Abstract
The MPR1 gene, which is found in the Σ1278b strain but is not present in the sequenced laboratory strain S288C, of the budding yeast Saccharomyces cerevisiae encodes a previously uncharacterized N-acetyltransferase that detoxifies the proline analogue azetidine-2-carboxylate (AZC). However, it is unlikely that AZC is a natural substrate of Mpr1 because AZC is found only in some plant species. In our search for the physiological function of Mpr1, we found that mpr1-disrupted cells were hypersensitive to oxidative stresses and contained increased levels of reactive oxygen species (ROS). In contrast, overexpression of MPR1 leads to an increase in cell viability and a decrease in ROS level after oxidative treatments. These results indicate that Mpr1 can reduce intracellular oxidation levels. Because put2-disrupted yeast cells lacking Δ1-pyrroline-5-carboxylate (P5C) dehydrogenase have increased ROS, we examined the role of Mpr1 in put2-disrupted strains. When grown on media containing urea and proline as the nitrogen source, put2-distrupted cells did not grow as well as WT cells and accumulated intracellular levels of P5C that were first detected in yeast cells and ROS. On the other hand, put2-disrupted cells that overexpressed MPR1 had considerably lower ROS levels. In vitro studies with bacterially expressed Mpr1 demonstrated that Mpr1 can acetylate P5C, or, more likely, its equilibrium compound glutamtate-γ-semialdehyde, at neutral pH. These results suggest that the proline catabolism intermediate P5C is toxic to yeast cells because of the formation of ROS, and Mpr1 regulates the ROS level under P5C-induced oxidative stress.
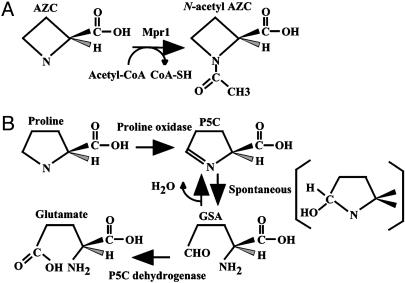
We discovered, on the chromosome of budding yeast Saccharomyces cerevisiae Σ1278b, previously uncharacterized genes required for resistance to the proline analogue azetidine-2-carboxylate (AZC) (1). Intriguingly, the genes MPR1 and MPR2 (sigma 1278b gene for proline-analogue resistance) were present on chromosomes XIV and X, respectively, of the Σ1278b background strains but were absent in S. cerevisiae strain S288C, which was used to determine the genomic sequence (1). Although one amino acid change at position 85 occurred between MPR1 and MPR2, both genes are expressed in S. cerevisiae and play similar roles in AZC resistance (1). Gene expression in Escherichia coli and enzymatic analysis showed that MPR1 encodes an AZC acetyltransferase, by which proline itself and other proline analogues are not acetylated (2). The MPR1-encoded protein (Mpr1) is a member of the N-acetyltransferase superfamily (3). AZC is transported into the cells via proline transporters (4–6). There it causes misfolding of proteins into which it is incorporated competitively with proline, thus inhibiting cell growth in both prokaryotic and eukaryotic cells (7–9). We believe that Mpr1 converts AZC into N-acetyl AZC (Fig. 1A), and consequently that N-acetyl AZC does not replace proline during the biosynthesis of protein (2).
Fig. 1.
Metabolism of AZC and proline. (A) Proposed scheme for the AZC N-acetyltransferase reaction by Mpr1. (B) Degradation of proline in S. cerevisiae. A possible transition state between GSA and P5C, which may be the substrate for Mpr1, is shown in parentheses.
A homology search detected MPR1 homologue genes in the genomes of Saccharomyces paradoxus (Spa MPR1) (10) and fission yeast Schizosaccharomyces pombe (ppr1+) (11). These genes were also shown to be required for AZC resistance in each strain and to encode a similar acetyltransferase (10, 11). Further, genomic PCR analysis showed that most of the S. cerevisiae complex species including Saccharomyces bayanus and Saccharomyces pastorianus have sequences highly homologous to MPR1 (10). We recently found by a blast search of protein databases that Saccharomyces mikatae, Saccharomyces kudriavzevii, and Saccharomyces kluyveri contain a DNA fragment (AABZ01000076.1, AACI01000538.1, and AACE01000067.1, respectively) that is highly homologous to MPR1. These results suggest that MPR1 is the “yeast-specific gene” that is widely present in yeast strains and probably derived from a common ancestral gene. However, the question arises as to why the yeast strains possess this acetyltransferase. Because AZC, a rare toxic imino acid, occurs only in plants belonging to the Lilaceae family, it is not likely a natural substrate of Mpr1 (12, 13). The Mpr1 enzyme may have a physiologically conserved function in addition to AZC detoxification. The natural substrate(s) of this enzyme in yeast cells might be unknown four-membered ring compounds.
In this study, we found that Mpr1 is involved in yeast cell growth and reduces levels of intracellular reactive oxygen species (ROS) under oxidative stresses, including H2O2 and heat-shock treatments. We then focused on proline catabolism intermediate Δ1-pyrroline-5-carboxylate (P5C) and its equilibrium compound, glutamate-γ-semialdehyde (GSA), because ROS was detected in a yeast mutant lacking P5C dehydrogenase (14). In S. cerevisiae, proline is converted to glutamate within mitochondria by the action of two mitochondrial enzymes: proline oxidase (the PUT1 gene product) and P5C dehydrogenase (the PUT2 gene product) (Fig. 1B) (15–17). Genetic and enzymatic analyses showed that Mpr1 regulates the ROS level by acetylating P5C or, more likely, GSA, under P5C-induced oxidative stress. In addition, we propose a possible function of Mpr1 under oxidative stress that produces ROS.
Materials and Methods
Strains and Plasmids. The yeast haploid strains used in this study are listed in Table 1. The S. cerevisiae strains with a Σ1278b background were the WT strain L5685 supplied by G. Fink (Massachusetts Institute of Technology, Cambridge) and the mpr-disrupted strain LD1014 (2). Strains L5685Dput1 and L5685Dput2 were constructed from strain L5685. Strain LD1014Dput2 was constructed from strain LD1014ura3, which was isolated from the 5-fluoroorotic acid-resistant colonies of strain LD1014. We also used the S. cerevisiae strain CKY8 with a S288C background lacking MPR1 supplied by C. Kaiser (Massachusetts Institute of Technology), and strain CKY8Dput2 was constructed from strain CKY8. The 2μ-based high-copynumber plasmid pAD-MPR (11), which contains LEU2, was used to express MPR1 under control of the ADH1 promoter. The centromere-based low-copy-number plasmid pRS-MPR, which contains LEU2, was constructed by cloning the 3.7-kb SacI–SacI fragment of MPR1 from pMH1 (1) into the SacI site of pRS415 (Stratagene). Plasmid YEp24 (18) harboring URA3 was used for disruption of PUT1 and PUT2. Plasmids pRS414 and pRS416 (Stratagene) harboring TRP1 and URA3, respectively, were used for complementing the auxotrophic markers.
Table 1. Yeast strains used in this study.
| Strain | Genotype | Background and/or description |
|---|---|---|
| L5685 | a ura3-52 trp1 MPR1 MPR2 | Σ1278b, WT |
| L5685Dput1 | a ura3-52 trp1 MPR1 MPR2 put1::URA3 | L5685, put1 disruptant |
| L5685Dput2 | a ura3-52 trp1 MPR1 MPR2 put2::URA3 | L5685, put2 disruptant |
| LD1014 | a ura3-52 trp1 mpr1::URA3 mpr2::TRP1 | L5685, mpr1 mpr2 double disruptant |
| LD1014ura3 | a ura3-52 trp1 mpr1::URA3 mpr2::TRP1 ura3 | LD1014, 5-fluoroorotic acid-resistant mutant |
| LD1014Dput2 | a ura3-52 trp1 mpr1::URA3 mpr2::TRP1 put2::URA3 | LD1014ura3, put2 disruptant |
| CKY8 | α ura3-52 leu2-3,112 | S288C, WT |
| CKY8Dput2 | α ura3-52 leu2-3,112 put2::URA3 | CKY8, put2 disruptant |
An E. coli strain JM109 (19) and the isopropyl-β-d-thiogalactopyranoside-inducible plasmid pQE-MPR (11) were used for expression of MPR1 in E. coli. Plasmid pBluescript II SK+ (Toyobo Biochemicals, Osaka) was used to subclone PUT1 and PUT2.
Culture Media. The media used for growth of S. cerevisiae were synthetic dextrose (SD) (2% glucose and 0.67% Bacto yeast nitrogen base without amino acids and ammonia sulfate) (Difco) and yeast extract/peptone/dextrose (2% glucose, 1% yeast extract, and 2% peptone). SD medium contains 10 mM ammonium sulfate (SD+Am) or 16.7 mM urea plus 100 mM proline (SD+Urea+Pro) as the nitrogen source. When appropriate, required amino acids were added to the media for auxotrophic strains. The E. coli recombinant cells were grown in M9 medium (19) containing 2% casamino acids and 50 μg/ml ampicillin. If necessary, 2% agar was added to solidify the medium.
Oxidative Stress Tolerance Test. Yeast cells were cultured to the exponential growth phase in SD+Am medium at 30°C with shaking, washed with 0.9% NaCl, and concentrated to 2 × 106 cells per ml (optical density at 600 nm of 0.1) for H2O2 treatment or to 2 × 108 cells per ml (optical density at 600 nm of 10) for heat-shock treatment. Cells were then exposed to 3 mM H2O2 or to heat shock at 50°C. Before exposure to oxidative stress and at intervals thereafter, 0.1 ml of the culture was removed and diluted in distilled water, and aliquots were plated on yeast extract/peptone/dextrose plates. After incubation at 30°C for 2 days, the survival rates were expressed as percentages, calculated as follows: (no. of colonies after exposure to oxidative stress/no. of colonies before exposure to oxidative stress) × 100.
Northern Blot Analysis. Northern blot analysis was performed as described in ref. 2.
Disruption of PUT1 and PUT2. The DNA fragments of PUT1 and PUT2 were prepared by PCR as described in ref. 27. The plasmids harboring PUT1 and PUT2 were designated pPUT1 and pPUT2, respectively. Plasmids pPUT1U and pPUT2U were then constructed by deleting the HindIII fragment in PUT1 from plasmid pPUT1 and inserting the 1.2-kb HindIII fragment containing URA3 of plasmid YEp24 or the AatII–HindIII fragment in PUT2 from plasmid pPUT2 and inserting the 1.1-kb SmaI–HindIII fragment containing URA3 of plasmid YEp24, respectively, by ligation. For put1 or put2 disruption, the 2.0-kb SalI–SacI fragment containing put1::URA3 of pPUT1U or the 2.0-kb SalI–SacI fragment containing put2::URA3 of pPUT2U was integrated into the PUT1 or PUT2 locus in strains L5685, LD1014ura3, and CKY8 by transformation. The Ura+ phenotype was selected, and the correct disruption event was confirmed by chromosomal PCR. We also tested the growth of these disruptants on SD medium containing 0.1% monosodium glutamate or proline as the sole nitrogen source. These disruptants failed to grow on proline-containing plates.
Intracellular Contents of Proline and P5C. Yeast cells were cultured in SD+Urea+Pro medium. After cultivation for 72 h at 30°C, 5 ml of cell suspension was removed, washed twice with 0.9% NaCl, and suspended in 0.5 ml of distilled water. The 1.5-ml microcentrifuge tube containing cells was transferred to a boiling water bath, and intracellular amino acids were extracted by boiling for 10 min. After centrifugation (5 min at 15,000 × g), proline in each supernatant was subsequently quantified with an amino acid analyzer (L-8500A, Hitachi, Tokyo). P5C in each supernatant was measured by monitoring the amount of the P5C-o-aminobenzaldehyde complex with its extinction coefficient (2,710 M–1·cm–1) as described in refs. 20 and 21. The contents of proline and P5C were expressed as a percentage of the dry weight.
Measurement of Intracellular Oxidation Level. The level of intracellular oxidation was measured with the oxidant-sensitive probe 2′,7′-dichlorof luorescin diacetate (DCFDA) (Molecular Probes) (22). For heat-shock treatment, exponential yeast cells were incubated at 50°C for 30 and 60 min in the presence of 10 μM DCFDA. In the put2-disrupted strain, 10 ml of cell culture grown in SD+Urea+Pro medium at 30°C for 72 h was removed, resuspended in 10 ml of distilled water containing 10 μM DCFDA, and incubated at 30°C for 15 min. In both experiments, cells were then washed, resuspended in 300 μl of distilled water, and disrupted with glass beads by vortex mixer for 5 min. Cell extracts (50 μl) were mixed in 450 μl of distilled water, and fluorescence was measured with λEX = 490 nm and λEM = 524 nm by using a fluorescence spectrophotometer (F4500, Hitachi). The value of λEM = 524 nm was normalized by protein in the mixture.
Expression and Purification of Mpr1 in E. coli. Recombinant Mpr1 was expressed and purified from the cell-free extracts of E. coli strain JM109 harboring pQE-MPR as described in ref. 11.
Assay of Acetyltransferase Activity. The acetyltransferase activity was assayed at 30°C by monitoring the decrease in absorbance at 232 nm, which occurs as a result of the cleavage of the thioester bond of acetyl-CoA, as described by Denk and Böck in ref. 23. The reaction mixture (final volume 1.5 ml) contained 50 mM Tris·HCl buffer (pH 8.5) or 50 mM 2-morpholinoethanesulfonate (pH 7.0), 1 mM AZC (Sigma) or P5C (Sigma), 0.1 mM acetyl-CoA, and enzyme solution. The reaction rate was calculated with an extinction coefficient for acetyl-CoA of 6,500 M–1·cm–1. The kinetic parameters kcat and Km were obtained from initial rate measurements by monitoring the absorbance decrease at 232 nm with a DU-640 spectrophotometer (Beckman Coulter). When the apparent Km value for AZC or P5C was determined, the AZC or P5C concentration was varied from 0.0625 to 1 mM in the presence of a fixed concentration of acetyl-CoA (0.1 mM). Protein concentrations were determined with a Bio-Rad protein assay kit. BSA was used as the standard protein.
Results
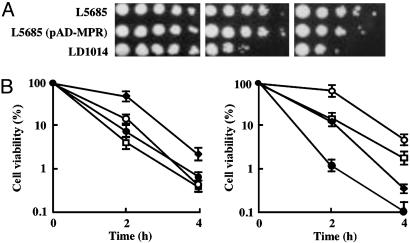
Mpr1 Plays a Crucial Role in Protecting Yeast Cells Under Oxidative Stress. To examine the physiological function of Mpr1 in addition to its detoxification of AZC, we compared the growth phenotype of the WT (MPR1 MPR2) and the mpr1 mpr2 (mpr)-disrupted strains with a Σ1278b background under various culture conditions. Interestingly, when yeast cells were exposed to oxidative stresses such as H2O2 and heat-shock treatments on SD+Am agar medium (Fig. 2A), the mpr-disrupted strain LD1014 was much more sensitive than the WT strain L5685. Overexpression of MPR1 had no influence on the growth of yeast cells under these conditions.
Fig. 2.
Effect of Mpr1 on yeast cells under oxidative stress. (A) About 106 cells of each strain and serial dilutions of 10–1 to 10–4 were spotted onto SD+Am medium (Left), 1 mM H2O2-containing SD+Am medium (Center), or SD+Am medium after heat-shock treatment (50°C for 4 h) (Right). The plates were incubated at 30°C for 3 days. (B) Time course of the cell viability of each strain in SD+Am liquid medium after addition of 3 mM H2O2 (Left) and heat shock at 50°C(Right). Detailed conditions for experiments are described in Materials and Methods. The S. cerevisiae strains with a Σ1278b background used are the WT L5685 (filled diamonds) and the mpr disruptant LD1014 (filled circles). The strains with a S288C background used are the WT CKY8 (open squares) and CKY8 carrying pAD-MPR (open circles). Results indicate the mean and standard deviation from three independent experiments.
We further determined the viability of S. cerevisiae cells exposed to the same oxidative stresses in a liquid medium. As shown in Fig. 2B, the cell viability of the mpr disruptant after the addition of 3 mM H2O2 fell to 18% of that of the WT strain. On the other hand, when high-copy-number plasmid pAD-MPR harboring MPR1 was introduced into strain CKY8 with a S288C background lacking MPR1, the transformant showed a 3-fold higher survival rate after exposure to H2O2 for 2 h as compared with that of strain CKY8 carrying only the vector. Similar results were obtained with heat-shock treatment at 50°C (Fig. 2B). The survival rates of the mpr-disrupted cells after exposure to heat shock were 6–10 times lower than those observed in the WT cells. In contrast, overexpression of MPR1 in strain CKY8 showed a 3-fold higher cell viability compared with that of strain CKY8 carrying only the vector against heat shock. These results indicate that Mpr1 has an important function in cell survival during exposure to oxidative stress.
Mpr1 Reduces Levels of Intracellular Oxidation. Lethal heating at 50°C is known to cause oxidative stress in the cell by producing oxygen-free radicals and other ROS that could attack vulnerable nucleic acids, proteins, lipids, carbohydrates, and other cellular components (24, 25). The oxidant-sensitive probe 2′,7′-dichlorofluorescin diacetate was used to measure the levels of intracellular oxidation produced during heating at 50°C. This probe is trapped inside the cells after cleavage of the diacetates by an intracellular esterase (22). It then becomes susceptible to attack by radical species, producing a more fluorescent compound (26).
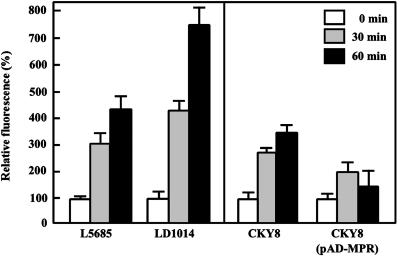
As shown in Fig. 3, crude extracts from strain L5685 with a Σ1278b background showed a 3- to 4-fold increase in fluorescence in response to incubation at 50°C, suggesting that the heat-shock stress represents toxicity through intracellular ROS generation. Further, the level of oxidation was increased by 30–80% in the mpr-disrupted strain. The same experiment was performed with strain CKY8 with a S288C background lacking MPR1 (Fig. 3). The mean fluorescence in CKY8 cells was significantly increased (almost 3-fold) after exposure to heat shock for 60 min. At that time point, however, we observed a 2-fold reduction in the fluorescence rates of MPR1-overexpressing cells. These findings are consistent with those of the survival experiments (Fig. 2). Northern blot analysis showed quantitatively similar amounts of MPR1 transcripts in strains L5685 and CKY8 (pMH1) in the presence or absence of H2O2 or heat shock (data not shown). These results indicate that Mpr1 reduces the intracellular level of ROS and protects yeast cells from damage by oxidative stress.
Fig. 3.
Effect of Mpr1 on the intracellular oxidation levels after heat-shock treatment. Detailed conditions for the experiments are described in Materials and Methods. Intensity of fluorescence of the WT strains L5685 and CKY8 before heat-shock treatment (0 min) was relatively taken as 100%. Results indicate the mean and standard deviation from three independent experiments for the Σ1278b (Left) and S288C (Right) backgrounds.
The put2 Disruptant Inhibits Growth Because of Accumulation of P5C and ROS. It is possible that Mpr1 would acetylate a toxic intermediate involved in the generation of ROS via some metabolic pathway. Deuschle et al. (14) recently reported that excess proline inhibits growth and forms ROS in a put2-disrupted yeast lacking P5C dehydrogenase. We also found that the put2 disruptant is hypersensitive to the freeze–thaw process (27), which causes oxidative stress to cellular components by ROS (28). Taking into account these observations, we assume that mitochondrial accumulation of P5C or GSA, which are in spontaneous equilibrium, in the put2-disrupted cells impairs mitochondrial function and the subsequent production of ROS.
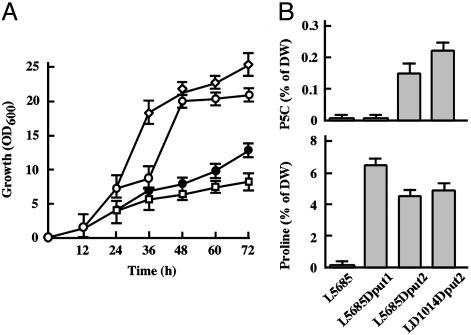
To first examine the effect of proline and its catabolism intermediate P5C on yeast cell growth, we constructed put1- and put2-disrupted strains with a Σ1278b background and tested their growth (Fig. 4A). When urea plus proline was the nitrogen source, the growth of the put2 disruptant was significantly inhibited, whereas little inhibition was observed in the put1-disrupted strain. Growth inhibition of the put2 disruptant occurred at a proline concentration of 100 mM and became more obvious as the concentration increased (data not shown). It should be noted that, as shown in Fig. 4B, the put2 disruptant accumulated higher P5C levels (0.15% of the dry weight) than did the WT and put1-disrupted strains (0.01% in both) after 72 h cultivation. On the other hand, proline accumulated significantly in both the put1 and put2 disruptants (5–6%) (Fig. 4B). In S. cerevisiae, the PRO3-encoded P5C reductase, which converts P5C to proline in the cytoplasm, is constitutively expressed (29). Therefore, there was significant accumulation of proline even in the put2-disrupted strain. The level of oxidation accompanied by P5C accumulation was measured (Fig. 5). A 2- to 3-fold increase in fluorescence evolved after 72 h cultivation in cells lacking P5C dehydrogenase with both Σ1278b and S288C strains. We also found that the intracellular ROS level in the put1-disrupted strain L5685Dput1 was 55% lower than that of the WT strain L5685, probably because of the proline accumulation. These observations demonstrate that P5C, not proline, is toxic to yeast cells, through intracellular ROS production.
Fig. 4.
P5C accumulation caused by disruption of PUT2. (A) The growth curve of S. cerevisiae strains with Σ1278b background. The S. cerevisiae strains with a Σ1278b background used are the WT L5685 (open diamonds), the put1 disruptant L5685Dput1 (open circles), the put2 disruptant L5685Dput2 (filled circles) and the mpr put2 disruptant LD1014Dput2 (open squares). (B) Intracellular contents of P5C (Upper) and proline (Lower) of S. cerevisiae strains with a Σ1278b background. Detailed conditions for experiments are described in Materials and Methods. All results represent the mean and standard deviation from three independent experiments. DW, dry weight.
Fig. 5.
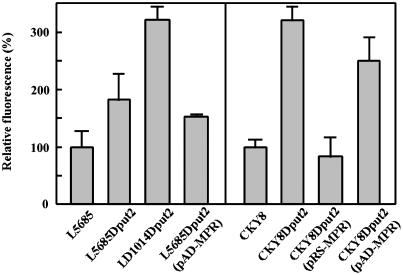
Effect of Mpr1 on the intracellular oxidation levels under P5C-induced oxidative stress. Detailed conditions for experiments are described in Materials and Methods. Intensity of fluorescence of the WT strains L5685 and CKY8 was relatively taken as 100%. Results represent the mean and standard deviation from three independent experiments for the Σ1278b (Left) and S288C (Right) backgrounds.
Mpr1 Regulates the Intracellular Level of ROS in put2-Disrupted Cells. As shown in Fig. 3, Mpr1 can reduce the level of intracellular ROS under oxidative stress conditions. We therefore analyzed the role of Mpr1 in P5C-induced growth inhibition and oxidative damage. Interestingly, disruption of MPR in the put2-disrupted strain severely inhibited growth (Fig. 4A) and increased P5C content by 50% (Fig. 4B) as compared with the single put2 disruptant. With respect to the oxidation of the fluorescent probe after 72 h cultivation, an ≈2-fold increase was observed in the mpr put2 disruptant compared with that of the single put2 disruptant (Fig. 5). In contrast, overexpression of MPR1 in the put2 disruptant significantly reduced the intracellular ROS level (Fig. 5). Similar results were observed for strain CKY8 with a S288C background lacking MPR1 (Fig. 5). Low expression of MPR1 in the put2 disruptant was more effective at decreasing intracellular oxidation than was overexpression, which may cause the misfolding or mislocalization of Mpr1. When MPR1 is overexpressed in the WT strains L5685 and CKY8, the intracellular ROS levels were virtually unchanged from those of the strains carrying only the vector (data not shown). Two copies (MPR1 and MPR2) in Σ1278b strain or the low-level expression in S288C strain would be adequate to protect yeast cells from oxidative stress. These results suggest that Mpr1 regulates the intracellular ROS level under P5C-induced oxidative stress, probably by controlling P5C content.
Mpr1 Acetylates P5C/GSA Involved in ROS Production. We attempted to identify the cellular substrate for Mpr1 under oxidative stress caused by put2 disruption. Consideration of GSA's structure suggests that, under a weak acidic condition, it could be converted to a cyclic-like structure because of the instability of the aldehyde group and that the amino group can be N-acetylated. We purified recombinant Mpr1 from E. coli cells and examined its acetyltransferase activity for the commercially available P5C, which is more stable than GSA (Table 2). It is noteworthy that the acetyltransferase activity of Mpr1 for P5C was clearly detected at neutral pH. The optimal pH of Mpr1 was ≈8.5–9.0 for AZC (11) but ≈6.5–7.0 for P5C. At the optimal pH, the kcat/Km value was virtually unchanged for both AZC and P5C, although kinetic constants kcat and Km values were higher for P5C than for AZC. These results suggest that P5C, or, more likely its equilibrium compound GSA, is one of the cellular substrates of Mpr1.
Table 2. Specific activities and kinetic constants of Mpr1 toward AZC and P5C.
| Substrate | Specific activity, milliunits/mg | Kcat, sec-1 | Km, mM | Kcat/Km, sec-1·mM-1 |
|---|---|---|---|---|
| AZC | 24.0 ± 1.49 | 17.9 ± 0.64 | 0.90 ± 0.07 | 19.8 ± 0.69 |
| P5C | 14.8 ± 1.97 | 121 ± 8.80 | 7.01 ± 0.84 | 17.2 ± 1.80 |
Assays were performed in 50 mM Tris·HCl (pH 8.5) for AZC and in 50 mM 2-morpholinoethanesulfonate (pH 7.0) for P5C at 30°C with 0.29 μg of purified enzyme. The values are mean ± standard deviation from three independent experiments.
Discussion
How Does Mpr1 Protect Yeast Cells Under Oxidative Stress? During the normal course of respiratory metabolism in all aerobic organisms, several ROS are produced as byproducts and damage the cell, e.g., superoxide anion (O–2), hydrogen peroxide (H2O2), and hydroxyl radical (OH). ROS production has been described as a critical feature of programmed cell death in yeast, animals, and plants (30, 31). In response to the destructive nature of ROS, aerobic organisms have evolved cellular antioxidant systems capable of detoxifying ROS.
In S. cerevisiae, oxidative stress, such as exposure to lethal H2O2 or heat shock, elicits the induction of a variety of antioxidant enzyme genes, including catalase (CTT1), superoxide dismutase (SOD), thioredoxin peroxidase (TSA), glutathione peroxidase (GPX), and cytochrome c peroxidase (CCP1) (32–35), as well as heat-shock response genes, including HSP104 and HSP70. Several transcriptional factors are also crucial for oxidative-stress response. Yap1 (yeast AP-1) regulates the expression of many antioxidant genes, including GSH1, GLR1, TRX2, and TRR1 (34, 36). Despite the multiple defense mechanisms against oxidative damage in yeast, the function of MPR1 under oxidative stress is particularly intriguing. The antioxidant enzymes found so far localize in the cytoplasm or mitochondria and catalyze the direct disproportion, decomposition, or reduction of ROS. However, Mpr1 catalyzes the N-acetylation reaction (11) and is a member of the N-acetyltransferase superfamily, which includes histone acetyltransferases and transcriptional regulators (3). In the case of Yap1, the localization changes dramatically in the response to oxidative stress. Yap1 exists in both the cytoplasm and the nucleus under nonstress conditions, whereas under oxidative stress it is concentrated in the nucleus (34). On the other hand, Mpr1 contains no amino acid sequences that are similar to the DNA binding motifs and the amino-terminal basic domain found in other yeast transcriptional factors (37, 38) or the carboxyl-terminal cysteine-rich domain, which is important for intracellular localization of Yap1 (36). It is thus unlikely that Mpr1 is a transcriptional regulator; such a regulator would induce the genes involved in detoxification of ROS. Like P5C, we consider that Mpr1 would acetylate some unknown toxic intermediates accumulated in the cytoplasm via the oxidative event. It is also possible that Mpr1 acts as a backup system for antioxidant enzymes.
P5C Accumulation Was Detectable in the put2-Disrupted Yeast Cells. P5C has certain effects on gene expression. It selectively inhibits the initiation of translation of mammalian RNAs (39) and selectively enhances the accumulation of P1450 mRNA in mice (40). Iyer and Caplan (41) described the induction of stress-related genes by P5C in rice. P5C is assumed to play a role in cell death in animals and plants if it is not metabolized. Treatment with P5C induced apoptosis in human tumor cell lines (42). Hellman et al. (43) reported that an external supply of P5C was lethal for Arabidopsis plants. Genetic approaches, such as overexpression of proline oxidase and the proline-hypersensitive mutant that is affected in P5C dehydrogenase activity, were also applied to elucidate P5C's toxicity (42, 43).
In S. cerevisiae, the growth of a mutant deficient in P5C dehydrogenase was inhibited by a high concentration of proline, probably because of the degradation of proline by proline oxidase and the subsequent accumulation of toxic amounts of P5C (14). The determination of intracellular P5C was problematic because of its chemical instability (44), so there has been no report on the cellular content of P5C. Through a proline oxidase assay (26, 45), we have succeeded in detecting and quantifying P5C in yeast cells. This discovery was accomplished by reacting P5C with o-aminobenzaldehyde to form, presumably, a dihydroquinazolium derivative with an absorbancy in the visible range that is proportional to the concentration of the product (21). As we would expect, the put2 disruptant caused a significant accumulation of P5C, compared with the WT and put1-disrupted strains (Fig. 4B).
Toxic Accumulation of P5C Causes Oxidative Damage in Yeast Cells. The growth inhibition caused by 100 mM proline in the put2-disrupted cells was related to the production of ROS (Fig. 5). This result agrees with the qualitative analysis of Deuschle et al. (14), in which dihydrorhodamine 123 was used to detect H2O2. Proline is believed to function as a compatible solute and as a free-radical scavenger to protect macromolecular structures form damage during osmotic and oxidative stresses (46, 47). We also found that intracellular proline in yeast has an important role in reducing the oxidative stress induced during freezing and thawing or during exposure to H2O2 (26, 48–50). It is therefore suggested that the toxic accumulation of P5C, not proline, triggers oxidative damage in yeast cells by reacting with various cellular compounds.
However, the mechanism underlying P5C's toxicity can hardly be explained yet. The degradation of proline to P5C takes place in mitochondria. Because P5C and GSA are in spontaneous equilibrium, it is unclear which of these compounds is the active substance. In higher eukaryotes, mitochondria participate in apoptosis by activating caspases through the release of cytochrome c into the cytosol (51). The mitochondrial proline oxidase is involved in the transfer of redox potential across the mitochondrial membrane through the proline–P5C pathway (52). In yeast, P5C accumulation could cause the shuttling of redox potential, thus inducing oxidative stress. P5C/GSA might prevent the opening of mitochondrial permeability transition pores, or they might have a signal-molecule activating process related to oxidative stress. It remains unclear whether the detrimental effects of P5C/GSA are a direct result of their intrinsic activities or an indirect consequence associated with oxidative stress.
Mpr1 Protects Yeast Cells from P5C-Induced Oxidative Stress by Reducing ROS. Under P5C-induced oxidative stress, Mpr1 was shown to regulate the intracellular ROS level, probably by controlling P5C content. The amino terminus of Mpr1 has no signal sequence to be imported into the mitochondria, unlike the case with proline oxidase and P5C dehydrogenase (53). The question arises as to why the cytoplasmic enzyme Mpr1 can acetylate the mitochondrial P5C/GSA. One simple explanation is that the mitochondrial permeability is impaired by excess P5C/GSA because of its chemical reactivity, resulting in its leakage into the cytoplasm. Another possible explanation is that there is a system for transporting excess P5C/GSA from the mitochondria to the cytoplasm. It is likely that P5C/GSA that have accumulated not in the cytoplasm but in the mitochondria are highly toxic to yeast cells. Therefore, our results indicate that Mpr1 is crucial in preventing the formation of ROS induced by P5C accumulation, which by unknown mechanisms could directly or indirectly trigger oxidative stress.
GSA or Its Transition State May Be One of the Cellular Substrates of Mpr1. The recombinant Mpr1 apparently has acetyltransferase activity for P5C in addition to AZC (Table 2). This reaction was not expected because cyclic P5C does not contain any amino or imino group to be acetylated. We consider two possibilities to explain this finding. Because it contains an aldehyde group, GSA is thought to be chemically more reactive than P5C. It is probable that, under a weak acidic condition, GSA is converted to a cyclic-like structure and that the amino group can be N-acetylated by Mpr1. Another possibility is that amino alcohol is formed as a transition state between GSA and P5C (Fig. 1B). In such a case, Mpr1 is supposed to N-acetylate the imino group of the amino alcohol. It should also be noted that Mpr1 exhibited larger kcat and Km values for the metabolic intermediate P5C than for AZC. The structural determinations of N-acetyl P5C (GSA) and Mpr1 are needed to prove our hypothesis.
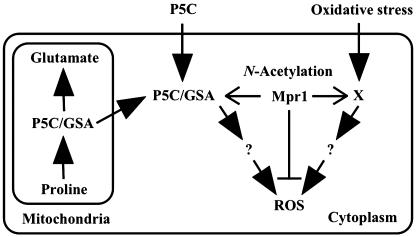
In summary, the null mutant of the MPR genes displayed hypersensitivity to oxidative stress, and the expression of the MPR1 gene conferred stress resistance. As an antioxidant mechanism, Mpr1 is supposed to reduce the intracellular oxidation level by acetylating the toxic metabolite(s) involved in ROS production, such as P5C (Fig. 6). The proposed role of Mpr1 against oxidative stress could be promising for breeding stress-resistant yeast strains.
Fig. 6.
Proposed model for Mpr1-regulated ROS under oxidative stress conditions. Mpr1 is considered to acetylate P5C/GSA that is excreted into the cytoplasm in yeast cells lacking P5C dehydrogenase. Oxidative stress conditions, such as exposure to H2O2 and heat shock, probably lead to accumulate an identified intermediate X, which may be a cellular substrate of Mpr1. As a result, Mpr1 reduces the intracellular level of ROS, although the pathway that P5C/GSA or X induces ROS production is unknown (?). The addition of P5C is also toxic in animals and plants (42, 43).
Acknowledgments
We thank S. Nakamori, M. Takahashi, M. Wada, Y. Hamano, and J. Oda for discussions on this work; G. R. Fink and C. A. Kaiser for providing yeast strains; and K. Matsuura and X. Du for technical assistance. This work was supported by Grant-in-Aid for Scientific Research 15380076 from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to H.T.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: AZC, azetidine-2-carboxylic acid; GSA, glutamate-γ-semialdehyde; ROS, reactive oxygen species; P5C, Δ1-pyrroline-5-carboxylate; SD, synthetic dextrose.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AB031349, M18107, and M10029).
References
- 1.Takagi, H., Shichiri, M., Takemura, M., Mohri, M. & Nakamori, S. (2000) J. Bacteriol. 182, 4249–4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shichiri, M., Hoshikawa, C., Nakamori, S. & Takagi, H. (2001) J. Biol. Chem. 276, 41998–42002. [DOI] [PubMed] [Google Scholar]
- 3.Neuwald, A. F. & Landsman, D. (1997) Trends Biochem. Sci. 22, 154–155. [DOI] [PubMed] [Google Scholar]
- 4.Helliwell, S. B., Losko, S. & Kaiser, C. A. (2001) J. Cell Biol. 153, 649–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoshikawa, C., Shichri, M., Nakamori, S. & Takagi, H. (2003) Proc. Natl. Acad. Sci. USA 100, 11505–11510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andréasson, C., Neve, E. P. A. & Ljungdahl, P. O. (2004) Yeast 21, 193–199. [DOI] [PubMed] [Google Scholar]
- 7.Mizzen, L. A. & Welch, W. J. (1988) J. Cell Biol. 106, 1105–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yokota, S., Yanagi, H., Yura, T. & Kubota, H. (2000) Eur. J. Biochem. 267, 1658–1664. [DOI] [PubMed] [Google Scholar]
- 9.Trotter, E. W., Kao, C. M.-F., Berenfeld, L., Botstein, D., Petsko, G. A. & Gray, J. V. (2002) J. Biol. Chem. 277, 44817–44825. [DOI] [PubMed] [Google Scholar]
- 10.Kimura, Y., Nakamori, S. & Takagi, H. (2002) Yeast 19, 1437–1445. [DOI] [PubMed] [Google Scholar]
- 11.Nomura, M., Nakamori, S. & Takagi, H. (2003) J. Biochem. (Tokyo) 133, 67–74. [DOI] [PubMed] [Google Scholar]
- 12.Peterson, P. J. & Fowden, L. (1963) Nature 200, 148–151. [Google Scholar]
- 13.Leete, E. (1964) J. Am. Chem. Soc. 86, 3162. [Google Scholar]
- 14.Deuschle, K., Funck, D., Hellmann, H., Däschner, K., Binde, S. & Frommer, W. B. (2001) Plant J. 27, 345–355. [DOI] [PubMed] [Google Scholar]
- 15.Brandriss, M. C. & Magasanik, B. (1980) J. Bacteriol. 143, 1403–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brandriss, M. C. & Magasanik, B. (1981) J. Bacteriol. 145, 1359–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tomenchok, D. M. & Brandriss, M. C. (1987) J. Bacteriol. 169, 5364–5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carlson, M. & Botstein, D. (1982) Cell 28, 145–154. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook, J. & Russell, D. W. (2001) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY), 3rd Ed.
- 20.Brandriss, M. C. & Magasanik, B. (1979) J. Bacteriol. 140, 498–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strecker, H. J. (1971) Methods Enzymol. 17, 251–254. [Google Scholar]
- 22.Royall, J. A. & Ischiropoulos, H. (1993) Arch. Biochem. Biophys. 302, 348–355. [DOI] [PubMed] [Google Scholar]
- 23.Denk, D. & Böck, A. (1987) J. Gen. Microbiol. 133, 515–525. [DOI] [PubMed] [Google Scholar]
- 24.Halliwell, B. & Gutteridge, J. M. C. (1984) Biochem. J. 219, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Longo, V. D., Liou, L. L., Valentine, J. S. & Gralla, E. B. (1999) Arch. Biochem. Biophys. 365, 131–142. [DOI] [PubMed] [Google Scholar]
- 26.Tsuchiya, M. & Suematsu, M. (1994) Methods Enzymol. 233, 128–140. [DOI] [PubMed] [Google Scholar]
- 27.Morita, Y., Nakamori, S. & Takagi, H. (2001) J. Biosci. Bioeng. 94, 390–394. [DOI] [PubMed] [Google Scholar]
- 28.Park, J.-I., Grant, C. M., Davies, M. J. & Dawes, I. W. (1997) J. Biol. Chem. 273, 22921–22928. [DOI] [PubMed] [Google Scholar]
- 29.Brandriss, M. C. & Falvey, D. A. (1992) J. Bacteriol. 174, 3782–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jabs, T. (1999) Biochem. Pharmacol. 57, 231–245. [DOI] [PubMed] [Google Scholar]
- 31.Madeo, F., Fröhlich, E., Ligr, M., Grey, M., Sigrist, S. J., Wolf, D. H. & Fröhlich, K. U. (1999) J. Cell Biol. 145, 757–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Collinson, L. P. & Dawes, I. W. (1992) J. Gen. Microbiol. 138, 329–335. [DOI] [PubMed] [Google Scholar]
- 33.Godon, C., Lagniel, G., Lee, J., Buhler, J. M., Kieffer, S., Perrot, M., Boucherie, H., Toledano, M. B. & Labarre, J. (1998) J. Biol. Chem. 273, 22480–22489. [DOI] [PubMed] [Google Scholar]
- 34.Inoue, Y., Matsuda, T., Sugiyama, K., Izawa, S. & Kimura, A. (1999) J. Biol. Chem. 274, 27002–27009. [DOI] [PubMed] [Google Scholar]
- 35.Davidson, J. F., Whyte, B., Bissinger, P. H. & Schiestl, R. H. (1996) Proc. Natl. Acad. Sci. USA 93, 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuge, S., Jones, N. & Nomoto, A. (1997) EMBO J. 16, 1710–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Natsoulis, G., Winston, F. & Boeke, J. D. (1994) Genetics 136, 93–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dollard, C., Ricupero-Hovasse, G., Natsoulis, G., Boeke, J. D. & Winston, F. (1994) Mol. Cell. Biol. 14, 5223–5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mick, S. J., Thach, R. E. & Hagedorn, C. H. (1988) Biochem. Biophys. Res. Commun. 150, 296–303. [DOI] [PubMed] [Google Scholar]
- 40.Nemoto, N. & Sakurai, J. (1991) Jpn. J. Cancer Res. (GANN) 82, 901–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iyer, S. & Caplan, A. (1998) Plant Physiol. 116, 203–211. [PMC free article] [Google Scholar]
- 42.Maxwell, S. A. & Davis, G. E. (2000) Proc. Natl. Acad. Sci. USA 97, 13009–13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hellman, H., Funck, D., Rentsch, D. & Frommer, W. B. (2000) Plant Physiol. 123, 779–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mezl, V. A. & Knox, W. E. (1976) Anal. Biochem. 74, 430–440. [DOI] [PubMed] [Google Scholar]
- 45.Brandriss, M. C. & Magasanik, B. (1979) J. Bacteriol. 140, 498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hong, Z., Lakkineni, K., Zhang, Z. & Verma, D. P. S. (2000) Plant Physiol. 122, 1129–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nanjo, T., Kobayashi, M., Yoshida, Y., Sanada, Y., Wada, K., Tsukaya, H., Kakubari, Y., Yamaguchi-Shinozaki, K. & Shinozaki, K. (1999) Plant J. 18, 185–193. [DOI] [PubMed] [Google Scholar]
- 48.Takagi, H., Iwamoto, F. & Nakamori, S. (1997) Appl. Microbiol. Biotechnol. 47, 405–411. [DOI] [PubMed] [Google Scholar]
- 49.Morita, Y., Nakamori, S. & Takagi, H. (2003) Appl. Environ. Microbiol. 69, 212–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Terao, Y., Nakamori, S. & Takagi, H. (2003) Appl. Environ. Microbiol. 69, 6527–6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zou, H., Henzel, W. J., Liu, X., Lutschg, A. & Wang, X. (1997) Cell 90, 405–413. [DOI] [PubMed] [Google Scholar]
- 52.Herzfeld, A., Mezl, V. A. & Knox, W. E. (1977) Biochem. J. 166, 95–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krzywicki, K. A. & Brandriss, M. C. (1984) Mol. Cell. Biol. 4, 2837–2842. [DOI] [PMC free article] [PubMed] [Google Scholar]