Abstract
Due to the acidic nature of the stomach, enteric organisms must withstand extreme acid stress for colonization and pathogenesis. Escherichia coli contains several acid resistance systems that protect cells to pH 2. One acid resistance system, acid resistance system 2 (AR2), requires extracellular glutamate, while another (AR3) requires extracellular arginine. Little is known about how these systems protect cells from acid stress. AR2 and AR3 are thought to consume intracellular protons through amino acid decarboxylation. Antiport mechanisms then exchange decarboxylation products for new amino acid substrates. This form of proton consumption could maintain an internal pH (pHi) conducive to cell survival. The model was tested by estimating the pHi and transmembrane potential (ΔΨ) of cells acid stressed at pH 2.5. During acid challenge, glutamate- and arginine-dependent systems elevated pHi from 3.6 to 4.2 and 4.7, respectively. However, when pHi was manipulated to 4.0 in the presence or absence of glutamate, only cultures challenged in the presence of glutamate survived, indicating that a physiological parameter aside from pHi was also important. Measurements of ΔΨ indicated that amino acid-dependent acid resistance systems help convert membrane potential from an inside negative to inside positive charge, an established acidophile strategy used to survive extreme acidic environments. Thus, reversing ΔΨ may be a more important acid resistance strategy than maintaining a specific pHi value.
Enteric organisms that colonize and cause disease in the human intestine must first endure a transient but extreme acid challenge in the stomach. The normal human stomach presents an antimicrobial acid environment averaging pH 2, with an emptying time of approximately 2 h (53). As a result, acid-sensitive pathogens like Vibrio cholerae must be ingested in massive numbers (10 to 100 million) to increase the possibility that some will survive and enter the intestine. Other microbes, such as Escherichia coli and Shigella, can colonize or cause disease even when small numbers of cells (10 to 100) are ingested. These resilient microbes are equipped with potent acid resistance systems able to withstand pH 2 challenge for at least 2 h (11, 31, 32, 52). In fact, E. coli possesses a level of acid resistance rivaling that of the gastric pathogen Helicobacter pylori (39, 45, 50, 59).
It has now been shown that E. coli uses four inducible acid resistance systems to survive extreme acid environments. Acid resistance system 1 (AR1), also referred to as the oxidative or glucose-repressed system, is acid induced in stationary phase. Its expression requires the alternative sigma factor RpoS and the cyclic AMP receptor protein CRP (11). However, the structural components of AR1 as well as the mechanism by which it protects are still unknown. The second AR system, AR2, requires extracellular glutamate to work at pH 2.0 and is induced upon entry into stationary phase or by log-phase growth in acidic minimal medium (10). Known components of glutamate-dependent acid resistance include two isoforms of glutamate decarboxylase (GadA and GadB) and a putative glutamate:γ-aminobutyric acid (GABA) antiporter called GadC (11, 12, 19, 33, 46). The third system, AR3, is similar to AR2 except that AR3 only protects cells if extracellular arginine is present. AR3 is induced by low pH under anaerobic conditions and has only been demonstrated following growth in complex media. This arginine-dependent system is composed of the acid-inducible arginine decarboxylase AdiA and the AdiC antiporter, which exchanges extracellular arginine for the intracellular end product of decarboxylation, agmatine (11, 15, 22, 31). The last AR system was recently described as lysine dependent and probably involves the inducible lysine decarboxylase (22).
Although it is clear that these systems protect E. coli during transient exposure to pH 2, how they actually function has been subject to speculation. It is believed that AR2 and AR3 protect the cell from acid stress by consuming intracellular protons during each decarboxylation reaction (11, 14). The siphoning off of intracellular protons was proposed to enhance pH homeostasis and allow the cell to maintain an internal pH compatible with viability. This model suggests that a specific internal pH may be crucial for survival during exposure to extreme acid stress. If the cell's internal pH fell below that point, it would succumb. The data obtained in the present study indicate that maintenance of a specific internal pH may not be paramount to cell survival. Survival may depend on E. coli taking an approach used by acidophiles, which is to reverse the electrical membrane potential (ΔΨ) in the presence of extreme low pH. Converting membrane potential from negative inside to positive inside can repel protons and mitigate the excess proton motive force (PMF) that can form.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The strains used in this study included EK227, wild-type K-12; EK592, wild-type MG1655; EK590, ΔclcB ΔclcA (derived from MG1655 [24]); EF333, gadC::Tn10 (3); EF522, gadA::pRR10 (AP) gadB::Tn10 (3); and EF996, ΔuncB-C ilv::Tn10 (derived from EK227). Media included Luria-Bertani broth (LB) and brain heart infusion (BHI) medium containing 0.4% glucose (LBG and BHIG). LB broth, where indicated, was buffered with either 100 mM morpholinepropanesulfonic acid (MOPS; pH 8.0) or morpholineethanesulfonic acid (MES; pH 5.0). For internal pH measurements, these media also contained 25 mM sucrose to block nonspecific binding of radiolabeled sucrose (see below). Acid challenge medium was minimal EG (58) prepared at various pH values (adjusted with HCl). For the reasons noted above, EG also contained 25 mM sucrose when used for internal pH measurements. To test whether Na+- or K+-deficient medium was important, Milli-Q Ultrapure water was prepared at pH 2.5 either with or without 1 mM glutamic acid · HCl. In addition, a Na+- and K+-deficient medium (M63 K/Na-deficient) containing 15 mM (NH4)2SO4, 18 μM FeSO4 · 7H2O, 1 mM MgSO4, and 0.2% glucose was used. All chemicals used were ultrapure (Sigma or Fluka) or Suprapur (EM). Na+ and K+ measurements were made using sodium and potassium ion-specific combination electrodes (Thermo-Orion). Cultures were grown at 37°C with shaking at 220 rpm. Exponential-phase cultures were grown to an optical density at 600 nm of 0.4 (2 × 108 CFU/ml), while stationary-phase cultures were grown overnight (18 h).
Acid resistance assays.
Acid resistance assays were performed as described previously (11). Briefly, cells were grown overnight in LB MOPS and LB MES for AR1, LBG for AR2, or BHIG for AR3. LBG was used when studying the glutamate-dependent system, but since arginine decarboxylase is not induced well in LBG, BHIG was used to induce this system. Stationary-phase cultures were diluted 1:1,000 into prewarmed pH 2.5 EG medium without amino acid supplementation (for AR1), with 1.6 mM glutamate (for AR2), or with 1.0 mM arginine (AR3). At various time points, 10-μl aliquots were removed and serially diluted, and 10 μl of each dilution was plated on LB plates. CFU were determined, and percent survival was calculated relative to time zero.
Internal pH measurements.
Internal pH was estimated by measuring the distribution of a weak acid (radiolabeled salicylic acid) across the membrane (4, 8, 23). Salicylate has been used by us and others as a reliable indicator of internal pH (13, 24, 26, 34, 47). Control cultures were grown to log phase (2 × 108 CFU per ml) or stationary phase (109CFU per ml) in LBG containing 25 mM sucrose (final pH at time of assay, 6.9). Sucrose was added to prevent nonspecific binding of radiolabeled sucrose, used later for water space measurements. Cultures to test decarboxylase-dependent effects on internal pH were grown overnight to stationary phase in LBG containing 25 mM sucrose for AR2 measurements or in BHIG containing 25 mM sucrose for AR3 measurements. Cultures were then harvested by centrifugation and resuspended in pH 7 EG medium containing no additions or in pH 2.5 EG medium with and without 40 mM glutamate or arginine. Final cell density after resuspension was 6 × 109 CFU/ml. Two reactions were required for each assay. A total of 2,000 to 3,000 dpm of 3H2O/μl (0.1 μCi/μl) was added to each reaction mixture. Next, 2,000 to 3,000 dpm of [14C]salicylate (56.5 mCi/mmol) was added to one tube, and the same amount of [14C]sucrose (462 mCi/mmol) was added to the other at time zero. At specific times (30 or 60 min), 5 μl was taken from each tube for a direct isotope count, and 100-μl aliquots were centrifuged through equal amounts of dibutylphthalate (50 μl) and silicone oil (50 μl) to separate the supernatant from the cell pellet. The amount of [14C]sucrose, which does not penetrate the cytoplasmic membrane, was used to determine the extracellular and periplasmic water space remaining after centrifugation. Disintegrations per minute for [14C]salicylate and 3H2O were then used to determine the internal pH value by the following equation: pHi = log{([Ain]/[Aout]) (10pKa + 10pHout) − 10pKa}, where [A] is a measure of salicylic acid and the pKa is 3.0.
Ψ measurements.
ΔΨ was measured using radiolabeled lipophilic anions or cations (4, 23). Cells were grown as for internal pH measurements. Log-phase and stationary-phase cultures were harvested by centrifugation and resuspended in pH 2.5 EG medium containing 40 mM glutamate or 40 mM arginine. Final cell densities were approximately 6 × 109 CFU per ml. Each measurement involved two assays, one for charge distribution and one to determine water space. The general methodology was similar to that used to estimate internal pH. For charge distribution, 1,200 dpm of [14C]tetraphenylphosphonium bromide (TPP+; 5 mCi/mmol) or potassium [14C]thiocyanate (S14CN−; 54 mCi/mmol)/μl was added to one tube along with 2,000 to 3,000 dpm of 3H2O. Extracellular and intracellular water spaces were determined as above. Extracellular water space was used to correct for the carryover of extracellular radiolabeled lipophilic ion not removed during centrifugation. At 30 min, 5 μl was taken for total direct counts and 100 μl was centrifuged through dibutylphthalate-silicone oil. The accumulations of [14C]TPP+ (or S14CN−) and 3H2O in cell pellets were used to determine ΔΨ (23) by the following equation: ΔΨ = RT/zF · ln([Aout]/[Ain]), where R is the gas constant (8.28 J/K · mol), T is temperature (310.15 K), z is the charge of the molecule (+ or −), and F is the Faraday constant (96,485 J/V · mol).
Similar assays were done with butanol-treated cells (20% butanol) to determine nonspecific binding, which was subtracted as background from the experimental results. Background counts were no more than 10% of experimental counts.
Whole-cell decarboxylation and antiport measurements.
Transport and conversion of [3H]arginine and [3H]glutamate to [3H]agmatine or [3H]γ-aminobutyric acid, respectively, and the subsequent end product efflux were measured using intact and Triton X-100-permeabilized cells. Wild-type and gadA/B and gadC mutant cells were grown for 22 h in 3 ml of BHIG (for arginine decarboxylation) or LBG (for glutamate decarboxylation). Cells were harvested by centrifugation, washed twice with EG medium (pH 7.0), and resuspended to 108 cells/ml in 3.0 ml of prewarmed EG medium adjusted to pH 2.5 with HCl or to other pH values as indicated. The medium contained 1.0 mM arginine, including 4 μCi of [3H]arginine (61 Ci/mmol) per ml or a final concentration of 0.4 mM glutamate including 22,000 dpm of [3H]glutamate/μl. At timed intervals, 500-μl aliquots of cell-free supernatants were obtained by filtration (0.45-μm-pore-size filters) and adjusted to pH 7.5 with 5 N NaOH, and 30-μl samples were subjected to paper chromatographic separation as described previously (29). Marked bands were cut and counted for radioactivity. Percent conversion was calculated from total radioactivity on each strip.
Western blot analysis and cellular location of GadC.
Cells were grown overnight in 50 ml of EG pH 7.7 and EG pH 5.5. The cells were harvested by centrifugation at 4,500 × g for 10 min (4°C), resuspended in 5 ml of cold 10 mM HEPES buffer (pH 7.4), and passed twice through a French pressure cell (Aminco) at 16,000 lb/in2. Crude extracts were cleared of cell debris by centrifugation at 4,500 × g for 10 min (two times). The resulting cleared supernatant was ultracentrifuged at 90,000 × g (4°C) to separate membrane and soluble fractions. Membrane pellets were washed with 2.0 ml of 10 mM HEPES buffer (pH 7.4) to remove residual soluble proteins and resuspended in 300 μl of the same buffer. Soluble fractions were also centrifuged at 90,000 × g to remove residual membrane. Protein concentrations were measured using the Bio-Rad protein assay reagent.
Western blot analysis of these fractions was carried out using rabbit anti-GadC antibodies raised against a GadC-specific peptide (N′-CRARSPHYIVMNDKKH) by Genemed Synthesis, Inc. Membrane and soluble fractions (3 μg of protein) were separated on 10% polyacrylamide-sodium dodecyl sulfate gels (30). Proteins were transferred to Immobilon-P (polyvinylidene difluoride) membranes with a Semiphore transfer cell (Hoefer Scientific) at 100 mA for 2 h. The membranes were blocked with 5% nonfat milk in Tris-buffered saline (10 mM Tris [pH 8], 150 mM NaCl) containing 0.05% Tween 20 and incubated with rabbit primary (1:8,000) and mouse anti-rabbit secondary (1:8,000) antibodies for 1 h at room temperature. The blot was developed with ECL detection reagents (Amersham Pharmacia Biotech).
RESULTS
The role of Cl− transporters and the F0/F1 proton-translocating ATPase in acid resistance.
The goal of this study was to further define how the amino acid-dependent AR systems protect against acid stress. We initially wanted to determine how similar these systems were to the amino acid-independent system AR1, whose mechanism is also a mystery. An elegant study by Iyer et al. previously demonstrated that Cl− transporters encoded by the clc genes of E. coli were important for AR2 and AR3 function (1, 21). However, their potential role in system 1 was not explored. Consequently, we examined whether the Clc transporters also contributed to AR1.
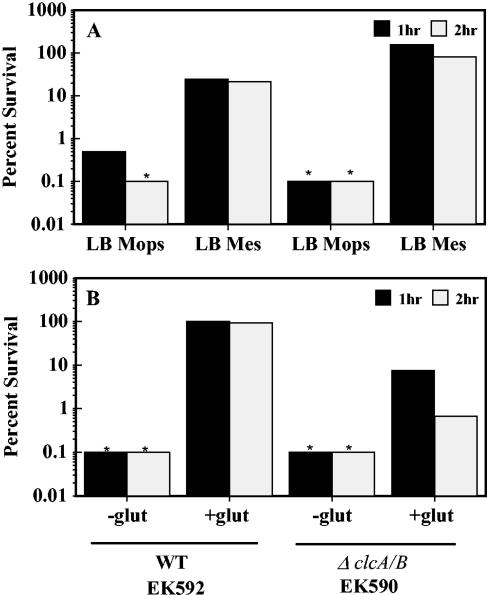
Wild-type and ΔclcA/B cells were grown to stationary phase in LB MES pH 5.5, an inducing condition for AR1, and then tested for survival in pH 2.5 minimal medium without amino acid supplementation (Fig. 1A). Both strains exhibited normal AR1 survival, indicating that the Clc Cl− transporters are not required for AR1. Our results also confirmed that the clc transporters are not essential but are important to both AR2 (Fig. 1B) and AR3 (data not shown) function. These data indicate that a fundamental mechanistic difference exists between the amino acid-dependent and -independent acid resistance mechanisms.
FIG. 1.
Clc Cl− transporters and acid resistance. (A) Cells (EK592 and EK590) were grown to stationary phase in LB MOPS (pH 8.0) and LB MES (pH 5.5) and diluted 1:1,000 into EG pH 2.5 without exogenous amino acids to test AR1. (B) Cells were grown in LBG to stationary phase and diluted 1:1,000 into EG pH 2.5 medium with and without 1.6 mM sodium glutamate to test AR2.
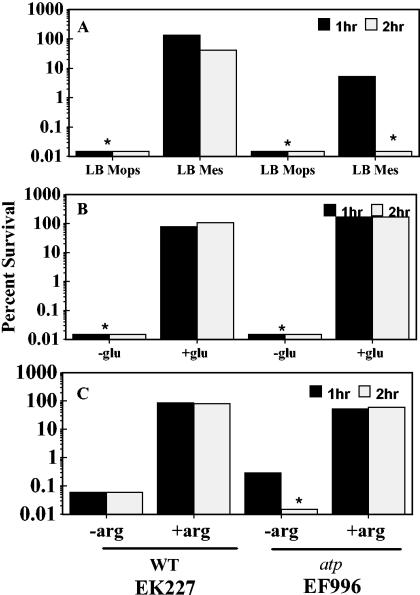
Another potential contributor to acid resistance is the F0/F1 H+-translocating ATPase. The F0/F1 ATPase is a well-established mover of protons across the cell membrane. This complex couples the energy released as protons move into the cell to the generation of ATP from ADP and Pi (56). The ATPase can also function in the opposite direction, hydrolyzing ATP to pump protons out of the cell (28, 60). Because the basic problem of acid stress is thought to be cytoplasmic acidification, a H+ pumping system like the F0/F1 ATPase has the potential to aid in resistance to extreme acid stress (26). To explore this possibility, each of the three acid resistance systems was tested for its dependence on the proton-translocating ATPase by comparing the pH 2.5 resistance of an atp mutant strain to that of the wild type. The data presented in Fig. 2A reveal that the ATPase was important for the protection provided by AR1. However, AR1 was not totally dependent on the ATPase, since residual survival was seen in the atp mutant. In contrast, the ATPase was not needed for AR2 or AR3 to function properly and did not function as a proton extruder in this situation. (Fig. 2B and C). These results and the chloride transporter results revealed fundamental differences between amino acid-dependent and -independent acid resistance mechanisms.
FIG. 2.
The F0/F1 proton-translocating ATPase is required by AR1. (A) Cells (EK227 and EF996) were grown in LB MOPS (pH 8.0) or LB MES (pH 5.5) to stationary phase and diluted 1:1,000 into EG pH 2.5 medium without exogenous amino acids (pH 5.5 induces AR1 function). (B) The same cells were grown in LBG to repress AR1 and diluted 1:1,000 into EG pH 2.5 medium, with or without 1.6 mM sodium glutamate. (C) Cells were grown in BHIG to repress AR1 and diluted 1:1,000 into EG pH 2.5 medium with or without 1 mM l-arginine. *, below the level of detection.
These results also addressed another question. Some amino acid decarboxylation reactions in other organisms are coupled to ATP synthesis carried out by the F0/F1 proton-translocating ATPase (2, 40, 44, 49). Knowing that the ATPase is not required for systems 2 and 3 indicates that any effect systems 2 or 3 may have on internal pH is most likely due to the glutamate and arginine decarboxylation reactions themselves and not due to an indirect effect on ATP synthesis.
AR2 and AR3 help acid-stressed cells maintain an elevated internal pH.
We then examined if the amino acid-dependent systems (AR2 and AR3) contribute to internal pH homeostasis as predicted. Internal pH measurements were made on cells suspended at various external pH values (Table 1). Control experiments using pH 7-grown exponential-phase cells gave an internal pH value (pH 7.8) comparable to that in previous reports (51, 57). Cells grown at pH 7 to stationary phase had an internal pH of 7.6.
TABLE 1.
Effects of AR2 and AR3 on Δψ during acid challenge of EK227
| Expt no. | Systema | Addition | % Survival | External pHb | Internal pHc | ΔpH | Δψc (mV) | PMF (mV) |
|---|---|---|---|---|---|---|---|---|
| 1 | Log phase | None | NDd | 6.9 ± 0.2e | 7.8 ± 0.1 | −53 ± 4 | −86 ± 3 | −139 |
| Stationary phase | None | ND | 6.8 ± 0.2 | 7.6 ± 0.1 | −44 ± 4 | −52 ± 6 | −96 | |
| 2 | AR2 | None | <0.1 | 2.3 ± 0.0 | 3.6 ± 0.1 | −73 ± 5 | −5 ± 4 | −78 |
| Glutamate | 100 | 2.5 ± 0.1 | 4.2 ± 0.1 | −108 ± 6 | 30 ± 6 | −78 | ||
| 3 | AR3 | None | <0.1 | 2.4 ± 0.1 | 3.7 ± 0.1 | −80 ± 7 | 4 ± 1 | −76 |
| Arginine | 75 | 2.6 ± 0.1 | 4.7 ± 0.1 | −128 ± 2 | 81 ± 17 | −47 |
Cells used in experiments 1 and 2 were grown in LBG medium to induce AR2. Cells used in experiment 3 were grown in BHIG to induce AR3.
Starting external pH values were the same with and without glutamate, but the decarboxylation of glutamate over 30 min raised external pH slightly.
Internal pH and Δψ were measured after 30 min of acid challenge.
ND, not determined.
Standard error of the mean.
When stationary-phase cells were examined during acid stress at external pH 2.5, differences in internal pH were observed depending on whether the cells were challenged in the presence or absence of glutamate or arginine. In the absence of amino acid supplementation, the internal pH was 3.6. When glutamate or arginine was added, internal pH rose to pH 4.2 and 4.7, respectively (Table 1). These results indicate that AR2 and AR3 do elevate internal pH. However, the internal pH achieved was lower than anticipated. Based on studies with Salmonella, we expected that internal pH levels lower than pH 5.5 would be lethal (13, 43). The internal pH gained by the arginine-dependent system was higher than with the glutamate-dependent system, yet survival was somewhat lower. The difference in internal pH attained by these different systems suggested that maintenance of internal pH is not the only element contributing to survival. AR1 and the lysine-dependent system were not tested for effects on internal pH because viability could not be maintained above 10% over the course of the experiments.
Internal pH levels achieved during acid stress correlate to the pH of maximal glutamate or arginine decarboxylation.
Since the internal pH measured in acid-resistant cells was lower than anticipated, we sought another method to confirm the readings. One approach used various forms of green fluorescent protein, whose fluorescence level changes with pH. The results indicated that the internal pH at external pH 2.5 was below the range of detection with this method (pH 5) (data not shown). This supported the earlier estimates. We then realized that the reported pH optima of the decarboxylases (pH 4 and 5 for glutamate and arginine decarboxylases, respectively) seemed remarkably similar to the internal pH measurements. If this correlation held, one would predict that decarboxylation by intact cells should be maximal at external pH 2.5, at which internal pH (pH 4 to 5) was near the pH optima of the decarboxylases.
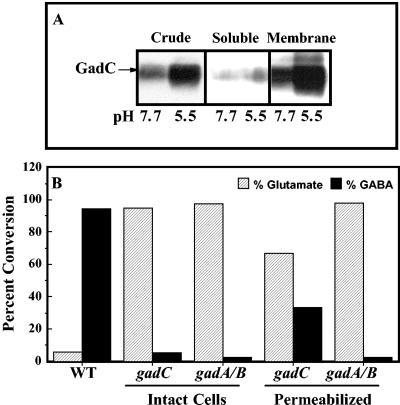
Before determining the pH at which intact cells maximally converted glutamate to GABA, we established that GadC serves as the antiporter for glutamate-dependent acid resistance. Previous work from our laboratory revealed that AdiC was the antiporter for the arginine-dependent acid resistance system (11, 15, 31). GadC, however, is only presumed to be the glutamate:GABA antiporter, based on computer sequence analysis. To provide evidence that GadC is the antiporter, we first established that this protein was membrane associated (Fig. 3A) and then demonstrated that gadC mutants would only decarboxylate glutamic acid if Triton X-100 were used to solubilize the membrane and bypass the transport requirement (Fig. 3B).
FIG. 3.
GadC is the antiporter for glutamate-dependent acid resistance. (A) Western blot assay. Cells were grown to stationary phase in minimal EG medium, and extracts were prepared with a French press. Crude extracts were separated into soluble and insoluble membrane pellets by ultracentrifugation. Samples of each fraction were electrophoresed through a 10% polyacrylamide gel electrophoresis gel and probed with anti-GadC antibody. (B) GadC is required for intact cells to convert glutamic acid to GABA. Cells were grown in LBG to stationary phase, washed in minimal EG pH 7.0 medium, and resuspended in EG at different pH values. Radiolabeled glutamic acid was added to suspensions of intact cells placed at pH 2.5 and to suspensions of cells permeabilized with 0.1% Triton X at pH 4.4, the reported internal pH of acid-challenged cells. Substrate (striped bars) and product (solid bars) present in filtered supernatants were separated by paper chromatography and identified by staining with 0.3% ninhydrin. WT, EK227; gadC, EF962; gadAB, EF522.
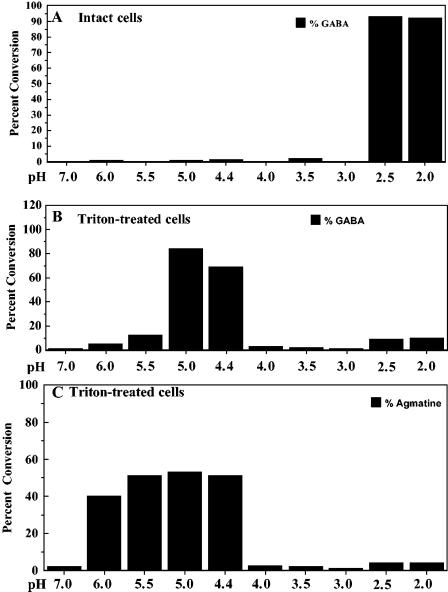
We then measured the extracellular pH at which intact cells carried out maximal transport and conversion of glutamate to GABA (or arginine to agmatine) and compared those values to what occurred with Triton X-100-permeabilized cells, where internal pH approximates the external pH. Conversion of substrate to product was determined using equal amounts of protein from intact and permeabilized cells, equal amounts of substrate, and equal reaction times as described in Materials and Methods. The decarboxylation of glutamate to GABA in permeabilized cells occurred maximally between pH 4.4 and 5 (Fig. 4B). Under similar conditions, maximal conversion of arginine to agmatine took place between pH 5 and 5.5 (Fig. 4C). These values were in agreement with published reports of pH optima using cell extracts (5, 55). However, the situation was very different using intact cells, where pH homeostasis mechanisms such as amino acid decarboxylation operate to alkalinize internal pH relative to external pH. Under these conditions, maximal conversion for both systems occurred around pH 2.5, as predicted (Fig. 4A) (11). The results suggest that optimal decarboxylation at external pH 2.5 by intact cells is consistent with an internal pH between pH 4 and 5 and that the different internal pH values generated using glutamate versus arginine might reflect the different pH optima of the two decarboxylases.
FIG. 4.
Intracellular pH optima of glutamate and arginine decarboxylases correlate to the internal pH of acid-stressed cells. EK227 cells were grown to stationary phase in LBG. Conversions of glutamate and arginine to GABA and agmatine, respectively, were carried out essentially as described in the legend for Fig. 3. Cells, either intact or Triton X solubilized, were resuspended at different pH values, and radiolabeled substrate was added.
Of course, for intact cells, the external pH allowing maximal conversion is factorial, reflecting separate pH optima of transport and decarboxylation. Although it has yet not been tested, it is possible that the antiporters only activate at external pH 2.5. Thus, the optimal pH of substrate-to-product conversion by intact cells may only reflect transport. However, once the amino acids enter the cell, it seems unlikely that the decarboxylases could cause internal pH to rise much above their pH optima.
A specific internal pH value is not required for extreme acid stress survival.
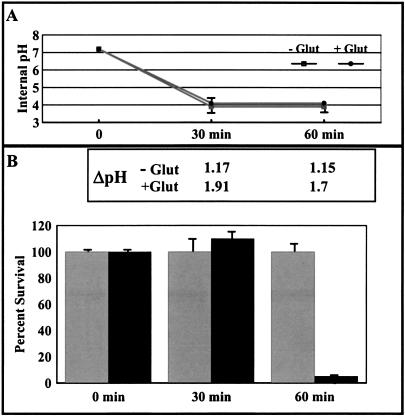
Taking into consideration that the internal pH values calculated for AR2 and AR3 were different from one another yet both systems protected cells from acid stress, an experiment was performed to determine if a specific internal pH was required for survival during acid stress. Cells were acid challenged with and without glutamate, but culture conditions were manipulated such that the internal pH of both cultures was equal. If a given pH were all that was required for survival, then both cultures should survive equally well. To conduct this experiment, the external pH values of the two cultures were adjusted independently so that the internal pH values were equivalent over the time of the experiment (Fig. 5A). When this was done, cells challenged in the presence of glutamate still survived better than cells challenged without glutamate (Fig. 5B, 60 min). This result supports the hypothesis that achieving a specific internal pH value is not the only goal of these acid resistance systems. Since cells challenged at pH 2.5 in the presence of glutamate or arginine still generated a greater ΔpH than cells challenged without amino acid, maintenance of ΔpH may be of greater importance to survival than achieving a specific internal pH value (Fig. 5B, inset).
FIG. 5.
A specific internal pH is not the only requirement for acid stress survival. Cells (EK227) were grown in LBG to stationary phase. A 1-ml aliquot of the culture was harvested and resuspended in 200 μl of EG pH 2.3 medium with 40 mM sodium glutamate or pH 2.7 medium without exogenous glutamate. (A) Internal pH measurements were made at 0, 30, and 60 min after acid challenge. (B) ΔpH (inset) and survival were measured in the presence (hatched bars) and absence (solid bars) of glutamate. ΔpH was calculated by subtracting the internal pH from the external pH.
Reversal of ΔΨ during survival under extreme acid stress.
Clues as to how E. coli handles pH 2 acid stress could come from acidophiles, organisms that naturally live and grow under extreme acid stress. Acidophiles use a novel approach to cope with the rigors of low pH. Neutralophiles like E. coli maintain an electrochemical gradient (ΔΨ) with a negative inside charge as part of generating PMF (16, 25). However, the ΔΨ of acidophiles growing at low pH is reversed, consisting of an inside positive charge (16, 35). It has been proposed that this strategy helps repel protons and maintain a higher internal pH. In acidophiles, this is thought to be achieved by an array of transporters and ion pumps that convert a negative ΔΨ to a positive ΔΨ under extremely acidic conditions (20, 35, 37).
We hypothesized that E. coli might carry out a similar feat as a result of the decarboxylation and antiport process. To address this possibility, ΔΨ values for E. coli under extreme acid stress conditions were determined. Log-phase cells at pH 7 exhibited an expected negative ΔΨ (−86 mV) and a PMF of −140 mV (Table 1). These values are somewhat lower than in some other reports (−160 to −180 mV) due to differences in growth media (LBG and BHIG here, versus minimal media), aeration, and the fact that these cells were fermenting rather than respiring. Stationary-phase cells at pH 7 had a somewhat lower ΔΨ (−52 mV) and a PMF of approximately −100 mV. However, under extreme acid stress at pH 2.5, E. coli changed its normally negative ΔΨ to a positive ΔΨ when glutamate or arginine was present (Table 1). Cells challenged at pH 2.5 for 30 min in the absence of glutamate or arginine had a ΔΨ near zero. In contrast, the addition of glutamate or arginine reversed the membrane potential, and ΔΨ became positive inside (+30 for glutamate and +80 for arginine). Confirmation of this reversal was obtained using TPP+, which was excluded from cells when the glutamate decarboxylase system was functioning (data not shown). The production of a positive ΔΨ in acid-stressed, living E. coli was a surprising finding, but it may explain the survival characteristics of the organism.
There has been controversy over similar studies performed using H. pylori, another neutralophilic organism that can survive extreme acid pH (36, 54). One report indicated that H. pylori inverts ΔΨ during extreme acid stress, similar to acidophiles and consistent with our results in E. coli (36). The second report found that the urease this organism produces will elevate internal pH when external pH is 1.2, but in contrast to acidophiles, they did not find Helicobacter inverted transmembrane electrical potential (54). In fact, the PMF measured was as high as −254 mV. The first study was criticized by the latter one for not taking into account nonspecific binding of radioactive probes. The results presented here with butanol-treated E. coli clearly accounted for nonspecific binding and still measured a positive inside electrical potential.
Neither potassium nor sodium ions are required for glutamate- or arginine-dependent acid resistance mechanisms.
Knowing that E. coli reverses its transmembrane potential raises the question of the source of the positive charges leading to positive inside ΔΨ. Two obvious candidates would be sodium or potassium ions, known to contribute to pH homeostasis under other circumstances (23, 41, 42). To address this question, cultures were grown to stationary phase in EG to induce the glutamate decarboxylase system. Cells were washed several times in MC buffer (10 mM MgCl2 and 5 mM CaCl2) to remove most residual Na+ and K+. The washed cells were then added to pH 2.5 water (Milli-Q Ultrapure) containing 1 mM glutamic acid · HCl or to M63 media containing different amounts of both ions. There were no significant survival differences in pH 2.5 media containing K+ concentrations ranging from 10 μM to 100 mM or Na+ concentrations ranging from 100 μM to 100 mM. After 2 h at pH 2.5, survival was maintained between 50 and 80% regardless of the Na+ or K+ concentration (data not shown). The results suggest that neither Na+ nor K+ ions were required for the system to work. The low intracellular pH (approximately 4.5) would likely prevent many, if not all, other housekeeping ion movement mechanisms from working efficiently. Thus, the accumulation of the positively charged decarboxylation product most likely accounts for the reversal of transmembrane potential.
DISCUSSION
E. coli is a remarkably acid-resistant neutralophilic organism that prefers growth near neutral pH but is able to withstand transient exposures to pH 2 environments for hours. Four systems contribute to this acid resistance. The two most robust systems use glutamate and arginine decarboxylases. Transcriptional controls regulating the synthesis of these systems have been heavily studied; however, the mechanisms by which they provide acid resistance have not been established. The prevailing hypothesis has been that protons entering the cell are consumed by the decarboxylase reaction via exchange with the amino acid α-carboxyl group. The decarboxylated product is returned to the exterior by antiport in exchange for more substrate (38). The ultimate goal of this cycle would be to raise internal pH to a level that protects sensitive cell constituents. Surprisingly, Na+:H+ and potassium:H+ antiport systems, thought to be important for pH homeostasis under pH conditions more suited for growth, appeared unimportant for survival under extreme acid stress (6, 7, 9, 27, 61). The results of this work indicate that the amino acid-dependent AR systems increase internal pH and reverse transmembrane potential. Whether one feature is more important than the other is not known.
The CO2 that evolved as a result of decarboxylation did not appear to contribute to pH homeostasis at this acid extreme. Carbonic anhydrase essentially adds water, not free protons, to CO2 to make carbonic acid (H2CO3); carbonic acid can then dissociate to HCO3− and H+. Because the pKa of this reaction is 6.5, an internal pH that is estimated to be between 4.2 and 4.7 will prevent this dissociation (18). Another reason carbonic anhydrase probably does not contribute to pH homeostasis at this extreme is that the protein has an alkaline pH optimum, making it unlikely that the enzymatic formation of carbonic acid will occur at pH 4 to 5. However, the system might contribute to pH homeostasis in the vicinity of the pKa of carbonic acid.
Although glutamate and arginine decarboxylases were shown to raise internal pH at external pH 2.5, the cytoplasm remained remarkably acidic (pH 4.2 to 4.7). Ordinarily, E. coli prefers to keep internal pH slightly alkaline (pH 7.8) during growth. This surprising finding was supported by two other results. First, besides using the radiolabel distribution assay, efforts to use green fluorescent protein derivatives for internal pH measurements indicated that the internal pH during pH 2.5 acid stress was below pH 5, the limit of detection by this method. Second, if internal pH were considerably higher than our measurements indicated, then other amino acid decarboxylases with higher pH optima, such as ornithine decarboxylase (pH optimum 7.0), might be expected to function as effective acid resistance systems. However, ornithine decarboxylase does not protect cells at pH 2.5, a finding we attribute to the high pH optimum limiting enzymatic function at the acidic internal pH reported here (3, 22). The situation with lysine decarboxylase (CadA) further supports the hypothesis. This enzyme has a pH optimum of pH 5.7, which is more acidic than ornithine decarboxylase but less acidic than the arginine or glutamate enzymes (48). Consistent with its intermediate pH optimum, there is a lysine-dependent acid resistance mechanism, but it is much less effective than the glutamate or arginine systems (22).
A major advance in our understanding of acid resistance came with the discovery that the chloride transport proteins of E. coli (ClcA and ClcB) are important (but not essential) to acid resistance (21). In their discussion, Iyer et al. proposed that at pH 2.5, protons cross the membrane and enter the cell in the form of uncharged HCl molecules that dissociate intracellularly into H+ and Cl−. At the high KCl concentration used in their experiments (40 mM), that is certainly possible. The intracellular protons are then consumed by decarboxylation, and the excess Cl− (or other anions) is thought to be removed via the Clc chloride transporters, which are thought to be channels. They proposed that these “channels” would provide an electrical shunt to prevent excessive inner membrane polarization predicted to occur, in the case of the arginine-dependent system, during the exchange of the intracellular decarboxylation product agmatine (+2) for the extracellular substrate arginine (+1). In their model, excessive charge, negative inside, would build in the absence of the Clc channels. Cl− exit through the channels would prevent the cell interior from becoming too negatively charged. Although not explicitly stated, their model implied that when all the systems are functioning, the resulting ΔΨ would still be negative inside when external pH is 2.5. The discovery that the Clc products are actually H+:Cl− antiporters suggests that the original model of Iyer et al. requires revision (1, 21).
We have carried out the internal pH and ΔΨ measurements needed to test the model, and we found that E. coli does not hyperpolarize but reverses membrane potential during acid stress, mimicking the strategy of an acidophile. We propose that the reversal of transmembrane charge by AR2 and AR3 is the direct result of proton influx combined with the production of positively charged decarboxylation products (GABA or agmatine) inside the cell. At low Cl− ion concentrations (no added KCl), it is generally perceived that protons, under pH 2.5 acidic conditions, enter the cell without an associated chloride ion (Fig. 6). If true, then proton intrusion alone would add considerably to the positive charges inside the cell. For instance, when the external pH is 2.5, internal pH moves from roughly 7.5 to 4.5, meaning the number of protons inside the cell (starting at 10 protons per cell at pH 7.5) increases 1,000-fold (to approximately 10,000 protons per cell). This may be enough to dissipate the normally negative interior charge of stationary-phase cells. Movement of about 10,000 H+ ions across the membrane can change the calculated ΔΨ by 60 mV (17). Of course, more than 10,000 protons must enter the cell to lower pH, because many protons become buffered by cell constituents, which can also add to the positive charge.
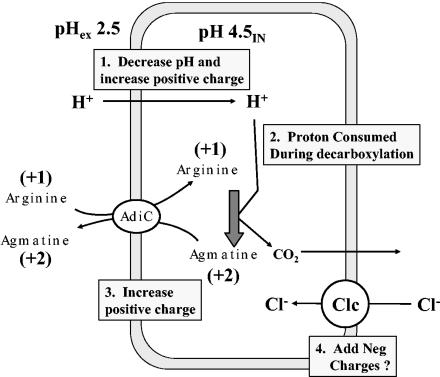
FIG. 6.
Model for amino acid-dependent acid resistance. Acid stress at pH 2.5 results in illicit entry of H+, which decreases pH and increases positive charge. As pHi drops to around 5, arginine decarboxylase will start to consume protons and convert +1-charged arginine to +2-charged agmatine, further increasing the positive charge. An antiporter will not completely drain agmatine from the cell, as it is continually being made during decarboxylation. In this model, the evolution of CO2 does not contribute toward internal pH or charge since (i) the proton donor to make carbonic acid is water, not a proton; (ii) carbonic anhydrase will not function at pH 4.5; and (iii) at this internal pH bicarbonate will not form (pKa = 6.1). The role of the Clc H+:Cl− antiporter is unknown but may help expel H+, limit excess internal positive charge, and aid in returning the cell to an inside negative charge as external pH returns to neutrality.
At steady state in the presence of glutamate or arginine, additional protons that enter are effectively consumed by the decarboxylation reaction, which keeps internal pH near 4.5. This consumption removes the proton from pH consideration, but the associated positive charge remains in the end product (GABA or agmatine). Although the charge is eventually removed by what is probably electrogenic antiport, there must always be a pool of end product in the cell to continuously drive antiport. Thus, the intracellular pool of agmatine at any one instant could be as high as half of the original intracellular arginine pool (approximately 5 to 10 mM). This scenario is consistent with our findings.
The precise role of the Clc antiporters in this system remains unclear. Eliminating the antiporters clearly impairs acid resistance, whether high-Cl− or low-Cl− media are used (15) (Fig. 1B). It would make sense if these antiporters allowed entry of Cl− ions to counter the positive inside ΔΨ produced as a result of proton influx and decarboxylation, so that when the inside charge is positive, the Clc antiporters could allow entry of negative charges and prevent membrane hyperpolarization in the direction opposite to that predicted by Iyer et al. (21). This would also allow excess H+ ions to be expelled from the cytoplasm in exchange for Cl−.
Calculations by Iyer et al. established the conversion rate of glutamate to GABA to be approximately three times that of arginine to agmatine within minutes of exposure to pH 2.5 (104 versus 3 × 103 molecules per min per cell, respectively) (21). This does not contradict the differences we observed in the ΔΨ measurements, where the arginine system generated 2.7 times more positive charge than glutamate. The apparent contradiction can be resolved by considering the different ionizable groups on glutamate and arginine.
The α-carboxyl groups of both glutamate and arginine have pKa values of 2.1 and, thus, they will be less than 50% protonated at external pH 2.5. Upon entering what is probably a less-acidic GadC antiporter channel, we predict the remaining H+ will be released to the periplasm, and so the group on both amino acids would enter as COO−. However, glutamate, but not arginine, also has a side chain carboxyl group with a dissociation constant of 4.3. If the pH of the mouth of the antiporter is near external pH 2.5 (pH 3 for instance), then this carboxyl group will be mostly protonated as it enters the cell. Once inside the cytoplasm, about half of those side chain protons will dissociate at the higher internal pH (4.2) and then be consumed again during decarboxylation to form GABA. The result would be a futile proton cycle as well as an electroneutral conversion. Thus, the higher reported conversion of glutamate to GABA would not generate as much internal positive charge as the slower arginine system.
How might the cell recover from an inverted ΔΨ? In our model, as acid stress is reduced (i.e., as external pH becomes less acidic) proton intrusion stops, but the decarboxylases continue to remove internal protons and allow internal pH to rise. The excess positive charges leading to the inverted ΔΨ are now in the form of decarboxylation products that can be removed by the antiporters. In the case of arginine and agmatine, a +2 is extruded in exchange for a +1 charge. As long as no more H+ flows into the cell, this exchange combined with the import of anions by the Clc antiporters (or other means) are predicted to restore a negative inside electrical potential. Finally, normal homeostatic mechanisms take over, and the cell can resume growing.
How might an inside positive membrane potential aid survival at extreme acid pH? One hypothesis is that converting membrane potential from negative inside to positive inside may be a way to repel protons. While it is true that internal pH falls to 4.2 to 4.7, the drop would be more severe without the repelling force of a positive inside charge. However, in the experiment where internal pH was maintained at 4.0 with and without glutamate, the cells with glutamate survived better, arguing that the primary role of a positive ΔΨ would not be to repel protons to change internal pH. Alternatively, a positive ΔΨ might mitigate excess PMF that can form when ΔpH is large.
An internal pH of 4.5 and a positive inside membrane potential would be disastrous to growing E. coli cells. We propose these conditions pose only minor problems to a cell under extreme acid stress. At external pH 2.5, the internal pH of the cell is so low that very few metabolic reactions can even take place. For instance, protein synthesis is undetectable under these conditions, and the cells clearly are not growing. As a result of metabolic stasis, the cell could also survive a relatively short period of reversed transmembrane potential. The overall result may actually protect the cell from inadvertent, self-inflicted damage.
In sum, the results presented indicate that even though E. coli cannot grow under acidophilic pH conditions, the organism may have learned to traverse the gastric acid barrier by adopting part of the acidophile survival strategy. The decarboxylase systems may protect against severe acid stress in two ways. Consuming protons by decarboxylation would produce a less acidic internal pH and generate an inside positive potential that could help repel protons and/or prevent excessive PMF.
Acknowledgments
This work was supported by National Institutes of Health award R01-GM61147.
We thank Patricia Couling for help in preparing the manuscript.
REFERENCES
- 1.Accardi, A., and C. Miller. 2004. Secondary active transport mediated by a prokaryotic homologue of ClC Cl− channels. Nature 427:803-807. [DOI] [PubMed] [Google Scholar]
- 2.Anantharam, V., M. J. Allison, and P. C. Maloney. 1989. Oxalate:formate exchange. The basis for energy coupling in Oxalobacter. J. Biol. Chem. 264:7244-7250. [PubMed] [Google Scholar]
- 3.Applebaum, D., D. L. Sabo, E. H. Fischer, and D. R. Morris. 1975. Biodegradative ornithine decarboxylase of Escherichia coli. Purification, properties, and pyridoxal 5′-phosphate binding site. Biochemistry 14:3675-3681. [DOI] [PubMed] [Google Scholar]
- 4.Atkinson, W. H., and H. H. Winkler. 1981. A centrifugal filtration method for the study of transport of nicotinamide adenine and pyruvate by Rickettsia prowazekii, p. 411-420. In W. Burgerdorfer and R. L. Anacker (ed.), Rickettsiae and rickettsial disease. Academic Press, Inc., New York, N.Y.
- 5.Boeker, E. A., and E. E. Snell. 1972. Amino acid decarboxylases. Enzymes 6:217-250. [Google Scholar]
- 6.Booth, I. R. 1985. Regulation of cytoplasmic pH in bacteria. Microbiol. Rev. 49:359-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Booth, I. R. 1999. The regulation of intracellular pH in bacteria, p. 19-27. In R. Poole (ed.), Bacterial responses to pH. John Wiley & Sons, Ltd., Chichester, England.
- 8.Booth, I. R., W. J. Mitchell, and W. A. Hamilton. 1979. Quantitative analysis of proton-linked transport systems. The lactose permease of Escherichia coli. Biochem. J. 182:687-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brey, R. N., B. P. Rosen, and E. N. Sorensen. 1980. Cation/proton antiport systems in Escherichia coli. Properties of the potassium/proton antiporter. J. Biol. Chem. 255:39-44. [PubMed] [Google Scholar]
- 10.Castanie-Cornet, M. P., and J. W. Foster. 2001. Escherichia coli acid resistance: cAMP receptor protein and a 20 bp cis-acting sequence control pH and stationary phase expression of the gadA and gadBC glutamate decarboxylase genes. Microbiology 147:709-715. [DOI] [PubMed] [Google Scholar]
- 11.Castanie-Cornet, M. P., T. A. Penfound, D. Smith, J. F. Elliott, and J. W. Foster. 1999. Control of acid resistance in Escherichia coli. J. Bacteriol. 181:3525-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Biase, D., A. Tramonti, F. Bossa, and P. Visca. 1999. The response to stationary-phase stress conditions in Escherichia coli: role and regulation of the glutamic acid decarboxylase system. Mol. Microbiol. 32:1198-1211. [DOI] [PubMed] [Google Scholar]
- 13.Foster, J. W., and H. K. Hall. 1991. Inducible pH homeostasis and the acid tolerance response of Salmonella typhimurium. J. Bacteriol. 173:5129-5135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foster, J. W., and M. Moreno. 1999. Inducible acid tolerance mechanisms in enteric bacteria. Novartis Found. Symp. 221:55-69. [DOI] [PubMed] [Google Scholar]
- 15.Gong, S., H. Richard, and J. W. Foster. 2003. YjdE (AdiC) is the arginine:agmatine antiporter essential for arginine-dependent acid resistance in Escherichia coli. J. Bacteriol. 185:4402-4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goulbourne, E., Jr., M. Matin, E. Zychlinsky, and A. Matin. 1986. Mechanism of ΔpH maintenance in active and inactive cells of an obligately acidophilic bacterium. J. Bacteriol. 166:59-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gould, J. M., and W. A. Cramer. 1977. Relationship between oxygen-induced proton efflux and membrane energization in cells of Escherichia coli. J. Biol. Chem. 252:5875-5882. [PubMed] [Google Scholar]
- 18.Hakansson, K., M. Carlsson, L. A. Svensson, and A. Liljas. 1992. Structure of native and apo carbonic anhydrase II and structure of some of its anion-ligand complexes. J. Mol. Biol. 227:1192-1204. [DOI] [PubMed] [Google Scholar]
- 19.Hersh, B. M., F. T. Farooq, D. N. Barstad, D. L. Blankenshorn, and J. L. Slonczewski. 1996. A glutamate-dependent acid resistance gene in Escherichia coli. J. Bacteriol. 178:3978-3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsung, J. C., and A. Haug. 1977. Membrane potential of Thermoplasma acidophila. FEBS Lett. 73:47-50. [DOI] [PubMed] [Google Scholar]
- 21.Iyer, R., T. M. Iverson, A. Accardi, and C. Miller. 2002. A biological role for prokaryotic ClC chloride channels. Nature 419:715-718. [DOI] [PubMed] [Google Scholar]
- 22.Iyer, R., C. Williams, and C. Miller. 2003. Arginine-agmatine antiporter in extreme acid resistance in Escherichia coli. J. Bacteriol. 185:6556-6561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kashket, E. R. 1985. Effects of K+ and Na+ on the proton motive force of respiring Escherichia coli at alkaline pH. J. Bacteriol. 163:423-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kashket, E. R. 1985. The proton motive force in bacteria: a critical assessment of methods. Annu. Rev. Microbiol. 39:219-242. [DOI] [PubMed] [Google Scholar]
- 25.Khan, S., and R. Macnab. 1980. Proton chemical potential, proton electrical potential and bacterial motility. J. Mol. Biol. 138:599-614. [DOI] [PubMed] [Google Scholar]
- 26.Kobayashi, H. 1985. A proton-translocating ATPase regulates pH of the bacterial cytoplasm. J. Biol. Chem. 260:72-76. [PubMed] [Google Scholar]
- 27.Kroll, R. G., and I. R. Booth. 1983. The relationship between intracellular pH, the pH gradient and potassium transport in Escherichia coli. J. Biochem. 216:706-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krulwich, T. A. 1986. Bioenergetics of alkalophilic bacteria. J. Membr. Biol. 89:113-125. [DOI] [PubMed] [Google Scholar]
- 29.Kumar, S., N. S. Punekar, V. SatyaNarayan, and K. V. Venkatesh. 2000. Metabolic fate of glutamate and evaluation of flux through the 4-aminobutyrate (GABA) shunt in Aspergillus niger. Biotechnol. Bioeng. 67:575-584. [PubMed] [Google Scholar]
- 30.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 31.Lin, J., I. S. Lee, J. Frey, J. L. Slonczewski, and J. W. Foster. 1995. Comparative analysis of extreme acid survival in Salmonella typhimurium, Shigella flexneri, and Escherichia coli. J. Bacteriol. 177:4097-4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin, J., M. P. Smith, K. C. Chapin, H. S. Baik, G. N. Bennett, and J. W. Foster. 1996. Mechanisms of acid resistance in enterohemorrhagic Escherichia coli. Appl. Environ. Microbiol. 62:3094-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malashkevich, V. N., D. De Biase, Z. Markovic-Housley, M. P. Schlunegger, F. Bossa, and J. N. Jansonius. 1998. Crystallization and preliminary X-ray analysis of the beta-isoform of glutamate decarboxylase from Escherichia coli. Acta Crystallogr. D 54:1020-1022. [DOI] [PubMed] [Google Scholar]
- 34.Maloney, P. C. 1978. Coupling between H+ entry and ATP formation in Escherichia coli. Biochem. Biophys. Res. Commun. 83:1496-1501. [DOI] [PubMed] [Google Scholar]
- 35.Matin, A., B. Wilson, E. Zychlinsky, and M. Matin. 1982. Proton motive force and the physiological basis of delta pH maintenance in Thiobacillus acidophilus. J. Bacteriol. 150:582-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matin, A., E. Zychlinsky, M. Keyhan, and G. Sachs. 1996. Capacity of Helicobacter pylori to generate ionic gradients at low pH is similar to that of bacteria which grow under strongly acidic conditions. Infect. Immun. 64:1434-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Michels, M., and E. P. Bakker. 1985. Generation of a large, protonophore-sensitive proton motive force and pH difference in the acidophilic bacteria Thermoplasma acidophilum and Bacillus acidocaldarius. J. Bacteriol. 161:231-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Molenaar, D., J. S. Bosscher, B. ten Brink, A. J. Driessen, and W. N. Konings. 1993. Generation of a proton motive force by histidine decarboxylation and electrogenic histidine/histamine antiport in Lactobacillus buchneri. J. Bacteriol. 175:2864-2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mooney, C., D. J. Munster, P. F. Bagshow, and R. A. Allardyck. 1993. Helicobacter pylori acid resistance. Lancet 335:1232. [DOI] [PubMed] [Google Scholar]
- 40.Olsen, E. B., J. B. Russell, and T. Henick-Kling. 1991. Electrogenic l-malate transport by Lactobacillus plantarum: a basis for energy derivation from malolactic fermentation. J. Bacteriol. 173:6199-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Padan, E., and T. A. Krulwich. 2000. Sodium stress. ASM Press, Washington, D.C.
- 42.Padan, E., and S. Schuldiner. 1996. Bacterial Na+/K+ antiporters: molecular biology, biochemistry, and physiology, vol. 2. Elsevier Science, Amsterdam, The Netherlands.
- 43.Park, Y. K., B. Bearson, S. H. Bang, I. S. Bang, and J. W. Foster. 1996. Internal pH crisis, lysine decarboxylase and the acid tolerance response of Salmonella typhimurium. Mol. Microbiol. 20:605-611. [DOI] [PubMed] [Google Scholar]
- 44.Poolman, B., D. Molenaar, E. J. Smid, T. Ubbink, T. Abee, P. P. Renault, and W. N. Konings. 1991. Malolactic fermentation: electrogenic malate uptake and malate/lactate antiport generate metabolic energy. J. Bacteriol. 173:6030-6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rektorschek, M., A. Buhmann, D. Weeks, D. Schwan, K. W. Bensch, S. Eskandari, D. Scott, G. Sachs, and K. Melchers. 2000. Acid resistance of Helicobacter pylori depends on the UreI membrane protein and an inner membrane proton barrier. Mol. Microbiol. 36:141-152. [DOI] [PubMed] [Google Scholar]
- 46.Richard, H. T., and J. W. Foster. 2003. Acid resistance in Escherichia coli. Adv. Appl. Microbiol. 52:167-186. [DOI] [PubMed] [Google Scholar]
- 47.Rottenberg, H. 1979. The measurement of membrane potential and ΔpH in cells, organelles, and vesicles. Methods Enzymol. 55:547-569. [DOI] [PubMed] [Google Scholar]
- 48.Sabo, D. L., E. A. Boeker, B. Byers, H. Waron, and E. H. Fischer. 1974. Purification and physical properties of inducible Escherichia coli lysine decarboxylase. J. Biochem. 13:662-670. [DOI] [PubMed] [Google Scholar]
- 49.Salema, M., B. Poolman, J. S. Lolkema, M. C. Dias, and W. N. Konings. 1994. Uniport of monoanionic l-malate in membrane vesicles from Leuconostoc oenos. Eur. J. Biochem. 225:289-295. [DOI] [PubMed] [Google Scholar]
- 50.Scott, D. R., D. Weeks, C. Hong, S. Postius, K. Melchers, and G. Sachs. 1998. The role of internal urease in acid resistance of Helicobacter pylori. Gastroenterology 114:58-70. [DOI] [PubMed] [Google Scholar]
- 51.Slonczewski, J. L., B. P. Rosen, J. R. Alger, and R. M. Macnab. 1981. pH homeostasis in Escherichia coli: measurements by 31P nuclear magnetic resonance of methylphosphonate and phosphate. Proc. Natl. Acad. Sci. USA 78:6271-6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Small, P., D. Blankenhorn, D. Welty, E. Zinser, and J. L. Slonczewski. 1994. Acid and base resistance in Escherichia coli and Shigella flexneri: role of rpoS and growth pH. J. Bacteriol. 176:1729-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith, J. L. 2003. The role of gastric acid in preventing foodborne disease and how bacteria overcome acid conditions. J. Food Prot. 66:1292-1303. [DOI] [PubMed] [Google Scholar]
- 54.Stingl, K., E. M. Uhlemann, R. Schmid, K. Altendorf, and E. P. Bakker. 2002. Energetics of Helicobacter pylori and its implications for the mechanism of urease-dependent acid tolerance at pH 1. J. Bacteriol. 184:3053-3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sukhareva, B. S., A. S. Tikhonenko, and E. L. Darii. 1994. Study of the quaternary structure of glutamate carboxylase from Escherichia coli. Mol. Biol. (Moscow) 28:1407-1411. [PubMed] [Google Scholar]
- 56.Tanabe, M., K. Nishio, Y. Iko, Y. Sambongi, A. Iwamoto-Kihara, Y. Wada, and M. Futai. 2001. Rotation of a complex of the γ subunit and c ring of Escherichia coli ATP synthase: the rotor and stator are interchangeable. J. Biol. Chem. 276:15269-15274. [DOI] [PubMed] [Google Scholar]
- 57.Thoma, W. J., J. G. Steiert, R. L. Crawford, and K. Ugurbil. 1986. pH measurements by 31P NMR in bacterial suspensions using phenyl phosphonate as a probe. Biochem. Biophys. Res. Commun. 138:1106-1109. [DOI] [PubMed] [Google Scholar]
- 58.Vogel, H. J., and D. M. Bonner. 1956. Acetylornithase of Escherichia coli: partial purification and some properties. J. Biol. Chem. 218:97-106. [PubMed] [Google Scholar]
- 59.Weeks, D. L., S. Eskandari, D. R. Scott, and G. Sachs. 2000. A H+-gated urea channel: the link between Helicobacter pylori urease and gastric colonization. Science 287:482-485. [DOI] [PubMed] [Google Scholar]
- 60.Winkler, H. H., and T. H. Wilson. 1966. The role of energy coupling in the transport of beta-galactosides by Escherichia coli. J. Biol. Chem. 241:2200-2211. [PubMed] [Google Scholar]
- 61.Zilberstein, D., V. Agmon, S. Schuldiner, and E. Padan. 1982. The sodium/proton antiporter is part of the pH homeostasis mechanism in Escherichia coli. J. Biol. Chem. 257:3687-3691. [PubMed] [Google Scholar]