Abstract
The PrfA protein of Listeria monocytogenes functions as a key regulatory factor for the coordinated expression of many virulence genes during bacterial infection of host cells. PrfA activity is controlled by multiple regulatory mechanisms, including an apparent requirement for either the presence of a cofactor or some form of posttranslational modification that regulates the activation of PrfA. In this study, we describe the identification and characterization of a novel PrfA mutation that results in constitutive activation of the PrfA protein. The PrfA L140F mutation was found to confer high-level expression of PrfA-regulated genes and to be functionally dominant over the wild-type allele. The presence of the PrfA L140F mutation resulted in the aggregation of L. monocytogenes in broth culture and, unlike previously described prfA mutations, appeared to be slightly toxic to the bacteria. High-level PrfA-dependent gene expression showed no additional increase in L. monocytogenes strains containing an additional copy of prfA L140F despite a >4-fold increase in PrfA protein levels. In contrast, the introduction of multiple copies of the wild-type prfA allele to L. monocytogenes resulted in a corresponding increase in PrfA-dependent gene expression, although overall expression levels remained far below those observed for PrfA L140F strains. These results suggest a hierarchy of PrfA regulation, such that the relative levels of PrfA protein present within the cell correlate with the levels of PrfA-dependent gene expression when the protein is not in its fully activated state; however, saturating levels of the protein are then quickly reached when PrfA is converted to its active form. Regulation of the PrfA activation status must be an important facet of L. monocytogenes survival, as mutations that result in constitutive PrfA activation may have deleterious consequences for bacterial physiology.
Listeria monocytogenes is a ubiquitous gram-positive bacterial pathogen that can cause serious food-borne infections in immunocompromised individuals, pregnant women, and neonates (15, 18). This facultative intracellular pathogen is able to invade a wide variety of host cells, gain entry to the cytosol, and subsequently replicate and spread to adjacent host cells (6, 7, 9, 10, 23, 24, 32, 34, 35, 46). Several virulence factors important for the different steps of L. monocytogenes infection have been identified (reviewed in references 26, 41, and 48). The majority of these virulence determinants are located within a 10-kb chromosomal region and are regulated by the positive transcriptional regulator PrfA. The prfA gene is also located within this cluster, which is commonly referred to as the PrfA regulon (29, 30). The prfA gene product is a key factor for L. monocytogenes pathogenesis, and bacterial strains lacking functional PrfA are essentially avirulent in mouse models of infection (13, 30, 31). PrfA is a 27-kDa site-specific DNA-binding protein that recognizes a 14-bp palindrome (PrfA box) within the −40 region of PrfA-dependent promoters (1, 8, 13, 40).
Multiple mechanisms exist to regulate prfA expression and protein activity. Three promoters contribute to the transcriptional regulation of prfA. The promoters prfAp1 and prfAp2 are located immediately upstream of the prfA coding region and are important for providing the initial levels of PrfA protein required for the activation of gene products essential for bacterial escape from host cell vacuoles (12, 13). These promoters are functionally redundant in vivo and appear to contribute to both positive and negative regulation of prfA expression (12, 13, 19). The third promoter contributing to regulation of prfA is located upstream of the plcA gene and increases prfA expression via the generation of a bicistronic plcA-prfA transcript; this promoter is PrfA dependent and represents a positive feedback loop for prfA expression (5, 13). The 5′ untranslated region of prfA mRNA has recently been shown to function as a thermosensor that regulates the translation of prfA mRNA in response to changes in temperature (22).
In addition to the regulation of prfA expression and mRNA translation, the activation state of PrfA appears to be regulated either through the binding of a cofactor or through some mode of posttranslational modification. The PrfA protein is a member of the Crp/Fnr family of transcriptional activators (25, 27, 41) and has significant structural and functional homology to the Escherichia coli cyclic AMP (cAMP) receptor protein (Crp) (17, 39, 41). Crp requires the binding of the cofactor cAMP for full activity, and cAMP-independent, constitutively active Crp* mutants have been described (14). A PrfA mutant analogous to Crp* has been identified in L. monocytogenes (37); a glycine-to-serine substitution at position 145 in PrfA resulted in constitutively high expression of all PrfA-regulated genes (37). The phenotype of the PrfA G145S mutant suggests that PrfA, like Crp, requires cofactor binding or modification for full activity (37). Two additional mutations in PrfA leading to activation of the protein have recently been identified (PrfA E77K and PrfA G155S); both were found to result in increased levels of PrfA-dependent gene expression, but strains containing the mutant alleles were phenotypically distinct, suggesting that the mutations altered different aspects of PrfA function (43).
In this study, we describe a fourth PrfA mutation (PrfA L140F) that results in the constitutive activation of PrfA but is also functionally distinct from the three previously described constitutive PrfA mutations.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains used in this study are listed in Table 1. E. coli DH5α was used as a host strain for recombinant plasmids. E. coli strain SM10 (44) was used as the donor strain for conjugation of pPL2 plasmid constructs. L. monocytogenes 10403S (serotype 1/2) is resistant to streptomycin and has a 50% lethal dose for mice of 2 × 104 CFU/ml (13). L. monocytogenes wild-type (WT) strain 10403S containing a chromosomal actA-gus-plcB transcriptional fusion (NF-L476) and NF-L758, a derivative of NF-L476 containing an erythromycin resistance gene (erm) inserted between orfZ and orfB downstream of the PrfA regulon, have been described (43). A 10403S L. monocytogenes strain containing a 339-bp in-frame deletion in prfA was generously provided by Hao Shen (University of Pennsylvania) and Jeff Miller (UCLA); this strain was transduced with a phage U153 lysate (20) obtained from NF-L758, and the resulting erythromycin-resistant transductant was designated NF-L1003. NF-L1003 therefore contains an in-frame deletion of prfA with a downstream copy of erm located between orfZ and orfB in the presence of a chromosomal actA-gus transcriptional fusion. L. monocytogenes strain NF-L879 was isolated as a mutant with high in vitro actA-gus expression following chemical mutagenesis of NF-L476 with ethylene methylsulfonate (EMS) (43). The integration vector pPL2 has been described (28) and was used for the construction of the recombinant pPL2prfA constructs listed in Table 1 (described below). L. monocytogenes and E. coli were grown in brain heart infusion (BHI) broth (Difco Laboratories, Detroit, Mich.) and Luria broth (LB) (Invitrogen Corp., Grand Island, N.Y.), respectively. For selection of pPL2 or its recombinant plasmids, chloramphenicol at 25 μg/ml was used for E. coli; chloramphenicol at 7.5 μg/ml was used for L. monocytogenes, and streptomycin at 200 μg/ml was used to select for L. monocytogenes following conjugation. For solid media, 2.5% (wt/vol) agar was included.
TABLE 1.
Bacterial strains used in this study
| Strain | Relevant descriptiona | Source or reference |
|---|---|---|
| DH5α | E. coli strain for constructing recombinant plasmids | |
| SM10 | E. coli strain as conjugation donor for pPL2 plasmid or constructs | 44 |
| NF-L476 | L. monocytogenes WT strain 10403S with actA-gus-plcB transcriptional fusion | 42 |
| NF-L758 | NF-L476 with an erythromycin resistance gene (erm) inserted downstream of orfXYZ at the 3′ end of the PrfA regulon | 43 |
| NF-L879 | NF-L476 EMS mutant with increased level of in vitro actA expression | 43 |
| NF-L1003 | NF-L476 ΔprfA with erythromycin resistance gene inserted downstream of the PrfA regulon | This work |
| NF-L1009 | NF-L1003 tRNAArg::pPL2 (ΔprfA + pPL2i) | This work |
| NF-L1041 | NF-L1003 tRNAArg::pPL2-prfA476 (ΔprfA + WTi) | This work |
| NF-L1042 | NF-L1003 tRNAArg::pPL2-prfAL147P (ΔprfA + L147Pi) | This work |
| NF-L1011 | NF-L1003 tRNAArg::pPL2-prfAL140F (ΔprfA + L140Fi) | This work |
| NF-L1006 | NF-L476 tRNAArg::pPL2 (WT + pPL2i) | This work |
| NF-L1039 | NF-L476 tRNAArg::pPL2-prfA476 (WT + WTi) | This work |
| NF-L1040 | NF-L476 tRNAArg::pPL2-prfAL147P (WT + L147Pi) | This work |
| NF-L1008 | NF-L476 tRNAArg::pPL2-prfAL140F (WT + L140Fi) | This work |
| NF-L1067 | NF-L879 tRNAArg::pPL2-prfAL140F (L140F + L140Fi) | This work |
pPL2i is the integration of pPL2 at tRNAArg-attBB′ of the chromosome. i indicates the integration of the particular prfA allele at tRNAArg-attBB′ of the chromosome.
Preparation of bacteriophage U153 lysates.
High-titer U153 lysates were prepared as described previously with minor modifications (20). Dilutions of a starting lysate were prepared in TM buffer (10 mM Tris-HCl, pH 7.5, 10 mM MgSO4), and 100 μl of diluted lysate was added to sterile 13- by 100-mm glass tubes containing 100 μl of mid-log-phase L. monocytogenes grown in LB with shaking at 30°C. Bacterial cultures were incubated with U153 lysate dilutions at room temperature (RT) for 40 min, and then 3 ml of molten LB plus 0.75% [wt/vol] agar-10 mM MgSO4-10 mM CaCl2 was added to each tube, followed by gentle mixing and immediate pouring of the tube contents onto LB-10 mM MgSO4-10 mM CaCl2 plates, which were then incubated at RT overnight. Plates containing U153 plaques just touching one another (giving the lawn of L. monocytogenes a lacy appearance) were selected for the preparation of fresh bacteriophage lysates. TM buffer (2 ml) was added to the selected plates, and the plates were incubated for 20 min at RT. The soft top agar layer was then broken up with a sterile glass spreader, and the agar and liquid were transferred into a sterile centrifuge tube and vortexed. The supernatant was recovered after centrifugation at 7,000 × g for 10 min, and chloroform was added to 10% (vol/vol) final volume. After the chloroform was mixed in and allowed to settle, bacteriophage lysates in the supernatant were transferred to sterile tubes and mixed with equal volumes of 100% glycerol. The lysates obtained were generally on the order of 108 to 109 PFU/ml.
U153 bacteriophage-mediated transduction.
High-titer U153 lysates (50 to 200 μl) prepared from strain NF-L758 (NF-L476 with an erm insertion downstream of the PrfA regulon) were mixed with 200 μl of mid-exponential-phase L. monocytogenes mutant strain NF-L879 (grown at 30°C with shaking) in the presence of 10 mM MgSO4 and 10 mM CaCl2, and the mixture was incubated at RT for 1 h with occasional gentle mixing. An aliquot of 3 ml of warm BHISA (BHI plus 0.75% [wt/vol] agar-10 mM sodium citrate, pH 7.5) containing 50 μg of 5-bromo-4-chloro-3-indolyl-β-d-glucuronide salt (XG; Inalco)/ml was added with gentle mixing to the bacteriophage-L. monocytogenes suspension, erythromycin at 1.3 μg/ml was included to induce erm expression, and the mixture was immediately poured onto BHI plates containing 10 mM sodium citrate (pH 7.5) and 50 μg of XG/ml. After incubation at 37°C for 2 h, another layer of BHISA containing 50 μg of XG/ml and 13 μg of erythromycin/ml was poured onto the plates, which were again incubated at 37°C. Bacterial transductants were usually visible within 1 to 2 days, and Ermr transductants were scored for blue or white colony color. Mutants containing mutations closely linked to the erm gene produced white transductants on indicator plates at a frequency that correlated with the distance of the mutation from the erm marker [distance = (1 − the cube root of the transduction frequency) × phage size (40.8 kb for U153)] (20). Transductants isolated after phage infection of mutant NF-L879 were either dark blue or white. The dark-blue ones retained the mutation that conferred a high level of in vitro actA-gus expression, whereas the white colonies contained the WT copy of prfA from NF-L758.
Construction of pPL2 recombinant plasmids.
Genomic DNA was isolated from the L. monocytogenes WT strain NF-L476 and the EMS mutant strain NF-L879 for splicing by overlap extension PCR (21) to clone the WT or mutant prfA genes with the prfA and plcA promoters and their PrfA-binding palindromes into the integrative vector pPL2. The primers 5′SacI-pPlcA (5′-GGGAATTCGGTATCAAATAAAACG-3′; the added SacI site is underlined) and 3′SalI prfA tx term (5′-AATCGCTTTCTTTACTGCAGGA-3′; the added SalI site is underlined) (sequences of both primers were provided by D. Higgins, Harvard Medical School, Boston, Mass.) were used as the external primer pair for splicing by overlap extension PCR with the internal primer pairs delplcAR (5′-TACTTTGTTGTTTAATGCTGCATTAAAATAAATTGG-3′) and delplcAF (5′-CCAATTTATTTTAATGCAGCATTAAACAACAAAGTA-3′). The internal primers generated a 453-bp in-frame deletion in plcA. An ∼500-bp product was obtained from PCR with 5′SacI-pPlcA and delplcAR, and a 1.2-kb fragment was amplified with delplcAF and 3′SalI prfA tx term. The two overlapping products formed were purified with a QIAquick gel extraction kit (Qiagen Inc., Valencia, Calif.) and subjected to a second PCR containing only the two external primers. The final product (∼1.7 kb) was gel purified, digested with SacI and SalI, and ligated to pPL2 cut with the same enzymes for transformation into E. coli DH5α. After the inserted fragments were checked by sequencing, each of the recombinant plasmids was electroporated into E. coli SM10 for conjugation into recipient L. monocytogenes strains.
Conjugation and plasmid integration.
Conjugation of the pPL2 recombinant plasmids from E. coli SM10 into recipient strains was performed as previously described (28) with slight modifications. Briefly, the bacterial strains were grown at 30°C with shaking to mid-log phase (optical density at 600 nm [OD600], ∼0.6). E. coli donor strains were grown in LB containing 25 μg of chloramphenicol/ml, and L. monocytogenes recipient strains were grown in BHI. The donor culture (250 μl) was mixed with 150 μl of recipient culture, put onto a 0.45-μm-pore-size HA-type filter (Millipore, Billerica, Mass.) on a BHI plate without antibiotics, and incubated at 30°C for 2 h. The bacterial cells were then resuspended in 2 ml of BHI, and aliquots (50 to 200 μl) were mixed with 3 ml of LB top agar and overlaid onto BHI plates with 7.5 μg of chloramphenicol/ml and 200 μg of streptomycin/ml. The plates were then incubated at 30°C overnight and shifted to 37°C for 2 to 3 days. Individual colonies were screened by PCR. The colonies were picked with sterile toothpicks, streaked onto a fresh selection plate, and then boiled at 100°C for 10 min in 75 μl of sterile water. PCR was performed with 5 μl of the boiled bacterial suspension. The primer pair NC16 (5′-GTCAAAACATACGCTCTTATC-3′) and PL95 (5′-ACATAATCAGTCCAAAGTAGATGC-3′) amplified a 499-bp product in strains that are lysogenic or contain the integration vector at tRNAArg-attBB′ (28). The primer pairs 5′SacI-pPlcA and 3′SalI prfA tx term amplified a single 2.1-kb product from NF-L476 or when the vector alone (pPL2) was integrated into the chromosome; a single 1.8-kb fragment was amplified from NF-L1003 (ΔprfA) and its pPL2 integrant. An additional band at ∼1.7-kb was obtained if a pPL2 recombinant plasmid carrying a prfA allele was integrated into the chromosome. Potential mutant insertion strains were confirmed by PCR using isolated genomic DNA.
Determination of hemolytic activity.
Overnight cultures of L. monocytogenes grown without shaking in BHI at 37°C were diluted 1:10 in fresh BHI broth and grown with shaking at 37°C. The culture supernatants were assayed for hemolytic activity with sheep erythrocytes as previously described (4). Hemolytic activity was determined as the reciprocal of the supernatant dilution at which 50% lysis of erythrocytes was observed and then normalized to the OD595 of the bacterial cultures.
Measurement of GUS activity.
Overnight cultures of L. monocytogenes grown without shaking in BHI at 37°C were diluted 1:20 into fresh BHI and grown with shaking at 37°C. Chloramphenicol at 7.5 μg/ml was included in the medium for L. monocytogenes strains integrated with the pPL2 vector or a pPL2 recombinant plasmid. At various time points, the OD595 was determined for each culture by using a spectrophotometer (UV-1201 UV-VIS spectrophotometer; Shimadzu Scientific Instruments, Inc., Columbia, Md.). Bacterial pellets were harvested from 1-ml culture suspensions by centrifugation, washed once with ABT buffer (1 M potassium phosphate [pH 7.0], 0.1 M NaCl, 1% Triton), and resuspended in 100 μl or 1 ml of the same buffer. β-Glucuronidase (GUS) activity was measured as described by Youngman (51) with the substitution of 4-methylumbelliferyl-β-d-glucuronide (Sigma, St. Louis, Mo.) in place of 4-methylumbelliferyl-β-d-galactoside.
Motility assays.
Swimming motility was evaluated on semisolid (0.3% [wt/vol] agar) BHI medium treated with 0.2% [wt/vol] activated charcoal as previously described (43). The plates were inoculated with 2 μl of mid-log-phase (OD600, ∼0.7) bacterial cultures grown in BHI at 37°C; chloramphenicol at 7.5 μg/ml was included in the media for strains with the pPL2 vector or a pPL2 recombinant plasmid integrated into the chromosome. The plates were incubated overnight at 37°C. Motility was quantified as the diameter of the swimming colony minus the diameter of a nonmotile ΔflaA L. monocytogenes deletion mutant (43). Results were obtained from duplicate samples of two independent experiments for each strain.
Plaque formation in murine L2 fibroblasts.
Plaque assays were performed with murine L2 fibroblasts as described previously (45), with a multiplicity of infection of ∼1:3. Plaque size was measured using a comparator (Finescale, Orange, Calif.). Chloramphenicol at 5 μg/ml was included in the tissue culture media for L2 cells infected by L. monocytogenes strains integrated with the pPL2 vector or a pPL2 recombinant plasmid. The average diameter of 10 plaques from at least three independent experiments was determined for each strain.
PlcB activity assay.
PlcB activities were assayed by streaking bacterial colonies onto BHI medium overlaid with molten agar containing activated-charcoal-treated (0.2% [wt/vol]) BHI and a 5% ([vol/vol]) concentration of a 1:1 egg yolk-phosphate-buffered saline (PBS) solution. The plates were then incubated at 37°C or room temperature for 1 to 2 days.
PrfA monoclonal antibodies.
Monoclonal antibody MAB-prfA55 derived against recombinant PrfA protein containing an N-terminal six-residue histidine tag was generated by the Hope Heart Institute (Seattle, Wash.).
SDS-PAGE and Western immunoblotting.
To isolate cell lysates, overnight cultures of L. monocytogenes strains were diluted 1:20 in 30 ml of fresh BHI broth and grown to an OD595 of ∼0.7 (chloramphenicol at 7.5 μg/ml was included in the media for strains with the pPL2 vector or a pPL2 recombinant plasmid integrated into the chromosome). Each culture was centrifuged at ∼5,000 × g for 10 min, washed in 1× PBS (Invitrogen, Grand Island, N.Y.), and centrifuged again. Each pellet was resuspended in 400 μl of lysis buffer (50 mM Tris-HCl, pH 7.5, 1 mM dithiothreitol, 0.1% [vol/vol] Triton X-100). Bacterial suspensions were mixed with 200 mg of glass beads (Sigma) and disrupted using a Mini-Beadbeater (Biospec Products, Barttesville, Okla.) at maximum speed for 20 s. Samples were kept on ice and then centrifuged at top speed in a microcentrifuge for 1 min to recover the supernatants. Protein concentrations were determined using the DC Protein Assay kit (Bio-Rad Laboratories, Hercules, Calif.) following instructions provided by the manufacturer. Equivalent amounts of total protein from each culture lysate were mixed with 4× sample buffer (0.25 mM Tris-HCl, pH 6.8, 8% [wt/vol] sodium dodecyl sulfate [SDS], 40% [vol/vol] glycerol, 4% [vol/vol] 2-mercaptoethanol, 0.04% bromophenol blue). Samples were heated to 100°C for 5 min prior to SDS- polyacrylamide gel electrophoresis (SDS-PAGE) on 15% polyacrylamide gels. The proteins were transferred to nitrocellulose membranes at 100 V for 1.5 h. Immunobloting for PrfA or PrfA derivatives was performed at RT. The membranes were briefly washed with 1× PBS, blocked with 5% (wt/vol) nonfat dry milk in 1× PBS for 1 h, and then incubated overnight with a 1:100 dilution of the monoclonal antibody MAB-prfA55 in 1× PBS with 5% (wt/vol) nonfat dry milk and 0.05% (vol/vol) Tween 20. After three 10-min washes in 1× PBS, the membranes were incubated with a 1:2,000 dilution of an alkaline phosphatase-conjugated goat anti-mouse antibody (Sigma, St. Louis, Mo.) for 30 min. Following three 10-min PBS washes and equilibration of the membranes for 5 min in detection buffer (100 mM Tris-HCl, 100 mM NaCl, 50 mM MgCl2, pH 9.5), PrfA proteins were detected by using Sigma Fast 5-bromo-4-chloro-3-indolyl phosphate-nitroblue tetrazolium tablets, a colorimetric alkaline phosphatase substrate. For quantitative comparison of PrfA protein levels visualized by Western analysis, serial dilutions of protein extracts were compared to determine the relative amounts of PrfA produced by different strains.
Intracellular growth in macrophage-like J774 cells.
To examine intracellular growth, cell monolayers were grown on circular coverslips (12-mm diameter) in tissue culture dishes. Bacterial cultures were grown overnight without shaking in BHI at 37°C; chloramphenicol at 7.5 μg/ml was included in the media for strains with the pPL2 vector or a pPL2 recombinant plasmid integrated into the chromosome. The cultures were washed with PBS and then used to infect J774 cells at a multiplicity of infection of ∼1:20. After infection for 30 min, the monolayers were washed three times with 37°C PBS prior to the addition of 5 ml of 37°C medium, and gentamicin was added to a final concentration of 10 μg/ml at 1 h postinfection to kill any remaining extracellular bacteria. At various time points, the coverslips were removed to determine the number of intracellular bacteria. The cell monolayers on each coverslip were lysed by vortexing them for 10 s in 5 ml of sterile water, and dilutions of the cell lysate were plated on LB plates. Bacterial CFU were determined for each strain in triplicate after overnight incubation at 37°C on LB plates.
RESULTS
Identification of a novel prfA mutation that results in constitutive activation of PrfA.
Previous experiments using EMS chemical mutagenesis resulted in the isolation of 43 L. monocytogenes mutants that exhibited high-level in vitro expression of gene products normally induced within the cytosol of infected host cells (43). These mutants were identified by screening for isolates exhibiting increased expression of actA, a gene normally expressed at low levels in bacteria grown outside of host cells, through the use of a transcriptional actA-gus reporter gene fusion located within the bacterial chromosome. Mutants with increased GUS activity were identified by the dark-blue color of the colonies on solid media containing the indicator substrate XG. One mutant, NF-L879, exhibited stable high-level in vitro actA expression based on the dark-blue color of the colonies and was selected for further study.
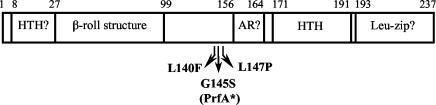
To determine whether the mutation in NF-L879 was associated with a gene located within or near the prfA locus, the L. monocytogenes-transducing bacteriophage U153 (20) was used to transduce an erythromycin resistance (erm) gene marker linked to the PrfA regulon from strain NF-L758 to the NF-L879 mutant strain. NF-L758 is a derivative of the WT strain NF-L476 (actA gus), which contains a copy of erm inserted downstream of orfXYZ at the 3′ end of the PrfA regulon. If the mutation in NF-L879 was closely linked to the PrfA regulon, then transduction of NF-L879 with a phage lysate derived from NF-L758 would result in a population of white transductants with WT levels of actA-gus expression on XG medium, as well as a population of dark-blue transductants that retained the NF-L879 mutation. The frequency of white versus blue transductants would reflect how closely linked the NF-L879 mutation was to the erm marker. If the mutation in NF-L879 was not linked to the PrfA regulon, then transduction of NF-L879 with the NF-L758-derived phage lysate would result in only dark-blue transductants. Transduction of NF-L879 resulted in the isolation of white colonies on XG medium at a frequency of 47% (31 white colonies identified from a total of 66 transductants), indicating that the mutation in NF-L879 conferring high-level in vitro actA expression was closely linked (within ∼9 kb) to the PrfA regulon. DNA sequence analysis of NF-L879 prfA revealed the presence of a CTT-to-TTT mutation within the prfA coding region, which would result in the replacement of a leucine with a phenylalanine at amino acid 140 of the protein sequence (PrfA L140F) (Fig. 1). This mutation maps near the site of another prfA mutation that has been described (37), PrfA G145S (or PrfA*), which is believed to lock the protein into a constitutively activated state (37, 49). No other mutations were found within the 9-kb region encompassing the prfA regulon of NF-L879.
FIG. 1.
Locations of PrfA mutations with respect to predicted protein secondary-structure motifs and functional regions. The previously described PrfA* mutation (G145S) is included for comparison (37). AR, activation region that may form contacts with RNA polymerase; Leu-zip, leucine zipper-like motif. Depictions of structural motifs and functional regions are adapted from Goebel et al. (17).
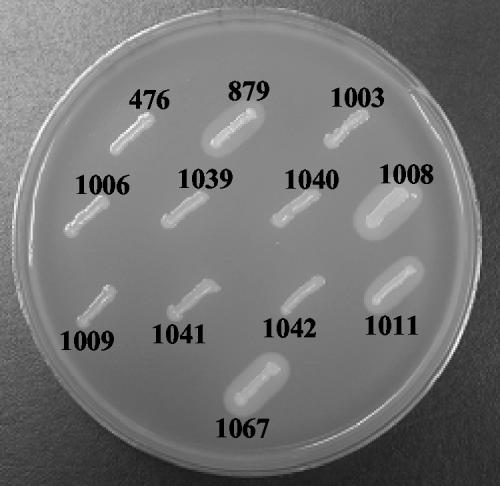
Several attempts were made to confirm that the PrfA mutation was responsible for the high actA expression phenotype via the introduction of the PrfA L140F mutation into the parent strain, NF-L476, by allelic replacement. These attempts proved unsuccessful, despite the fact that other PrfA mutations resulting in increased virulence gene expression (PrfA G145S, PrfA E77K, and PrfA G155S) have been successfully introduced into NF-L476 using this approach (43 and data not shown). The NF-L879 mutant had no obvious growth defect in comparison to the WT parent strain, NF-L476 (data not shown), but interestingly, cultures of NF-L879 grown overnight in broth culture without shaking were repeatedly found to settle to the bottoms of the culture tubes rather than to remain in suspension (Fig. 2), suggesting that the mutant cells possessed altered surface structure or cell density. A comparative Gram stain of WT and mutant bacteria taken from settled broth cultures revealed no obvious changes in cell septation or cell shape (data not shown); thus, the bacteria were apparently not settling as a result of cell filamentation.
FIG. 2.
Overnight cultures of L. monocytogenes strains grown in BHI broth at 37°C without shaking. The strain numbers and relevant genotypes are shown underneath the respective culture tubes. 476, NF-L476 (WT); 879, NF-L879 (EMS L140F mutant); 1041, NF-L1041 (ΔprfA + WTi); 1011, NF-L1011 (ΔprfA + L140Fi); 1067, NF-L1067 (L140F + L140Fi); 1006, NF-L1006 (WT + pPL2i); 1008, NF-L1008 (WT + L140Fi).
As an alternative approach to introduction of the PrfA L140F mutation by allelic exchange, an L. monocytogenes site-specific phage integration vector, pPL2 (28), was used to introduce a single copy of a WT or mutant prfA allele into strain NF-L1003, which contains an in-frame deletion within prfA in addition to the actA-gus reporter gene fusion. Recombinant pPL2 plasmids integrated at the phage attachment site of the PSA prophage within the tRNAArg gene that is present only once in the genome of L. monocytogenes (16, 28). To provide all promoters known to contribute to prfA expression, coding sequences for WT PrfA or PrfA L140F were introduced in the presence of both of the prfAp1 and prfAp2 promoters, as well as the upstream plcA promoter. During the cloning process, another PrfA mutation (PrfA L147P) (Fig. 1) was fortuitously generated, and this mutation was also independently introduced for comparison into pPL2 for integration into the chromosome of NF-L1003. The three integrants generated were NF-L1041 (containing an integrated copy of WT prfA [WTi]), NF-L1011 (containing an integrated copy of prfA L140F [L140Fi]), and NF-L1042 (containing an integrated copy of prfA L147P [L147Pi]) (Table 1). The L140Fi integrants were isolated at the same frequency and with the same colony growth characteristics as prfA, L147Pi, and pPL2 integrants on agar plates (data not shown); however, unshaken overnight cultures of NF-L1011 (ΔprfA + L140Fi) settled to the bottoms of tubes in a manner identical to that of the original EMS mutant strain, NF-L879 (Fig. 2). In addition, in contrast to NF-L1041 (ΔprfA + WTi) or NF-L1042 (ΔprfA + L147Pi), the NF-L1011 (ΔprfA + L140Fi) integrant was found to be unstable in the absence of drug selection when grown in BHI broth culture (data not shown), suggesting that the presence of the L140Fi allele may adversely affect some aspect of L. monocytogenes growth.
Analysis of PrfA protein levels in ΔprfA strains containing integrated copies of WT or mutant prfA.
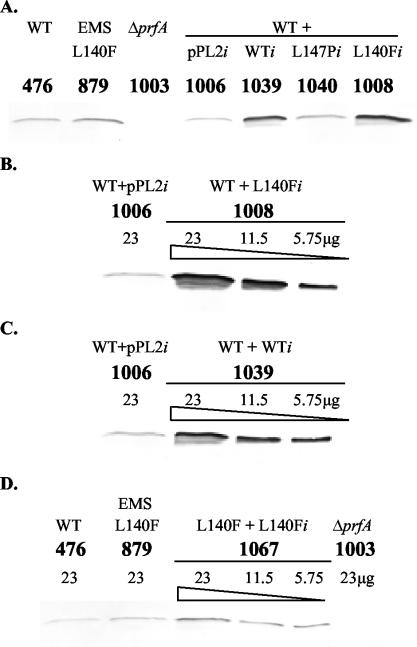
To compare the levels of PrfA present in prfA integrant strains, total cell extracts derived from broth cultures grown to similar mid-log-phase densities were examined by Western analysis using a mouse monoclonal antibody directed against PrfA. No PrfA was detected in the ΔprfA parent strain NF-L1003 or in the vector control integrant NF-L1009 (ΔprfA + pPL2i) (Fig. 3A). The EMS mutant NF-L879 produced increased levels of PrfA protein in comparison to the WT strain NF-L476 (Fig. 3A). Similarly, larger amounts of PrfA protein were detected in the integrant strain NF-L1011 (ΔprfA + L140Fi) than in the control strain NF-L1041 (ΔprfA + WTi) (Fig. 3A). Interestingly, no PrfA was detected from the PrfA L147P integrant NF-L1042 (Fig. 3A), even when a rabbit polyclonal antibody against PrfA was used for detection (data not shown). The PrfA L147P mutation therefore appeared to result in an unstable, rapidly degraded form of the protein. For quantitative comparison of PrfA protein levels, serial dilutions of protein extracts were compared (Fig. 3B and C). The EMS mutant NF-L879 produced about two to four times more PrfA protein than the WT strain NF-L476 (Fig. 3B), and the integrant strain NF-L1011 (ΔprfA + L140Fi) produced at least four times more PrfA protein than the control strain NF-L1041 (ΔprfA + WTi) (Fig. 3C). These results demonstrated that the introduction of the integrated prfA mutant and WT alleles into L. monocytogenes ΔprfA strains resulted in levels of PrfA protein that were comparable to WT and mutant protein levels produced by NF-L476 and NF-L879 strains.
FIG. 3.
Western analysis of PrfA protein levels produced by the various L. monocytogenes strains. Soluble bacterial whole-cell lysates were prepared from mid-log-phase cultures. The strain numbers and relevant genotypes are shown above the lanes. PrfA was detected by using a monoclonal antibody against PrfA and an alkaline phosphatase-conjugated goat anti-mouse secondary antibody. (A) PrfA protein levels from L. monocytogenes integrant strains in the NF-L1003 (ΔprfA) background. Equal amounts of total protein (23 μg) solubilized in SDS-PAGE sample buffer were loaded for each sample. (B) Quantitative comparison of PrfA protein levels from WT strain NF-L476 (476) and original EMS mutant strain NF-L879 (879), with the amount of total proteins loaded in each lane indicated. (C) Quantitative comparison of PrfA protein levels from integrant strains NF-L1041 (ΔprfA + WTi) (1041) and NF-L1011 (ΔprfA + L140Fi) (1011), with the amount of total proteins loaded in each lane indicated.
In vitro expression of virulence genes by prfA integrant strains.
PrfA is the major regulatory element controlling the transcription of many L. monocytogenes virulence genes (30, 31); thus, the effects of the PrfA mutations on the expression of several PrfA-dependent genes were examined. Listeriolysin O (LLO), encoded by hly, is important for L. monocytogenes to escape from primary host cell vacuoles and is normally expressed during bacterial growth in standard broth culture (3, 4, 11, 33). LLO activity can be readily detected in culture supernatants. In contrast to hly, actA is expressed at low-to-undetectable levels in broth-grown cultures but is highly induced following entry of L. monocytogenes into the host cell cytosol (2, 3, 11, 24, 33). Following bacterial spread into adjacent host cells, the activity of the phospholipase PlcB is important for bacterial escape from double-membrane vacuoles, and similar to actA, plcB is normally expressed at low-to-undetectable levels in bacteria grown in standard broth culture (38).
Relative amounts of secreted LLO were determined by measuring bacterial supernatants derived from mid-log-phase cultures for hemolytic activity against sheep red blood cells. Supernatants derived from the ΔprfA strain NF-L1003, the vector control integrant NF-L1009 (ΔprfA + pPL2i), and the PrfA L147P integrant strain NF-L1042 (ΔprfA + L147Pi) exhibited very little hemolytic activity (Table 2). The EMS mutant strain NF-L879 produced significantly larger amounts of LLO activity than the WT strain NF-L476 (Table 2), indicating that the PrfA L140F mutation is capable of elevating the expression of PrfA-dependent gene products distinct from actA. The integrated WTi allele in NF-L1041 (ΔprfA + WTi) complemented the prfA deletion in the NF-L1003 background, and similarly, the integrated L140Fi allele conferred levels of hemolytic activity on NF-L1011 (ΔprfA + L140Fi) that were comparable to those observed for NF-L879 (Table 2).
TABLE 2.
Phenotypic analysis of L. monocytogenes strains with prfA mutations
| Strain | PrfA genotype | Hemolytic activitya | Plaque sizeb | Motilityc |
|---|---|---|---|---|
| NF-L476 | WT | 100 | 100d | 100 |
| NF-L879 | L140F | 461.5 ± 225.3 | 78.9 ± 2.7d | 0.8 ± 28.5 |
| NF-L1003 | ΔprfA | 20.8 ± 22.7 | 0 | 114.2 ± 14.1 |
| NF-L1009 | ΔprfA + pPL2i | 16.3 ± 19.1 | 0 | 121.4 ± 14.7 |
| NF-L1041 | ΔprfA + WTi | 75.6 ± 28.4 | 100e | 148.6 ± 24.0 |
| NF-L1042 | ΔprfA + L147Pi | 24.5 ± 10.3 | 0 | 145.1 ± 28.7 |
| NF-L1011 | ΔprfA + L140Fi | 564.0 ± 164.8 | 90.4 ± 2.2e | 46.4 ± 22.5 |
| NF-L1006 | WT + pPL2i | 86.2 ± 27.6 | 100f | 85.6 ± 11.8 |
| NF-L1039 | WT + WTi | 187.7 ± 54.4 | 109.1 ± 4.3f | 111.0 ± 13.4 |
| NF-L1040 | WT + L147Pi | 82.2 ± 20.7 | 107.0 ± 5.4f | 130.1 ± 22.8 |
| NF-L1008 | WT + L140Fi | 884.1 ± 598.7 | 104.1 ± 8.9f | 7.2 ± 26.7 |
| NF-L1067 | L140F + L140Fi | 312.9 ± 35.9 | 100.6 ± 4.9e | 4.3 ± 21.1 |
Determined after 5 h of growth in BHI broth at 37°C; expressed as percentage ± standard deviation from at least three independent experiments, with WT set at 100%.
Expressed as percentage ± standard deviation from four independent experiments.
Soft agar (BHI-activated charcoal with 0.3% [wt/vol] agar) was inoculated with mid-log-phase bacterial cultures and incubated overnight at 37°C. Motility was measured as the diameter of the swimming colony minus that of a ΔflaA mutant and is expressed as percentage ± standard deviation from duplicate samples from two experiments, with WT set at 100%.
Percent ± standard deviation with WT (NF-L476) set at 100%.
Percent ± standard deviation with NF-L1041 set at 100%.
Percent ± standard deviation with NF-L1006 set at 100%.
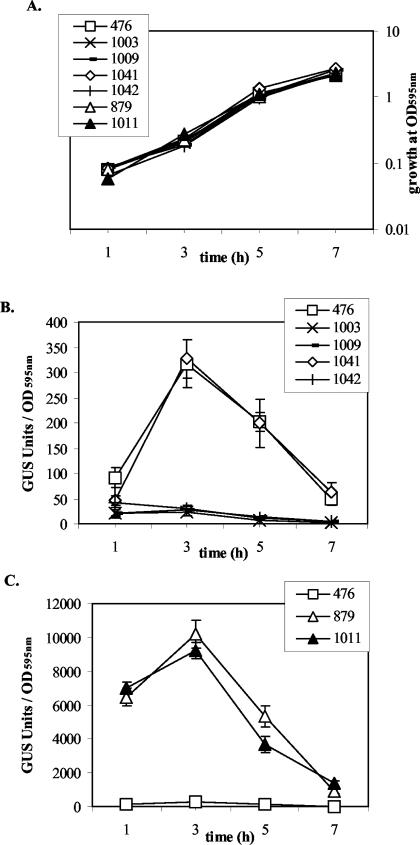
The PrfA L140F mutation was originally identified based on its apparent ability to confer high levels of actA expression in bacteria grown outside of host cells. Quantitative analysis of levels of actA expression was performed for prfA WT and mutant strains by measuring levels of GUS activity in strains containing actA-gus transcriptional fusions. As mentioned previously, the PrfA L140F integration appeared to be unstable under nonselective conditions, and all integrant strains were therefore grown in the presence of chloramphenicol. The inclusion of selective concentrations of chloramphenicol did not affect the growth rates of integrant strains compared to the rate observed for NF-L476, NF-L879, or NF-L1003 (ΔprfA) grown in BHI alone (Fig. 4A). The ΔprfA control strains NF-L1003 and NF-L1009 (ΔprfA + pPL2i) had almost undetectable levels of GUS activity, and the same was observed for the integrant NF-L1042 (ΔprfA + L147Pi) (Fig. 4B); all of these strains did not produce detectable PrfA protein as observed by Western analysis (Fig. 3A). The integration of a WTi copy into the prfA deletion strain NF-L1003 completely restored actA expression levels in NF-L1041 (ΔprfA + WTi) to those observed for the WT strain NF-L476 (Fig. 4B). actA expression of the original EMS mutant NF-L879 was observed to peak during exponential growth, and levels of expression were up to 37-fold higher than the expression levels observed for strains containing a single copy of the WT prfA allele (NF-L476 and NF-L1041) (Fig. 4C). The actA expression profile of integrant NF-L1011 (ΔprfA + L140Fi) was very similar to that of NF-L879 (Fig. 4C), indicating that the L140F mutation in PrfA was responsible for the high in vitro actA induction in the original EMS mutant. NF-L1011 (ΔprfA + L140Fi) produced increased levels of PrfA protein in comparison to the original NF-L879 mutant strain (Fig. 3A); the similar levels of actA expression observed for NF-L879 and NF-L1011, therefore, suggest that the mutant PrfA L140F reached a level of PrfA saturation beyond which there was no further increase in the induction of actA expression despite increased production of PrfA.
FIG. 4.
actA expression of PrfA mutant strains and corresponding control strains during growth in broth culture. Units of GUS activity were determined at the indicated time intervals as described in Materials and Methods and were normalized for bacterial culture OD595 (51), using 4-methylumbilliferyl-β-d-glucuronide as the substrate. The data shown are from duplicate samples and are representative of at least three independent experiments, expressed as average ± standard error. (A) Growth of WT and PrfA mutant strains in BHI broth at 37°C as measured by OD595 of cultures. (B) actA expression from strains NF-L476 (WT), NF-L1003 (ΔprfA), NF-L1009 (ΔprfA + pPL2i), NF-L1041 (ΔprfA + WTi), and NF-L1042 (ΔprfA + L147Pi). (C) actA expression from strains NF-L476 (WT), NF-L879 (EMS L140F), and NF-L1011 (ΔprfA + L140Fi).
The expression of plcB is coregulated with that of actA (42). PlcB activity was not observed for the ΔprfA control strains NF-L1003 and NF-L1009 (ΔprfA + pPL2i) or for the integrant NF-L1042 (ΔprfA + L147Pi) (Fig. 5). No PlcB activity was observed for WT strain NF-L476 and the integrant NF-L1041 (ΔprfA + WTi) (Fig. 5). In contrast, NF-L1011 (ΔprfA + L140Fi) exhibited increased levels of PlcB activity similar to that observed for the original EMS mutant NF-L879 (Fig. 5), even in media not treated with activated charcoal (treatment of BHI broth with activated charcoal has been reported to induce the expression of plcB [38]) (data not shown).
FIG. 5.
PlcB (lecithinase) phenotypes of L. monocytogenes mutants and corresponding control strains on BHI medium overlaid with molten agar containing activated-charcoal-treated (0.2% [wt/vol]) BHI and 5% (vol/vol) 1:1 egg yolk-PBS solution. The plates were incubated at 37°C overnight. 476, NF-L476 (WT); 879, NF-L879 (EMS L140F mutant); 1003, NF-L1003 (ΔprfA); 1006, NF-L1006 (WT + pPL2i); 1039, NF-L1039 (WT + WTi); 1040, NF-L1040 (WT + L147Pi); 1008, NF-L1008 (WT + L140Fi); 1009, NF-L1009 (ΔprfA + pPL2i); 1041, NF-L1041 (ΔprfA + WTi); 1042, NF-L1042 (ΔprfA + L147Pi); 1011, NF-L1011 (ΔprfA + L140Fi); 1067, NF-L1067 (L140F + L140Fi).
In summary, the introduction of a WT copy of prfA via vector integration at the tRNAArg-attBB′ site was sufficient to fully complement PrfA-dependent gene expression in the prfA deletion strain NF-L1003. The introduction of the PrfA L140F mutation demonstrated that this mutation was sufficient for generating the high-level in vitro actA-gus expression observed for the EMS mutant NF-L879; production of LLO and PlcB was also increased in both strains. In contrast, the introduction of the PrfA L147P mutation did not lead to the induction of PrfA-dependent gene expression, indicating that this mutation results in an inactive or unstable form of the protein.
The PrfA L140F mutation is phenotypically dominant to WT PrfA.
The PrfA protein is thought to be active as a multimer based on its similarity to E. coli Crp and on the recently solved PrfA crystal structure (27, 49; http://www.rcsb.org/pdb/). It seemed possible, therefore, that strains producing both the PrfA WT and PrfA L140F proteins might exhibit altered patterns of PrfA-dependent gene expression as a result of PrfA heterodimer formation. The PrfA L140F and PrfA L147P mutations, with the two prfA promoters and the plcA promoter, were therefore introduced into the chromosome of WT strain NF-L476 to examine the potential effects on PrfA-dependent gene expression. prfA merodiploid integrants NF-L1008 (WT + L140Fi) and NF-L1040 (WT + L147Pi) were generated, along with strain NF-L1006 with an integrated vector alone (WT + pPL2i) and a WTi merodiploid strain, NF-L1039 (WT + WTi), as controls. All strains exhibited apparently normal growth characteristics in broth cultures (data not shown). Similar to the EMS mutant NF-L879 and the NF-L1011 (ΔprfA + L140Fi) integrant, unshaken overnight cultures of NF-L1008 (WT + L140Fi) were observed to settle to the bottoms of culture tubes (Fig. 1).
Western analysis of PrfA protein levels in the vector control integrant NF-L1006 (WT + pPL2i) and the merodiploid NF-L1040 (WT + L147Pi) demonstrated levels of PrfA protein comparable to those of the WT strain NF-L476 (Fig. 6A). This result suggested that mutant PrfA L147P protein was not produced in NF-L1040 (WT + L147Pi) and that the presence of the mutant allele did not affect the expression or stability of WT PrfA. Increased amounts of PrfA protein were detected in extracts derived from NF-L1008 (WT + L140Fi) (Fig. 6A), with protein levels at least fourfold higher than in the vector control integrant NF-L1006 (WT + pPL2i) (Fig. 6B). Two copies of the WTi allele in NF-L1039 (WT + WTi) also led to a >4-fold increase in PrfA synthesis (Fig. 6A and C).
FIG. 6.
Western analysis of PrfA protein levels produced by the various L. monocytogenes strains. Soluble bacterial whole-cell lysates were prepared from mid-log-phase cultures. The strain numbers and relevant genotypes are shown above the lanes. PrfA was detected by using a monoclonal antibody against PrfA and an alkaline phosphatase-conjugated goat anti-mouse secondary antibody. (A) PrfA protein levels from L. monocytogenes integrant strains in the WT strain NF-L476 background. Equal amounts of total protein (23 μg) solubilized in SDS-PAGE sample buffer were loaded for each sample. (B) Quantitative comparison of PrfA protein levels from integrant strains NF-L1006 (WT + pPL2i) (1006) and NF-L1008 (WT + L140Fi) (1008). (C) Quantitative comparison of PrfA protein levels from integrant strains NF-L1006 (WT + pPL2i) and NF-L1039 (WT + WTi) (1039). (D) Quantitative comparison of PrfA protein levels from EMS mutant strain NF-L879 (879), NF-L1003 (ΔprfA) (1003), and integrant strain NF-L1067 (L140F + L140Fi) (1067). The amounts of total proteins loaded in each lane in panels B, C, and D are indicated.
The various prfA merodiploid strains were then examined for PrfA-dependent gene expression. The vector control integrant NF-L1006 (WT + pPL2i) had actA expression levels similar to those observed for WT strain NF-L476, as did the NF-L1040 (WT + L147Pi) integrant (Fig. 7B). Despite the high level of PrfA protein produced by NF-L1039 (WT + WTi) (Fig. 6A and C), this strain had only a twofold increase in actA expression in comparison to WT strains at both 3 and 5 h of growth (Fig. 7B). actA expression levels from the NF-L1008 (WT + L140Fi) integrant were the same as those from the original EMS mutant NF-L879, indicating the apparent dominance of PrfA L140F over WT PrfA (Fig. 7C).
FIG. 7.
actA expression of PrfA mutant strains and corresponding control strains during growth in broth culture. Units of GUS activity were determined at the indicated time intervals as described in Materials and Methods and were normalized for bacterial culture OD595 (51) using 4-methylumbilliferyl-β-d-glucuronide as the substrate. The data shown are from duplicate samples and are representative of at least three independent experiments, expressed as average ± standarderror. (A) Growth of WT and PrfA mutant strains in BHI broth at 37°C as measured by OD595 of cultures. (B) actA expression from strains NF-L476 (WT), NF-L1003 (ΔprfA), NF-L1006 (WT + pPL2i), NF-L1039 (WT + WTi), and NF-L1040 (WT + L147Pi). (C) actA expression from strains NF-L476 (WT), NF-L879 (EMS L140F), and NF-L1008 (WT + L140Fi). (D) actA expression from strains NF-L476 (WT), NF-L879 (EMS L140F), and NF-L1067 (L140F + L140Fi).
Similar results were obtained when the strains were examined for LLO activity. NF-L1006 (WT + pPL2i) and NF-L1040 (WT + L147Pi) produced levels of LLO that were the same as those produced by WT NF-L476, and NF-L1039 (WT + WTi) LLO activity was approximately twofold higher than that of strains with only one copy of WTi (Table 2). Integrant NF-L1008 (WT + L140Fi) retained high hemolytic activity, confirming the dominance of the PrfA L140F mutation over the WT (Table 2). An increase in PlcB activity was observed only for the NF-L1008 strain (WT + L140Fi) (Fig. 5). If there was increased PlcB activity in NF-L1039 (WT + WTi), it was too low to be detected using this assay.
Construction of an L. monocytogenes strain containing two copies of L140Fi.
To examine the potential effects on PrfA-dependent gene expression that would result from the presence of multiple copies of the PrfA L140F mutation, the L140Fi allele was introduced via integration into the EMS mutant NF-L879 to generate NF-L1067 (L140F + L140Fi). Unshaken overnight cultures of NF-L1067 were observed to settle to the bottoms of culture tubes (Fig. 1), although NF-L1067 had no obvious growth defects in broth culture (data not shown). Approximately two- to fourfold more PrfA protein was produced in NF-L1067 than in its EMS mutant parent strain, NF-L879 (Fig. 6D), but hly, actA, and plcB expression levels were not found to differ significantly between the two strains (Table 2 and Fig. 5 and 7D). These data indicate that the induction of PrfA-dependent gene expression was already saturated with the amount of activated PrfA protein synthesized from one L140Fi allele, and therefore, the introduction of a second copy resulted in no additional increase.
Intracellular growth and cell-to-cell spread of prfA mutant strains in tissue culture cells.
The capacity of L. monocytogenes to replicate within the cytosol and spread to adjacent cells can be assessed by visualizing zones of cell clearing (plaques) in monolayers of infected tissue culture cells (45). Strains that are deficient in intracellular growth and/or cell-to-cell spread fail to form plaques or form plaques of reduced size. It has been demonstrated that strains lacking functional PrfA do not escape from host cell vacuoles and are unable to replicate within cells or spread to adjacent cells; these strains do not form plaques in monolayers of mouse L2 fibroblasts (13, 30). As anticipated, strains NF-L1003 (ΔprfA), NF-L1009 (ΔprfA + pPL2i), and NF-L1042 (ΔprfA + L147Pi) failed to form plaques in monolayers of L2 fibroblasts (Table 2). The EMS mutant NF-L879 formed plaques that were ∼79% the size of those formed by WT NF-L476 (Table 2). In the presence of chloramphenicol, integrant strain NF-L1041 (ΔprfA + WTi) formed plaques smaller than those formed by NF-L476 (63.5% ± 6.4% [average ± standard deviation with NF-L476 set at 100%]); however, in the absence of the antibiotic, plaque formation was comparable to that of the WT (data not shown). Chloramphenicol therefore appeared to reduce the efficiency of plaque formation, despite the presence of the cat gene in the pPL2 integrant strains. The inclusion of chloramphenicol was necessary to ensure the stability of the integrated L140Fi allele; therefore, the drug was included for all pPL2 integrants, and the plaque size of NF-L1041 (ΔprfA + WTi) was set at 100% for comparison with mutant prfA integrant strains. The integrant strain NF-L1011 (ΔprfA + L140Fi) was still found to form reproducibly smaller plaques than strain NF-L1041 (ΔprfA + WTi); however, the plaques were only slightly (∼10%) smaller than those formed in the presence of the WTi allele (Table 2). L. monocytogenes strains containing the L140Fi allele may therefore be slightly deficient in invasion, intracellular replication, and/or cell-to-cell spread. No significant differences were observed in plaque size or frequency for any of the various prfA integrant strains in the WT background or, interestingly, for NF-L1067 containing two copies of L140Fi (Table 2). Experiments examining bacterial infection of the macrophage-like cell line J774 indicated that all integrant strains had similar intracellular growth for at least 7 h postinfection (data not shown).
L. monocytogenes strains containing L140Fi exhibit decreased swimming motility.
Previous work has demonstrated that two strains of L. monocytogenes that contained constitutively activated forms of PrfA protein (PrfA E77K and PrfA G155S) exhibited decreased swimming motility in soft agar (43). We therefore examined the swimming motility of the PrfA L140F strains. The EMS mutant NF-L879 and integrants NF-L1008 (WT + L140Fi), NF-L1011 (ΔprfA + L140Fi), and NF-L1067 (L140F + L140Fi) were all strikingly deficient in swimming motility, while the other integrants were similar in motility to WT strain NF-L476 (Table 2). Strains containing constitutively activated forms of PrfA (such as PrfA L140F) may thus be deficient in nutrient acquisition and exhibit decreased fitness for survival in environments outside of host cells.
DISCUSSION
The transcriptional regulator PrfA of L. monocytogenes tightly coordinates the expression of many virulence determinants to facilitate bacterial survival both inside and outside of host cells. Regulation of prfA expression and PrfA activity occurs on multiple levels and includes transcriptional, posttranscriptional, and posttranslational regulatory mechanisms, all of which are necessary for pathogenesis (5, 12, 13, 19, 22). In this study, we have identified a novel mutation within PrfA (PrfA L140F) that results in a constitutively activated form of the protein and high-level virulence gene expression in bacteria grown outside of host cells. The discovery of the PrfA L140F mutation brings the total number of PrfA-activating mutations defined to four, and each mutation appears to have its own distinguishable effect on PrfA function. Deciphering how specific structural alterations in the PrfA protein alter distinct functional aspects of this key virulence regulator will further illuminate how L. monocytogenes controls virulence gene expression in response to environmental cues.
In contrast to the three previously described mutations within PrfA that lead to the protein's constitutive activation (PrfA G145S, PrfA G155S, and PrfA E77K), the PrfA L140F mutation conferred some degree of toxicity upon bacterial cells. Multiple attempts to reconstruct the PrfA L140F mutation in WT L. monocytogenes by allelic replacement proved unsuccessful. In addition, when the mutant allele was integrated into an ectopic location on the L. monocytogenes chromosome (as in strain NF-L1011 [ΔprfA + L140Fi]), the L140Fi allele could not be stably maintained in the absence of selective pressure (unlike the WT allele, which was stable without selection). The observation that unshaken overnight cultures of L. monocytogenes strains harboring the L140Fi allele settled to the bottoms of culture tubes suggests that the presence of PrfA L140F results in an altered bacterial surface structure or cell density (Fig. 1). Altered bacterial cell physiology or morphology probably reflects changes in the spectrum of gene products normally produced by the bacteria during growth in broth culture; these products are likely to be either directly or indirectly regulated by PrfA. Preliminary analysis of L. monocytogenes membranes and secreted proteins of PrfA L140F strains using denaturing PAGE indicates readily detectable changes in the abundance of several polypeptide species (K. K. Y. Wong, unpublished data). The PrfA-directed alterations in L. monocytogenes bacterial surfaces or cell physiology may function to promote bacterial survival in the cytosol of infected host cells, an environment demonstrated to induce PrfA activation (42). It is notable that, in contrast to the pPL2-prfA L140F reconstructed L. monocytogenes strains, the PrfA L140F mutation is stably maintained in the original NF-L879 mutant isolate; it is possible that this strain contains additional mutation(s) that alleviate the toxicity associated with this PrfA mutant allele. NF-L879, however, formed plaques in fibroblast monolayers that were significantly smaller than those formed by WT strains and smaller than those formed by the isogenic NF-L1011 (ΔprfA + L140Fi) mutant (79% ± 3% for NF-L879 versus 90% ± 2% for NF-L1011). It is possible, therefore, that the mutation in NF-L879 that compensates for the presence of the L140Fi allele adversely affects the ability of L. monocytogenes to spread to adjacent cells. Alternatively, NF-L879 may contain an independent mutation, unrelated to the presence of L140Fi, that reduces cell-to-cell spread. We do not believe that the L140Fi suppressor mutation is present in either NF-L1011 or NF-L1008, as these strains do not stably maintain the L140Fi allele in the absence of drug selection, and multiple L. monocytogenes isolates obtained from independent pPL2-prfA L140F vector transformations were indistinguishable from NF-L1011 and NF-L1008 in independent assays (K. K. Y. Wong, unpublished data). Additionally, L. monocytogenes integrants containing the pPL2-prfA L140F plasmid were isolated at the same frequency and with the same colony growth characteristics on agar plates as integrants containing the pPL2-prfA or pPL2 vector, a result that suggests that the toxicity associated with the presence of L140Fi is not severe.
The relative levels of PrfA protein have been shown to be important for optimal induction of virulence gene expression (5, 12, 13, 36). PrfA synthesis increases upon entry of L. monocytogenes into host cells (36), and this increase is necessary to direct the expression of gene products required for bacterial spread to adjacent cells (5, 12, 13). The data presented here indicate that saturating levels of virulence gene expression are more rapidly achieved by activated forms of PrfA. For example, the introduction of a second WT copy of prfA into L. monocytogenes doubled the production of LLO and ActA (Table 2 and Fig. 7). However, no significant increase in either actA or hly expression was observed for L. monocytogenes strains containing two L140Fi alleles (NF-L1067 [L140F + L140Fi]) versus those with one copy (NF-L879 and NF-L1011 [ΔprfA + L140Fi]) (Table 2 and Fig. 4 and 7), even though the presence of a second copy increased the levels of PrfA protein produced in these strains (Fig. 6). These results suggest that the expression of PrfA-dependent genes is most sensitive to variations in PrfA protein levels when PrfA is present in its nonactivated state.
The apparent phenotypic dominance of the L140Fi allele over the WT allele is interesting in light of predictions that PrfA is active as a multimer. These predictions are based upon the homology PrfA shows with Crp and upon the recently solved crystal structure of PrfA (37, 49; http://www.rcsb.org/pdb/). The merodiploid integrant NF-L1008 (WT + L140Fi) presumably produces both WT PrfA and PrfA L140F, but this has not been definitely demonstrated. The PrfA L140F protein would be predicted to activate expression not only from its own upstream plcA promoter but also from that of the WT copy, thus leading to high-level expression of both proteins. It is possible that PrfA L140F has sufficiently high promoter binding affinity to outcompete the WT protein at all promoters examined; it has been reported that WT PrfA could compete and neutralize the activation of virulence genes by an activated PrfA form (PrfA G145S) if the WT PrfA was produced in trans from a multicopy plasmid (37). We favor the possibility that PrfA L140F may induce conformational changes in the WT protein upon multimerization that lead to activation of both PrfA molecules. Alternatively, the PrfA L140F mutation may serve to stabilize the formation of active PrfA dimers.
As mentioned above, the crystal structure of PrfA has recently been solved and released (Protein Data Bank accession code 1OMI [http://www.rcsb.org/pdb/]). The structure obtained indicates that PrfA is structurally similar to the cAMP receptor protein (Crp) from E. coli (50). Figure 8 depicts the locations of the PrfA L140F and the PrfA L147P mutations within the WT structure. Also depicted is the original PrfA G145S mutation, the first mutation identified that resulted in a constitutively active form of PrfA (PrfA*) (37). The PrfA L140F and L147P mutations are located very close to the DNA-binding helix-turn-helix (HTH) domain of PrfA. The phenylalanine side chain of PrfA L140F, based on its location within the WT structure, might possibly alter the positioning of the HTH domain to facilitate the interaction of this region with DNA; confirmation of this hypothesis awaits further structural and functional analyses of PrfA L140F. In contrast, proline substitutions often introduce dramatic turns into protein chains, and thus, the PrfA L147P substitution might result in improper folding and protein degradation, consistent with the failure of this strain to produce detectable PrfA protein.
FIG. 8.
Locations of PrfA mutations in the crystal structure of PrfA (http://www.rcsb.org/pdb/). The two diagrams illustrate two different views of the PrfA dimer. The blue and yellow traces are the Cα traces of the two monomers of PrfA. The HTH motifs are shown in red. The previously described PrfA* mutation (G145S) is included for comparison (37).
Finally, it has been proposed that L. monocytogenes is a bacterium that must mediate a balance between virulence for mammalian hosts and survival in environments outside of host cells (43). L. monocytogenes strains containing the prfA E77K or prfA G155S allele are fully virulent (or, in the case of PrfA G155S, hypervirulent) in murine models of infection; however, these mutants are severely compromised in swimming motility, a behavior that is likely to be important for nutrient acquisition outside of host cells (43). Similarly, strains containing the PrfA L140F mutation were found to be defective for swimming motility (Table 2). Elucidating how the activation status of PrfA contributes to the survival of L. monocytogenes in its varied environments will aid our understanding of how L. monocytogenes is able to adapt and flourish in such a wide diversity of habitats.
Acknowledgments
We thank Darren Higgins for the pPL2 plasmid, the SM10/pPL2 strain, and the sequences for primers 5′SacI-pPlcA and 3′SalI prfA tx term. We thank Hao Shen and Jeff Miller for the gift of the L. monocytogenes prfA deletion strain and Daniel Portnoy and Richard Calendar for the gift of the bacteriophage U153. We also thank members of the Freitag laboratory for helpful discussions and the reviewers of the manuscript for insightful comments.
This work was supported by Public Health Service grant AI41816 (N.E.F.) from the National Institutes of Health and by the M. J. Murdock Charitable Trust.
REFERENCES
- 1.Böckmann, R., C. Dickneite, B. Middendorf, W. Goebel, and Z. Sokolovic. 1996. Specific binding of the Listeria monocytogenes transcriptional regulator PrfA to target sequences requires additional factor(s) and is influenced by iron. Mol. Microbiol. 22:643-653. [DOI] [PubMed] [Google Scholar]
- 2.Brundage, R. A., G. A. Smith, A. Camilli, J. A. Theriot, and D. A. Portnoy. 1993. Expression and phosphorylation of the Listeria monocytogenes ActA protein in mammalian cells. Proc. Natl. Acad. Sci. USA 90:11890-11894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bubert, A., Z. Sokolovic, S. K. Chun, L. Papatheodorou, A. Simm, and W. Goebel. 1999. Differential expression of Listeria monocytogenes virulence genes in mammalian host cells. Mol. Gen. Genet. 261:323-336. [DOI] [PubMed] [Google Scholar]
- 4.Camilli, A., C. R. Paynton, and D. A. Portnoy. 1989. Intracellular methicillin selection of Listeria monocytogenes mutants unable to replicate in a macrophage cell line. Proc. Natl. Acad. Sci. USA 86:5522-5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Camilli, A., L. G. Tilney, and D. A. Portnoy. 1993. Dual roles of plcA in Listeria monocytogenes pathogenesis. Mol. Microbiol. 8:143-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cossart, P., M. F. Vicente, J. Mengaud, F. Baquero, J. C. Perez-Diaz, and P. Berche. 1989. Listeriolysin O is essential for virulence of Listeria monocytogenes: direct evidence obtained by gene complementation. Infect. Immun. 57:3629-3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dabiri, G. A., J. M. Sanger, D. A. Portnoy, and F. S. Southwick. 1990. Listeria monocytogenes moves rapidly through the host-cell cytoplasm by inducing directional actin assembly. Proc. Natl. Acad. Sci. USA 87:6068-6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dickneite, C., R. Böckmann, A. Spory, W. Goebel, and Z. Sokolovic. 1998. Differential interaction of the transcription factor PrfA and the PrfA-activating factor (Paf) of Listeria monocytogenes with target sequences. Mol. Microbiol. 27:915-928. [DOI] [PubMed] [Google Scholar]
- 9.Domann, E., and T. Chakraborty. 1989. Nucleotide sequence of the listeriolysin gene from a Listeria monocytogenes serotype 1/2a strain. Nucleic Acids Res. 17:6406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Domann, E., J. Wehland, M. Rohde, S. Pistor, M. Hartl, W. Goebel, M. Leimeister-Wachter, M. Wuenscher, and T. Chakraborty. 1992. A novel bacterial virulence gene in Listeria monocytogenes required for host cell microfilament interaction with homology to the proline-rich region of vinculin. EMBO J. 11:1981-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freitag, N. E., and K. E. Jacobs. 1999. Examination of Listeria monocytogenes intracellular gene expression by using the green fluorescent protein of Aequorea victoria. Infect. Immun. 67:1844-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freitag, N. E., and D. A. Portnoy. 1994. Dual promoters of the Listeria monocytogenes prfA transcriptional activator appear essential in vitro but are redundant in vivo. Mol. Microbiol. 12:845-853. [DOI] [PubMed] [Google Scholar]
- 13.Freitag, N. E., L. Rong, and D. A. Portnoy. 1993. Regulation of the prfA transcriptional activator of Listeria monocytogenes: multiple promoter elements contribute to intracellular growth and cell-to-cell spread. Infect. Immun. 61:2537-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garges, S., and S. Adhya. 1988. Cyclic AMP-induced conformational change of cyclic AMP receptor protein (CRP): intragenic suppressors of cyclic AMP-independent CRP mutations. J. Bacteriol. 170:1417-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gellin, B. G., and C. V. Broome. 1989. Listeriosis. JAMA 261:1313-1320. [PubMed] [Google Scholar]
- 16.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. G. Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 17.Goebel, W., J. Kreft, and R. Böckmann. 2000. Regulation of virulence genes in pathogenic Listeria, p. 499-506. In V. A. Fischetti et al. (ed.), Gram-positive pathogens. American Society for Microbiology, Washington, D.C.
- 18.Gray, M. L., and A. H. Killinger. 1966. Listeria monocytogenes and listeric infections. Bacteriol. Rev. 30:309-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greene, S. L., and N. E. Freitag. 2003. Negative regulation of PrfA, the key activator of Listeria monocytogenes virulence gene expression, is dispensable for bacterial pathogenesis. Microbiology 149:111-120. [DOI] [PubMed] [Google Scholar]
- 20.Hodgson, D. A. 2000. Generalized transduction of serotype 1/2 and serotype 4b strains of Listeria monocytogenes. Mol. Microbiol. 35:312-323. [DOI] [PubMed] [Google Scholar]
- 21.Horton, R. 1993. In vitro recombination and mutagenesis of DNA, p. 251-260. In B. White (ed.), PCR protocols: current methods and applications. Humana Press Inc., Totowa, N.J.
- 22.Johansson, J., P. Mandin, A. Renzoni, C. Chiaruttini, M. Springer, and P. Cossart. 2002. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell 110:551-561. [DOI] [PubMed] [Google Scholar]
- 23.Kathariou, S., P. Metz, H. Hof, and W. Goebel. 1987. Tn916-induced mutations in the hemolysin determinant affecting virulence of Listeria monocytogenes. J. Bacteriol. 169:1291-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kocks, C., E. Gouin, M. Tabouret, P. Berche, H. Ohayon, and P. Cossart. 1992. L. monocytogenes-induced actin assembly requires the actA gene product, a surface protein. Cell 68:521-531. [DOI] [PubMed] [Google Scholar]
- 25.Körner, H., H. J. Sofia, and W. G. Zumft. 2003. Phylogeny of the bacterial superfamily of Crp-Fnr transcription regulators: exploiting the metabolic spectrum by controlling alternative gene programs. FEMS Microbiol. Rev. 792:1-34. [DOI] [PubMed] [Google Scholar]
- 26.Kreft, J., and J. A. Vazquez-Boland. 2001. Regulation of virulence genes in Listeria. Int. J. Med. Microbiol. 291:145-157. [DOI] [PubMed] [Google Scholar]
- 27.Lampidis, R., R. Gross, Z. Sokolovic, W. Goebel, and J. Kreft. 1994. The virulence regulator protein of Listeria ivanovii is highly homologous to PrfA from Listeria monocytogenes and both belong to the Crp-Fnr family of transcription regulators. Mol. Microbiol. 13:141-151. [DOI] [PubMed] [Google Scholar]
- 28.Lauer, P., M. Y. Chow, M. J. Loessner, D. A. Portnoy, and R. Calendar. 2002. Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J. Bacteriol. 184:4177-4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leimeister-Wachter, M., W. Goebel, and T. Chakraborty. 1989. Mutations affecting hemolysin production in Listeria monocytogenes located outside the listeriolysin gene. FEMS Microbiol. Lett. 53:23-29. [DOI] [PubMed] [Google Scholar]
- 30.Leimeister-Wachter, M., C. Haffner, E. Domann, W. Goebel, and T. Chakraborty. 1990. Identification of a gene that positively regulates expression of listeriolysin, the major virulence factor of Listeria monocytogenes. Proc. Natl. Acad. Sci. USA 87:8336-8340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mengaud, J., S. Dramsi, E. Gouin, J. A. Vazquez-Boland, G. Milon, and P. Cossart. 1991. Pleiotropic control of Listeria monocytogenes virulence factors by a gene that is autoregulated. Mol. Microbiol. 5:2273-2283. [DOI] [PubMed] [Google Scholar]
- 32.Mengaud, J., M. F. Vicente, J. Chenevert, J. M. Pereira, C. Geoffroy, B. Gicquel-Sanzey, F. Baquero, J. C. Perez-Diaz, and P. Cossart. 1988. Expression in Escherichia coli and sequence analysis of the listeriolysin O determinant of Listeria monocytogenes. Infect. Immun. 56:766-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moors, M. A., B. Levitt, P. Youngman, and D. A. Portnoy. 1999. Expression of listeriolysin O and ActA by intracellular and extracellular Listeria monocytogenes. Infect. Immun. 67:131-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mounier, J., A. Ryter, M. Coquis-Rondon, and P. J. Sansonetti. 1990. Intracellular and cell-to-cell spread of Listeria monocytogenes involves interaction with F-actin in the enterocytelike cell line Caco-2. Infect. Immun. 58:1048-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Portnoy, D. A., T. Chakraborty, W. Goebel, and P. Cossart. 1992. Molecular determinants of Listeria monocytogenes pathogenesis. Infect. Immun. 60:1263-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Renzoni, A., P. Cossart, and S. Dramsi. 1999. PrfA, the transcriptional activator of virulence genes, is upregulated during interaction of Listeria monocytogenes with mammalian cells and in eukaryotic cell extracts. Mol. Microbiol. 34:552-561. [DOI] [PubMed] [Google Scholar]
- 37.Ripio, M. T., G. Dominguez-Bernal, M. Lara, M. Suarez, and J. A. Vazquez-Boland. 1997. A Gly145Ser substitution in the transcriptional activator PrfA causes constitutive overexpression of virulence factors in Listeria monocytogenes. J. Bacteriol. 179:1533-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ripio, M. T., G. Dominguez-Bernal, M. Suarez, K. Brehm, P. Berche, and J. A. Vazquez-Boland. 1996. Transcriptional activation of virulence genes in wild-type strains of Listeria monocytogenes in response to a change in the extracellular medium composition. Res. Microbiol. 147:371-384. [DOI] [PubMed] [Google Scholar]
- 39.Sheehan, B., A. Klarsfeld, R. Ebright, and P. Cossart. 1996. A single substitution in the putative helix-turn-helix motif of the pleiotropic activator PrfA attenuates Listeria monocytogenes virulence. Mol. Microbiol. 20:785-797. [DOI] [PubMed] [Google Scholar]
- 40.Sheehan, B., A. Klarsfeld, T. Msadek, and P. Cossart. 1995. Differential activation of virulence gene expression by PrfA, the Listeria monocytogenes virulence regulator. J. Bacteriol. 177:6469-6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sheehan, B., C. Kocks, S. Dramsi, E. Gouin, A. D. Klarsfeld, J. Mengaud, and P. Cossart. 1994. Molecular and genetic determinants of the Listeria monocytogenes infectious process. Curr. Top. Microbiol. 192:187-216. [DOI] [PubMed] [Google Scholar]
- 42.Shetron-Rama, L. M., H. Marquis, H. G. Bouwer, and N. E. Freitag. 2002. Intracellular induction of Listeria monocytogenes actA expression. Infect. Immun. 70:1087-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shetron-Rama, L. M., K. Mueller, J. M. Bravo, H. G. Bouwer, S. S. Way, and N. E. Freitag. 2003. Isolation of Listeria monocytogenes mutants with high-level in vitro expression of host cytosol-induced gene products. Mol. Microbiol. 48:1537-1551. [DOI] [PubMed] [Google Scholar]
- 44.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vitro genetic engineering: transponson mutagenesis in Gram negative bacteria. Bio/Technol. 1:784-791. [Google Scholar]
- 45.Sun, A. N., A. Camilli, and D. A. Portnoy. 1990. Isolation of Listeria monocytogenes small-plaque mutants defective for intracellular growth and cell-to-cell spread. Infect. Immun. 58:3770-3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tilney, L. G., P. S. Connelly, and D. A. Portnoy. 1990. Actin filament nucleation by the bacterial pathogen, Listeria monocytogenes. J. Cell Biol. 111:2979-2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tilney, L. G., and D. A. Portnoy. 1989. Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite, Listeria monocytogenes. J. Cell Biol. 109:1597-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vazquez-Boland, J. A., M. Kuhn, P. Berche, T. Chakraborty, G. Dominguez-Bernal, W. Goebel, B. Gonzalez-Zorn, J. Wehland, and J. Kreft. 2001. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14:584-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vega, Y., C. Dickneite, M. T. Ripio, R. Böckmann, B. Gonzalez-Zorn, S. Novella, G. Dominguez-Bernal, W. Goebel, and J. A. Vazquez-Boland. 1998. Functional similarities between the Listeria monocytogenes virulence regulator PrfA and cyclic AMP receptor protein: the PrfA* (Gly145Ser) mutation increases binding affinity for target DNA. J. Bacteriol. 180:6655-6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weber, I. T., and T. A. Steitz. 1987. Structure of a complex of catabolite gene activator protein and cyclic AMP refined at 2.5 Å resolution. J. Mol. Biol. 198:311-326. [DOI] [PubMed] [Google Scholar]
- 51.Youngman, P. 1987. Plasmid vectors for recovering and exploiting Tn917 transpositions in Bacillus and other gram-positive bacteria, p. 79-103. In K. Hardy (ed.), Plasmids: a practical approach. IRL Press, Oxford, England.