Abstract
Inherited and somatic mutations in the adenomatous polyposis coli occur in most colon cancers, leading to activation of β-catenin-responsive genes. To identify small molecule antagonists of this pathway, we challenged transformed colorectal cells with a secondary structure-templated chemical library, looking for compounds that inhibit a β-catenin-responsive reporter. We identified ICG-001, a small molecule that down-regulates β-catenin/T cell factor signaling by specifically binding to cyclic AMP response element-binding protein. ICG-001 selectively induces apoptosis in transformed cells but not in normal colon cells, reduces in vitro growth of colon carcinoma cells, and is efficacious in the Min mouse and nude mouse xenograft models of colon cancer.
Keywords: coactivators, T cell factor, Wnt pathway
The Wnt/β-catenin pathway initiates a signaling cascade critical in both normal development and the initiation and progression of cancer (1-4). The hallmark of this pathway is that it activates the transcriptional role of the multifunctional protein β-catenin. In its diverse roles as a mediator of cell adhesion at the plasma membrane and as a transcriptional activator, β-catenin interacts with a host of proteins, the majority of which, despite a lack of significant sequence homology, compete for the same armadillo repeats of β-catenin (5, 6).
Canonical Wnt signaling inactivates GSK-3β, preventing β-catenin phosphorylation. This leads to accumulation of β-catenin in the cytoplasm and subsequent translocation to the nucleus (7). A key step in the activation of target genes is the formation of a complex between β-catenin and members of the T cell factor (TCF)/lymphoid enhancer factor family of transcription factors (8). To generate a transcriptionally active complex, β-catenin recruits the transcriptional coactivators, cyclic AMP response element-binding protein (CBP) or its closely related homolog, p300 (9, 10), as well as other components of the basal transcription machinery.
The Wnt/β-catenin pathway normally regulates expression of a range of genes involved in promoting proliferation and differentiation. In >85% of colon cancers, one of the components of the destruction complex, adenomatous polyposis coli (APC), and/or β-catenin itself is mutated, leading to an increase in nuclear β-catenin and constitutive expression of target genes (11). Many of these genes, including cyclin D1 (12, 13) and c-myc (14), which play critical roles in cell growth, proliferation, and differentiation, are inappropriately activated in colon cancer.
Given that the majority of colorectal cancers involve activation of the β-catenin signaling pathway, and given that multiple mutations lead to this activation, there is a clear need for drugs that attenuate the nuclear functions of β-catenin (15). Here, we report the discovery of a selective low molecular-weight inhibitor (ICG-001), which antagonizes β-catenin/TCF-mediated transcription. We show that ICG-001 specifically down-regulates the expression of a subset of β-catenin/TCF-responsive genes. We demonstrate that ICG-001 binds specifically to CBP but not the related transcriptional coactivator p300, thereby disrupting the interaction of CBP with β-catenin. We show that treatment with ICG-001 induces apoptosis in colon carcinoma cells but not in normal colonic epithelial cells. We also demonstrate that ICG-001 is efficacious in both the Min mouse and nude mouse SW620 xenograft models of cancer. Taken together, these data suggest that this small molecule inhibitor of β-catenin/TCF-mediated transcription has significant therapeutic potential for the treatment of cancer.
Materials and Methods
Plasmids. Optimized TOPFLASH, FOPFLASH reporter plasmids (16), β-catenin, and Wnt1 expression vectors were provided by R.T.M.
Cell Culture. Human colon carcinoma cell lines SW480, SW620, and HCT116, normal colonic epithelial cell line CCD-841Co, and Jurkat, PC12, and C2C12 myoblasts (American Type Culture Collection) were maintained according to recommendations.
Transfection and Luciferase Assays. Cells were transfected with Fugene6 (Roche Molecular Biochemicals). Transfection efficiencies were normalized with pRL-null luciferase plasmid. Luciferase assays were performed by using the DUAL-Luciferase Reporter Assay System (Promega). Data represent the mean of two independent experiments, performed in duplicate.
Affinity Purification. Cells were lysed in protein-binding buffer [PBB, 20 mM Hepes, pH 7.9/100 mM NaCl/0.5 mM EDTA/0.5% Nonidet P-40/6 mM MgCl2/5 mM 2-mercaptoethanol/one tablet of Complete protease inhibitor mixture (Roche Molecular Biochemicals)]. Biotinylated ICG-002 was bound overnight at room temperature to a 50% slurry of streptavidin-agarose beads (Amersham Pharmacia) in buffer containing 50% DMSO and 50% PBB. Beads were washed to remove unbound ICG-002 and then incubated with whole-cell lysates. Proteins eluted, either specifically with 100 μM ICG-001 or by boiling in SDS, were immunoblotted and silver stained.
Immunoblotting. Lysates from cultured cells and tissues were immunoblotted by using polyclonal CBP A-22, polyclonal p300 N-15, β-catenin H102 (polyclonal), monoclonal cyclin-D1 HD11 (Santa Cruz Biotechnology); survivin 6E4 (monoclonal, Cell Signaling Technology); and α-tubulin Ab-1 (monoclonal, EMD Biosciences, Madison, WI). Immune complexes were visualized by using enhanced chemiluminescence detection (Amersham Pharmacia).
RNA Extraction and Real-Time RT-PCR. Total RNA was extracted (RNeasy Maxi kit; Qiagen, Valencia, CA), and cDNA synthesized (TaqMan RT, Roche Molecular Biochemicals).
Real-time RT-PCR (SYBR Green PCR Master Mix; Roche Molecular Biochemicals) was performed by using the following: survivin forward primer: 5′-AGCCCTTTCTCAAGGACCAC-3′, reverse primer: 5′-GCACTTTCTTCG CAGTTTCC-3′; β-actin forward primer: 5′-ATCTGGCACCACACCTTCTACAATGAGCTGCG-3′, reverse primer: 5′-CGTCATACTCCTCCTTGCYGATCCACATCTGC-3′.
Min Mouse Model. Seven-week-old male C57BL/6J-ApcMin/+ (The Jackson Laboratory) and WT C57BL/6J mice were treated orally for 9 weeks with ICG-001a (300 mg/kg per day) or vehicle (1% carboxymethylcellulose), once daily, six times per week. Sulindac (Sigma) was administered in drinking water (160 ppm, dissolved in 8 mM Na2PO4 buffer, pH 7.6). At 16 weeks, the polyp number was counted manually by using a dissecting microscope (Zeiss).
SW620 Xenograft Model. The antitumor effect (150 mg/kg, i.v.) was investigated in the SW620 mouse xenograft model, as described (17).
CBP and p300 siRNA. Small interfering RNAs (siRNAs) with a stretch of two thymine residues overhanging at the 3′ end were synthesized by using 2′-deprotected oligos, duplexed, desalted, and purified (Proligo, Boulder, CO). Human CBP and p300 siRNAs were CBP sense strand: UCAACAGCCGCCAUCUUGUtt; antisense strand: ACAAGAUGGCGGCUGUUGAtt; and p300 sense strand: GCGGCCUAAACUCUCAUCUtt; antisense strand: AGAUGAGAGUUUAGGCCGCtt. siRNA duplexes were transfected into SW480. Twenty-four hours posttransfection, cells were treated with ICG-001 or DMSO control.
Chromatin Immunoprecipitation (ChIP) Assay. ChIP assays in SW480 cells were performed as described (18). Primers for PCR for the cyclin D1 promoter are cyclin D1 forward primer, 5′-TGCTTAACAACAGTAACGT-3′; reverse primer, 5′-GGGGCTCTTCCTGGGCAGC-3′.
Caspase-3/7 Activity and in Vitro Cytotoxicity Assays. Caspase 3/7 activity (Apo-One Homogeneous, Promega) and MTS cytotoxicity assays (Promega) were performed according to the manufacturer's instructions.
Results
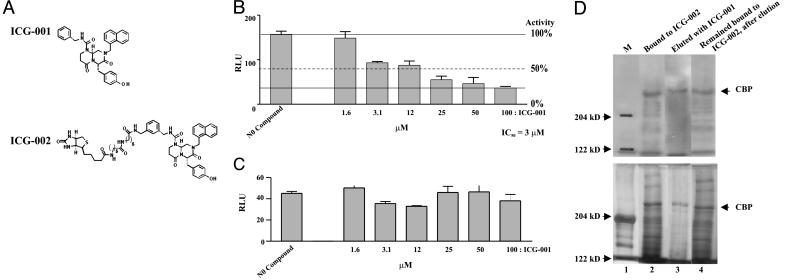
ICG-001 Antagonizes β-Catenin/TCF Transcription. Due to mutations in APC, SW480 colon carcinoma cells exhibit constitutive translocation of β-catenin to the nucleus, and thus high basal β-catenin/TCF transcription as assessed by the TOPFLASH reporter system (16). Using this reporter assay, we screened a secondary structure-templated small molecule library of 5,000 compounds (19, 20) for inhibitors of β-catenin/TCF-mediated transcription. We have developed a series of privileged templates that mimic protein secondary structure elements and incorporate multiple sites of diversity (19-21). From the initial screen, there were three closely related compounds, from which we selected the most potent one for further investigation. ICG-001 (molecular weight 548) (Fig. 1A) had an IC50 of 3 μM (Fig. 1B) against TOPFLASH and had no effect on the related reporter construct, FOPFLASH, which contains mutated TCF sites (Fig. 1C).
Fig. 1.
ICG-001 binds CBP and inhibits β-catenin/TCF-mediated transcription. (A) Chemical structures of ICG-001 and ICG-002. (B and C) ICG-001 selectively inhibits the β-catenin/TCF reporter construct TOPFLASH but not FOPFLASH. (D) CBP is the molecular target of ICG-001. Nuclear extracts of SW480 cells were incubated with streptavidin-agarose beads coated with ICG-002. Bound proteins were eluted and immunoblotted (4% gel) with anti-CBP antibody (Upper) or silver-stained (Lower).
To determine the molecular target(s) of ICG-001, we synthesized a biotinylated derivative of ICG-001, termed ICG-002 (Fig. 1A), for use as an affinity reagent. Nuclear extracts were prepared from SW480 cells after pretreatment with ICG-001 or DMSO and then incubated with ICG-002. The complexes were separated on streptavidin-agarose beads, gel electrophoresed, followed by immunoblotting (Fig. 1D Upper) and silver staining (Fig. 1D Lower). The only major band retained on the ICG-002 affinity column that was specifically eluted by ICG-001 had an apparent molecular mass of 225 kDa and was identified by immunoblotting as the transcriptional coactivator CBP (Fig. 1D, lane 3). The antibody used was specific for CBP and does not crossreact with p300. Immunoblotting with anti-p300, anti-β-catenin, or anti-TCF4 antibodies did not detect any bands on the gel (data not shown).
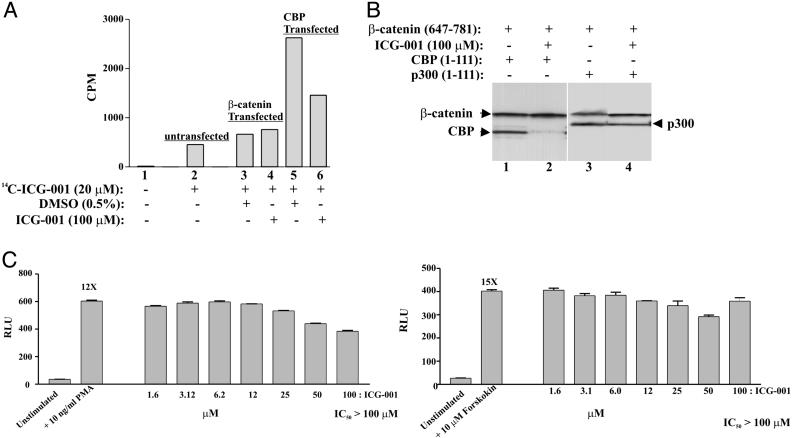
ICG-001 Specifically Interacts with CBP. We next performed a series of independent investigations to validate that ICG-001 binds CBP. We synthesized a 14C-labeled version of ICG-001. Nuclear lysates of SW480 cells, untransfected or transfected with either β-catenin or CBP expression vectors, were treated with 14C-labeled ICG-001 alone or with cold ICG-001 overnight. Unbound 14C-labeled ICG-001 was removed by using a G-25 column, and the incorporation of radioactivity was measured. The nuclear lysates transfected with CBP had an ≈4- to 6-fold increased incorporation of 14C-labeled ICG-001 compared to control (Fig. 2A, compare lanes 2-4 with lane 5), which was competed away by excess cold ICG-001 (Fig. 2A, compare lane 5 with lane 6).
Fig. 2.
ICG-001 binds CBP and selectively disrupts the β-catenin/CBP interaction. (A) SW480 cells were transfected with full-length β-catenin (2.2 μg) or full-length CBP (1.1 μg). Nuclear lysates (50 μg) were treated with 20 μM 14C-labeled-ICG-001 alone (14C-ICG-001) or with 100 μM cold ICG-001. Unbound 14C-labeled-ICG-001 was removed, and 14C-labeled ICG-001 incorporation was measured. (B) ICG-001 specifically disrupts the CBP/β-catenin interaction without disrupting the p300/β-catenin. CBP (amino acids 1-111), p300 (amino acids 1-111), and β-catenin (amino acids 647-781) were expressed in E. coli and purified. β-catenin (5 μg) was bound to protein A-agarose beads coated with β-catenin-specific antibody (2.5 μg) and incubated with either CBP (5 μg) or p300 (5 μg). Unbound proteins were washed away, and specific interactions between these proteins were challenged by using 100 μM ICG-001. The complexes were washed and immunoblotted by using an anti-His antibody. (C) ICG-001 has no effect on AP1 and CRE reporter constructs. RLU, relative light units.
To exclude the possibility of an indirect association between CBP and ICG-001 mediated by another cellular component and to confirm that ICG-001 inhibits the interaction of β-catenin with CBP, we used recombinant proteins expressed in Escherichia coli. We first mapped the minimal binding domain of ICG-001 to amino acids 1-111 of CBP (data not shown). We next mapped the minimum interacting C-terminal region of β-catenin to the same 1-111 amino acids of both CBP and p300 (M.K., unpublished data) (9, 10). Using purified CBP (1-111), p300 (1-111), and β-catenin (647-781), we demonstrated that both CBP and p300 bind to the C-terminal region of β-catenin in the absence of ICG-001 (Fig. 2B, lanes 1 and 3). As anticipated, the CBP/β-catenin interaction but not the p300/β-catenin was disrupted by treatment with ICG-001 (Fig. 2B, compare lane 2 with lane 4). These data confirm the direct association between CBP and ICG-001 and the ability of ICG-001 to selectively disrupt the CBP/β-catenin interaction.
To further examine the selectivity of ICG-001, we used additional reporter gene assays. ICG-001 did not inhibit other CBP-dependent reporters, including AP-1 and CRE, both of which exhibit IC50>100 μM (Fig. 2C). We conclude that ICG-001 specifically inhibits the TCF/β-catenin reporter by binding to CBP.
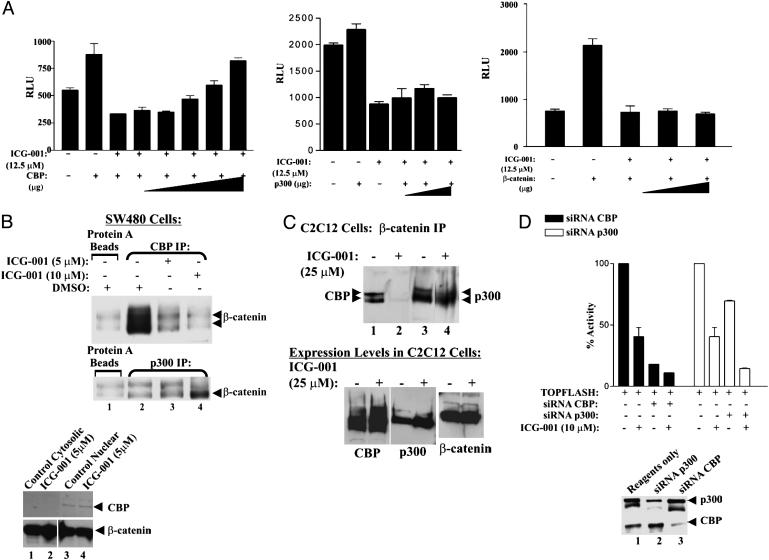
ICG-001 Competes with β-Catenin for CBP. CBP is a rate-limiting factor in many transcriptional events; therefore, we anticipated that increased levels of CBP, but not p300, would raise the IC50 of the inhibitor. As seen in Fig. 3A, transfection of increasing amounts of CBP (Fig. 3A Left), but not p300 (Center) or β-catenin (Right), overrode the inhibition of ICG-001 in a dose-dependent fashion.
Fig. 3.
ICG-001 is specific for CBP but not p300. (A) Full-length CBP, p300, or β-catenin plasmids were cotransfected with TOPFLASH in SW480 cells. Luciferase assays were performed 24 h after ICG-001 treatment. (B) ICG-001 competes with β-catenin for CBP. SW480 cells were treated with ICG-001 or DMSO. Nuclear lysates were coimmunoprecipitated with anti-CBP or anti-p300 antibody and immunoblotted for β-catenin. (C) Nuclear lysates of treated C2C12 myoblasts (25 μM ICG-001, 24 h) were immunoprecipitated with anti-β-catenin antibody and immunoblotted with anti-CBP antibody (Upper, lanes 1 and 2) or anti-p300 antibody (lanes 3 and 4). ICG-001 does not affect levels of CBP, p300, or β-catenin (Lower). (D) SW480 cells were cotransfected with TOPFLASH and duplex siRNA against either CBP or p300. Twenty-four hours after transfection, the cells were treated with 10 μM ICG-001 or DMSO. Luciferase assays were performed 24 h later (Upper). Nuclear lysates from siRNA-transfected cells were immunoblotted (Lower).
We next performed immunoprecipitation assays in SW480 cells to further confirm that ICG-001 selectively disrupts β-catenin binding to CBP, but not p300 (Fig. 3B Top and Middle). Immunoprecipitation of β-catenin with CBP was inhibited by ICG-001 in a dose-dependent manner [Fig. 3B Top, CBP immunoprecipitation (IP), compare lane 2 with lanes 3 and 4; lane 1 control, protein A beads only]. As shown in Fig. 3B Top, two forms of differentially posttranslationally modified β-catenin coimmunoprecipitate with CBP. The binding of both forms of β-catenin to CBP is completely inhibited by ICG-001 (Fig. 3B Top, lane 4). The minimal interaction seen in the SW480 cells between β-catenin and p300 was not blocked by ICG-001, despite the fact that CBP and p300 are highly homologous (Fig. 3B Middle, p300 IP, compare lane 2 with lanes 3 and 4). In fact, treatment with 10 μM ICG-001 increases the amount of β-catenin coimmunoprecipitated with p300 (Fig. 3B Middle, p300 IP, compare lanes 2 and 4). We interpret these data in the following manner: Treatment with ICG-001 dissociates β-catenin from its primary target CBP, thereby resulting in a pool of transcriptionally competent free β-catenin in the nucleus that can now bind to p300. CBP and β-catenin levels, in both the cytoplasm and nucleus of ICG-001-treated cells, remained essentially unchanged with ICG-001 treatment (Fig. 3B Bottom).
To further confirm the specific inhibition of the β-catenin/CBP interaction by ICG-001, we used C2C12 myoblasts in which, unlike SW480 cells, the levels of CBP and p300 associated with β-catenin are approximately equivalent. ICG-001 blocked the β-catenin/CBP interaction without interfering with the interaction between β-catenin and p300 (Fig. 3C Top). The coimmunoprecipitation of CBP with β-catenin was completely inhibited by ICG-001 (25 μM) treatment (Fig. 3C Top, compare lanes 1 and 2). Strikingly, the coimmunoprecipitation of p300 was unaffected by ICG-001 (Fig. 3C Top, compare lanes 3 and 4). CBP, p300, and β-catenin levels remained essentially unchanged (Fig. 3C Bottom).
Further confirmation of CBP as the molecular target of ICG-001 was obtained by using siRNA. We designed siRNA sequences for CBP and p300 that selectively knocked down the targeted coactivator protein (Fig. 3D Bottom). These coactivator-specific siRNAs were cotransfected with TOPFLASH into SW480 cells. Specific down-regulation of CBP protein levels dramatically reduced the inhibitory effects of ICG-001 on TOPFLASH activity (Fig. 3D Top). As anticipated, down-regulation of p300 protein levels had no effect on the inhibitory potency of ICG-001 (Fig. 3D Top). This demonstrates that depletion of CBP, the molecular target of ICG-001, selectively affects ICG-001 inhibition of TCF/β-catenin-mediated transcription.
ICG-001 Treatment Reduces the Steady-State Levels of Survivin and Cyclin D1 RNA and Protein. To further address the selectivity of ICG-001, we performed cDNA microarray analysis by using the Clontech Atlas Human Cancer 1.2 Array (no. 7851-1) (M.K., unpublished data). The data demonstrated that ICG-001 had a very selective effect on global gene transcription. After 8 h of treatment of SW480 cells with 25 μM ICG-001, ≈2% of the genes analyzed were up-regulated >2-fold, whereas only ≈0.3% of the genes were down-regulated by >50%. Interestingly, two of the genes down-regulated are S100A4 and survivin, the number one and four mRNAs up-regulated in cancer cells, respectively (22).
Survivin belongs to the Inhibitor of Apoptosis gene family that counteracts cell death and controls mitotic progression (23). Survivin is present during embryonic development but is virtually undetectable in terminally differentiated adult tissues (23, 24). Overexpression of survivin has been reported in many types of cancers, including colorectal, stomach, lung, pancreatic, prostate, and breast, and is generally associated with a poor prognosis (25-31).
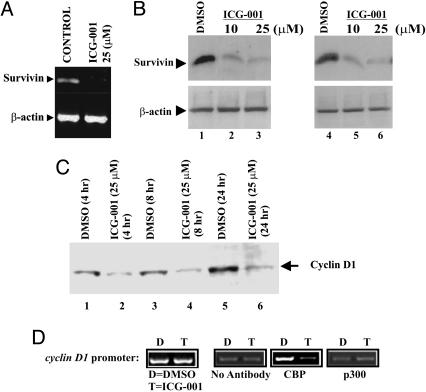
It has been shown recently that TCF/β-catenin stimulated a 6- to 12-fold increase in survivin gene expression in colorectal cancer cells (28, 29), and mutagenesis experiments demonstrated the importance of TCF-binding elements in the survivin promoter (28). In particular, we were interested in the down-regulation of the survivin message, because survivin has been shown to inhibit caspase activation (31). To confirm the microarray results, we investigated the endogenous mRNA (Fig. 4A) and protein levels of survivin (Fig. 4B). As shown in Fig. 4 A and B, ICG-001 potently reduced the steady-state levels of survivin mRNA and protein in treated colorectal cancer cells.
Fig. 4.
ICG-001 inhibits the expression of the TCF/β-catenin-mediated genes survivin and cyclin D1. (A) Inhibition of survivin gene transcription was measured by semiquantitative RT-PCR in SW480 cells treated with ICG-001 (25 μM, 24 h) or DMSO. (B) Survivin immunoblots from SW480 and HCT116 cells treated with ICG-001 (10 and 25 μM, 24 h) or DMSO. (C) Cyclin D1 immunoblots from SW480 cells treated with 25 μM ICG-001 or DMSO. (D) Chromatin immunoprecipitation assay using the cyclin D1 promoter. After 8-h treatment with ICG-001 (25 μM) or DMSO, promoter occupancy was evaluated by using CBP-(AC-22) or p300-(C-20) specific antibodies.
The cyclin D1 gene is inappropriately expressed in many different tumor types and is known to be a direct target of the TCF/β-catenin pathway (12, 13). To determine the effect of ICG-001 on the expression of another direct target gene of the TCF/β-catenin pathway, we evaluated cyclin D1 protein levels in cells treated with ICG-001. Whole-cell lysates of treated and untreated SW480 cells were immunoblotted, as shown in Fig. 4C, and there was a clear reduction in cyclin D1 upon treatment with ICG-001 (25 μM) as early as 4 h posttreatment. Using the chromatin immunoprecipitation assay, we also show that ICG-001 decreases cyclin D1 message levels via selective inhibition of the β-catenin/CBP interaction at the endogenous cyclin D1 promoter (Fig. 4D). We conclude that ICG-001 specifically interferes with TCF/β-catenin mediated expression of both survivin and cyclin D1.
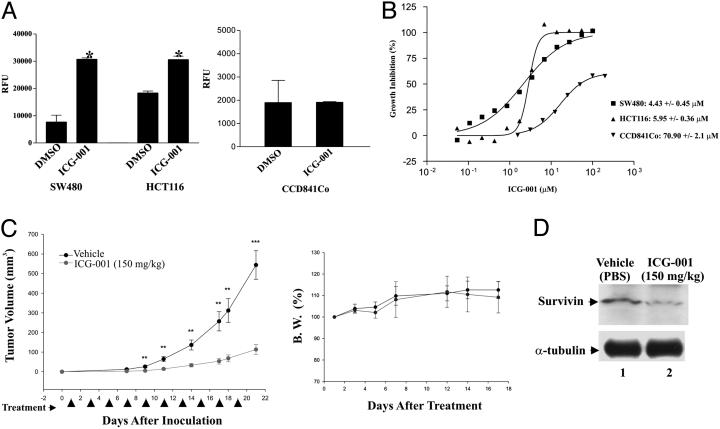
Therapeutic Potential of ICG-001. We have shown that ICG-001 down-regulate survivin, a known inhibitor of caspase activation (31). Caspases are cysteine proteases that are activated in cells undergoing apoptosis. We compared caspase activation in SW480 and HCT116 cells, which carry mutations in APC or β-catenin, respectively, to CCD-841Co normal colonic epithelial cells. All cells were treated with either ICG-001 (25 μM) or DMSO. Twenty-four hours posttreatment, caspase-3/7 activity was measured. ICG-001 significantly increased caspase activity in both SW480 and HCT116 cell lines but not in CCD-841Co (Fig. 5A).
Fig. 5.
ICG-001 is selectively cytotoxic to colon carcinoma cell lines and effective in vivo. (A) SW480, HCT116 cells and the normal colonic epithelial cells CCD-841Co were treated with ICG-001 (25 μM) or DMSO. Caspase 3/7 enzymatic activity was measured. Error bars represent mean ± SD. *, P < 0.05. (B) SW480 (▪), HCT116 (▴), and CCD-841Co (▾) cells were treated with increasing amounts of ICG-001, followed by MTS cell viability assay. (C) i.v administration (150 mg/kg) or vehicle (PBS) into 7-week-old female BALB/c nude mice for 22 days. Tumor growth inhibition (TGI) (Right) was calculated by using the following equation: % TGI = 100 × [1-(mean final tumor volume of treated group/mean fi-nal volume of vehicle group)]. Effect of treatment on body weight (Left). Error bars are mean ± SD; n = 10 for each treatment group; *, P < 0.05; **, P < 0.01; ***, P < 0.001. (D) Treated or vehicle-treated tumors were removed from xenografted animals and immunoblotted for Survivin.
To be considered a lead compound for therapeutic development for colorectal cancer, ICG-001 should demonstrate growth inhibitory effects and selective cytotoxicity in colon carcinoma cells. We tested the efficacy of ICG-001 in SW480, HCT116, and CCD-841Co cells. The results demonstrate the selective growth inhibitory effects of ICG-001 in cancer cells (Fig. 5B). We conclude that ICG-001 is cytostatic at lower concentrations and at higher concentrations increases caspase activity and is selectively cytotoxic in colon carcinoma cells.
We next evaluated a water-soluble analog of ICG-001 in vivo in two mouse models of cancer. The Min mouse, which has a germ-line mutation in one allele of the APC tumor suppressor gene, is a well characterized model for human familial adenomatous polyposis (32). Administration of the analog for 9 weeks reduced the formation of colon and small intestinal polyps by 42% as effectively as the nonsteroidal antiinflammatory agent Sulindac (Table 1), which has consistently demonstrated efficacy in this model (33). No overt toxicity was detected throughout the course of treatment. In the SW620 nude mouse xenograft model of tumor regression (17), 150 mg/kg, i.v. of the analog demonstrated a dramatic reduction in tumor volume over the 19-day course of treatment (Fig. 5C Left), with no mortality or weight loss (Fig. 5C Right). We conclude that ICG-001 is efficacious in vivo.
Table 1. Min mouse model.
| Polyp number (mean ± SE)
|
||||||
|---|---|---|---|---|---|---|
| Group | Number of mice | Small intestine | Colon | Total | P (total vs. VH) | % inhibition vs. VH |
| Wild type | 8 | 0.0 ± 0.0 | 0.0 ± 0.0 | 0.0 ± 0.0 | — | — |
| Vehicle | 10 | 67.4 ± 5.0 | 1.8 ± 0.5 | 69.2 ± 4.8 | — | — |
| ICG-001a (300 mg/kg) | 8 | 39.4 ± 3.5 | 1.1 ± 0.4 | 40.3 ± 3.6 | <0.001 | 42 |
| Sulindac (160 ppm) | 6 | 37.8 ± 3.7 | 0.4 ± 0.2 | 38.3 ± 3.5 | <0.001 | 45 |
Polyp numbers were scored after treatment with the water-soluble analog or Sulindac. Data are shown as mean ± standard error. Differences were considered significant if P < 0.005.
To demonstrate the molecular mechanism in vivo, we evaluated survivin levels in the SW620 xenografted tumors taken from the treated mice. Real-time RT-PCR demonstrated a decrease in survivin message (ΔΔCT = +0.56). The Survivin protein level was also dramatically decreased, as judged by immunoblotting (Fig. 5D). We conclude that the in vivo efficacy correlates with the inhibition of TCF/β-catenin-mediated expression of survivin.
Discussion
There is compelling evidence that misregulation of the β-catenin pathway is involved in the development and progression of many cancers (2-4). Here, we describe the discovery of ICG-001, a small molecule that selectively inhibits TCF/β-catenin transcription in a CBP-dependent fashion. ICG-001 selectively blocked the β-catenin/CBP interaction without interfering with the β-catenin/p300 interaction. Using siRNA against CBP and p300, we demonstrated that depletion of CBP, the molecular target of ICG-001, selectively affects ICG-001 inhibition of TCF/β-catenin-mediated transcription.
Binding studies using ICG-001 led to the discovery of a minimal region of interaction at the NH2 terminus of CBP (amino acids 1-111). ICG-001 did not bind to the homologous sequence in p300. ICG-001 selectively blocked the interaction between CBP (1-111) and β-catenin, without interfering with the p300 (1-111)/β-catenin interaction (Fig. 2B). Our mapping studies demonstrated that the interacting domains of both CBP and p300 with the C terminus of β-catenin reside within the N-terminal 110 residues of the coactivators. Interestingly, this region also contains the CBP/p300-binding site for retinoic acid (RA) receptors, RXR/RAR (34). It has been shown that RA treatment inhibits TCF/β-catenin-mediated transcription (35).
Finally, ICG-001 increases caspase activity in colon carcinoma cell lines (SW480 or HCT116) but not in normal colonic epithelial cells (CCD-841Co). The increased caspase activity is manifested in selective cytotoxicity in colorectal cancer cells. cDNA microarray analysis demonstrated that ICG-001 had a very selective effect on gene transcription. Interestingly, two of the most highly up-regulated mRNAs in cancer cells (survivin and S100A4) (22) are down-regulated by ICG-001. Down-regulation of survivin is notable, because survivin has been shown to inhibit caspase activation (31). We also demonstrated that ICG-001 reduces endogenous survivin levels in a TCF/β-catenin fashion, both in vitro (Fig. 4 A and B) and in vivo (Fig. 5D). We believe that the observed decrease in survivin is responsible for the induction of apoptosis of the cancer cells, leading to tumor reduction in vivo.
The exquisite selectivity of ICG-001 is at first surprising, given that it interacts with CBP, a coactivator protein used by a wide array of transcription factors (36). We attribute this selectivity to two elements, the interaction of the small molecule ICG-001 (Mr 548) with the enormous coactivator protein CBP (≈300 kDa) blocks only a very small percentage of the CBP surface. The selectivity of ICG-001 is further enhanced by its specific binding to CBP, but not the highly homologous coactivator p300 (63% identity). The functions of CBP and p300 have been described as redundant in several studies (36), and their expression pattern during mouse development is almost identical (37). However, it is becoming increasingly clear from knockout studies that these highly homologous coactivators are not completely redundant under physiological conditions and are responsible for distinct transcriptional programs (38-41). In this regard, ICG-001 and related p300 selective analogs are providing unique chemogenomic tools to investigate gene selective coactivator usage (M.K., unpublished results).
Elegant structural studies of the interactions between β-catenin and TCF (42-44) offered an a priori attractive mode for inhibition of this pathway. Recently, this molecular interaction was successfully targeted in a small molecule screen to provide a series of inhibitors of TCF/β-catenin-mediated transcription (45). However, concerns arise about the development of specific inhibitors due to the diverse partners besides TCF (e.g., APC and E-cadherin) that also bind to the central Arm repeats of β-catenin (6, 45). The elegant selectivity of ICG-001, through its specific inhibition of the β-catenin/CBP interaction via binding to CBP, provides an alternative mechanism to inhibit a subset of TCF/β-catenin-mediated transcription. The specificity of ICG-001, its ability to2 inhibit survivin expression, thereby selectively activating apoptotic caspases in cancerous but not normal colonic epithelial cells, and its activity in both the Min and nude mouse xenograft models (17, 33), are all very positive signs for its potential therapeutic use in colon cancer. We anticipate that aberrant activation of β-catenin/CBP transcription is a critical event in the development of a broad array of cancers, and that its inhibition offers a previously unappreciated therapeutic approach.
Acknowledgments
We gratefully acknowledge Drs. Richard Goodman (Vollum Institute, Oregon Health Science Center, Portland, OR) and David Livingston (Dana-Farber Cancer Institute, Boston) for generous gifts of CBP and p300 expression vectors.
Abbreviations: CBP, cyclic AMP response element-binding protein; TCF, T cell factor; APC, adenomatous polyposis coli; CRE, cyclic AMP-response element; siRNA, small interfering RNA.
References
- 1.Wodraz, A. & Nusse, R. (1998) Annu. Rev. Cell Dev. Biol. 14, 59-88. [DOI] [PubMed] [Google Scholar]
- 2.Morin, P. J. (1999) BioEssays 21, 1021-1030. [DOI] [PubMed] [Google Scholar]
- 3.Oving, I. M. & Clevers, H. C. (2002) Eur. J. Clin. Invest. 32, 448-457. [DOI] [PubMed] [Google Scholar]
- 4.Moon, R. T., Bowerman, B., Boutros, M. & Perrimon, N. (2002) Science 296, 1644-1646. [DOI] [PubMed] [Google Scholar]
- 5.Gottardi, C. J. & Gumbiner, B. M. (2001) Curr. Biol. 11, R792-R794. [DOI] [PubMed] [Google Scholar]
- 6.Huber, A. H. & Weis, W. I. (2001) Cell 105, 391-402. [DOI] [PubMed] [Google Scholar]
- 7.Behrens, J. (2000) Ann. N.Y. Acad. Sci. 910, 21-33. [DOI] [PubMed] [Google Scholar]
- 8.Brantjes H., Barker, N., van Es, J. & Clevers, H. (2002) Biol. Chem. 383, 255-261. [DOI] [PubMed] [Google Scholar]
- 9.Hecht, A., Vleminckx, K., Stemmler, M.P., van Roy, F. & Kemler, R. (2000) EMBO J. 19, 1839-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takemaru, K. I. & Moon, R. T. (2000) J. Cell Biol. 149, 249-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fearnhead, N. S., Wilding, J. L. & Bodmer, W. F. (2002) Br. Med. Bull. 64, 27-43. [DOI] [PubMed] [Google Scholar]
- 12.Shtutman, M., Zhurinsky, J., Simcha, I., Albanese, C., D'Amico, M., Pestell, R. & Ben-Ze'ev, A. (1999) Proc. Natl. Acad. Sci. USA 96, 5522-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tetsu, O. & McCormick, F. (1999) Nature 398, 422-426. [DOI] [PubMed] [Google Scholar]
- 14.He, T. C., Sparks, A. B., Rago, C., Hermeking, H., Zawel, L., da Costa, L. T., Morin, P. J., Vogelstein, B. & Kinzler, K. W. (1998) Science 281, 1509-1512. [DOI] [PubMed] [Google Scholar]
- 15.Kim, J. S., Crooks, H., Foxworth, A. & Waldman, T. (2002) Mol. Cancer Ther. 1, 1355-1359. [PubMed] [Google Scholar]
- 16.Korinek, V., Barker, N., Morin, P.J., van Wichen, D., de Weger, R., Kinzler, K.W., Vogelstein, B. & Clevers, H. (1997) Science 275, 1784-1787. [DOI] [PubMed] [Google Scholar]
- 17.Lee, C. W., Hong, D. H., Sang, B. H., Jung, S. H., Kim, H. C., Fine, R. L., Lee, S. H. & Kim, H. M. (2002) Biochem. Pharmacol. 64, 473-480. [DOI] [PubMed] [Google Scholar]
- 18.Spencer, V. A., Sun, J.-M., Li, L. & Davie, J. R. (2003) Methods 31, 67-75. [DOI] [PubMed] [Google Scholar]
- 19.Ogbu, C. O., Qabar, M. N., Boatman, P. D., Urban, J., Meara, J. P., Ferguson, M. D., Tulinsky, J., Lum, C., Babu, S., Blaskovich, M. A., et al. (1998) Bioorg. Med. Chem. Lett. 8, 2321-2326. [DOI] [PubMed] [Google Scholar]
- 20.Eguchi, M., Lee, M. S., Nakanishi, H., Stasiak, M., Lovell, S. & Kahn, M. (1999) Am. Chem. Soc. 102, 22031-22032. [Google Scholar]
- 21.Kim, H. O. & Kahn, M. (2000) Res. Adv. Organ. Chem. 1, 43-59. [Google Scholar]
- 22.Velculescu, V. E., Madden, S. L., Zhang, L., Lash, A. E., Yu, J., Rago, C., Lal, A., Wang, C. J., Beaudry, G. A., Ciriello, K. M., et al. (1999) Nat. Genet. 23, 387-388. [DOI] [PubMed] [Google Scholar]
- 23.Chiou, S. K., Jones, M. K. & Tarnawski, A. S. (2003) Med. Sci. Mon. 9, PI25-PI29. [PubMed] [Google Scholar]
- 24.Altieri, D. C. (2001) Trends Mol Med. 7, 542-547. [DOI] [PubMed] [Google Scholar]
- 25.Tanaka, K., Iwamoto, S., Gon, G., Nohara, T., Iwamoto, M. & Tanigawa, N. (2000) Clin. Cancer Res. 6, 127-134. [PubMed] [Google Scholar]
- 26.Lu, C.-D., Altieri, D. C. & Tanigawa, N. (1998) Cancer Res. 58, 1808-1812. [PubMed] [Google Scholar]
- 27.Ambrosini, G., Adida, C. & Altieri, D. C. (1997) Nat. Med. 3, 917-921. [DOI] [PubMed] [Google Scholar]
- 28.Kim, P. J., Plescia, J., Clevers, H., Fearon, E. R. & Altieri, D. C. (2003) Lancet 362, 205-209. [DOI] [PubMed] [Google Scholar]
- 29.Kawasaki, H., Altieri, D. C., Lu, C. D., Toyoda, M., Tenjo, T. & Tanigawa, N. (1998) Cancer Res. 58, 5071-5074. [PubMed] [Google Scholar]
- 30.Adida, C., Recher, C., Raffoux, E., Daniel, M. T., Taksin, A. L., Rousselot, P., Sigaux, F., Degos, L., Altieri, D. C. & Dombret, H. (2000) Blood 96, 1921-1925. [DOI] [PubMed] [Google Scholar]
- 31.Tamm, I., Wang, Y., Sausville, E., Scudiero, D. A., Vigna, N., Oltersdorf, T. & Reed, J. C. (1998) Cancer Res. 58, 5315-5320. [PubMed] [Google Scholar]
- 32.Moser A. R., Luongo, C., Gould, K. A., McNeley, M. K., Shoemaker, A. R. & Dove, W. F. (1995) Eur. J. Cancer A, 1061-1064. [DOI] [PubMed] [Google Scholar]
- 33.Corpet, D. E. & Pierre, F. (2003) Cancer Epidemiol. Biomarkers Prev. 12, 391-400. [PMC free article] [PubMed] [Google Scholar]
- 34.Easwaran, V., Pishvaian, M., Salimuddin & Byers, S. (1999) Curr. Biol. 2, 1415-1418. [DOI] [PubMed] [Google Scholar]
- 35.Minucci, S. & Pelicci, P. G. (1999) Semin. Cell Dev. Biol. 10, 215-225. [DOI] [PubMed] [Google Scholar]
- 36.Goodman, R. H. & Smolik, S. (2000) Genes Dev. 14, 1553-1577. [PubMed] [Google Scholar]
- 37.Patanen, A., Motoyama, J. & Hui, C. C. (1999) Int. J. Dev. Biol. 43, 487-494. [PubMed] [Google Scholar]
- 38.Kung, A. L., Rebel, V. I., Bronson, R. T., Ch'ng, L. E., Sieff, C. A., Livingston, D. M. & Yao, T. P. (2000) Genes Dev. 14, 272-277. [PMC free article] [PubMed] [Google Scholar]
- 39.Eid, J. E., Kung, A. L., Scully, R. & Livingston, D. M. (2000) Cell 102, 839-848. [DOI] [PubMed] [Google Scholar]
- 40.Rebel, V. I., Kung, A. L., Tanner, E. A., Yang, H., Bronson, R. T. & Livingston, D. M. (2002) Proc. Natl. Acad. Sci USA 99, 14879-14894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roth, J. F., Shikama, N., Henzen, C., Desbaillets, I., Lutz, W., Marino, S., Wittwer, J., Schorle, H., Gassmann, M. & Eckner, R. (2003) EMBO J. 19, 5186-5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Graham, T. A., Weaver, C., Mao, F., Kimelman, D. & Xu, W. (2000) Cell 103, 885-896. [DOI] [PubMed] [Google Scholar]
- 43.Graham, T. A., Ferkey, D. M., Mao, F., Kimelman, D. & Xu, W. (2001) Nat. Struct. Biol. 8, 1048-1052. [DOI] [PubMed] [Google Scholar]
- 44.Poy, F., Lepourcelet, M., Shivdasani, R. A. & Eck, M. .J. (2001) Nat. Struct. Biol. 8, 1053-1057. [DOI] [PubMed] [Google Scholar]
- 45.Lepourcelet, M., Chen, Y. N., France, D. S., Wang, H., Crews, P., Petersen, F., Bruseo, C., Wood, A. V. & Shivdasani, R. A. (2004) Cancer Cell 5, 91-102. [DOI] [PubMed] [Google Scholar]