Abstract
Nine voltage-gated sodium channels are expressed in complex patterns in mammalian nerve and muscle. Three channels, Nav1.7, Nav1.8, and Nav1.9, are expressed selectively in peripheral damage-sensing neurons. Because there are no selective blockers of these channels, we used gene ablation in mice to examine the function of Nav1.7 (PN1) in pain pathways. A global Nav1.7-null mutant was found to die shortly after birth. We therefore used the Cre-loxP system to generate nociceptor-specific knockouts. Nav1.8 is only expressed in peripheral, mainly nociceptive, sensory neurons. We knocked Cre recombinase into the Nav1.8 locus to generate heterozygous mice expressing Cre recombinase in Nav1.8-positive sensory neurons. Crossing these animals with mice where Nav1.7 exons 14 and 15 were flanked by loxP sites produced nociceptor-specific knockout mice that were viable and apparently normal. These animals showed increased mechanical and thermal pain thresholds. Remarkably, all inflammatory pain responses evoked by a range of stimuli, such as formalin, carrageenan, complete Freund's adjuvant, or nerve growth factor, were reduced or abolished. A congenital pain syndrome in humans recently has been mapped to the Nav1.7 gene, SCN9A. Dominant Nav1.7 mutations lead to edema, redness, warmth, and bilateral pain in human erythermalgia patients, confirming an important role for Nav1.7 in inflammatory pain. Nociceptor-specific gene ablation should prove useful in understanding the role of other broadly expressed genes in pain pathways.
Voltage-gated sodium channels underlie the electrical excitability demonstrated by mammalian nerve and muscle (1). The complexity of voltage-gated sodium channel subtype expression suggests that particular isoforms may have specialized roles in different physiological systems. In the absence of subtype-specific blockers, analysis of null mutant mice has proved informative (2). So far, four α-subunits and two different β-subunits have been deleted in transgenic mice. Nax, a non-voltage-gated sodium channel homologue, has been shown to play an important role in salt homeostasis (3), whereas Nav1.8 seems to play a specialized role in pain pathways (4). Nav1.2 deletion leads to massive brainstem neuronal apoptosis and perinatal death (5), and loss of Nav1.6 in natural “med” mutants has been shown to lead to ataxia, dystonia, and paralysis (6). The deletion of β-1 or β-2 subunits leads to a complex phenotype resulting in seizures and epileptogenic activity (7). Naturally occurring cardiac Nav1.5 mutations are associated with epilepsy and sudden death in humans with Brugada syndrome (8).
Nav1.7 was first cloned from the pheochromocytoma PC12 cell line (9, 10). Its presence at high levels in the growth cones of small-diameter neurons (9, 11) suggests that it is likely to play some role in the transmission of nociceptive information. Immunochemical studies of sensory neurons in guinea pigs supports the view that Nav1.7 is associated with nociceptors (12). Recently, a human heritable pain condition, primary erythermalgia, has been shown to map to Nav1.7 (13). We therefore examined the role of Nav1.7 in pain pathways by generating null mutant mice. The Cre-recombinase-loxP system allows tissue-specific and/or inducible gene deletion (14). By driving Cre with a tissue-specific promoter, gene deletion can be effected in particular tissues. This refinement of null mutant technology has proved useful for functional studies (15-17). To elucidate the physiological roles of Nav1.7, we generated “floxed” mice in which loxP sites were inserted in introns flanking exons 14 and 15, which encode most of domain II. Deletions of S4 voltage sensors have been shown to abolish sodium channel function in Nav1.8-null mutants (4). To ablate Nav1.7 globally, we used a general deletor mouse in which Cre recombinase is expressed in all tissues (18). Next, we used a nociceptor-specific Cre mouse to delete Nav1.7 only in damage-sensing sensory neurons. Nociceptor-specific Cre recombinase expression was achieved by driving this gene with the Nav1.8 promoter. The Nav1.8 gene is expressed predominantly in nociceptive sensory neurons and is completely absent in tissue other than sensory neurons (19). Heterozygous null mutant Nav1.8 mice are identical to WT mice (4), suggesting that “knocking in” Cre recombinase to the Nav1.8 locus is unlikely to have deleterious effects in heterozygous mice that express single alleles of Nav1.8 and Cre. These mice were constructed and analyzed and showed no phenotypic deficits, although expressing Cre recombinase in an identical pattern to Nav1.8 (20). Crossing these mice with floxed Nav1.7 mice resulted in viable animals that showed specific deletion of Nav1.7 only in a subset of sensory neurons. Here we present an analysis of the consequences of deleting Nav1.7 on pain behavior in mice.
Methods
Gene Targeting: Nav1.8 Cre Construct. A rat cDNA probe encoding Nav1.8 exons 10-12 was used to identify genomic clones from a RCPI-22 129S6/SvEvTac mouse bacterial artificial chromosome (BAC) library. The targeting vector was derived from two adjacent BamHI flanked fragments, 6.5 and 8.5 kb in size, containing exons 1-3 of the Nav1.8 gene. A PCR product encoding ≈400 bp upstream of the Nav1.8 translational start site and the first 45 bp of the Cre gene was obtained by PCR using the 8.5-kb fragment as a template (primers 5′-AGCAAGAGGCAAATCATAGTCAGC-3′ and 5′-TCGACCGGTAATGCAGGCAAATTTTGGTGTACGGTCAGTAAATTGGACATCTTCTCATTCTTCTTGGGGAAGGATTTACA-3′). This product was cloned into a modified pBSSK-vector by using XmaI and AgeI. The remainder of the Cre gene, derived from pBS500 (1), was cloned by using AgeI and MluI. The remainder of the 5′ arm of homology was cloned by using XmaI followed by BstEII digestion and religation. A FRT-site flanked (flrted) neomycin-resistance (neor) cassette, for positive selection, was cloned in a separate modified pBSSK-vector by using SacI and SalI. Two polyadenylation signals were cloned by using NsiI and HindIII. The 3′-UTR was derived by PCR from a genomic BAC clone (primers 5′-AGGGATCC AACTCGAGAGACACTCCAGCATGCACGGGGCG-3′ and 5′-CGCGGATCCGGCCGACCCTCAGGTATTGTCCGG-3′) and cloned by using BamHI. The 3′ arm of homology was excised from the 6.5-kb Nav1.8 genomic subclone and cloned by using BamHI and XmaI.
The two fragments were cloned into a herpes simplex virus thymidine kinase-containing vector (4) by using NotI and SacII. The vector was linearized by using NotI and electroporated into 129-derived embryonic stem cells. A total of 192 clones survived G418 selection, and they were screened for homologous recombination by using Southern blotting. DNA was digested with BamHI and screened with 3′ and Cre probes. Seven clones were correctly targeted, and they were injected into C57BL/6J blastocysts to generate chimeras. F1 heterozygotes were crossed to FLPe deletor animals to excise the positive selection marker.
Nav1.7 Construct. An RCPI-22 129S6/SvEvTac mouse BAC library was screened with a probe representing a 900-bp BglI-BamHI fragment from Nav1.7 intron 7. Four positive BACs were analyzed further with probe for exons 14 and 15, and only one was positive. Two exon-14-15-containing genomic fragments were then subcloned from the positive BAC into pBSSK-, a 7.5-kb EcoRI, and an 8.2-kb ApaI fragment. The targeting vector was constructed from three genomic regions obtained from the subclones. The 5′ arm is a 4.7-kb fragment from the SacI site after exon 11 to the HindIII before exon 14. The floxed sequence is a 2.9-kb HindIII to HindIII fragment containing exons 14 and 15. The 3′ arm is a 4.5-kb fragment from the HindIII site after exon 15 to the NsiI site before exon 16.
The loxP site was isolated as a 111-bp EcoRI-PstI fragment from vector Pl2-neor (a gift from R. Schoepfer, University College London) and cloned in a modified pBSSK-. The 2.9-kb HindIII to HindIII fragment containing exons 14 and 15 was inserted in the HindIII site of the vector-containing loxP site. The correct orientation then was determined by an EcoRI digest, and the product was called pPN1415lox. A loxP site and the frt-flanked neor cassette were joined in a modified pBSSK-. The loxP- neor fragment was then fused to the 3-kb 1415lox fragment in pBSSK-. The product was named pPN1415loxneolox.
A pBSSK- with a modified polylinker was made by inserting the oligos MAN-PL2s (GCGGCCGCCCGGGAATTCAATTGAAGCTTGGGCCCGCTAGCACTAGTAT) and MANPL2a (CGATACTAGTGCTAGCGGGCCCAAGCTTCAATTGAATTCCCGGG CGGCCGCAGCT) into the SacI ClaI sites; the product is called pMANPL2. The 5′ Nav1.7 homology arm was generated in pMANPL2 from two genomic fragments. These are the 1-kb HindIII → MfeI and the 3.9-kb MfeI → XmaI fragments; the product is called pPN15′arm. The part of exon 11 present in pPN15′arm was deleted by digesting with SacI-XmaI followed by T4 blunting; the product is called pPN15′armΔ. The XhoI → SpeI fragment from p1415loxneolox was inserted into XhoI-NheI of pPN15′armΔ. The product is called pPN15′-Neo. The 3′ Nav1.7 homology arm was generated in pBSSK- from two genomic fragments from the EcoRI subclone. These are the 1-kb HindIII → ApaI and the 3.7-kb ApaI → NsiI fragments. They were inserted into the HindIII → PstI of pBSSK-. The product is called pPN13′arm. Part of the pBSSK- polylinker in pPN13′arm was then deleted. pPN13′arm was digested with NotI-XmaI, followed by blunting with T4. This product was named pPN13′armΔ. The final construct was put together from three fragments, which are as follows: (i) backbone, a modified pBSSK- containing the herpes simplex virus thymidine kinase cassette digested with SacII+NotI; (ii) pPN13′armΔ, 4.5-kb SacII → XhoI fragment; and (iii) pPN15-1415loxneolox, 9-kb XhoI → NotI fragment.
The 3′ probe, a deletion clone, was generated from the EcoRI subclone. Digesting it with PstI+NsiI followed by ligation left only a 700-bp genomic insert. A 600-bp probe was cut out by using XmaI+MfeI.
The I12 internal probe was a 450-bp PacI-KpnI fragment from the ApaI subclone.
The complete targeting vector was linearized by using NotI and electroporated into 129-derived embryonic stem cells. The 192 clones that survived G418 selection were screened for homologous recombination by using Southern blotting. Five positive clones were identified by using EcoRI digestion and a 3′ probe. The clones were further analyzed with ApaI digests and an internal probe to intron 12. Two clones were correctly targeted and injected into C57BL/6J blastocysts to generate chimeras. F1 heterozygotes were crossed to FLPe deletor animals to excise the positive selection marker (22).
Electrophysiological and Immunochemical Studies. These studies were carried out as described previously (20) by using voltage-clamp recordings from single dorsal root ganglion (DRG) neurons of up to 25 μm in diameter in culture by using the whole cell variant of the patch-clamp technique. Measurements of electrical input into wide dynamic range neurons in spinal cord neurons were carried out as described in ref. 4. Polyclonal antisera to Nav1.8 and antibodies to other sensory neurons markers were used as described in refs. 19 and 20.
Behavioral Analysis. All tests were approved by the United Kingdom Home Office Animals (Scientific Procedures) Act 1986 and were carried out as described in ref. 4. Mice were between 10 and 20 weeks old when tested.
Results
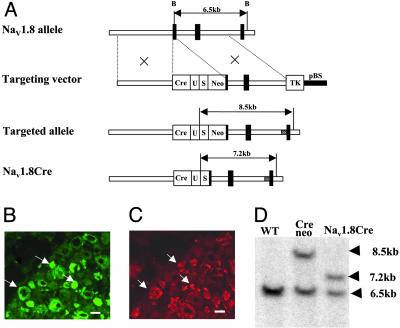
Generation of Transgenic Mice. The strategy for generating the Cre targeting construct is shown in Fig. 1. Cre recombinase with the 3′-UTR of Nav1.8 was knocked into the Nav1.8 initiator methionine so that the translational start-site of Nav1.8 was substituted by Cre recombinase (21). After screening for homologous recombination in 129/Sv embryonic stem cells selected for the presence of the neor cassette, a single positive cell line was used to generate transgenic mice, which were analyzed for germ-line transmission by Southern blotting with probes shown in Fig. 1B. BamHI genomic digests distinguish the WT allele (6.5 kb) from the targeted allele (8.5 kb). After a mouse line had been established expressing Cre at the Nav1.8 locus, the neor cassette was excised by crossing the mice with the universal Flp deletor mouse before analysis of the expression of the Cre recombinase (22). BamHI restriction fragments of 7.2 kb were then detected (Fig. 1).
Fig. 1.
Generation of Nav1.8Cre mice. (A) Diagram of the native Nav1.8 allele, Nav1.8Cre targeting construct, and Nav1.8Cre-targeted allele before and after excision of neor. (B and C) Histological localization of Nav1.8Cre-mediated β-galactosidase activity. Serial 10-μm sections from 4% paraformaldehydefixed Nav1.8Cre+/-,R26R+/- animals were stained with antibodies to β-galactosidase (B) and with anti-Nav1.8 polyclonal antisera (C). Arrows highlight double-labeled cells. (Scale bars, 50 μm.) (D) Southern blot with BamHI and external probe confirms correct targeting and excision of neor. The 6.5-kb WT band is seen in all lanes, and the 8.5- and 7.3-kb bands represent the targeted allele before (+neor) and after (-neor) excision of neor, respectively.
Analysis of the expression pattern of functional Cre recombinase used ROSA26 reporter mice in which β-galactosidase is expressed as a result of Cre activity (23). Functional Cre was expressed in a pattern identical to that of Nav1.8. Expression began at embryonic day 14 in small-diameter neurons in dorsal root, trigeminal, and nodose ganglia but was absent in nonneuronal and CNS tissues. This pattern of expression was unaltered in adult animals.
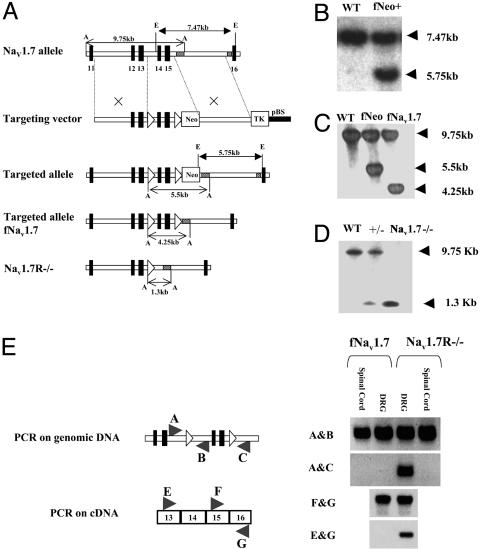
Mice with floxed Nav1.7 (fNav1.7) were constructed by inserting loxP sites at HindIII sites in introns flanking exons 14 and 15 with a neor cassette downstream of the 3′ loxP site. Homologous recombination in 129/Sv embryonic stem cells used a fragment from the SacI site 5′ of exon 11 to the NsiI site before exon 16. Transgenic mice were screened by Southern blotting with EcoRI digests and an external probe as shown in Fig. 2. The neor cassette was excised by crossing with a Flp-deletor strain and again analyzed by Southern blotting with ApaI digests and an internal probe as shown in Fig. 2C.
Fig. 2.
Generation and analysis of Nav1.7 floxed mice. (A) Structure of the native Nav1.7 allele, Nav1.7 targeting construct, fNav1.7 allele, fNav1.7 allele after excision of neor, and Nav1.7 knockout allele. (B) Southern blotting with EcoRI and the external probe confirms correct targeting. (C) Southern blotting with ApaI and internal probe confirms the removal of neor cassette. (D) Southern blotting confirms the deletion of exons 14 and 15 in Nav1.7-/-. (E) PCR was used to detect exon 14-15 deletion in genomic and cDNA from DRG but not spinal cord in Nav1.7R-/-.
Nav1.7-Null Mutants. The universal Cre-deletor mouse was used to generate conventional Nav1.7-/- mice by crossing with fNav1.7 mice. Heterozygous Nav1.7 matings were carried out to compare global nulls and WT mice. Southern blots demonstrated the deletion of exons 14 and 15 in genomic DNA. Of 92 pups that survived, 72% were heterozygotes and the rest were Nav1.7 WTs. Nav1.7-/- pups all died shortly after birth, apparently because of a failure to feed. Thus, deleting Nav1.7 in all sensory and sympathetic neurons causes a lethal perinatal phenotype.
We sequentially crossed fNav1.7 mice with Nav1.8Cre mice to eventually generate animals that were homozygous for the fNav1.7 allele and heterozygous for Nav1.8Cre. By further crossing these animals with homozygous fNav1.7 mice, litters comprising approximately equal numbers of fNav1.7 mice and tissue-restricted Nav1.7-/- (Nav1.7R-/-) mice were generated. In this way, we could compare the behavior of DRG-null animals (Nav1.7R-/-) with littermate control animals (fNav1.7). We assessed the deletion of Nav1.7 in two ways. First, we used PCR primers to examine genomic DNA from Nav1.7R-/- mice and fNav1.7 littermates. A PCR product showed that adjacent exons 13 and 16 were present in DNA from Nav1.7R-/- DRG but not spinal cord. fNav1.7 DRG genomic DNA did not generate such a product. cDNA copied from Nav1.7R-/- DRG mRNA also showed a similar product, which was absent in fNav1.7 DRG mRNA. Excision of Nav1.7 exons 14 and 15 thus occurred only in DRG.
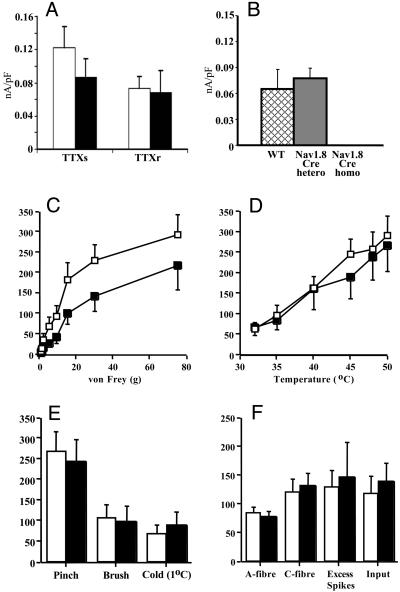
We examined the functional consequences of deleting Nav1.7 on sodium channel expression in DRG neurons in culture by using electrophysiological recording (20, 24). A small effect on peak Tetrodotoxin (TTX)-sensitive current, consistent with residual high levels of Nav1.1 and Nav1.6 in DRG neurons, was observed in the Nav1.7R-/- knockout. TTX-resistant currents, accounted for by Nav1.8 expression, were normal (Fig. 3). Thus, in contrast to the Nav1.8-null mutant (4), there were no reciprocal changes in TTX-resistant sodium channel expression as a result of deleting Nav1.7. This finding was confirmed by examination of the electrical properties of sensory neurons by using a spinal cord preparation. Again, in contrast to the Nav1.8-null mutant (4), there were no changes in electrical thresholds of activation of C or A fibers in Nav1.7R-/- mice.
Fig. 3.
Electrophysiology of Nav1.7-/-.(A) TTX-sensitive and -resistant peak sodium current density in Nav1.7R-/- (black) (n = 7) and fNav1.7 (white) (n = 14) mice. (B) TTX-resistant currents in WT (hatched) (n = 22), Nav1.8Cre heterozygotes (filled) (n = 22), and homozygous (n = 26) mice. (C-F) Responses of lamina V dorsal horn neurons (L3 and L4) receiving input from the hindpaw to peripheral stimuli in Nav1.7 R-/- (black) (n = 14) and littermate fNav1.7 (white) (n = 18) mice. (C) Evoked responses to von Frey hairs showed a mechanical deficit in Nav1.7R-/- mice (P = 0.048). Temperature (water jet) (D) and pinch, brush, and noxious cold (E) applied to the peripheral receptive field over 10 s were normal. Data are expressed as mean spikes (±SEM) evoked over 10 s. (F) Response to transcutaneous electrical stimulation of the receptive field (train of 16, 2-ms-wide electrical pulses, at 0.5 Hz, at three times C-fiber threshold). Spikes evoked between 0 and 50 ms were classified as caused by A-fiber input, and those evoked between 50 and 250 ms were classified as caused by C-fiber input. “Input” is the number of spikes evoked by the first stimulus of the train (50-800 ms) and reflects nonpotentiated C-fiber-evoked dorsal horn neuron response. Excess spikes, additional spikes recorded above the predicted constant baseline response, are a measure of “wind-up,” defined as increased neuronal excitability to repeated constant stimulation.
We examined the evoked responses of single spinal cord lamina V wide dynamic range neurons by using in vivo extracellular recording techniques (25). The application of a range of nonnoxious and noxious thermal stimuli to the peripheral receptive field over the hindpaws of Nav1.7 R-/- and littermate fNav1.7 mice revealed similar patterns of neuronal coding. However, there was a clear deficit in the responses evoked by noxious mechanical input into the spinal cord. By using von Frey filaments, the dorsal horn responses were reduced in the Nav1.7 R-/- mice compared with littermate controls in the noxious range, whereas low-threshold mechanical input was unaffected. No difference was observed between the knockout and control mice in the levels of ongoing spontaneous activity, neuronal activity to pinch or brush, or noxious cold stimuli. Likewise, in response to transcutaneous electrical stimulation, Nav1.7R-/- and fNav1.7 mice displayed similar thresholds for activation of A- and C-fiber input into the spinal cord and the evoked neuronal responses via these afferents after the stimulus train (Fig. 3).
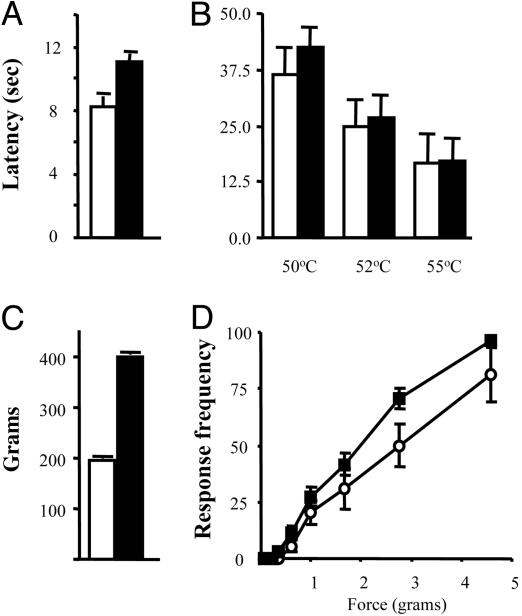
Nav1.7R-/- Pain Behavior. We examined the behavior of Nav1.7R-/- mice (26). Motor activity was normal as measured with a rotarod (fNav1.7 87.8 ± 20.1 s; null 69 ± 10.4 s), and there were no obvious differences in weight (fNav1.7 male, 30.6 ± 0.92 g; Nav1.7R-/-, 29.3 ± 0.58 g; fNav1.7 female, 24 ± 0.63 g; and Nav1.7R-/-, 22.9 ± 0.94 g) or sex ratios between Nav1.7R-/- mice and fNav1.7 controls. Pain behavior induced on a hot plate at 50°C, 52°C, or 55°C was identical. However, withdrawal latencies obtained by using thermal stimuli applied with a Hargreave's apparatus showed that Nav1.7R-/- mice were ≈20% less sensitive to noxious thermal stimulation than controls (Fig. 4). In addition, noxious mechanosensation was dramatically reduced. Although responses to von Frey hairs were similar, application of noxious pressure with a Randall-Selitto apparatus revealed a pronounced analgesia in the Nav1.7R-/- mice. These observations are consistent with the peripheral deficit demonstrated in the electrophysiological analysis of spinal cord input (Fig. 3).
Fig. 4.
Acute pain behavior of Nav1.7R-/- mice. (A) Noxious thermal stimulation of fNav1.7 (white) and Nav1.7R-/- (black) mice by using Hargreave's apparatus. The latency of hindpaw withdrawal was significantly increased in Nav1.7R-/- mice (11.01 ± 0.47, n = 14, and 8.23 ± 0.67, n = 12, respectively) (P = 0.003, t test). (B) Response to noxious thermal stimulation by using the hotplate apparatus was not significantly different in fNav1.7 (n = 12) and Nav1.7R-/- (n = 17) mice (P = 0.27, 0.11, and 0.79, t test). (C)Nav1.7R-/- mice (396.4 ± 4.0, n = 19) showed profound analgesia to noxious mechanical pressure when using the Randall-Selitto apparatus compared with fNav1.7 mice (195.5 ± 4.8, n = 18) (P < 0.0001, t test). (D) Response to mechanical stimulation when using von Frey hairs was not significantly different in fNav1.7 (n = 11) and Nav1.7R-/- (n = 19) mice.
The Nav1.7R-/- and fNav1.7 controls differ not only in terms of deletion of Nav1.7 but also in the expression of Cre recombinase. We therefore assessed Nav1.8Cre heterozygotes in all of the above acute pain tests and showed that they were indistinguishable from C57Bl6 mice (20). Thus, the behavioral differences that we see between Nav1.7R-/- mice and fNav1.7 mice are due to the ablation of the Nav1.7 sodium channel rather than the differential expression of Cre recombinase.
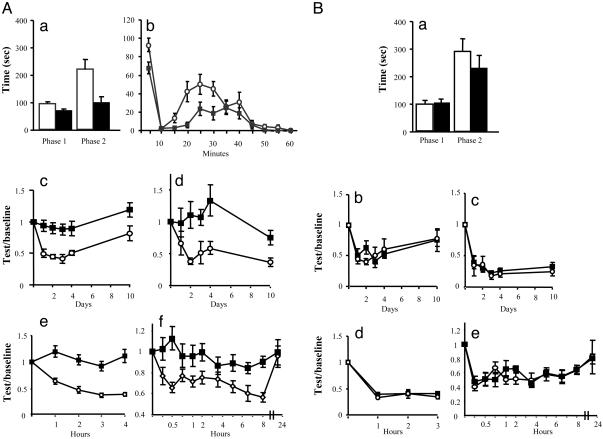
Nav1.7 and Inflammatory Pain. There were striking deficits in the development of inflammatory pain in Nav1.7R-/- mice. The behavior of mice in response to intraplantar injection of formalin showed a reduction in the first phase of the response and much reduction and a delay in onset of the second phase (Fig. 5). Longer-term inflammation induced by the injection of complete Freund's adjuvant (CFA) caused substantial thermal and mechanical hyperalgesia in fNav1.7 mice, but the Nav1.7R-/- mice were essentially unaffected at all time points. In contrast, there was no difference in edema evoked by CFA between the two mouse strains (weight injected/noninjected paw = 1.59 ± 0.04 for fNav1.7 and 1.62 ± 0.06 for Nav1.7R-/-).
Fig. 5.
Inflammatory pain in Nav1.7R-/-. (A) Analysis of behavior of fNav1.7 (white) and Nav1.7R-/- (black) mice in models of inflammatory pain. (Aa) Pain behavior of fNav1.7 (n = 14) and Nav1.7R-/- (n = 17) mice after intraplantar injection of 20 μl of 5% formalin. Time spent licking/biting the injected hindpaw in phase I (1-10 min) and phase II (10-55 min) was recorded. There is a reduction in phase I (P = 0.03, t test) (fNav1.7, 93.1 ± 7.2; Nav1.7R-/-, 70.4 ± 6.6). In phase II, Nav1.7R-/- mice also showed a significant reduction in pain behavior (P = 0.004, t test) (101.3 ± 18.9 and 220.9 ± 34.8 respectively). (Ab) Time course of the formalin response (P < 0.05 at 5-, 20-, 25-, and 30-min time points). (Ac) Thermal hyperalgesia after intraplantar injection of 20 μl of CFA. fNav1.7 mice developed pronounced hyperalgesia (0.56 ± 0.1, n = 9) from day 1, whereas Nav1.7R-/- mice did not (0.97 ± 0.06, n = 14) (P < 0.001, ANOVA; P < 0.05 at all time points). (Ad) Mechanical allodynia induced by intraplantar injection of 20 μl of CFA. fNav1.7 mice developed pronounced allodynia from day 1 (0.50 ± 0.16, n = 8), whereas the Nav1.7R-/- mice did not (1.05 ± 0.12, n = 13) (P = 0.012, ANOVA; P < 0.05 at days 2, 3, 4, and 10). (Ae) Thermal hyperalgesia after intraplantar injection of 20 μl of 2% carrageenan. fNav1.7 mice developed pronounced hyperalgesia (0.47 ± 0.03, n = 7) within an hour, whereas Nav1.7R-/- mice did not (1.07 ± 0.04, n = 5) (P < 0.001, ANOVA; P < 0.05 at all time points). (Af) Thermal hyperalgesia after intraplantar injection of 500 ng of NGF. fNav1.7 mice (n = 15) developed thermal hyperalgesia in a biphasic pattern, whereas Nav1.7R-/- mice (n = 8) showed no phase I (first hour) and a reduced phase II (P < 0.05 at 0.25, 0.5, 1, 2, 6.5, and 8 h, ANOVA). (B) Inflammatory pain is normal in Nav1.8Cre mice. Analysis of behavior of Nav1.8Cre+/- (black) and C57BL/6 (white) mice in models of inflammatory pain. (Ba) Behavior of Nav1.8Cre+/- (n = 6) and C57BL/6 (n = 6) mice after intraplantar injection of 20 μl of 5% formalin was not different between groups in phase I (107 ± 12.6 s and 96.1 ± 10.3 s, respectively; P = 0.51) and phase II (230 ± 47.7 and 290 ± 45.4 s, respectively; P = 0.38, t test). (Bb) Thermal hyperalgesia after intraplantar injection of 20 μl of CFA was not different in Nav1.8Cre+/- (0.58 ± 0.05, n = 6) and WT (0.55 ± 0.05, n = 6) littermates (P = 0.79, ANOVA). (Bc) Mechanical allodynia after intraplantar injection of 20 μl of CFA was not different in Nav1.8Cre+/- (0.30 ± 0.16, n = 6) and WT (0.27 ± 0.04, n = 6) littermates (P = 0.74, ANOVA). (Bd) Thermal hyperalgesia induced by intraplantar injection of 20 μl of 2% carrageenan was not different in Nav1.8Cre+/- (0.40 ± 0.26, n = 9) and C57BL/6 (0.36 ± 0.03, n = 7) mice (P = 0.31, two-way ANOVA). (Be) Thermal hyperalgesia induced by intraplantar injection of 500 ng of NGF was not different in Nav1.8Cre+/- (0.58 ± 0.02) (n = 6) and C57BL/6 (0.56 ± 0.03) (n = 6) mice (P = 0.83, two-way ANOVA).
Two other forms of inflammation are commonly studied in rodents. Carrageenan induces a short-term inflammation involving prostanoids (27). Nerve growth factor (NGF) acting through TrkA receptors also causes hyperalgesia (28). In both of these models of inflammatory pain, the development of thermal hyperalgesia was absent in Nav1.7R-/- mice.
Once again, to confirm that this phenotype was not the result of the presence of Cre recombinase in Nav1.7R-/- mice, we compared C57/Bl6 mice with heterozygous Nav1.8Cre mice. There was no difference in inflammatory pain behavior in these control experiments, confirming that the lack of inflammatory hyperalgesia can be ascribed to the absence of Nav1.7.
The crucial role of Nav1.7 in setting inflammatory pain thresholds may be explained by three possible mechanisms. First, the channel could form an essential component of a complex containing kinases, scaffolding proteins, and ion channels that function to alter neuronal excitability. Second, the intrinsic properties of Nav1.7 could be altered by posttranslational modifications such as phosphorylation to increase peak current density in a mechanism analogous to the mechanism that works with Nav1.8. Finally, the trafficking of the channel into the membrane could be up-regulated by inflammatory mediators to cause increased neuronal excitability.
We tested the first possibility by examining the effect of PGE2 on Nav1.8 TTX-resistant currents in DRG neurons lacking Nav1.7. Both control (35.2 ± 4.7%; n = 3) and null neurons (69 ± 26%; n = 3) showed a dramatic increase in peak Nav1.8 currents minutes after micromolar PGE2 application, suggesting that posttranslational regulation of other channels was normal in the conditional null mutant.
The second possibility can be discounted because both protein kinase A- and C-mediated phosphorylation diminish peak Nav1.7 currents in an oocyte expression system (29).
In contrast, the third possibility, enhanced trafficking of Nav1.7 into a functional form induced by inflammatory mediators, is supported by experimental data (30). Nav1.7 contributes a small proportion of the total TTX-sensitive current in DRG neurons, and sensory neuron terminals are difficult to isolate for electrophysiological studies (4). However, it is practicable to examine PC12 and adrenal chromaffin cells that only express Nav1.7, and, here, strong evidence for enhanced trafficking of Nav1.7 in response to elevated cAMP or NGF levels has been obtained. Both tritiated saxitoxin binding and functional studies demonstrate an increased current density of Nav1.7 of 50% to several-fold (30). This increase in current density correlates well with the increased peripheral excitability induced by inflammatory mediators.
Discussion
Nav1.7 is expressed in peripheral, mainly small-diameter sensory and sympathetic neurons. It is coexpressed in sensory neurons with many other sodium channel isoforms including, at high levels, Nav1.1, -1.2, and -1.6, and the TTX-resistant channels Nav1.8 and -1.9. As yet, there are no specific blockers of individual sodium channel subtypes, although TTX distinguishes between subsets of sodium channels (1). Nav1.7 falls into the TTX-sensitive category, showing an IC50 of 2 nM for TTX. There has been a resurgence of interest recently in sodium channels as potential analgesic drug targets based on antisense and null mutant mice studies (31, 32). Interpreting null mutant phenotypes can be problematic, because developmental compensatory mechanisms may occur, and, occasionally, embryonic or perinatal death precludes behavioral studies. In Nav1.7, global gene deletion leads to death shortly after birth, apparently as a consequence of failure to feed. This effect may reflect central, autonomic, or enteric sensory neuron dysfunction.
Deleting the gene in a subset of sensory neurons that are predominantly nociceptive demonstrates that Nav1.7 plays an important role in pain mechanisms, especially in the development of inflammatory pain. The specific deficits in acute mechanosensation, rather than thermal sensitivity associated with the Nav1.7 deletion, are striking. However, because TTX does not block mechanically activated currents in sensory neurons (33), Nav1.7 must play a downstream role in mechanotransduction. Nav1.8-null mice are also refractory to Randall-Selitto-induced mechanical stimuli (4). TrpV4, a potential mechanosensory primary transducer, shows a similar null phenotype (34). These data suggest that Nav1.7 and mechanosensitive channels are associated at nociceptor terminals, and the generator potential produced by activating mechanosensors activates Nav1.7.
The roles of Nav1.7 and -1.8 in pain pathways may be related. In the Nav1.8-null mouse, NGF-induced thermal hyperalgesia is diminished, and there are small deficits in other inflammatory pain models (4, 35). However, in the Nav1.8-null mouse, an up-regulation of Nav1.7 mRNA has been noted, as well as an increase in TTX-sensitive sodium current density (4). Thus, Nav1.7 may partially compensate for the depletion of Nav1.8. In contrast, deletion of Nav1.7 is not compensated for by increased TTX-resistant current activity (Fig. 3) and leads to a dramatic phenotype in terms of inflammatory pain.
Peripheral changes in pain thresholds seem to involve several mechanisms, including channel phosphorylation. NGF acting through TrkA-mediated activation of phospholipase C relieves phosphatidylinositol-4,5-bisphosphate [PtdIns(4,5)P2] channel block of TRPV1; p38 mitogen-activated protein kinase also plays a role in NGF-induced hyperalgesia (36, 37). Carrageenan increases prostanoid levels that, acting through EP receptors, cause sodium channel phosphorylation involving protein kinase A (38). CFA induces longer-term changes that are partially blocked by aspirin-like drugs (39). Remarkably, all of these forms of inflammatory hyperalgesia are attenuated by deletion of Nav1.7, possibly as a result of deficits in Nav1.7 trafficking.
The recent discovery that primary erythermalgia, a dominant human disease associated with recurrent episodes of severe pain, maps to SCN9A confirms an important role for Nav1.7 in pain pathways in both mice and humans (40). Strikingly, this condition is associated with chronic inflammation involving edema, redness, and bilateral pain, particularly in the extremities, that can be induced by standing or exercise. These symptoms are the converse of the deficits found in the nociceptor-specific Nav1.7-null mutant.
These studies have demonstrated a role for Nav1.7 in inflammatory pain that would be impossible to define pharmacologically. Like Nav1.7, a number of proteins expressed in both sensory neurons and other cell types have been suggested to play an important role in pain pathways (1, 41, 42). The deletion of broadly expressed genes only in nociceptors by using Nav1.8Cre mice is likely to prove useful in understanding mechanisms in nociception and pain.
Acknowledgments
We thank Gail Mandel, Monica Mendelson, Liam Drew, Tom Jessell, and Kenji Okuse for helpful comments. We thank the Medical Research Council and The Wellcome Trust for invaluable support. E.A.M. and M.A.N. are now supported by The Wellcome Trust-funded London Pain Consortium.
Abbreviations: BAC, bacterial artificial chromosome; CFA, complete Freund's adjuvant; DRG, dorsal root ganglion; fNav1.7, floxed Nav1.7; Nav1.7R-/-, tissue-restricted Nav1.7-/-; neor, neomycin-resistance; NGF, nerve growth factor; TTX, tetrodotoxin.
References
- 1.Catterall, W. A. (2000) Neuron 26, 13-25. [DOI] [PubMed] [Google Scholar]
- 2.Wood, J. N., Akopian, A. N., Baker, M., Ding, Y., Geoghegan, F., Nassar, M., Malik-Hall, M., Okuse, K., Poon, L., Ravenall, S., et al. (2002) in Sodium Channels and Neuronal Hyperexcitability, Novartis Foundation Symposium (Wiley, West Sussex, U.K.), Vol. 241, pp. 159-68. [PubMed] [Google Scholar]
- 3.Watanabe, E., Fujikawa A., Matsunaga, H., Yasoshima, Y., Sako, N., Yamamoto, T., Saegusa, C. & Noda, M. (2000) J. Neurosci. 20, 7743-7751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akopian, A. N., Souslova, V., England, S., Okuse, K., Ogata, N., Ure, J., Smith, A., Kerr, B. J., McMahon, S. B., Boyce, S., et al. (1999) Nat. Neurosci. 2, 541-548. [DOI] [PubMed] [Google Scholar]
- 5.Planells-Cases, R., Caprini, M., Zhang, J., Rockenstein, E. M., Rivera, R. R., Murre, C., Masliah, E. & Montal, M. (2000) Biophys. J. 78, 2878-2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burgess, D. L., Kohrman, D. C., Galt, J., Plummer, N. W., Jones, J. M., Spear, B. & Meisler, M. H. (1995) Nat. Genet. 10, 461-465. [DOI] [PubMed] [Google Scholar]
- 7.Chen, C. C., Zimmer, A., Sun, W. H., Hall, J., Brownstein, M. J. & Zimmer, A. (2002) Proc. Natl. Acad. Sci. USA 99, 17072-17077.12481039 [Google Scholar]
- 8.Takahata, T., Yasui-Furukori, N., Sasaki, S., Igarashi, T., Okumura, K., Munakata, A. & Tateishi, T. (2003) Life Sci. 72, 2391-2399. [DOI] [PubMed] [Google Scholar]
- 9.Toledo-Aral, J. J., Moss, B. L., He, Z. J., Koszowski, A. G., Whisenand, T., Levinson, S. R., Wolf, J. J., Silos-Santiago, I., Halegoua, S. & Mandel, G. (1997) Proc. Natl. Acad. Sci. USA 94, 1527-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sangameswaran, L., Delgado, S. G., Fish, L. M., Koch, B. D., Jakeman, L. B., Stewart, G. R., Sze, P., Hunter, J. C., Eglen, R. M. & Herman, R. C. (1997) J. Biol. Chem. 272, 14805-14809. [DOI] [PubMed] [Google Scholar]
- 11.Kretschmer, T., Happel, L. T., England, J. D., Nguyen, D. H., Tiel, R. L., Beuerman, R. W. & Kline, D. G. (2002) Acta Neurochir. 8, 803-810. [DOI] [PubMed] [Google Scholar]
- 12.Djouhri, L., Newton, R., Levinson, S. R., Berry, C. M., Carruthers, B. & Lawson, S. N. (2003) J. Physiol. 546, 565-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang, Y., Wang, Y., Li, S., Xu, Z., Li, H., Ma, L., Fan, J., Bu, D., Liu, B., Fan, Z., et al. (2004) J. Med. Genet. 41, 171-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Le, Y. & Sauer, B. (2001) Mol. Biotechnol. 17, 269-275. [DOI] [PubMed] [Google Scholar]
- 15.Gagneten, S., Le, Y., Miller, J. & Sauer, B. (1997) Nucleic Acids Res. 25, 3326-3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsien, J. Z., Chen, D. F., Gerber, D., Tom, C., Mercer, E. H., Anderson, D. J., Mayford, M., Kandel, E. R. & Tonegawa, S. (1996) Cell 87, 1317-1326. [DOI] [PubMed] [Google Scholar]
- 17.Hayashi, S. & McMahon, A. P. (2002) Dev. Biol. 244, 305-318. [DOI] [PubMed] [Google Scholar]
- 18.Schwenk, F., Baron, U. & Rajewsky, K. (1995) Nucleic Acids Res. 23, 5080-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Djouhri, L., Fang, X., Okuse, K., Wood, J. N., Berry, C. M. & Lawson, S. N. (2003) J. Physiol. 550, 739-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stirling, L. C., Nassar, M. A., Forlani, G., Baker, M. D. & Wood, J. N. (2003) J. Physiol. 555P, C53. [Google Scholar]
- 21.Souslova, V. A., Fox, M., Wood, J. N. & Akopian, A. N. (1997) Genomics 41, 201-209. [DOI] [PubMed] [Google Scholar]
- 22.Farley, F. W., Soriano, P., Steffen, L. S. & Dymecki, S. M. (2000) Genesis 28, 106-110. [PubMed] [Google Scholar]
- 23.Soriano, P. (1999) Nat. Genet. 21, 70-71. [DOI] [PubMed] [Google Scholar]
- 24.Okuse, K., Malik-Hall, M., Baker, M., Poon, L., Kong, H., Chao, M. & Wood, J. N. (2002) Nature 417, 653-656. [DOI] [PubMed] [Google Scholar]
- 25.Suzuki, R., Morcuende, S., Webber, M., Hunt, S. P. & Dickenson, A. H. (2002) Nat. Neurosci. 5, 1319-1326. [DOI] [PubMed] [Google Scholar]
- 26.Crawley, J. N. (2000) What's Wrong with My Mouse? Behavioral Phenotyping of Transgenic and Knockout Mice (Wiley, New York).
- 27.Ibuki, T., Matsumura, K., Yamazaki, Y., Nozaki, T., Tanaka, Y. & Kobayashi, S. (2003) J. Neurochem. 86, 318-328. [DOI] [PubMed] [Google Scholar]
- 28.Chao, M. V. (2003) Nat. Rev. Neurosci. 4, 299-309. [DOI] [PubMed] [Google Scholar]
- 29.Vijayaragavan, K., Boutjdir, M. & Chahine, M. (2004) J. Neurophysiol. 91, 1556-1569. [DOI] [PubMed] [Google Scholar]
- 30.Wada, A., Yanagita, T., Yokoo, H. & Kobayashi, H. (2004) Front. Biosci. 1, 1954-1966. [DOI] [PubMed] [Google Scholar]
- 31.Waxman, S. G., Cummins, T. R., Black, J. A. & Dib-Hajj, S. (2002) in Sodium Channels and Neuronal Hyperexcitability, Novartis Foundation Symposium (Wiley, West Sussex, U.K.), Vol. 241, pp. 34-51. [PubMed] [Google Scholar]
- 32.Baker, M. D. & Wood, J. N. (2001) Trends Pharmacol. Sci. 22, 27-31. [DOI] [PubMed] [Google Scholar]
- 33.Drew, L. J., Rohrer, D. K., Price, M. P., Blaver, K. E., Cockayne, D. A., Cesare, P. & Wood, J. N. (2004) J. Physiol. 556, 691-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suzuki, M., Mizuno, A., Kodaira, K. & Imai, M. (2003) J. Biol. Chem. 278, 22664-22668. [DOI] [PubMed] [Google Scholar]
- 35.Kerr, B. J., Souslova, V., McMahon, S. B. & Wood, J. N. (2001) NeuroReport 12, 3077-3080. [DOI] [PubMed] [Google Scholar]
- 36.Chuang, H. H., Prescott, E. D., Kong, H., Shields, S., Jordt, S. E., Basbaum, A. I., Chao, M. V. & Julius, D. (2001) Nature 411, 957-962. [DOI] [PubMed] [Google Scholar]
- 37.Ji, R. R., Samad, T. A., Jin, S. X., Schmoll, R. & Woolf, C. J. (2002) Neuron 26, 57-68. [DOI] [PubMed] [Google Scholar]
- 38.Dina, O. A., McCarter G. C., de Coupade, C. & Levine, J. D. (2003) Neuron 39, 613-624. [DOI] [PubMed] [Google Scholar]
- 39.Hutchins, B., Patel, H. & Spears, R. (2002) J. Orofac. Pain 16, 312-316. [PubMed] [Google Scholar]
- 40.Drenth, J. P., Finley, W. H., Breedveld, G. J., Testers, L., Michiels, J. J., Guillet, G., Taieb, A., Kirby, R. L. & Heutink, P. (2001) Am. J. Hum. Genet. 68, 1277-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo, Z. D., Calcutt, N. A., Higuera, E. S., Valder, C. R., Song, Y. H., Svensson, C. I. & Myers, R. R. (2002) J. Pharmacol. Exp. Ther. 303, 1199-205. [DOI] [PubMed] [Google Scholar]
- 42.Thompson, S. W., Bennett, D. L., Kerr, B. J., Bradbury, E. J. & McMahon, S. B. (1999) Proc. Natl. Acad. Sci. USA 96, 7714-7718. [DOI] [PMC free article] [PubMed] [Google Scholar]