Abstract
How olfactory sensory neurons converge on spatially invariant glomeruli in the olfactory bulb is largely unknown. In one model, olfactory sensory neurons interact with spatially restricted guidance cues in the bulb that orient and guide them to their target. Identifying differentially expressed molecules in the olfactory bulb has been extremely difficult, however, hindering a molecular analysis of convergence. Here, we describe several such genes that have been identified in a screen that compiled microarray data to create a three-dimensional model of gene expression within the mouse olfactory bulb. The expression patterns of these identified genes form the basis of a nascent spatial map of differential gene expression in the bulb.
In the olfactory system, a major challenge has been to identify the mechanisms responsible for guiding olfactory sensory neurons (OSNs) to specific targets in the olfactory bulb. OSNs expressing a given odorant receptor are distributed throughout large zones in the olfactory epithelium but ultimately converge on common targets in the bulb, which are called glomeruli. Remarkably, these glomerular positions are spatially invariant from animal to animal, consistent with the presence of a hard-wired map of connectivity in the peripheral olfactory system (1).
How are OSNs able to identify their glomerular targets? The stereotypic nature of this innervation suggests that regions of the bulb, perhaps the glomeruli, are molecularly distinct. These spatially distributed cues would then function to guide OSNs expressing complementary receptors. In this model, the collective expression patterns of these guidance cues would form a spatial map within the bulb, molecularly distinguishing potential paths and substrates encountered by OSNs. However, although it has become increasingly clear that a number of molecules have been identified that are differentially expressed by OSNs [for e.g., along the path that OSNs traverse to the bulb (2-4) and by ensheathing cells (5-7)], efforts to identify spatially restricted molecules within the bulb itself have been generally unsuccessful (8, 9), greatly impeding our understanding of how olfactory axons select their partner glomeruli. Indeed, the most plausible models of olfactory development assume that spatially restricted cues exist within the olfactory bulb to guide the sensory axons to their targets (1, 10). Yet, despite this widely held assumption, it has been remarkably difficult to identify such guidance and targeting cues.
Does a spatial map of differential gene expression exist within the bulb? We decided to address this issue directly by employing a molecular screen using microarrays to reconstruct, in three dimensions, patterns of gene expression within the mouse olfactory bulb. This statistical representation of the bulb was then scanned for genes with differential expression patterns, which were validated by RNA in situ hybridization. A number of genes known to be involved in neural development and pattern formation were shown to be expressed in restricted patterns, consistent with their possible roles in imparting spatial information within the developing olfactory bulb.
Materials and Methods
RNA Purification and Amplification. Olfactory bulbs were dissected from postnatal day 0 CD-1 mice, and slices were obtained by manual dissection, which was done approximately in thirds along each principal axis. For each aspect [anterior (A), dorsal (D), lateral (L), medial (M), posterior (P), and ventral (V)], samples from ≈10 mice were pooled and total RNA was isolated by using the TRIzol reagent. We amplified 3 μg of RNA once by using an optimized T7 RNA polymerase amplification protocol (D.M.L., P. Luu, T. Serafini, and J.N., unpublished data), incorporating an oligo(dT)-T7 primer (5′-ATCGATTCGAACTTCTGATAGACTTCGAAATTAATACGACTCACTATAGGGAGACCACAT21-3′) to generate cDNA. Amplified RNA was then obtained in an in vitro transcription reaction (1× transcription buffer/2 mM each NTP/1 unit of RNAsin/100 units of T7 RNA polymerase), and 5 μg of amplified RNA was used per labeling reaction.
Microarrays. We generated microarrays by using the RIKEN 19K full-length mouse cDNA set (11). Microarrays were hybridized with Cy3- and Cy5-labeled cDNAs by using protocols essentially as described by the Microarrays.org public protocols distributor (www.microarrays.org/protocols.html).
Normalization and MA Plot. Raw data from each scanned slide were processed by spot (12), with foreground seeds set at a 5 × 5 pixels square. Within- and between-slide normalization were performed on all data (13). An MA plot (14) was used to represent the data, where M = log2 (red intensity/green intensity) and 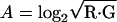 .
.
Estimation of Contrasts. For each gene, a linear model was used to estimate each of the 15 contrasts [AL, DM, VP, etc.; for details, see Supporting Materials and Methods and Figs. 5 and 6, which are published as supporting information on the PNAS web site]. For visualization purposes, we estimated the contrast of a single effect vs. an average of all six [e.g., ã = a - ⅙(a + p + d + v + m + l)], in effect, recreating the pooled bulb reference (a + p + d + v + m + l) in silico. Note that this estimate does not represent the absolute expression “profile” but rather the relative expression between a portion of the bulb to the pooled reference.
Two-Stage Cluster Analysis. The top 100 genes from each contrast were taken (a total of 614 unique genes), and hierarchical clustering of these genes was performed by building a dendrogram based on a modified Mahalanobis distance and Ward agglomeration (see Supporting Materials and Methods). The metric used to measure the similarity between the bulb expression profile for gene x and gene y is defined as δxy = (x - y)′ (X′X)(x - y), where X is the design matrix of our experiment. We considered all 614 clusters that consist of more than two genes in the tree, measured the heterogeneity h by calculating the 15 SDs across the cluster for each of the pairwise effects, and took the largest. By choosing a score of s = 0.45 and taking all maximal disjoint clusters with h < s, we obtained 16 clusters. For each cluster, we ranked genes based on their Mahalanobis distance between the bulb expression profile of the gene and the origin, which is written as δx = (x)′(X′X)(x). Genes with a high δx score were selected for further verification. To further understand the relationship between these clusters, we carried out a second hierarchical clustering by using the average profiles of the disjoint clusters that were obtained as described above. A dendrogram based on correlation metric and Ward agglomeration was then built. A complete list of the 614 genes can be found in Table 1, which is published as supporting information on the PNAS web site.
Spatial Map. Patterns obtained from in situ hybridization experiments were traced and digitized by using the neurolucida program. Up to 18 sections, taken from the entire AP axis of the bulb, were used per reconstruction. The location of NT3 expressing cells was estimated from individual slides.
In Situ Hybridization. In situ hybridizations were performed as described in ref. 15 by using 20-μm-thick coronal fresh-frozen sections of olfactory bulbs from postnatal day 0 mice. Expression patterns of candidate genes were verified with digoxigenin and/or 33P-labeled probes.
Results
Given the historical difficulty in determining where within the bulb differentially expressed molecules might exist, we decided to sample cell types from throughout the bulb by comparing relatively large domains (i.e., the A, P, D, V, M, and L aspects) (Fig. 1a). Because the bulb is composed of a series of concentric layers, these aspects will each contain all of the different cell types and layers within this structure.
Fig. 1.
Experimental design and profile generation. (a) Olfactory bulbs were dissected individually into three slices along each axis: AP (Left), DV (Middle), and ML (Right). Slices shown in red represent the six aspects that were used in this analysis. (b) All pairwise comparisons between the six aspects were made. This design permits gene expression to be measured directly (e.g., AD) and indirectly (APD, AVD, etc.). By convention, the sample to which the arrow points was labeled with Cy5. (c) For each gene (e.g., jagged, gene no. 15,228), 15 contrasts (MD, LD, etc.) are estimated and assigned values (average, estimated contrast value). By convention, the first letter in each contrast determines the sign of the y axis value (e.g., in the MD contrast for jagged, the positive MD value indicates that M has relatively higher expression than D).
Gene expression within the olfactory bulb fragments was assayed by using DNA microarrays, and spatial patterns were identified from these data by using statistical approaches. Previous attempts that used microarrays to compare grossly different regions of the brain against one another (e.g., bulb vs. amygdala) identified a surprisingly limited number of gene-expression differences (16, 17). By comparison, our intended approach relies on recreating patterns of expression that exist within a tissue of the brain, comparing highly similar subregions that are composed of morphologically equivalent populations of cells and layers.
To define three-dimensional patterns of gene expression in the olfactory bulb, we designed our experiments to incorporate all 15 possible comparisons among each of the six aspects (Fig. 1b). Next, we considered how to integrate this information to determine the three-dimensional expression pattern for each gene on the array. In essence, we wished to produce a “bulb profile” of expression for each gene that could then be surveyed to identify those patterns that indicate differential expression. To generate three-dimensional profiles, data from multiple hybridizations must be combined in a manner that maintains the spatial nature of each aspect used in the hybridization. Therefore, we expanded on a previous approach (14) and used fixed effects linear models to analyze our data (see Materials and Methods and Supporting Materials and Methods).
For each of the 15 possible pairwise comparisons (e.g., A vs. L), robust multiple regression was used to combine all measurements of gene expression (see Materials and Methods). Measurements obtained from direct measurements (e.g., A→L) and indirect measurements (e.g., A→D→L) were weighted and combined to produce a contrast value for that comparison. Each gene would, therefore, be associated with 15 contrast values. Thus, a gene may possess an AL contrast estimate of zero (indicating, on average, equal expression in a comparison of the A and L aspects) but a positive PM contrast estimate, indicating relatively higher P expression as compared with M expression. Together, the 15 contrast values for each gene represent for each gene a profile of expression within the bulb. Fig. 1c shows such a bulb profile for gene no. 15,228, which corresponds to jagged, a ligand for Notch (18).
For ease of visualization, we simplified these 15 contrast profiles and reduced each to a six-contrast representation, in which each effect (a, p, d, v, m, and l) is compared with a computed in silico pooled reference to generate an average estimate (ã, p̃, d̃, ṽ, m̃, and l̃) of overall expression (see Materials and Methods and Fig. 2a). In this simplified six-way profile, positive or negative values indicate relatively increased or decreased expression, respectively, for a given effect as compared with the computed pooled reference. Thus, it is more apparent that jagged is expressed relatively strongly in the D and P aspects of the bulb and comparatively evenly expressed across the other four aspects (Fig. 2 a and b). These relative increases in expression do not preclude expression in other regions of the bulb because our predictions depend on the aggregate total of mRNA that is present within each fragment used in these experiments.
Fig. 2.
Hierarchical clusting of bulb expression profiles. (a) The 15 contrasts can be simplified to produce an in silico estimate of gene expression along each axis relative to the pooled reference. This permits visualization of the predicted expression pattern of jagged (gene no. 15,228), which has relatively higher D and P expression (positive average value) as compared with the pooled reference. (b) The predicted jagged expression pattern can be superimposed on a model of the olfactory bulb, which is shown as a cube. On average, higher D and P expression is predicted for this gene within the bulb. (c) We generated 16 clusters, and the corresponding dendrogram shows the relationship among the clusters. The individual bulb profiles of all genes contained within a cluster were averaged to generate a six-way cluster profile, which is shown below the dendrogram. Red, green, and black indicate increased, decreased, and unchanged relative expression, respectively, as compared with the computed whole-bulb average. (d) A six-way cluster profile for cluster 2 shown as both a red-and-green map and as a bar diagram. Compare with the individual profile of jagged, which is shown in a.
Clustering Bulb Profiles. Our linear model analysis produced ≈19,000 bulb profiles, with each profile comprising 15 contrasts (e.g., Fig. 1c). To identify bulb profiles within this set that possess spatially restricted expression patterns, we considered different approaches. One possibility would be simply to rank genes according to their contrast values. A weakness of such an approach, however, is that genes with certain profiles may dominate other genes showing more subtle, but potentially interesting, patterns. We, therefore, chose to cluster genes with similar profiles to identify patterns that are prominently represented in our data.
In this approach, it is difficult to determine a priori how many clusters (or “groups”) should be represented within our data. To produce an experimentally manageable number, we generated 16 clusters by using a two-stage hierarchical clustering procedure (see Materials and Methods and Supporting Materials and Methods); the resulting dendrogram is shown in Fig. 2c, and the list of 614 genes, sorted by cluster, is given in Table 1. It is important to note that the number of clusters and the membership or rank of an individual gene within a cluster can be changed without affecting the profile of these genes. Thus, hierarchical clustering is used here to organize the data and not necessarily to infer mechanistic relationships between genes within a cluster.
The individual profiles of all genes contained within a cluster were averaged to generate corresponding six-way cluster profiles, shown below the dendrogram as a red-and-green profile map, where red, green, and black correspond to increased, decreased, and unchanged expression, respectively, as compared with the in silico pooled reference (Fig. 2d). We performed a randomization analysis to test the null hypothesis that the data and patterns that we observed have no biological basis (and, thus, are what we would expect from random data without structure). The null hypothesis was convincingly rejected in an analysis comprising 500 sets of randomized data, each containing all ≈19,000 genes (P < 0.01; see Supporting Materials and Methods).
An examination of the six-way cluster profiles reveals interesting differences among the various patterns, which we organized into four general categories. The first category (category 1) includes groups whose six-way cluster profiles indicate differences in expression along any one axis of the bulb (groups 2, 5, and 12). For example, the cluster profile of group 2 indicates reduced expression in the A bulb and high relative expression in the P bulb (low A, high P). This type of pattern, in which a bias is observed from one pole of the bulb relative to the opposite pole, is observed also in group 5 (low A, high P) and group 12 (low D, high V). Category 2 includes groups with cluster profiles showing differential expression within a single aspect of the bulb (groups 3, 9, and 14-16). For example, the cluster profile of group 3 shows high relative expression in the P and D aspects (high P and D) but minimal differences in the opposing A or V aspects, respectively. This pattern holds as well for group 9 (low A or M, no P or L changes), and groups 14-16 (all high M). Category 3 includes most of the groups (groups 1, 4, 6-8, 11, and 13), and it comprises cluster profiles that show dramatic and correlated changes along at least two axes of the bulb. For example, in group 1, there is elevated expression in both the A and P aspects (high A, high P) and reduced M and L expression (low M, low L). In group 11, expression is high in both the D and V aspects of the bulb (high D, high V) but is reduced in the M and L bulb (low M, low L). Such patterns of high expression along one axis but low expression along another axis suggest a discontinuity of expression within the bulb consistent with layer-specific, but not three-dimensionally restricted expression. Finally, category 4 contains a single member (group 10), which exhibits minimal differences between each aspect as compared with the computed whole-bulb average.
Validation. The qualitative categorization of cluster profiles provided a preliminary filter for genes showing potentially interesting expression patterns. Next, we used in situ hybridization to validate differentially expressed genes within these clusters, focusing on category 1 (groups 2, 5, and 12) and category 2 (groups 3, 14, and 15). Genes from categories 3 and 4 were not considered further because the nature of their predicted profiles did not suggest any obvious three-dimensional restrictions within the olfactory bulb. We disregarded group 16 (category 2) because it is composed predominantly of hemoglobin genes, which is likely an artifact of our dissection technique.
To select genes for in situ hybridizations, we typically assessed the individual profiles of the 10 top-ranked genes from each cluster and selected three to four genes showing the most promising differences (i.e., of a magnitude or pattern likely to be detected in an in situ hybridization). In some cases, however, some genes (e.g., NT3) were chosen based on their known function in the nervous system and not on their final ranking within a group. We successfully identified nine genes that indeed are differentially expressed within the olfactory bulb.
Category 1. Group 2. The cluster profile of group 2 (Fig. 2d), which contains three genes (Table 1), suggests differential expression along both the AP and DV axes. The individual profile (Fig. 3a) of jagged indicates relatively high D and P expression as compared with the remainder of the bulb. To determine the actual pattern of expression, we hybridized digoxigenin-labeled jagged probes to tissue sections of the olfactory bulb (Fig. 3b). As a control, we used the glutamate receptor (subunit 1), which labels cells in the periglomerular, mitral, and granule layers of the bulb (Fig. 3 c and d). For jagged, we detected significant reactivity in the accessory olfactory bulb, located on the DP surface of the main olfactory bulb. Weaker signal could be detected in other layers (data not shown). Thus, the in situ hybridization results for jagged are fully consistent with the expression pattern predicted from our three-dimensional statistical profile. Adhesion protein RA175C, another member of group 2, is expressed in a similar pattern, and is present in the accessory olfactory bulb and other cell layers of the main olfactory bulb although at higher levels in the mitral layer than jagged (see Figs. 5-9, which are published as supporting information on the PNAS web site). Its pattern of expression also confirms the individual six-way profile. However, the top-ranked member of this group (an EST) displayed only weak mitral expression and no obvious restriction in expression (data not shown).
Fig. 3.
In situ hybridization patterns of differentially expressed genes in the bulb. All tissue sections shown are coronal sections, oriented so that M is to the right and D is up. (a) Individual six-way profile for jagged. (b) In situ hybridization of jagged probe to a caudal olfactory bulb section. Note strong reactivity in the accessory olfactory bulb (arrow) on the D-caudal surface of the main olfactory bulb. (c) Control hybridization with a glutamate receptor probe (GluR1) to a caudal section. (d) Control hybridization of GluR1 probe to a more rostral bulb section. Strong reactivity is observed in the periglomerular (arrowhead), mitral (white arrow), and granule (black arrow) cell layers. (e) Individual six-way profile for ptd2.(f) In situ hybridization using a ptd2 probe reveals a strong signal in the leptomeninges (arrow). Note the absence of VL signal in this section (arrowheads). (g) Individual six-way profile for IGF-2.(h) In situ hybridization using an IGF-2 probe reveals similar expression to that of ptd2 in the A bulb. Expression is localized to the meningeal layer (arrow), with weak or absent V and L expression (arrowheads). (i) Individual six-way profile for pcVα2.1. (j) Expression of pcVα2 is located immediately external to the glomerular layer (arrow). Note higher expression on M and V aspects of the bulb as compared with the D and L aspects. (k) Individual six-way profile for cadherin-11.(l) Cadherin-11 probe labels cells in the mitral layer on the M aspect of the olfactory bulb (arrows). Signal is also detected in the granule layer on the L aspect of the bulb (arrowheads). (m) Individual six-way profile for vitronectin.(n) In situ hybridization with vitronectin reveals a similar pattern to that of pcVα2, with high M and V expression (arrow). Expression is reduced in the D and L aspects (arrowheads). (o) Individual six-way profile for NT3.(p)A 33P-labeled NT3 probe hybridized to tissue sections indicates expression in a subset of mitral cells (arrows). Scale bar indicates 300 μm.
Group 5. Of the top three genes tested from group 5, two are expressed in restricted patterns in the bulb. Prostaglandin D2 synthase (ptd2) is predicted to have relatively high P expression and reduced L expression (Fig. 3e). In situ hybridization shows expression of ptd2 in the leptomeninges of the bulb. This expression is not uniform, however, and is absent from the AVL surface of the bulb (Fig. 3f; compare with GluR1, Fig. 3d; see also Fig. 8), consistent with the prediction that was made from the DNA microarray analysis. Insulin-like growth factor 2 (IGF-2) exhibited a similar predicted pattern and was also validated by in situ hybridization (Fig. 3 g and h; see also Fig. 9). The top-ranked clone in this cluster (ZX00001L15) possessed strong mitral layer expression but no obvious differential expression in the bulb (data not shown).
Group 12. Four genes were tested from group 12. The top-ranked genes in this cluster, clones ZA00006A12 (EST, similar to dynein heavy chain) and ZX00032G15 (keratin complex 2, basic, gene 1), showed weak or no signal in the bulb (data not shown). However, the fourth- and fifth-ranked genes in this cluster, NAD(P)H menadione oxidoreductase 1 (MO 1) and procollagen Vα2 (pcVα2), were found to be expressed in similarly restricted patterns in the bulb. For both of these genes, expression is biased to the V portion of the bulb in a layer just external to the glomeruli, corresponding possibly to a subset of periglomerular cells (for pcVα2, see Fig. 3 i and j, and for MO 1, see Fig. 7 C and D). Thus, the in situ hybridization patterns confirm the individual six-way profiles, which predicted high V, low D expression.
In summary, of the 10 category 1 genes tested, 6 are spatially restricted in expression in the bulb (jagged, RA175C, ptd2, IGF-2, pcVα2, and MO 1), 2 are not restricted in their expression, and 2 gave a weak or not visible signal.
Category 2. We next tested genes belonging to clusters belonging to category 2 (groups 3, 14, and 15). These groups exhibited patterns predicting up-regulation in one aspect of the bulb but no complementary differences along the same axis in the opposite aspect (e.g., high P, unchanged A).
Group 3. We selected the first two clones in group 3, beta-site APP cleaving enzyme, which gave no signal, and clone ZX00035C13 (an EST), which showed uniform expression in the mitral cell layer (data not shown).
Group 14. In this cluster, neither ERK3 protein kinase nor clone ZX00028A22 (an EST), the 3rd- and 4th-ranked genes, exhibit restricted expression patterns. However, group 14 contains the cadherin-11 gene (19), a member of the cadherin superfamily, whose functions have been widely studied in the nervous system (20). The bulb profile for cadherin-11, ranked 5th in this cluster, predicts it to be relatively highly expressed along the AP axis, weakly along the DV axis, and somewhat higher in the M aspect (Fig. 3k). In situ hybridization shows high expression in the mitral cell layer of the bulb, particularly along the M aspect. Surprisingly, a subset of granule neurons (or possibly cells belonging to the A olfactory nucleus) along the L aspect of the bulb also labeled with this probe (Fig. 3l; compare with Fig. 3c, which shows the unrestricted labeling of all granule cells by using the glutamate receptor probe). This restriction of cadherin-11 was not predicted by its six-way bulb profile (Fig. 3k), probably because the L bias within the granule cell layer is overshadowed by a complementary M bias in the mitral cell layer. A similar pattern of protein expression has been published for ephrin A2 (Elf-1 in Table 1), the top-ranked gene in this cluster (21); however, we could not confirm this pattern by in situ hybridization using a probe for ephrin A2.
Group 15. Five clones were tested from group 15. The first two, ZX00001F10 (an EST) and ZX00032I10 (an EST) showed no restriction, and the 5th-ranked gene, ZX00001F09 (EST) gave no detectable signal. However, the 13th-ranked gene, vitronectin, is expressed in the extraglomerular layer (similar to pcVα2 and MO 1), with a three-dimensional restriction to the M and V aspects of the bulb (Fig. 3n), consistent with the predicted pattern (Fig. 3m). Cluster 15 also contains the neurotrophin-3 (NT3) gene, which is predicted to have relatively high M expression and lower D and V expression (Fig. 3o). Previous studies have been unable to determine where within the bulb NT3 is expressed (22, 23). We were unable to detect any signal by using digoxigenin-labeled probes. However, by using 33P-labeled probes, we detected hybridization within a subset of mitral cells located along the M surface of the bulb (Fig. 3p; compare with Fig. 3c). This band of NT3 positive cells extends more ventrally within this layer at more rostral positions (see Fig. 4).
Fig. 4.
A nascent spatial map of differential gene expression in the olfactory bulb. Major domains of expression were retained for each gene to simplify presentation. In this representation, A is toward the lower-right corner. Dark blue, outline of olfactory bulb and cortex; yellow, ptd2 expression; light blue; jagged expression; red, cadherin-11 expression; purple, NT3 expression.
In summary, of the 11 genes tested from category 2, 3 are differentially expressed within the bulb (cadherin-11, vitronectin, and NT3), 6 are uniformly expressed, and 2 gave no detectable signal.
A Nascent Spatial Map of Gene Expression in the Olfactory Bulb. The results of the in situ hybridization patterns paint a surprising and unexpected picture of differential gene expression in the olfactory bulb. Jagged and Adhesion protein RA175C are preferentially expressed in the accessory olfactory bulb; cadherin-11 labels a subpopulation of granule neurons; ptd2 and IGF-2 are absent in large areas of the leptomeninges; NT3 labels a small proportion of previously undistinguished mitral cells; and vitronectin, pcVα2, and MO 1 are expressed in the periglomerular region. To obtain a more global view of these spatial distributions, we reconstructed from multiple tissue sections the three-dimensional expression patterns for a subset of genes. Major domains of expression were then superimposed on a model of the olfactory bulb (Fig. 4). The resulting map reveals how different regions of the bulb can be distinguished from one another based on their profiles of gene expression. For example, the M aspect of the olfactory bulb, which expresses both ptd2 and NT3, is molecularly distinct from the L portion of the AV surface, which appears to express neither gene.
Discussion
Spatial maps of differential gene expression can be found throughout development. Such maps are particularly evident from studies on pattern formation, where complex spatial patterns have been identified in the Drosophila embryo (24), the mammalian telencephalon (25), and in the spinal cord (26). Similarly, spatial maps are thought to be used in the formation of topographic connections in the nervous system (27). However, although guidance cues have been identified, a comprehensive spatial map of these cues has yet to be produced. Such a map would begin to address how these multiple signals distinguish among potential targets by identifying overlapping patterns of expression. The function of these individual genes could then be tested for their role in this process (28, 29).
The identification of a spatial map in the olfactory system and its potential function has been particularly difficult to address. Although differentially expressed cues have been found in the periphery, only a limited number have been described in the bulb. In this article, we identify molecules with restricted patterns of expression in the bulb by using a statistical approach for DNA microarray-based expression profiling, and demonstrate the existence of a spatial map in this structure. The nascent map formed by these patterns allows us to begin to describe, on a global level, how regions of the bulb can be differentiated from one another at the molecular level. It should be noted, however, that our analysis provides only a “snapshot” of olfactory bulb development. Thus, it is possible that the patterns of gene expression we observe here are a result of differential developmental maturation across the olfactory bulb, and not reflective of spatial restrictions per se. Indeed, it has been reported that glomeruli form in a spatiotemporal wave, with rostral glomeruli maturing before caudal glomeruli (30). However, several of the patterns identified here describe gene-expression differences across axes of the bulb orthogonal to the spatiotemporal axis, indicating that such restrictions are not a secondary result of temporal waves of cellular maturation, but instead represent bona fide asymmetries within this tissue.
Despite the relatively large size of the tissue fragments used in our analysis, our estimate in general successfully predicted the location of expression patterns associated with individual genes. Note that in all of our validated cases, expected average differences in any one aspect of the bulb as compared with the pooled reference are <2-fold (Fig. 3), underscoring the ability of our approach to identify subtle changes in gene expression within the bulb and distinguish them from background noise. By clustering genes with similar patterns together, other, weaker patterns that may potentially be buried within the noise could be identified and considered for further analysis.
Several of the genes that were uncovered in this screen (jagged, IGF-2, cadherin-11, NT3, pcVα2, and vitronectin) have clearly been implicated in synapse formation and connectivity (18, 19, 22, 31-33). These genes, as well as other members of the families that they belong to, are excellent candidates for a more detailed analysis of how they might affect development and target selection within the bulb. For example, mutants in cadherin-11 have been obtained by employing transgenic techniques (19). In these mice, the olfactory bulbs are reduced significantly in size and can now be tested to determine whether there are alterations in convergence patterns. Thus, the genes uncovered in this work could possibly impact many aspects of olfactory bulb development and function.
The identification of differentially expressed genes in the olfactory system is a fundamental and necessary first step toward defining the mechanisms underlying the patterning of sensory input in the olfactory bulb. We anticipate that iterations of this nascent spatial map will incorporate progressively smaller olfactory bulb fragments, allowing finer resolution of gene expression patterns within the three-dimensional space of this structure. Ultimately, this information should provide a rational basis for multiple genetic alterations and interpretation of the subsequent phenotypes.
Supplementary Material
Acknowledgments
We thank K. van Fossen for technical assistance; J. Winer for use of the neurolucida software; A. Dittman for discussion of the manuscript; and S. Choksi, E. Diaz, A. Finn, P. Luu, L. Simirenko, D. Speca, and T. Alioto for help in generation of the microarrays. This work was supported by grants from the National Institutes of Health (to J.N. and T.P.S.) and by funds from the Department of Molecular Cell Biology and the Helen Wills Neuroscience Institute. D.M.L. was supported by the Whitehall Foundation, the National Science Foundation, the Sloan Foundation, and the Beckman Young Investigator Program. The 19K RIKEN cDNA set was supported in part by the Special Coordination Funds for Promoting Science and Technology and a research grant from the Science and Technology Agency of the Japanese government (to Y.H.).
Abbreviations: OSN, olfactory sensory neuron; A, anterior; D, dorsal; L, lateral; M, medial; P, posterior; V, ventral; ptd2, prostaglandin D2 synthase; IGF-2, insulin-like growth factor 2; pcVα2, procollagen Vα2.
References
- 1.Key, B. & St. John, J. (2002) Chem. Senses 27, 245-260. [DOI] [PubMed] [Google Scholar]
- 2.Cutforth, T., Moring, L., Mendelsohn, M., Nemes, A., Shah, N. M., Kim, M. M., Frisen, J. & Axel, R. (2003) Cell 114, 311-322. [DOI] [PubMed] [Google Scholar]
- 3.Norlin, E. M., Alenius, M., Gussing, F., Hagglund, M., Vedin, V. & Bohm, S. (2001) Mol. Cell. Neurosci. 18, 283-295. [DOI] [PubMed] [Google Scholar]
- 4.Yoshihara, Y., Kawasaki, M., Tamada, A., Fujita, H., Hayashi, H., Kagamiyama, H. & Mori, K. (1997) J. Neurosci. 17, 5830-5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gong, Q. & Shipley, M. T. (1996) J. Comp. Neurol. 366, 1-14. [DOI] [PubMed] [Google Scholar]
- 6.Crandall, J. E., Dibble, C., Butler, D., Pays, L., Ahmad, N., Kostek, C., Puschel, A. W. & Schwarting, G. A. (2000) J. Neurobiol. 45, 195-206. [DOI] [PubMed] [Google Scholar]
- 7.Schwarting, G. A., Kostek, C., Ahmad, N., Dibble, C., Pays, L. & Puschel, A. W. (2000) J. Neurosci. 20, 7691-7697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marillat, V., Cases, O., Nguyen-Ba-Charvet, K. T., Tessier-Lavigne, M., Sotelo, C. & Chedotal, A. (2002) J. Comp. Neurol. 442, 130-155. [DOI] [PubMed] [Google Scholar]
- 9.Otaki, J. M. & Firestein, S. (1999) NeuroReport 10, 2677-2680. [DOI] [PubMed] [Google Scholar]
- 10.Gogos, J. A., Osborne, J., Nemes, A., Mendelsohn, M. & Axel, R. (2000) Cell 103, 609-620. [DOI] [PubMed] [Google Scholar]
- 11.Miki, R., Kadota, K., Bono, H., Mizuno, Y., Tomaru, Y., Carninci, P., Itoh, M., Shibata, K., Kawai, J., Konno, H., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 2199-2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang, Y. H., Buckley, M. J., Dudoit, S. & Speed, T. P. (2002) J. Comput. Graph. Stat. 11, 108-136. [Google Scholar]
- 13.Yang, Y. H., Dudoit, S., Luu, P., Lin, D. M., Peng, V., Ngai, J. & Speed, T. P. (2002) Nucleic Acids Res. 30, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dudoit, S., Yang, Y. H., Callow, M. J. & Speed, T. P. (2002) Statistica Sinica 12, 111-139. [Google Scholar]
- 15.Speca, D. J., Lin, D. M., Sorensen, P. W., Isacoff, E. Y., Ngai, J. & Dittman, A. H. (1999) Neuron 23, 487-498. [DOI] [PubMed] [Google Scholar]
- 16.Sandberg, R., Yasuda, R., Pankratz, D. G., Carter, T. A., Del Rio, J. A., Wodicka, L., Mayford, M., Lockhart, D. J. & Barlow, C. (2000) Proc. Natl. Acad. Sci. USA 97, 11038-11043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zirlinger, M., Kreiman, G. & Anderson, D. J. (2001) Proc. Natl. Acad. Sci. USA 98, 5270-5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindsell, C. E., Shawber, C. J., Boulter, J. & Weinmaster, G. (1995) Cell 80, 909-917. [DOI] [PubMed] [Google Scholar]
- 19.Manabe, T., Togashi, H., Uchida, N., Suzuki, S. C., Hayakawa, Y., Yamamoto, M., Yoda, H., Miyakawa, T., Takeichi, M. & Chisaka, O. (2000) Mol. Cell. Neurosci. 15, 534-546. [DOI] [PubMed] [Google Scholar]
- 20.Yagi, T. & Takeichi, M. (2000) Genes Dev. 14, 1169-1180. [PubMed] [Google Scholar]
- 21.St. John, J. A., Pasquale, E. B. & Key, B. (2002) Brain Res. Dev. Brain Res. 138, 1-14. [DOI] [PubMed] [Google Scholar]
- 22.Nef, S., Lush, M. E., Shipman, T. E. & Parada, L. F. (2001) Dev. Biol. 234, 80-92. [DOI] [PubMed] [Google Scholar]
- 23.Guthrie, K. M. & Gall, C. M. (1991) J. Comp. Neurol. 313, 95-102. [DOI] [PubMed] [Google Scholar]
- 24.St Johnston, D. & Nusslein-Volhard, C. (1992) Cell 68, 201-219. [DOI] [PubMed] [Google Scholar]
- 25.Wilson, S. W. & Rubenstein, J. L. (2000) Neuron 28, 641-651. [DOI] [PubMed] [Google Scholar]
- 26.Price, S. R., De Marco Garcia, N. V., Ranscht, B. & Jessell, T. M. (2002) Cell 109, 205-216. [DOI] [PubMed] [Google Scholar]
- 27.Sperry, R. W. (1963) Proc. Natl. Acad. Sci. USA 50, 703-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Winberg, M. L., Mitchell, K. J. & Goodman, C. S. (1998) Cell 93, 581-591. [DOI] [PubMed] [Google Scholar]
- 29.Stein, E. & Tessier-Lavigne, M. (2001) Science 291, 1928-1938. [DOI] [PubMed] [Google Scholar]
- 30.Bailey, M. S., Puche, A. C. & Shipley, M. T. (1999) J. Comp. Neurol. 415, 423-448. [PubMed] [Google Scholar]
- 31.Chernousov, M. A. & Carey, D. J. (2000) Histol. Histopathol. 15, 593-601. [DOI] [PubMed] [Google Scholar]
- 32.Holland, S. J., Peles, E., Pawson, T. & Schlessinger, J. (1998) Curr. Opin. Neurobiol. 8, 117-127. [DOI] [PubMed] [Google Scholar]
- 33.Schvartz, I., Seger, D. & Shaltiel, S. (1999) Int. J. Biochem. Cell Biol. 31, 539-544. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.






