e.g., isochores (14)] in both coding and noncoding regions. The GC content in noncoding regions is correlated with the GC content at the third position of coding regions from the same isochore. Thus, codon biases observed in the human genome have been attributed to neutral processes [such as biased mutation (15) and gene conversion (16)] rather than to selection (17). [Early studies on cDNA clones derived from a diverse set of vertebrate genes failed to find evidence for tissue-specific or taxon-specific codon usage (18).]
Comparing Codon Usage Between Genes
The most common measure of codon bias, called the effective number of codons (ENC), is analogous to the effective number of alleles in population genetics. ENC does not describe the particulars of which codons are more frequent than others but rather measures the overall departure from random synonymous codon choice. As a result, two genes may exhibit the same degree of overall bias (ENC value) and yet differ dramatically in their particular choice of synonymous codons.
For this study, we desire a detailed measure of the “distance” between the synonymous codon usage of two genes. We are not concerned with degree of codon bias in the usual sense, that is, the departure from random synonymous codon choice, but rather with the degree to which genes differ in their encoding of amino acids. Given the coding sequences for a pair of genes, we compare their codon usage by first tabulating the absolute frequency of each codon in each gene. For each amino acid, we compute a two-tailed Fisher exact test (19) on the n × 2 contingency table given by the frequencies of the amino acid's synonymous codons (e.g., for Ala n = 4: GCC, GCG, GCA, and GCT). As a result, for each amino acid we obtain a P value indicating whether or not the genes use significantly different codons to encode that amino acid. Table 1 summarizes an example of this analysis by comparing the codon usage of two human genes.
Table 1. Comparison of codon usage between two human genes.
 |
For each codon, we report its absolute frequency of occurrence in each gene and its relative frequency compared with synonymous codons. The P value for each amino acid reflects whether or not the two genes differ in their encoding of the amino acid (Fisher exact test). A complete comparison of all 61 condons is given as Table 2, which is published as supporting information on the PNAS web site. The comparison between these genes is typical of comparisons between other genes from their respective tissues, testes and uterus. Gene A, testis-specific glycerol kinase (GI 516123); gene B, endometrial bleeding factor (GI 2058537).
The number of amino acids that exhibit a statistically different encoding is a biologically relevant metric of distance between the codon usage in two genes. All other things being equal (i.e., RNA folding, protein–RNA recognition, transport, etc.), for a fixed pool of tRNAs, this metric should naturally correlate with the difference in translation rates between the two genes. Unlike metrics such as “relative synonymous codon usage” (4), which are noisy when applied to individual genes, our measure of codon usage relies on the Fisher Exact test for small sample sizes, and it can be applied to genes that contain only a few examples of each amino acid.
Methods
The uterus- and testis-specific genes used in this study (Table 3, which is published as supporting information on the PNAS web site) were obtained directly from the tissue-specific lists compiled by Warrington et al. (1). The brain, liver, ovary, and vulva genes (Table 3) were taken from the online expression database of Hsiao et al. (2). A gene was considered to be brain-specific if, according to the Hsiao database (2), its mRNA transcript is present in brain but absent from all but at most two other tissue types tested by Hsiao et al. The criteria for tissue-specific consideration were the same for liver, ovary, and vulva.
Given a dendogram that represents the codon usage of genes in a pair of tissues (e.g., Fig. 1), we calculate a P value to test whether the observed clustering of genes is nonrandom. The P value is obtained by comparing the observed summed squared distances along the tree between genes of the same tissue against a null distribution produced by randomly permuting the labels of the leaves.
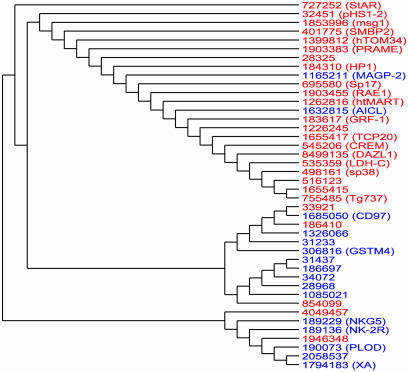
Fig. 1.
A dendogram reflecting the codon usage of 26 genes selectively expressed in human testis (red) and 16 genes selectively expressed in uterus (blue). Genes are denoted by their GI number. The pairwise distances underlying this tree reflect the degree to which the genes differ in their codon usage. As this tree demonstrates, testis-expressed genes can generally be distinguished from uterus-expressed genes purely on the basis of their synonymous codon usage. The observed separation between these two classes of genes would not have occurred by random chance (P = 0.0008)
For each of the 44 brain-specific genes, the corresponding mouse orthologs were obtained from the ensembl web-site by using ensmart, and they were aligned by using clustalw (20). The same procedure was used to produce orthologous alignments of the genes specific to ovary, testes, uterus, liver, and vulva.
Results on Tissue-Specific Codon Usage
On the basis of two extensive microarray mRNA expression studies (1, 2), we have identified genes that are selectively expressed in six adult healthy human tissues: testis (26 genes), uterus (16 genes), total brain (44 genes), liver (34 genes), ovary (36 genes), and vulva (42 genes). By analyzing expression patterns from only two studies, we limited ourselves to fewer data than are available in large compilations of many expression studies. On the other hand, the expression data we have used are comparable (both studies used the GeneChip HuGeneFL microarray), and they provide a consistent, unbiased method of assigning tissue-specificity. The total number of identified tissue-specific genes is smaller than in previous studies (21) because we use a conservative, stringent definition of tissue specificity (see Methods). The genes selectively expressed in each of these six tissues are distributed throughout the genome (Table 3), and they have similar distributions of gene sizes (the mean gene length within each tissue is well within one standard deviation of the means of all other tissues.)
We have compared codon usage between pairs of the six tissues. When comparing testis to uterus, for example, we calculate the distance between the codon usage of every pair of genes (including pairs from the same tissue), obtaining a 42-by-42 symmetric matrix of pairwise distances. The distance between two genes is given by the number of amino acids that exhibit significantly different (P < 0.01) codon usage, as defined above. Our results are not sensitive to the particular choice of a threshold P value within 0.001 and 0.05. By using the neighborjoining method (phylip v3.5), we produced a dendogram that graphically represents the measured pairwise distances between the codon usage in the study genes.
Fig. 1 shows the dendogram resulting from the codon usage in testis- and uterus-specific genes. Note that virtually all testis-associated genes are clustered in a separate clade from the uterus-associated genes. The observed clustering is the result of systematic differential codon usage between the testis- and uterus-specific genes. Fig. 1 indicates that we can generally discriminate between testis- and uterus-expressed genes on the basis of their codon usage alone.
The separation of testis and uterus genes seen in Fig. 1 would not have occurred by random chance (P < 0.0008, see Methods). Similarly, Fig. 2 indicates that brain-specific genes are easily distinguishable from liver-specific genes on the basis of their codon usage (P < 0.00018). We also find (trees not shown) that ovary-specific genes are distinguishable from vulva genes (P < 0.0032), brain genes are distinguishable from testis genes (P < 0.0044), brain genes are distinguishable from ovary genes (P < 0.00008), and vulva genes are distinguishable from testes genes (P < 0.0092). All but one of these results remain significant even after Bonferroni–Holm correction for multiple hypotheses.
Fig. 2.
A dendogram reflecting the codon usage of 44 brain-specific genes (red) and 34 liver-specific genes (blue). The observed separation between these two classes of genes would not have occurred by random chance (P = 0.00018).
Despite the results presented above, many pairs of tissue-specific gene sets do not exhibit significantly different codon usage (e.g., liver versus uterus). The evolutionary processes that produce differential codon usage between certain pairs of tissues but not others pose an intriguing question for further research.
Evolutionary Preservation of Codon Usage
It is tempting to hypothesize that the highly nonrandom, tissue-specific codon usage we have observed serves an adaptive function. Although we cannot impute an adaptive function, we can nevertheless demonstrate that the codon usage of brain-specific genes has been selectively preserved far more than expected by chance during the evolution of human and mouse from their common ancestor. For this analysis, we have identified and aligned mouse orthologs for the 44 brain-specific human genes (see Methods) and for the other study tissues.
We considered only those sites in the alignment of the human and mouse brain genes that exhibited either identical or synonymous codons. There are 31,050 such codons, which we concatenated into a single sequence for each organism. The resulting aligned mouse and human sequences are fairly similar in their codon usage. There are only two amino acids that have a significantly different encoding (P < 0.01) between the orthologous sequences.
The overall similarity of codon usage between the mouse and human brain-specific genes does not in itself imply that codon usage has been selectively preserved, because the human and mouse sequences are similar by descent. There are only 8,837 (synonymous) nucleotide mutations between the two sequences. We have applied a randomization test to compare the codon usage of the human and mouse sequences, controlling for their sequence similarity. In each randomization trial, we started with the mouse sequence, and we introduced in randomly chosen synonymous locations the observed number of nucleotide changes (preserving even the number of mutations of each type, A→C, A→T, A→G, C→A, etc.) to produce a randomized version of the human sequence. The resulting randomized sequence has the exact same amino acid and nucleotide composition as the observed human sequence. Moreover, the randomized human sequences contain virtually the same dinucleotide CpG content as the actual human sequence. The mean number of occurrences of CpG in the codons of the randomized sequences agrees with the actual number of CpGs in the observed human sequence (all randomization trials fall within 2% of the observed human CpG content).
Among 10,000 such randomization trials, there were on average 7.53 amino acids that exhibited significantly different encodings between the mouse sequence and the randomized human sequence. There were no examples in which the mouse sequence and the randomized human sequence exhibited fewer than four amino acids with different encodings. In other words, even when controlling for their amino acid compositions, their nucleotide compositions, and their CpG compositions, the human and mouse genes are far more similar in synonymous codon usage than expected by random chance (P < 10–4), given the mutations that have occurred between them. Although the aligned mouse and human sequences exhibit synonymous differences in 28% of their codons, these differences compensate in such a way so as to preserve the overall codon usage. This result suggests that there has been selection to preserve the codon usage of these brain-specific genes throughout the evolution of mouse and human from their common ancestor.
In addition to brain-specific genes, the genes associated with most of the other study tissues also show a highly significant degree of synonymous codon usage preservation compared with their mouse orthologs (P < 0.0032 each for liver, uterus, and vulva.) Notably, however, the synonymous codon usage in testes-specific and, to a lesser extent, ovary-specific genes do not show significant preservation between human and mouse (P = 0.48 and P = 0.058, respectively). This result is analogous to the well-established fact that the protein sequences of reproductive genes, particularly those related to spermatogenesis, have undergone rapid evolution in primates (22). Apparently, synonymous codon usage in testes-specific genes is also undergoing relatively more rapid divergence.
Discussion
Here we have reported a significant difference between genes that are selectively expressed in several human tissues: Such genes exhibit characteristic codon usage that, in many cases, distinguishes the genes expressed in one tissue from those expressed in another. Moreover, in most cases the tissue-specific codon usage has been selectively preserved throughout the evolution of human and mouse from their common ancestor. The biological mechanism and impact of this phenomenon certainly require further study. Nevertheless, our results suggest that synonymous codon usage in mammalian genes is not simply the result of neutral evolutionary processes or isochore structure.
Previous studies have explored GC content at the third position of coding sequences expressed in different human tissues (21). The GC3 content of the genes studied here does vary by tissue type, but the average GC3 content of one tissue is well within one standard deviation of another tissue's average: testes, 0.55 ± 0.059; uterus, 0.58 ± 0.014; brain, 0.56 ± 0.053; liver, 0.52 ± 0.053; ovary, 0.57 ± 0.19; vulva, 0.67 ± 0.15. As a result of the variation within each tissue, GC3 content alone is not powerful enough to reliably separate genes by tissue type. For example, a dendogram analogous to Fig. 1 based on pairwise GC3-content distances results in an insignificant separation of tissue-specific genes (P = 0.53). The tissue-specific genes that we have identified are characterized by differences in synonymous codon usage above and beyond their GC3 content.
Our results on differential, tissue-specific codon usage suggest several hypotheses about the mechanisms of protein regulation and tissue differentiation in humans. Differential codon usage can impact tissue-specific modulation of proteins at several levels. Codon usage in mammals is known to have dramatic effects on translation rate (11–13), especially during cell differentiation (23). The existence of systematic tissue-specific codon usages raises the important possibility that human tissues may differ in their relative tRNA abundances and that these differences may modulate the expression of the appropriate proteins. To our knowledge, detailed studies on relative tRNA abundances across human tissues have not yet been performed. Our results suggest that such studies may be important for understanding tissue differentiation.
Differential synonymous codon usage has further biological consequences because methylated C-residues in DNA frequently result in transcriptional silencing (24). mRNA modifications are also base-specific (e.g., pseudouridines). Furthermore, mRNA folding into secondary and tertiary structures is sensitive to the choice of synonymous codons (25). RNA transport, protein recognition of palendromic sequences, translational efficiency as modulated by either tRNA abundance or the secondary structure of the mRNA are all impacted by the differences in the codon usage of an mRNA. As a result, tissue-specific codon usage also has implications for the optimal design of gene therapies targeted at specific tissues.
Transcriptional control has been the primary focus of gene regulatory research, especially since the advent of mRNA expression arrays. Nevertheless, the level of mRNA expression alone is not directly important to cellular function. Instead, the level of protein activity, which is a complex result of transcriptional, posttranscriptional, translational, and transport processes, is most important to biological function. Our results suggest that synonymous differences in the encoding of genes may have been selected for and may be used at several levels of regulation to reinforce differential protein levels of tissue-specific genes.
Supplementary Material
Acknowledgments
We thank Jonathan Dushoff, Alice Chen, Hagar Barak, and Gyan Bhanot for their input throughout the preparation of this manuscript. J.B.P. acknowledges funding from the Harvard Society of Fellows.
References
- 1.Warrington, J., Nair, A., Mahadevappa, M. & Tsyganskya, M. (2000) Physiol. Genomics 2, 143–147. [DOI] [PubMed] [Google Scholar]
- 2.Hsiao, L., Dangond, F., Yoshida, T., Hong, R., Jensen, R., Misra, J., Dillon, W., Lee, K., Clark, K., Haverty, P., et al. (2001) Physiol. Genomics 7, 97–104. [DOI] [PubMed] [Google Scholar]
- 3.King, J. L. & Jukes, T. H. (1969) Science 164, 788. [DOI] [PubMed] [Google Scholar]
- 4.Sharp, P. M. & Li, W. H. (1987) Nucleic Acids Res. 15, 1281–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ikemura, T. (1985) Mol. Biol. Evol. 2, 13–34. [DOI] [PubMed] [Google Scholar]
- 6.Powell, J. R. & Moriyama, E. N. (1997) Proc. Natl. Acad. Sci. USA 94, 7784–7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zuckerkand, E. & Pauling, L. (1965) J. Theor. Biol. 8, 357. [DOI] [PubMed] [Google Scholar]
- 8.Ikemura, T. (1981) J. Mol. Biol. 146, 1–21. [DOI] [PubMed] [Google Scholar]
- 9.Debry, R. & Marzluff, W. F. (1994) Genetics 138, 191–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Srensen, M., Kurland, C. & Pedersen, S. (1989) J. Mol. Biol. 207, 365–377. [DOI] [PubMed] [Google Scholar]
- 11.Levy, J., Muldoon, R. R., Zolotukhin, S. & Link, C. J., Jr. (1996) Nat. Biotechnol. 14, 610–614. [DOI] [PubMed] [Google Scholar]
- 12.Zolotukhin, S., Potter, M., Hauswirth, W., Guy, J. & Muzyczka, N. (1996) J. Virol. 70, 4646–4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wells, K., Foster, J., Moore, K., Pursel, V. & Wall, R. (1999) Transgenic Res. 8, 371–381. [DOI] [PubMed] [Google Scholar]
- 14.Bernardi, G. (1995) Annu. Rev. Genet. 29, 445–476. [DOI] [PubMed] [Google Scholar]
- 15.Francino, M. P. & Ochman, H. (1999) Nature 400, 30–31. [DOI] [PubMed] [Google Scholar]
- 16.Galtier, N. (2003) Trends Genet. 19, 65–68. [DOI] [PubMed] [Google Scholar]
- 17.Urrutia, A. O. & Hurst, L. D. (2001) Genetics 159, 1191–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hastings, K. & Emerson, C. (1983) J. Mol. Evol. 19, 214–218. [DOI] [PubMed] [Google Scholar]
- 19.Agresti, A. (1992) Stat. Sci. 7, 131–153. [Google Scholar]
- 20.Higgins, D., Thompson, J. & Gibson, T. (1994) Nucl. Acid. Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vinogradov, A. (2003) Nucl. Acid. Res. 31, 5212–5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wyckoff, G., Wang, W. & Wu, C. I. (2000) Nature 403, 304–309. [DOI] [PubMed] [Google Scholar]
- 23.Calkhoven, C., Muller, C. & Leutz, A. (2002) Trends Mol. Med. 8, 577–583. [DOI] [PubMed] [Google Scholar]
- 24.Bird, A. & Wolffe, A. (1999) Cell 99, 451–454. [DOI] [PubMed] [Google Scholar]
- 25.Katz, L. & Burge, C. (2003) Genome Res. 9, 2042–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.