Abstract
We propose that chromosome function is governed by internal mechanical forces generated by programmed tendencies for expansion of the DNA/chromatin fiber against constraining features.
Chromosomal processes are generally considered to comprise the simple sum of a large number of individual biochemical changes. We present here a different idea in which internal mechanical forces play a governing role.
Spatial Patterning in Biological Systems Mimics Patterning Intrinsic to Mechanical Systems
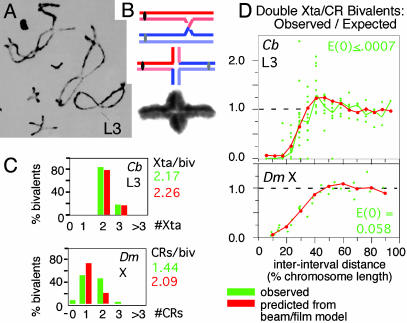
Distribution of Meiotic Crossovers/Chiasmata. During meiosis, homologous maternal and paternal chromosomes (“homologs”) become linked by cytologically observable connections, “chiasmata” (Fig. 1A). Each chiasma is the site of a DNA crossover between one sister of each homolog. A connection is created by this crossover, plus linkages between sister chromatids along their lengths (Fig. 1B). Crossovers/chiasmata occur in a strikingly nonrandomly distribution along the chromosomes (ref. 1 and Fig. 1 C and D).
Fig. 1.
Chiasmata and crossovers. (A) Diplotene bivalents of Chorthippus brunneus with homologs linked by chiasmata (1). (B) Differential staining of sister chromatids shows that each chiasma is the site of a reciprocal exchange between one sister of each homolog (2). (C and D) Numbers and interference distributions of crossovers (D. melanogaster X chromosome; 3) and chiasmata (Chorthippus L3 bivalent, marked in A; ref. 1). Experimental values (green) and values predicted by the beam/film model (red; details in Fig. 10). [A and B are reproduced with permission from, respectively, ref. 1 (Copyright 1984, Society for Experimental Biology) and ref. 2 (Copyright 1978, Nature Publishing Group).]
Along a given chromosome, events occur at different positions in different meiotic cells. Thus, the nonrandom distribution does not arise via unique specification of position.
Every pair of homologs always acquires at least one crossover/chiasma. This is biologically important, because at least one connection between homologs is required to direct them to opposite poles at the meiosis I division. Moreover, the number of events per chromosome is very low, usually two or three (e.g., Fig. 1C, green) and sometimes only one. Thus, the obligate event is not ensured by many events occurring randomly along the chromosomes.
When two or more events occur along a chromosome, they exhibit “interference”: if an event is present at one position, there is a reduced probability that another event will be found nearby. Interference is maximal at short distances and decreases progressively with increasing distance between positions. This pattern is seen by dividing a chromosome into intervals along its length, determining the frequencies of events occurring in each individual interval and then examining all pairs of intervals with respect to the observed frequency of chromosomes with events in both intervals (“double events”) as compared to that expected on the basis of independent occurrence (Fig. 1D, green). For intervals that are very close together, no double events are observed [observed/expected (“O/E”) = 0]. As interinterval distance increases, the probability of double events rises, eventually reaching that expected from independence (O/E = 1). At certain larger distance(s), double events are more probable than expected (O/E >1), implying that events tend to be evenly spaced at these particular distance(s).
A Mechanical Explanation. The phenomenon of crossover/chiasma interference implies communication along chromosomes. In physical systems, any local increase or decrease in mechanical stress at one position automatically tends to redistribute outward from that point. For example, a solid rubber rod that is bent and held firmly in the bent position experiences tensile (pulling) stress along its convex (“top”) surface and compressive (pushing) stress along its concave (“bottom”) surface, in proportion to the degree of curvature. If the rod is then cut partway through along its convex surface, the rod becomes straighter away from the cut but more sharply bent just under the cut point, with no rotation of one end relative to the other. The decrease in curvature away from the cut is accompanied by a relaxation of stress on both surfaces. The increase of curvature in the immediate vicinity of the cut is accompanied by stress intensification, including a local increase in compressive stress along the concave surface. Because redistribution of mechanical stress provides a built-in communication mechanism, crossover/chiasma distributions might be explained by a mechanical model.
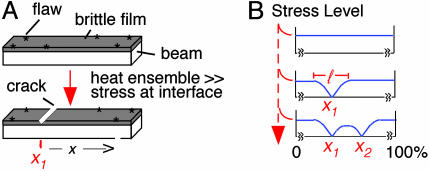
A convenient model for quantification of mechanical effects is as follows: Consider an elastic beam or plate (e.g., of metal) coated on one face by a thin brittle film (e.g., of ceramic) that contains flaws along its edges (Fig. 2A Upper; also Fig. 10, which is published as supporting information on the PNAS web site). If this ensemble is heated, the metal plate will have a greater tendency to expand than the overlaid ceramic film (due to higher and lower coefficients of thermal expansion of the two materials). If the two entities are tightly bonded, expansion of the metal plate will force the film to stretch (i.e., to expand beyond the extent dictated by its intrinsic thermal expansion). That is, “tensile stress” develops within the film. To some degree, the film resists this stretching tendency, thus generating a balancing compressive stress within the metal plate. However, if the film is very thin compared to the substrate, the amount of resistance and thus of compressive stress within the plate is very small. As a result, heating has an innocuous effect on the beam but gives rise to high tensile stress in the film that can trigger crack nucleation at the edge flaws. Once triggered, a crack extending down to the film–substrate interface propagates across the entire width of the ensemble from one edge to the other (Fig. 2A Lower).
Fig. 2.
The beam/film system. See also text and Fig. 10.
Formation of a crack results in local relief of stress on either side of the affected site (Fig. 2B Top to Middle). Because the beam is elastic, it participates in the redistribution process and sets the characteristic distance of stress relief beyond which the film stress is not influenced by the presence of the crack. The larger the stiffness (elastic modulus) of the beam material, the smaller the length of the region of stress relief. Put another way, local stress relief tends to spread (redistribute) from the crack outward in both directions along the beam. Further, because the beam is massive compared to the film, it can absorb the tensile load shed by the film at the crack and, through its elasticity, spread the stress relief in a manner that decreases gradually with distance from the site. The result is a graded spatial domain of stress relief (Fig. 2B Middle). Additional stress-promoted cracks may then occur. If so, they will tend to occur in regions outside of the “stress relief domains” of prior crack(s), where the stress level remains high (Fig. 2B Bottom). Occurrence of a crack at one position thus “interferes” with the occurrence of a crack nearby. As more and more cracks occur, they will tend to “fill in the holes” between prior cracks, resulting in a tendency for cracks to be evenly spaced along the beam.
The beam/film system, which can be simulated to compute the statistics of the distribution of crack locations, can be used to model experimentally observed meiotic crossovers/chiasmata distributions. Crossovers/chiasmata arise by stochastic selection of a few sites from a much larger array of undifferentiated precursor interaction sites. Thus, precursor interactions could be analogous to flaws, whereas designation of certain interactions to be crossovers/chiasmata could be a stress-promoted process corresponding to crack formation. When key parameters are set at optimal values, the numbers and distributions of crossovers (Drosophila X) or chiasmata (Chorthippus L3) can be matched very closely by the predicted numbers and distributions of cracks (Fig. 1 C and D; compare red and green; additional details are in Fig. 10C). Unique secondary features are also readily explained (Fig. 10C).
General Implications. Crack formation in the beam/film system illustrates three general properties that could be exhibited by any biological system governed by mechanical effects, including but not limited to chromosomes.
Ensuring an obligatory event. Occurrence of a (first) obligatory event is easily ensured by adequate stress or by adequate sensitivity to stress. The ensured event could occur at a unique site (e.g., activation of a particular promoter) or at a minimum of one among many potential sites (e.g., firing of at least one among several replication origins or occurrence of at least one crossover/chiasma at one of many precursor sites along a chromosome).
Self-limiting spatial domains. Occurrence of a local stress-mediated change sets up a defined spatial domain of stress relief (or stress). Such domains arise by nucleation and spreading. They are also self-limiting and thus can arise without unique genetic designation of boundaries. Such domains might explain chromosome compartments such as centromeres, imprinted regions, heterochromatic domains, and local domains of transcriptional activation, which often have these properties (e.g., refs. 4–6) or H2AX phosphorylation domains, which arise at sites of spontaneous double-strand breaks and spread outward within apparently disorganized interphase chromatin (7). When needed, specific boundaries could be provided by genetically positioned molecular “sinks” that absorb redistributing stress or stress relief, thus preventing spreading into adjacent regions.
Interference and even spacing. A particular type of change can occur at multiple sites along a chromosome, at different positions in different nuclei, but always with a tendency to be evenly spaced, as seen for meiotic crossovers/chiasmata (above). In fact, these properties are also exhibited by many or all basic chromosome organizational features, e.g., nucleosomes, chromatin loops, and axial chromosome coils, and by initiation of DNA replication (8, 9).
Summary. Chromosomal events exhibit spatial patterning. Many such patterns were not previously recognized as features to be explained and none is understood. All could be explained by a common mechanism if corresponding processes involve imposition and redistribution of mechanical stress and/or stress relief.
Mechanical Forces Within Chromosomes: Proposed Origin and Potential Predicted Effects
Spatial patterning effects suggest that physical forces are present and play important roles within chromosomes. How might such forces arise? And how might they exert their effects?
DNA/Chromatin Expansional Force. A segment of chromatin free in solution will tend to occupy a particular “envelope volume” (the volume of the smallest shell containing all of the chromatin). If the state of the chromatin changes appropriately, it may tend to occupy a larger envelope volume. In solution, such expansion can occur freely and without effect. However, inside the cell, such expansion may be constrained, or “caged,” either by external features (e.g., other chromatin) or by an internal meshwork of intersegment interlinks (e.g., nucleosome–nucleosome contacts plus interactions involving chromatin/chromosome structure molecules). In both cases, a tendency for expansion will cause the chromatin to push on the constraining components, which will push back. These pushing forces can do chromosomal work. Physically, chromatin expansion could result from any change in the DNA/chromatin fiber that causes it to become stiffer and/or more extended, i.e., to have a longer contour length. In eukaryotic cells, increased contour lengths are produced by histone modification, loss of nonhistone architectural components, or histone elimination.
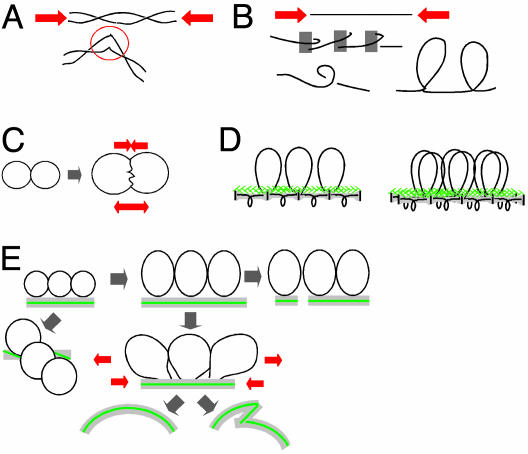
Predicted Effects. Compression stress along the DNA/chromatin fiber. Expansion of the DNA/chromatin fiber as constrained by a closepacked internal “meshwork” would cause it to come under compression stress along its length. Such stress would tend to destabilize the DNA duplex, causing discrete buckling or shearing/denaturation (Fig. 3A). It would also tend to promote conformational changes that place the fiber in a more compact shape, e.g., smooth bending and writhing as well as local buckling (Fig. 3B). Such tendencies could promote basic processes that involve access to Watson–Crick base pairs (initiation of transcription or DNA replication or DNA recombination) and organizational processes (wrapping of DNA around the nucleosome core, chromatin fiber folding, or development of programmed chromatin loops; Fig. 3B Bottom). Additionally, a single- or double-strand DNA interruption would lead to local and domainal relaxation, which could then be used to signal the presence of the lesion (e.g., refs. 7 and 10).
Fig. 3.
Stresses that could be generated by chromatin expansion against contraining features. (A and B) Compression stress along the DNA/chromatin fiber can destabilize DNA (A) or promote a more compact fiber configuration (B). (C) Interchromatin pushing produces opposing forces for separation and interface compression. (D) Linear/loop axis array (Left) and prophase configuration with cooriented sister arrays (Right). (E) Changes in axis status predicted from pushing forces between adjacent chromatin loops.
Chromatin pushing. If two touching chromatin masses expand, they will push against one another at their interface (“chromatin pressure”; ref. 11). Two opposing tendencies are created: the two masses will tend to push one another apart, thus providing a force for separation; concomitantly, they will tend to push into one another, thus providing a force for conjunction and intermingling (Fig. 3C). Either or both tendencies may come into play according to the state of the DNA/chromatin fiber. The separation force could drive, e.g., separation of sister chromatids or homologs, declustering of heterochromatic regions, and separation of unrelated chromosomes into distinct territories or, analogously, separation of chromatin from the nuclear envelope. The conjunction force could promote, e.g., intermingling of chromosome territories and linkages between different chromatin regions or chromatin and the nuclear envelope. Expansion forces will also cause different chromatin regions to push themselves through the nucleus into the “least-stressed” configuration, thus influencing local or global chromosome disposition (e.g., ref. 12).
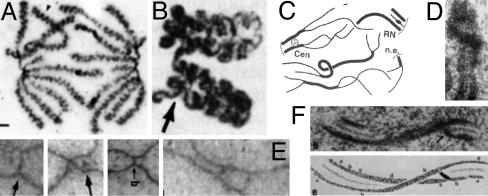
Axis destabilization. From prophase onward, the chromatin of each chromosome is organized into a linear array of loops, the bases of which are elaborated by proteins to form a geometric and structural axis (Fig. 3D). If adjacent loops expand, they cannot move apart because of tethering to their underlying axis; instead, the axis itself will come under stress (Fig. 3E). An immediate effect will be a tendency for axis extension and, ultimately, fracturing. Axes will also tend to twist, thus placing adjacent loops out of phase, where they have more room to expand. Finally, chromatin will tend to expand differentially in the periphery of the loops, where there is more room, rather than along the axis. This effect will cause the axis to bend, with stretching forces arising immediately above the axis and compression stress arising along the axis itself. Axial compression stress, in turn, can promote axis bending, writhing, or buckling. These various effects can explain axial coiling of late-stage chromosomes (Fig. 4A); twisting, bending, buckling, and fracturing seen along meiotic prophase chromosomes (e.g., Fig. 4 B–F); destabilization of axis components; and axis disassembly.
Fig. 4.
Observed changes in axis conformation. (A) Muntjac mitotic anaphase chromosomes with coiled axes (13). (B–F) Meiosis. (B) Coiled homologs of lily at diakinesis (14). (C) Axis fracturing and sister separation in late leptotene in Sordaria (15). (D) Twisting of a single leptotene homolog axis in Neotiella (16). (E)In Allium, coaligned homolog axes linked by bridges (Left) undergo apparent buckling plus ensuing nucleation of synaptonemal complex at late leptotene (1, 17). (F) Twisting synaptonemal complex (18). [Reproduced with permission from, respectively: (A) The Journal of Cell Biology, 1995, 131, pp. 7–17, by copyright permission of The Rockefeller University Press; (B) ref. 14 (Copyright 1991, NRC Research Press); (C) ref. 15 (Copyright 1999, Elsevier); (D) ref. 16 by courtesy of the Carlsberg Laboratory (Copyright 1985, The Carlsberg Laboratory, Copenhagen); (E) ref. 17 (Copyright 2004, Elsevier); and (F) ref. 18 (Copyright 1978, NRC Research Press).]
Additional effects. Chromatin expansion could drive and direct topoisomerase-mediated changes in DNA supercoiling and catenation/decatenation (11). Mechanical forces within chromatin/chromosomes could promote conformational changes in associated proteins or RNAs, altering structural or catalytic properties. Macromolecules that respond in this way would be molecular “stress sensors.” Onset of anaphase is known to be governed by regulatory sensing of tension on centromere/kinetochore regions arising from microtubule-mediated bipolar spindle forces (19). Diverse molecules might analogously sense mechanical stresses generated internally within the chromosomes, transducing that information to chromosomal or nonchromosomal components.
The Mitotic and Meiotic Programs Comprise Global Cycles of Chromatin Expansion and Contraction
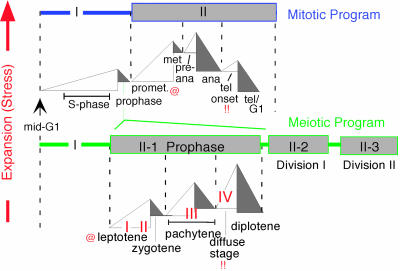
Our model requires dynamic changes in chromatin state. Inspection of eukaryotic chromosome literature suggests that chromatin cycles globally between expanded and contracted states (Fig. 5).
Fig. 5.
The mitotic and meiotic programs comprise related sequential cycles of global chromatin expansion and contraction, proposed to be cycles of expansional stress and stress relief. Four transitions of meiotic recombination (17) are indicated by I–IV. @ and!! symbolize analogies between prometaphase and telophase onset of the mitotic program and leptotene and diffuse stages of meiosis. @, appearance of silver-staining axis components along, and interaxis bridges between, sisters (mitosis, ref. 13) and homologs (meiosis, e.g., Fig. 4E) and chromosome destabilization in Sordaria spo76-1 (Fig. 14). (!!) Telophase onset and the diffuse stage both involve marked chromatin diffuseness and loss or destabilization of axial structures.
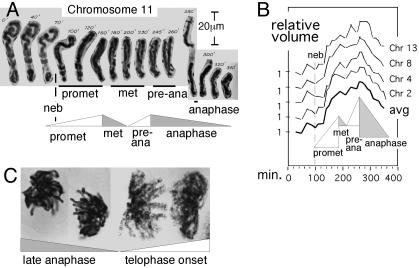
The Mitotic Program. The chromatin fiber is well folded at mid-G1 (20), unfolds and becomes diffuse during S phase and G2, and becomes compact again at prophase, as seen in the appearance of discrete compact individualized chromosomes. These events comprise a global chromatin expansion/contraction cycle. It is generally considered that, after prophase, chromosomes become more and more compact, as required for clean segregation to opposite poles at anaphase. However, actual measurements of chromosome volume, made from videomicrographs of living Haemanthus chromosomes, reveal this is not the case: chromosome volumes increase nearly 2-fold during prometaphase, decrease somewhat during metaphase, increase again to a maximum at preanaphase, and then decrease progressively through anaphase (ref. 21 and Fig. 6), as also seen in Tradescantia (22). The prophase/prometaphase/metaphase progression is also seen in ultrastructural chromatin fiber studies (23). These patterns imply two more global expansion/contraction cycles (Fig. 6 A and B). At telophase onset, chromosomes become expanded before completely disappearing (Fig. 6C Right), but by early G1, the chromatin is folded into compact fibers (above), thus implying a final cycle. Overall, the mitotic program would comprise four sequential cycles of global chromatin expansion and contraction (Fig. 5 Upper).
Fig. 6.
Chromatin expansion and contraction after prophase. (A and B) Individual Haemanthus chromosome volumes were determined from videomicrographs (e.g., A) (21). Shown are results for four chromosomes, normalized to 1 at t = 0 and their average. (C) At late anaphase, chromosomes are contracted, relaxed, and closely juxtaposed (Left); at telophase onset, chromosomes are expanded, distended, and separated (Right). [A and B are reproduced with permission from ref. 21 (Copyright 1959, Blackwell Publishing, Ltd.).]
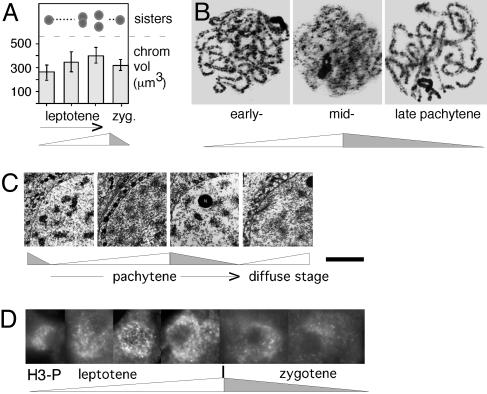
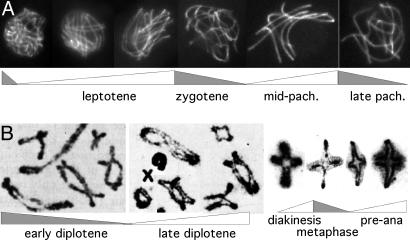
Meiosis. The meiotic program also initiates with chromatin expansion/contraction during G1-S/prophase. Meiotic prophase is then greatly prolonged as compared to its mitotic counterpart, to accommodate a complex program of interactions between homologs, and comprises three successive global chromatin expansion/contraction cycles that correlate with classical cytologically defined stages (Fig. 5). Direct measurements of chromatin volume, and analysis of chromatin diffuseness and compactness in wholemounted chromosomes spread nuclei or electron microscope thin sections, all show that: chromatin expands during leptotene, contracts at zygotene, expands at early–mid pachytene, contracts at late pachytene, expands at the “diffuse stage,” and contracts at diplotene (Figs. 5 Lower, 7 A–C, and Fig. 11, which is published as supporting information on the PNAS web site). Interestingly, the three meiotic prophase-specific expansion/contraction cycles appear to be directly analogous to the latter three cycles of the mitotic program (Fig. 5). Because prophase is followed by the two meiotic divisions that are closely analogous to their mitotic counterpart, meiosis may have arisen by programmatic triplication of the latter three cycles of the mitotic program (Fig. 5).
Fig. 7.
Chromatin expansion and contraction in meiotic prophase. (A) Chromatin volumes and sister chromatid cross-section morphologies seen by optical sectioning of maize leptotene and zygotene nuclei (25). (B) Pachytene chromosomes of Locusta migratoria (source, G.H.J). (C) Chromatin of human oocyte nuclei visualized by EM in ultrathin sections (26). At late zygotene/early pachytene, chromatin is highly condensed, leaving clear chromatin-free areas within the nucleus; at midpachytene, chromatin masses are less dense, and chromatin is more evenly distributed; compact chromatin returns at late pachytene; and less dense chromatin then returns at “the diffuse stage.” (D) Immunostaining of histone H3 Ser-10 phosphorylation at successive stages of meiotic leptotene/zygotene in Sordaria (source, D.Z.; Fig. 11). [A and C are reprinted with permission from, respectively, ref. 25 (Copyright 1994, Elsevier) and ref. 26 by courtesy of the Carlsberg Laboratory (Copyright 1970, The Carlsberg Laboratory, Copenhagen).]
Histone Phosphorylation Is Closely Correlated with Chromatin Expansion. Prometaphase (expansion) is characterized by dramatic global phosphorylation of histones H3 and H1, both of which promote a more extended chromatin conformation (e.g., Fig. 12, which is published as supporting information on the PNAS web site). We further find that histone H3 phosphorylation increases and decreases in tight temporal correlation with global chromatin expansion and contraction throughout the mitotic and meiotic programs (Fig. 7D and Fig. 12).
Predicted Effects of Expansional Stress and Stress Relief Occur in Temporal and Spatial Correlation with Increased and Decreased Chromatin Expansion
The Mitotic Program. By the above model, chromatin and chromosomes should tend to be active during periods of expansion, with regard to both “DNA events” and chromosome morphological changes, and should tend to be quiescent during intervening contraction periods. Correspondingly: There is a strong tendency for genes to be expressed differentially in G1/S and G2/M (prometaphase) in yeast and mammals (27, 28). DNA replication and development of basic organizational features (e.g., nucleosome installation, chromatin loops/axis arrays, and intersister connections) occur during S phase. Modulation and elimination of organizational features occur specifically during postprophase expansion periods: prominent sister separation and axis coiling at prometaphase, a second phase of sister chromatid separation at preanaphase, and axis disassembly at telophase onset. In contrast, periods of contraction exhibit less transcription and no basic changes in morphology.
Particular changes in chromosome morphology and disposition are also expected specifically during chromatin expansion or contraction phases due to increased or decreased “chromatin pushing.”
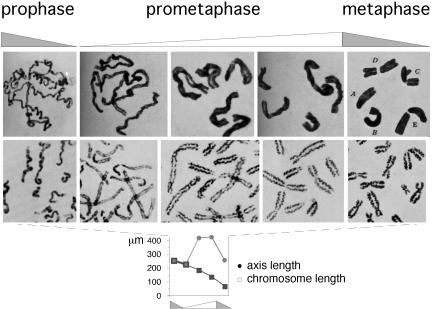
Axes should tend to be longer and/or destabilized in periods of expansion and shorter and/or stable in structure and conformation in periods of contraction. Correspondingly, chromosome axes coil during prometaphase (expansion), and coils remain present during metaphase (contraction) (Fig. 8 Top and Middle). Furthermore, whereas end-to-end chromosome lengths decrease smoothly from prometaphase through metaphase, inspection of axis lengths reveals exactly the predicted three-phase process: chromosome axis lengths increase at the onset of prometaphase, before the onset of coiling; remain constant while coiling develops; and then decrease at metaphase (Fig. 8 Middle and Bottom). Thus, chromatin expansion forces first promote axis extension; when no further extension is possible, compression stress develops along the axes and promotes axis coiling; when expansion forces are alleviated, axes contract back to their original lengths but now in their new expansional stress-promoted shape. The same progression was described for diakinesis coiling in higher plant meiosis, leading to a closely related model, 50 years ago (29, 31). Notably, also, mitotic prometaphase, an expansion phase (Fig. 5), and its meiotic counterpart diakinesis are both characterized not only by axis coiling but also by sister axis separation (e.g., Figs. 4B, 6, and 8).
Fig. 8.
Chromosomes at prometaphase/metaphase. (Top) Mitotic chromosomes of T. grandiflorum (acetocarmine preparations; figures 19–24 of ref. 29). (Middle and Bottom) Human chromosomes stained to reveal axes (30) and end-to-end and axis lengths for corresponding total chromosome complements (details in Supporting Text, which is published as supporting information on the PNAS web site). [Reproduced with permission from, respectively: (Top) ref. 29 (Copyright 1941, NRC Research Press); (Middle and Bottom) ref. 30 (Copyright 1968, Springer-Verlag GmbH & Co.).]
Different chromosomes should tend to be more separated in periods of expansion and more closely conjoined in periods of contraction. Correspondingly: In S phase/G1 (expansion), newly formed sisters are well separated (e.g., ref. 32). In prophase (contraction), sisters are conjoined into a single morphological unit, and all chromosomes are prominently coalesced (33). In prometaphase (expansion), sisters begin to separate (above). In metaphase (contraction), all chromosomes are closely juxtaposed on the spindle. In preanaphase (expansion), sisters “jump apart” further. At the end of anaphase (contraction), chromosomes are tightly coalesced at the two poles (e.g., Fig. 6C Left). In telophase (expansion), disorganizing chromosomes are more separated (Fig. 6C Right) and are concomitantly placed in individual territories, where they remain during the ensuing G1 (contraction).
Chromosomes should tend to be distended and stiff during periods of expansion versus relaxed and “floppy” during periods of contraction. Correspondingly: Classically prepared chromosomes are relaxed and “floppy” at prophase (contraction); straight and stiff at prometaphase (expansion) and also at metaphase, where volumes remain high; relaxed and floppy at anaphase (contraction); and distended at telophase onset (expansion) (Fig. 6C and Fig. 13, which is published as supporting information on the PNAS web site).
Meiotic Prophase. The morphological tendencies described above for mitotic chromosomes are also apparent in meiotic prophase chromosomes. During this period, the chromatin of each chromatid is organized into a linear array of chromatin loops, with sister chromatid linear loop arrays cooriented and with closely conjoined axes (Fig. 3D Right). This conformation undergoes cyclic modulations in correlation with global chromatin expansion and contraction. Homolog axes tend to be stiffer/straighter and/or longer during periods of expansion and more relaxed/floppier and/or shorter during periods of contraction (Fig. 9A and Fig. 14, which is published as supporting information on the PNAS web site). Changes in axis conformation (e.g., buckling and twisting) and loss of prominent axis-associated structures occur preferentially during expansion periods (Figs. 4 C–F and 14). Sister chromatids exhibit tendencies for separation at the chromatin and/or axis levels in each expansion period, preceded and followed by (re-)conjunction during flanking contraction periods (e.g., Figs. 9 B and C, 7A, and 14B). The same tendencies occur in exaggerated form in axial structure mutants (e.g., Fig. 14A). It appears that the same expansional stresses that would promote axis coiling, sister separation, and axis disassembly during postprophase stages are again present but are now severely constrained such that basic organization is never lost, except when constraining features are defective.
Fig. 9.
Modulations of meiotic prophase chromosome conformation in correlation with chromatin expansion/contraction cycles. (A) Sordaria chromosome axes stained with Spo76-GFP are stiffer or floppier in periods of expansion or contraction, respectively (source, D.Z.). (B) Whole nuclear complements of locust chromosomes at early (Left) and late (Right) diplotene exhibit conjoined and separated sister chromatids (34). (C) Individual grasshopper chromosomes showing stained chromatid axes (35) alternating between conjoined and separated configurations during periods ofcontraction and expansion, respectively. Additional examples are in Figs. 11 A and B and 13. [B is reproduced from ref. 34; C is reproduced with permission from ref. 35 (Copyright 1990, NRC Research Press).]
Relationships between homologs and among unrelated chromosomes also vary appropriately. Intermingling of different chromosomes, as required for pairing of homologs, and formation of initial links between homologs, occur in early leptotene (expansion). Maximal juxtaposition of homologs via the synaptonemal complex and concomitant global coalescence of all chromosomes occur during zygotene (contraction); redistribution of chromosomes and their envelope-associated ends occurs during early–mid pachytene (expansion); and global interhomolog links are lost during the diffuse stage (expansion).
Meiosis provides additional unique chromosome readouts, which match expectation. Changes of the chromatin fiber are manifested in the covalent DNA events of recombination. Crossovers arise via four discrete temporally distinct steps (17). Each step involves a covalent change in the DNA that could be promoted by compression stress along the DNA/chromatin fiber (a double-strand break, strand invasion/exchange at each of the two ends and resolution of a double Holliday junction); and each occurs during a period of chromatin expansion (Fig. 5 and ref. 17). Additionally, formation of a chiasma requires not only a DNA crossover but also an analogous exchange at the structural level, between the two involved underlying chromatid axes, as well as local separation of sister chromatids at the same position(s) (38). A simple explanation for such coordinate multilevel effects is as follows: Expansional stresses arise globally, at all necessary levels, i.e., along the DNA/chromatin fiber (for DNA recombination), along chromosome axes (for axis exchange), and between sister chromatids (for sister differentiation and separation). These stresses are constrained globally, by meiosis-specific axis and chromatin structure components; and they are targeted to the appropriate local positions via axis-associated recombination complexes that, suitably localized to the bases of their chromatin loops, would simultaneously create discontinuities (flaws) at all three levels (38). Local ensembles may then ultimately give way in response to imposed stresses, in specific, appropriately programmed ways, giving the desired molecular effects at all three levels, with coordination via direct mechanical linkage. During this process, crossover designation with accompanying interference can be explained by imposition, relief, and redistribution of compression stress and stress relief along chromosome axes (17).
Analogous Spatial Correlations. Mammalian and yeast chromosome arms consist of three basic types of compartments, R, G, and C bands, which exhibit higher, intermediate, and lower degrees of chromatin fiber extension, respectively. The three types of regions vary coordinately with respect to levels of gene expression and meiotic recombination (high, intermediate, low), timing of DNA replication during S phase (early, intermediate, and late), and axis status (36), all of which could be explained by higher, intermediate, and lower levels of chromatin expansional stress.
Additional Implications
Expanded Roles for DNA and Chromosomal Molecules. If molecular and chemical changes within chromosomes are governed by mechanical forces, the DNA and its associated chromosomal macromolecules acquire expanded roles in relation to those forces. DNA base sequence/structure, covalent DNA modifications (e.g., methylation, which favors melting), histone modifications, architectural proteins, and chromatin remodeling activities would be key determinants of basic expansion/contraction state. Other chromosomal components, including nucleosomes, bulk chromatin structure proteins, RNAs, and proteins with specialized functions, would be constrainers, transducers, or targeters of expansional stresses and/or sensors of expansional stress or stress relief.
Intrinsic Chromatin Stress Cycling Versus the Cell Cycle Engine. Global chromatin expansion/contraction cycling could be mediated by the cell cycle engine. Alternatively, chromosomes might have an intrinsic capacity to cycle between expanded and contracted conditions, e.g., via stress-sensitive chromatin modifying proteins. Information might then flow from the chromosomes to the cell cycle engine. Such “reverse information flow” could explain diverse powerful effects of altered histone modification on cell cycle progression and modulation of G1/S progression by chromosome structure proteins (15, 37). Intrinsic chromatin stress cycling could also explain situations where chromosome output varies cyclically irrespective of the cell cycle engine (38–40). Chromosome stress cycling might even have been the original cell cycle engine, mediated by DNA gyrase in eubacteria as a primordial cell cycle and then replaced by more complex mechanisms.
Diseases. If mechanical stress status governs chromosome function, conditions of chromosome dysfunction should be cases of “aberrant stress status.” Chromosomal molecules implicated in disease disorders could thus be assigned specific roles in modulating, constraining, sensing, or transducing mechanical stresses.
Beyond the Chromosomes. Chromosomes are in physical contact with nuclear architectural components and the nuclear envelope, which contact cytoplasmic and cytoskeletal components, which contact the cell surface, which contacts other cells. Information might therefore travel by direct mechanical linkage from the chromosomes outward and from external components inward to the chromosomes. Moreover, the types of effects described above for chromosomes might be involved in other cellular processes.
Conclusion
We propose that basic chromosome function is governed by internally generated mechanical forces generated by tendencies for DNA/chromatin expansion. An attractive feature of this model is that the DNA plays a governing role not only via its information content but also via its intrinsic mechanical properties.
Supplementary Material
Acknowledgments
We gratefully acknowledge the help of many colleagues. This research is supported by grants from the National Institutes of Health (GM44794 and GM25326, to N.K.), the Centre National de la Recherche Scientifique (UMR8126, to D.Z.); the National Science Foundation (DMR 0213805, to J.H.); and the United Kingdom–Biotechnology and Biological Sciences Research Council (6/G5097, to G.H.J.); and by European Molecular Biology Organization Fellowship ALTF 223-1998, a Medical Foundation Fellowship, and National Institutes of Health Grant HG03143 (to J.D.).
References
- 1.Jones, G. H. (1984) Symp. Soc. Exp. Biol. 38, 293–320. [PubMed] [Google Scholar]
- 2.Tease, C. (1978) Nature 272, 823–824. [DOI] [PubMed] [Google Scholar]
- 3.Charles, D. R. (1938) J. Genet. 36, 103–126. [Google Scholar]
- 4.Megee, P. C., Mistrot, C., Guacci, V. & Koshland, D. (1999) Mol. Cell 4, 445–450. [DOI] [PubMed] [Google Scholar]
- 5.Kageyama, Y., Mengus, G., Gilfillan, G., Kennedy, H. G., Stuckenholz, C., Kelley, R. L., Becker, P. B. & Kuroda, M. I. (2001) EMBO J. 20, 2236–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strehl, S., LaSalle, J. M. & Lalande, M. (1997) Mol. Cell. Biol. 17, 6157–6166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rogakou, E. P., Boon, C., Redon, C. & Bonner, W. M. (1999) J. Cell Biol. 146, 905–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brewer, B. J. & Fangman, W. L. (1993) Science 262, 1728–1831. [DOI] [PubMed] [Google Scholar]
- 9.Lucas, I., Chevrier-Miller, M., Sogo, J. M. & Hyrien, O. (2000) J. Mol. Biol. 296, 769–786. [DOI] [PubMed] [Google Scholar]
- 10.Bakkenist, C. J. & Kastan, M. B. (2003) Nature 421, 499–506. [DOI] [PubMed] [Google Scholar]
- 11.Marko, J. F. & Siggia, E. D. (1997) Mol. Biol. Cell 8, 2217–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Croft, J. A., Bridger, J. M., Boyle, S., Perry, P., Teague, P. & Bickmore, W. A. (1999) J. Cell Biol. 145, 1119–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gimenez-Abian, J. F., Clarke, D. J., Mullinger, A. M., Downes, C. S. & Johnson, R. T. (1995) J. Cell Biol. 131, 7–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stack, S. M. (1991) Genome 34, 900–908. [Google Scholar]
- 15.van Heemst, D., James, F., Poggeler, S., Berteaux-Lecellier, V. & Zickler, D. Cell. (1999) 98, 261–271. [DOI] [PubMed] [Google Scholar]
- 16.Westergaard, M. & von Wettstein, D. (1970) C. R. Trav. Lab. Carlsberg 37, 239–268. [PubMed] [Google Scholar]
- 17.Börner, G. V., Kleckner, N. & Hunter, N. (2004) Cell, 117, 29–45. [DOI] [PubMed] [Google Scholar]
- 18.Moens, P. B. (1978) Can. J. Genet. Cytol. 20, 567–579. [DOI] [PubMed] [Google Scholar]
- 19.Nicklas, R. B., Waters, J. C., Salmon, E. D. & Ward, S. C. (2001) J. Cell Biol. 114, 4173–4183. [DOI] [PubMed] [Google Scholar]
- 20.Belmont, A. S. & Bruce, K. (1994) J. Cell. Biol. 127, 287–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bajer, A. (1959) Hereditas 45, 579–596. [Google Scholar]
- 22.Wilson, G. B. & Coleman, P. G. (1952) Cytologia 17, 270–278. [Google Scholar]
- 23.Hao, S., Jiao, M., Zhao, J., Xing, M. & Huang, B. (1994) Chromosoma 103, 432–440. [DOI] [PubMed] [Google Scholar]
- 24.Rhoades, M. M. (1968) in Replication and Recombination of Gentic Material, eds. Peacock, W. J. & Brock, R. D. (Aust. Acad. Sci., Canberra, Australia), pp. 229–241.
- 25.Dawe, R. K., Sedat, J. W., Agard, D. A. & Cande, W. Z. (1994) Cell 76, 901–912. [DOI] [PubMed] [Google Scholar]
- 26.Bojko, M. (1985) Carlsberg Res. Commun. 50, 43–72. [Google Scholar]
- 27.Spellman, P. T., Sherlock, G., Zhang, M. Q., Iyer, V. R., Anders, K., Eisen, M. B., Brown, P. O., Botstein, D. & Futcher, B. (1998) Mol. Biol. Cell 9, 3273–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whitfield, M. L., Sherlock, G., Saldanha, A. J., Murray, J. I., Ball, C. A., Alexander, K. E., Matese, J. C., Perou, C. M., Hurt, M. M., Brown, P. O., et al. (2002) Mol. Biol. Cell 13, 1977–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sparrow, A. H., Huskins, C. L. & Wilson, G. B. (1941) Can. J. Res. C 19, 323–350. [Google Scholar]
- 30.Ohnuki, Y. (1968) Chromosoma 25, 402–428. [DOI] [PubMed] [Google Scholar]
- 31.Wilson, G. B. & Morrison, J. H. (1961) Cytology (Reinhold, New York).
- 32.Selig, S., Okumura, K., Ward, D. C. & Cedar, H. (1992) EMBO J. 11, 1217–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuwada, Y. (1939) Cytologia 10, 213–256. [Google Scholar]
- 34.Whitehouse, H. L. K. (1973) Towards an Understanding of the Mechanism of Heredity (St. Martin's Press, New York), 3rd Ed.
- 35.Suja, J. A., de la Torre, J., Gimenez-Abian, J. F., Garcia de la Vega C. & Rufas, J. S. (1990) Genome 34, 19–27. [DOI] [PubMed] [Google Scholar]
- 36.Blat, Y., Protacio, R., Hunter N. & Kleckner, N. (2002) Cell 111, 791–802. [DOI] [PubMed] [Google Scholar]
- 37.van Heemst, D., Kafer, E., John, T., Heyting, C., van Aalderen, M. & Zickler, D. (2001) Proc. Natl. Acad. Sci. USA. 98, 6267–6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haase, S. B. & Reed, S. I. (1999) Nature 401, 394–397. [DOI] [PubMed] [Google Scholar]
- 39.Mori, T. & Johnson, C. H. (2001) Semin. Cell Dev. Biol. 12, 271–278. [DOI] [PubMed] [Google Scholar]
- 40.Chan, R. C., Chan, A., Jeon, M., Wu, T. F., Pasqualone, D., Rougvie, A. E. & Meyer, B. J. (2003) Nature 424, 1002–1009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.