Abstract
During growth of the halophilic archaeon Haloarcula marismortui on d-xylose, a specific d-xylose dehydrogenase was induced. The enzyme was purified to homogeneity. It constitutes a homotetramer of about 175 kDa and catalyzed the oxidation of xylose with both NADP+ and NAD+ as cosubstrates with 10-fold higher affinity for NADP+. In addition to d-xylose, d-ribose was oxidized at similar kinetic constants, whereas d-glucose was used with about 70-fold lower catalytic efficiency (kcat/Km). With the N-terminal amino acid sequence of the subunit, an open reading frame (ORF)—coding for a 39.9-kDA protein—was identified in the partially sequenced genome of H. marismortui. The function of the ORF as the gene designated xdh and coding for xylose dehydrogenase was proven by its functional overexpression in Escherichia coli. The recombinant enzyme was reactivated from inclusion bodies following solubilization in urea and refolding in the presence of salts, reduced and oxidized glutathione, and substrates. Xylose dehydrogenase showed the highest sequence similarity to glucose-fructose oxidoreductase from Zymomonas mobilis and other putative bacterial and archaeal oxidoreductases. Activities of xylose isomerase and xylulose kinase, the initial reactions of xylose catabolism of most bacteria, could not be detected in xylose-grown cells of H. marismortui, and the genes that encode them, xylA and xylB, were not found in the genome of H. marismortui. Thus, we propose that this first characterized archaeal xylose dehydrogenase catalyzes the initial step in xylose degradation by H. marismortui.
The utilization of sugars, in particular of hexoses and hexose polymers and—to a lesser extent—of pentoses, has been reported for various species in the domain Archaea. So far, only the catabolic pathways of hexoses and glucose polymers (e.g., maltose and starch) have been studied in detail in particular in hyperthermophilic, thermoacidophilic, and extremely halophilic archaea. Comparative analyses of glucose degradation pathways in these organisms revealed that the classical Embden-Meyerhof- (EM) or Entner-Doudoroff- (ED) pathway found in bacteria is not operative in archaea; they use instead modified versions of these pathways as follows (for reviews, see references 31 and 41). In hyperthermophilic eury- and crenarchaeota, glucose degradation proceeds predominantly via modified EM pathways, which differ from the classical EM pathway by the presence of several unusual glucokinases (ADP or ATP dependent) and 6-phosphofructokinases (ADP, ATP, or PPi dependent), novel enzymes of glucose-6-phosphate isomerization and of glyceraldehyde-3-phosphate oxidation, and pyruvate kinases with reduced regulatory potential (15, 18, 41).
In thermoacidophilic archaea, Sulfolobus and Thermoplasma spp., glucose is degraded via a nonphosphorylated version of the ED pathway (22, 31, 41) by which glucose is oxidized to glycerate via the nonphosphorylated intermediates gluconate and 2-keto-3-deoxygluconate (KDG) involving glucose dehydrogenase, gluconate dehydratase, and KDG aldolase. Glycerate is then phosphorylated via a specific kinase to 2-phosphoglycerate, which is further converted to pyruvate via enolase and pyruvate kinase. In halophilic archaea, e.g., Halococcus, Haloarcula, and Haloferax spp., a modified, semiphosphorylated ED pathway is operative in which—as in thermoacidophiles—glucose is converted to KDG. However, KDG is then phosphorylated to 2-keto-3-deoxy-6-phosphogluconate by KDGkinase. Further degradation of 2-keto-3-deoxy-6-phosphogluconate proceeds via reactions of the conventional phosphorylated ED pathway found in bacteria (19, 45).
In contrast to hexose metabolism, the catabolic pathways of pentoses have not been studied in detail in the domain Archaea. The utilization of pentoses, e.g., xylose, ribose, and arabinose, has been reported for several halophiles, e.g., Halococcus, Haloarcula, and Halobacterium spp., and for Sulfolobus species (30, 40), rather than for the majority of hyperthermophiles. No studies of growth on pentoses or analyses of the enzymes involved in pentose degradation by these organisms have been reported.
In the domain Bacteria, the pathways for the degradation of pentoses, in particular, d-xylose, have been studied in detail in many species, including Escherichia coli, Salmonella enterica serovar Typhimurium, Lactobacillus pentosus, Lactococcus lactis, Bacillus spp., Staphylococcus xylosus, Bacteroides xylanolyticus, and Tetragenococcus halophilus. Degradation of xylose by these organisms, e.g., by E. coli, starts with its uptake via specific high- or low-affinity transport systems. Via xylose isomerase, xylose is then isomerized to xylulose, which is phosphorylated to xylulose-5-phosphate by the activity of xylulose kinase. The genes encoding xylose transporters, xylose isomerase (xylA gene), and xylulose kinase (xylB gene), which are arranged in an operon, are induced by xylose mediated by the transcriptional regulator XylR. Further degradation of xylulose-5-phosphate, proceeds—depending on the organism—either via the pentose phosphate cycle, the phosphoketolase pathway, or—as in Bacteroides spp. (4)—via a combination of both pentose phosphate and the EM pathway. Thus, the most common initial reactions of bacterial xylose catabolism involve xylose isomerase and xylulose kinase (4, 13, 23, 29, 36).
Xylose isomerase and the gene that encodes it, xylA, have been characterized in many bacteria, including the hyperthermophile Thermotoga maritima, as well as in eucarya. The enzymes from both domains are of significant industrial interest since they also catalyze, as a side activity, the isomerization of glucose to fructose, a reaction that constitutes the last step in the large-scale industrial process of the production of sweeteners from starch (3).
So far, activities of xylose isomerase and xylulose kinase have not been reported in any species of the archaeal domain. Further, homologs to the bacterial xylA and xylB genes could not be found in archaeal genomes. Thus, it might be speculated that the initial steps in xylose degradation by archaea are different from the common mechanism found in most bacteria.
In this communication, we report on studies of the growth of the halophilic archaeon Haloarcula marismortui on xylose. Evidence is presented that the first step in xylose degradation by this organism is oxidation of xylose to xylonate via a xylose-inducible NADP+-reducing d-xylose dehydrogenase. This first archaeal xylose dehydrogenase was purified, and the gene that encodes it, xdh, was identified in available sequenced contigs of H. marismortui. This enzyme represents a novel type of xylose dehydrogenase, showing high similarity to glucose-fructose oxidoreductase (GFOR) from Zymomonas mobilis.
MATERIALS AND METHODS
Growth of H. marismortui and preparation of cell extracts.
H. marismortui (DSM 3752) (24) was obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (Braunschweig, Germany). The organism was grown aerobically at 37°C in 500-ml Erlenmeyer flasks filled with 50 ml of medium containing 25 mM xylose, 0.05% yeast extract, 250 g of NaCl per liter, 20 g of MgSO4 · 7H2O per liter, 19.5 g of morpholineethanesulfonic acid (MES) per liter, 2 g of KCl per liter, 1 g of Na-glutamate per liter, and 3 g of Na-citrate per liter; 10 ml of the vitamin solution described by Staley (35); and 10 ml of a trace element solution containing (per liter) 1.5 g of EDTA, 0.01 g of Na2MoO4 · 2H2O, 0.5 g of MnSO4 · H2O, 0.1 g of FeSO4 · 7H2O, 0.1 g of CoCl2, 0.1 g of ZnSO4, and 0.01 g of CuSO4 · 5H2O. The pH was adjusted to 7.35 with 10 N NaOH. Growth was followed by measuring optical density at 578 nm (ΔOD578). During growth, samples were removed and centrifuged and the supernatants were analyzed for xylose and xylonate, as indicated. Extracts were prepared from late-log-phase cells by sonication as described previously (19), and enzyme activities were determined as described in the section on enzyme assays. Protein was determined by the biuret method with bovine serum albumin as the standard (5).
Induction of xylose dehydrogenase in H. marismortui was followed in 2,000-ml Erlenmeyer flasks filled with 400 ml of medium containing xylose (25 mM), yeast extract (2.5 g/liter), and Casamino Acids (5 g/liter); cells previously grown on glucose (see reference 19) were used as an inoculum (10%). At the times indicated, 60- to 80-ml samples were removed and centrifuged (2,600 × g, 10 min, 4°C) and the cell pellets were suspended in 1 ml of 0.1 M Tris-HCl, pH 7.5, containing 250 g of NaCl/liter. Cell extracts were prepared by sonication (19), followed by centrifugation at 12,000 × g for 10 min. The supernatants were analyzed for xylose dehydrogenase activities. The protein concentration of cell extracts was determined by the biuret method.
Purification of xylose dehydrogenase from H. marismortui.
Xylose dehydrogenase was purified from H. marismortui after growth of the organism in a medium (see above) containing 25 mM xylose and 0.1% yeast extract in a 10-liter Biostat fermentor. Extract was prepared from 4 g of cells, which were suspended in 100 mM Tris-HCl, pH 8.8, containing 2 M (NH4)2SO4 and 20 mM MgCl2 (buffer A). Cells were disrupted by passage through a French pressure cell at 1.3 × 108 Pa. Cell debris and unbroken cells were removed by centrifugation for 90 min at 100,000 × g at 4°C. The 100,000 × g supernatant was applied to a Sepharose CL 4B column (1.6 by 60 cm) that had been equilibrated with buffer A. Protein was eluted with a decreasing (NH4)2SO4 gradient from 2 to 0 M in buffer A. Fractions containing the highest xylose dehydrogenase activity [1.6 to 1.4 M (NH4)2SO4] were pooled, adjusted to 2 M (NH4)2SO4, and applied to a Phenyl Sepharose column (2.6 by 10 cm) equilibrated with buffer B [50 mM Tris-HCl, pH 8.5, containing 2 M (NH4)2SO4 and 20 mM MgCl2]. Protein was eluted with a linear gradient of buffer B to 50 mM Tris-HCl, pH 8.5, containing 20 mM MgCl2 and 10% glycerol. Fractions containing the highest xylose dehydrogenase activity [1.04 to 0.95 M (NH4)2SO4] were pooled and concentrated to 600 μl by ultrafiltration (cutoff, 20 kDa). The concentrated protein solution was applied to a Superdex 200 HiLoad gel filtration column (1.6 by 60 cm) that had been equilibrated with 50 mM Tris-HCl, pH 8.5, containing 20 mM MgCl2, 10% glycerol, and 100 mM NaCl. In this buffer, the enzyme was stable in the absence of a high salt (KCl or NaCl) concentration. Eluted fractions containing xylose dehydrogenase activity indicated essentially pure protein and were stored at −20°C. The purity of the preparations was checked by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in 12% gels in accordance with standard procedures (21). During the purification procedure, protein concentrations were determined by the Bradford method with bovine serum albumin as the standard (7).
Cloning and expression of xylose dehydrogenase from H. marismortui in E. coli.
On the basis of the N-terminal amino acid sequence, an open reading frame (ORF) was identified by a BLASTP search in contig 97 of the partially sequenced genome of H. marismortui (P. Zhang, W. V. Ng, and S. DasSarma, personal communication, 2003). The ORF was characterized as the xdh gene, encoding xylose dehydrogenase, by its functional overexpression in E. coli. The gene was amplified from genomic DNA of H. marismortui by PCR and cloned into pET17b (Novagen) via two restriction sites (NdeI and BamHI) created with the primers 5′-GACGACAGTCATATGAACGTTG-3′ and 5′-CAAAAAATCTGGATCCGGTTTC-3′ (restriction sites are underlined). The vector pET17b-xdh was transformed into E. coli BL21 codon plus(DE3)-RIL (Stratagene). For expression, cells were grown in Luria-Bertani medium at 37°C. Expression was initiated by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG; final concentration, 0.4 mM). After 18 h of further growth, cells were harvested by centrifugation.
Solubilization, refolding, and purification of recombinant xylose dehydrogenase.
Recombinant xylose dehydrogenase, which was expressed in inclusion bodies, was solubilized and refolded as described by Connaris et al. (11), with modifications. The E. coli cell pellets were suspended in 20 mM Tris-HCl, pH 7.5, containing 2 mM EDTA, 3 M KCl, and 10% glycerol (buffer C). The cell suspension was treated with 100 μg of lysozyme per ml and 0.1% (vol/vol) Triton X-100 and incubated at 30°C for 60 min, followed by incubation on ice for 15 min. The suspension was then sonicated and centrifuged at 40,000 × g for 30 min at 4°C. The insoluble fraction was washed twice in buffer C, yielding inclusion bodies and insoluble cell fragments. The insoluble fraction was dissolved in 20 mM Tris-HCl, pH 7.5, containing 2 mM EDTA, 8 M urea, and 50 mM dithioerythritol. Solubilization was carried out at 37°C for 1 h. Refolding was initiated by slowly diluting the suspension in 20 mM Tris-HCl, pH 7.5, containing 3 M KCl, 2 mM EDTA, 10% glycerol, 2 mM xylose, 0.1 mM NADP+, 3 mM reduced glutathione, and 0.3 mM oxidized glutathione to a final concentration of about 30 μg of protein per ml. After incubation for 6 days at 4°C, the renatured protein was concentrated 250-fold by ultrafiltration (cutoff, 30 kDa). The concentrated protein solution was applied to a Superdex 200 HiLoad 16/60 gel filtration column that had been equilibrated with 50 mM Tris-HCl, pH 8.5, containing 20 mM MgCl2, 10% glycerol, and 100 mM NaCl. Eluted fractions containing xylose dehydrogenase activity were pooled and applied to a Phenyl Resource column (1 ml), equilibrated with buffer B. Protein was eluted with a linear (NH4)2SO4 gradient to 0 M in 50 mM Tris-HCl, pH 8.5, containing 20 mM MgCl2 and 10% glycerol. Essentially pure enzyme was eluted at about 1 M (NH4)2SO4. The purity of the preparations was checked by SDS-PAGE, and protein concentrations were determined by the Bradford method (7).
Enzyme assays.
All enzyme assays were done at 37°C. One unit of enzyme activity corresponds to the conversion of 1 μmol of substrate consumed or product formed per min.
Xylose dehydrogenase activity (xylose + NADP+ → xylonate + NADPH + H+) was assayed by measuring the rate of reduction of NADP+ at 365 nm. The standard assay mixture contained 100 mM Tris-HCl (pH 8.3), 1.5 M KCl, 1 mM NADP+, 10 mM xylose, and protein.
Glucose dehydrogenase activity was tested as glucose-dependent reduction of NADP+. The assay mixture contained 100 mM Tris-HCl (pH 8.3), 1.5 M KCl, 10 mM glucose, 1 mM NADP+, and protein.
Xylose isomerase activity was tested as xylose-dependent formation of xylulose. The assay mixture contained 100 mM Tris-HCl (pH 8.5), 1 M KCl, 1 mM CoCl2, 5 mM MnSO4, 5 mM MgCl2, 10 to 100 mM xylose or glucose, and protein. During incubation (0 to 20 min), aliquots were taken and the reaction was stopped by addition of trichloroacetic acid to a final concentration of 10%. After centrifugation, xylulose was quantified by the cysteine-carbazole method (17). Crude extract from xylose-grown E. coli cells served as a positive control (12).
Xylulose-5-phosphate kinase activity was tested as the ATP-dependent decrease in xylulose. The assay mixture contained 100 mM Tris-HCl (pH 8.5), 1 M KCl, 10 mM MgCl2, 4 mM cysteine-HCl, 10 mM ATP, 6 mM xylulose, and protein. During incubation (0 to 30 min), aliquots were taken and the reaction was stopped by addition of trichloroacetic acid to a final concentration of 10%. After centrifugation, xylulose was quantified by the cysteine-carbazole method. Crude extract from xylose-grown E. coli cells served as a positive control.
Temperature and pH dependence, salt effects, and cation specificity.
The temperature dependence of xylose dehydrogenase was measured between 20 and 60°C in 50 mM Tris-HCl, pH 8.3, containing 1.5 M KCl, 1 mM NADP+, 10 mM xylose, and protein. The pH dependence of the enzyme was measured between pHs 4.4 and 10.3 at 37°C with either piperazine (pHs 4.9 to 6.0), bis-Tris (pHs 6.0 to 7.5), Tris-HCl (pHs 7.5 to 9.3), or piperazine (pHs 9.3 to 10.8), each at 20 mM, containing 1.5 M KCl, 1 mM NADP+, 10 mM xylose, and protein. The effects of salts (0 to 200 mM MgCl2, 0 to 3.5 M KCl, and 0 to 3.5 M NaCl) on xylose dehydrogenase activity were tested at 37°C in 20 mM Tris-HCl, pH 8.3, containing 1 mM NADP+, 10 mM xylose, and protein.
Substrate specificity.
The substrate specificity of xylose dehydrogenase was tested at 37°C in 20 mM Tris-HCl, pH 8.3, containing 1.5 M KCl with d isomers of the sugars in the presence of NADP+ at 10 mM each xylose and ribose; 1 mM NADP+; 100 mM glucose, 1 mM NADP+; 100 mM each galactose, fructose, and arabinose; and 2 mM NADP+. For the determination of apparent Km and Vmax values for sugars and the cosubstrates NADP+ and NAD+, the following concentrations were used: xylose or ribose, 0 to 10 mM with 1 mM NADP+; NADP+, 0 to 1 mM with 10 mM xylose or ribose; NAD+, 0 to 3 mM with 10 mM xylose or ribose; glucose, 0 to 100 mM with 1 mM NADP+; NADP+, 0 to 1 mM with 100 mM glucose.
Analytical assays.
Gel filtration chromatography was carried out with a flow rate of 1 ml/min on a Superdex 200 HiLoad column (1.6 by 60 cm). The column was equilibrated with 50 mM Tris-HCl, pH 8.5, containing 20 mM MgCl2, 10% glycerol, and 100 mM NaCl. HWM and LWM gel filtration calibration kits (Amersham Biosciences, Amersham, England) were used as the standards.
The concentration of xylose was determined by using the orcinol assay (8). The extinction coefficient (ɛ) at 546 nm was 4,800 M−1 cm−1. The concentration of xylonate was determined by high-performance liquid chromatography with an Aminex HPX87H column (Bio-Rad, Richmond, Calif.) operating at 37°C. Samples were diluted 1:5 in 5 mM H2SO4, boiled for 30 min, centrifuged, passed through a 0.2-μm-pore-size filter, and loaded onto the column. Xylonate was eluted with 5 mM H2SO4 at a flow rate of 0.6 ml/min and then monitored with a differential refractometer at 210 nm.
RESULTS
Growth of H. marismortui on xylose and induction of xylose dehydrogenase.
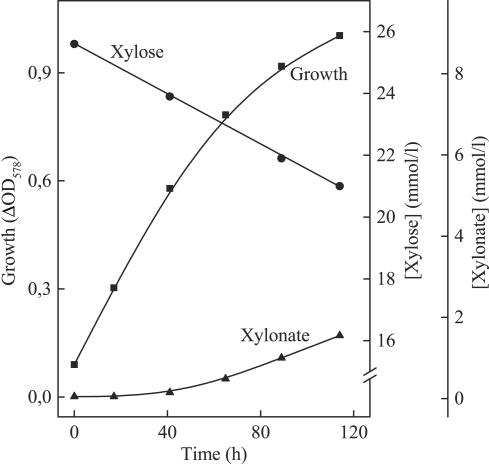
H. marismortui was grown on a medium containing 25 mM d-xylose and 0.05% yeast extract, with xylose-grown cells (10%) as the inoculum. The cells grew exponentially with a doubling time of about 20 h up to a ΔOD578 of about 1. During growth, xylose was consumed and small amounts of xylonate were formed (Fig. 1). In the absence of xylose, the cells grew with a doubling time of about 30 h up to a ΔOD578 of about 0.5 because of the yeast extract present in the medium (data not shown).
FIG. 1.
Growth of H. marismortui on 25 mM xylose and 0.05% yeast extract. Cultures were incubated at 37°C in 500-ml Erlenmeyer flasks filled with 50 ml of medium and shaken at 200 rpm. Growth (▪), xylose consumption (•), and xylonate formation (▴) were followed over time.
To identify the first enzymes of xylose degradation, extracts of xylose-grown H. marismortui cells were analyzed for xylose isomerase and xylulose kinase, the initial enzymes of xylose degradation by bacteria. Neither of these activities could be detected. As a control, both enzyme activities (xylose isomerase, 40 mU/mg; xylulose kinase, 420 mU/mg) were found in xylose-grown E. coli cells under conditions identical to those used for H. marismortui.
Since during growth on xylose small amounts of xylonate were formed, we looked for enzymes catalyzing the dehydrogenation of xylose. Indeed, extracts of xylose-grown cells catalyzed the NADP+-dependent conversion of xylose to xylonate at a specific activity of 0.15 U/mg with an apparent Km for xylose of 0.95 mM. Extracts of xylose-grown cells also catalyzed the oxidation of glucose with NADP+ to gluconate, however, at a 70-fold lower catalytic efficiency (apparent Vmax, 0.03 U/mg; Km, 15 mM), indicating that xylose-grown H. marismortui cells contain a specific xylose dehydrogenase different from glucose dehydrogenase. Glucose-grown cells of H. marismortui also contained xylose dehydrogenase activity, however, with about 10-fold lower catalytic efficiency (apparent Vmax, 0.03 U/mg; Km, 2 mM) compared to that of xylose-grown cells, suggesting that xylose dehydrogenase was induced during growth on xylose.
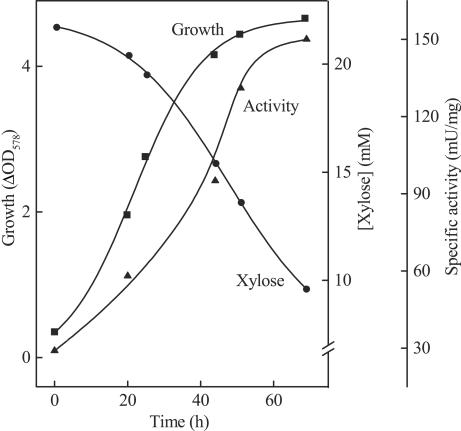
Induction of xylose dehydrogenase was demonstrated during growth of H. marismortui on xylose with cells pregrown on glucose as an inoculum. Because of the higher concentrations of yeast extracts and Casamino Acids present in the medium, the cells grew with a shorter doubling time (about 12 h) to significantly higher cell densities and the amount of xylose consumed increased (Fig. 2). During growth, the NADP+-dependent xylose dehydrogenase activity increased up to fivefold parallel to xylose consumption, indicating that the enzyme is induced by xylose and probably represents the first reaction of xylose catabolism in H. marismortui.
FIG. 2.
Induction of xylose dehydrogenase during growth of H. marismortui on xylose. A culture was incubated at 37°C in a 2,000-ml Erlenmeyer flask filled with 400 ml of medium containing xylose (25 mM), yeast extract (2.5 g/liter), and Casamino Acids (5 g/liter) and shaken at 200 rpm. Glucose-grown cells were used as the inoculum (10%). Growth (▪), xylose consumption (•), and the specific activity of xylose dehydrogenase (▴) were followed over time. Protein concentration was determined by the biuret method.
Purification of xylose dehydrogenase from H. marismortui.
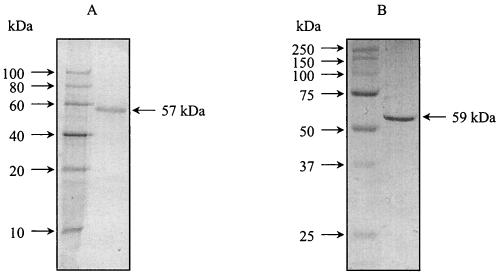
Xylose dehydrogenase was purified from cell extract of xylose-grown H. marismortui cells by only three chromatographic steps. The most efficient purification step was hydrophobic interaction chromatography on Phenyl Sepharose, resulting in 130-fold enrichment. With the entire procedure, the enzyme was purified about 210-fold, to a specific activity of 100 U/mg with a yield of 10% (Table 1). The purified protein was electrophoretically homogeneous as judged by denaturing SDS-PAGE (Fig. 3). Thus, xylose dehydrogenase represents about 0.5% of the cellular protein of H. marismortui.
TABLE 1.
Purification of xylose dehydrogenase from H. marismortui
| Purification step | Amt of proteinb (mg) | Enzyme activitya (U) | Sp act (U/mg) | Yield (%) | Purifi- cation (fold) |
|---|---|---|---|---|---|
| Cell extract | 260 | 125 | 0.48 | 100 | 1 |
| Sepharose CL 4B | 24 | 102 | 4.4 | 82 | 9 |
| Phenyl Sepharose | 0.57 | 37 | 66 | 30 | 137 |
| Superdex | 0.12 | 12 | 100 | 10 | 211 |
Enzyme activity was measured at 37°C as NADP+ and xylose-dependent xylonate formation. During the purification procedure, the assay mixture contained 100 mM Tris-HCl (pH 8.8), 1.5 M KCl, 1 mM NADP+, and 10 mM xylose.
Protein concentration was determined by the Bradford method (7).
FIG. 3.
Purified xylose dehydrogenase from H. marismortui (A) and recombinant xylose dehydrogenase from transformed E. coli (B) as analyzed by SDS-PAGE. (A) Lanes: 1, molecular mass standards; 2, native enzyme purified from H. marismortui. (B) Lanes: 1, molecular mass standards; 2, recombinant enzyme purified from E. coli.
Molecular and catalytic properties.
The apparent molecular mass of native xylose dehydrogenase determined by gel filtration on Superdex 200 was 175 ± 15 kDa. SDS-PAGE revealed only one subunit with an apparent molecular mass of 57 ± 3 kDa (Fig. 3). This value is significantly overestimated, as has been observed for various halophilic enzymes (see Discussion). Recombinant xylose dehydrogenase (see below), showed an apparent molecular mass on SDS-PAGE of 57 ± 3 kDa, although the calculated molecular mass is 39.9 kDa. We propose that native xylose dehydrogenase is a homotetrameric (α4) enzyme.
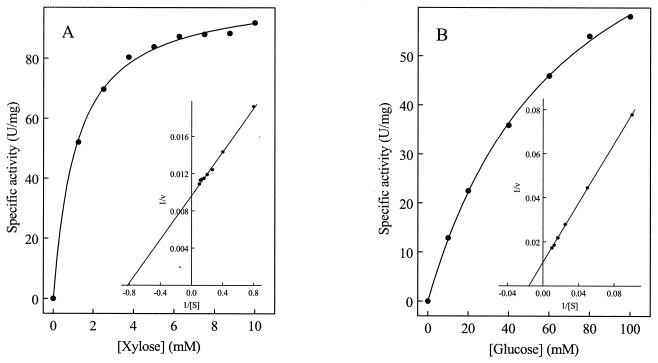
The purified enzyme catalyzed the oxidation of d-xylose with both NADP+ and NAD+. The rate dependence of the enzyme on xylose, NADP+, and NAD+ followed Michaelis-Menten kinetics with apparent Km values of 1.2, 0.15, and 0.9 mM, respectively. The corresponding Vmax values were about 100, 92, and 80 U/mg. The sixfold higher apparent Km for NAD+ compared to NADP+ indicates that NADP+ is the preferred electron acceptor. In addition to d-xylose, various other pentoses and hexoses (all d isomers) were tested as substrates for the dehydrogenase with NADP+ as the electron acceptor. The apparent Km values, Vmax, kcat values, and catalytic efficiencies (kcat/Km) are given in Table 2. The highest catalytic efficiency was obtained with xylose and NADP+. d-Ribose was also accepted by the enzyme at high efficiency, whereas d-arabinose was not oxidized at significant rates, indicating that the configuration change at C-2 significantly affects enzyme activity. d-Glucose was oxidized with a catalytic efficiency 70-fold lower than that with which xylose was oxidized, thus defining the enzyme as a specific xylose dehydrogenase (Fig. 4 A and B). Galactose oxidation was less efficient than glucose oxidation, and almost no activity was found with fructose (Table 2).
TABLE 2.
Kinetic parameters of purified xylose dehydrogenase from H. marismortuia
| Substrate | Vmax (U/mg) | Km (mM) | kcat (s−1) | kcat/Km (s−1, 102) |
|---|---|---|---|---|
| d-Xyloseb | 107 | 1.2 | 356 | 2.97 |
| d-Ribosec | 108 | 2.3 | 360 | 1.57 |
| d-Glucosed | 82 | 64 | 273 | 0.043 |
| d-Galactosee | 39 | NDh | ||
| d-Fructosef | 2 | ND | ||
| d-Arabinoseg | 1 | ND |
Enzyme activity was measured at 37°C with 0.1 M Tris-HCl (pH 8.3)-1.5 M KCl.
Concentrations under Vmax conditions: d-xylose, 10 mM; NADP+, 1 mM.
Concentrations under Vmax conditions: d-ribose, 10 mM; NADP+, 1 mM.
Concentrations under Vmax conditions: d-glucose, 100 mM; NADP+, 1 mM.
Concentrations under Vmax conditions: d-galactose, 100 mM; NADP+, 2 mM.
Concentrations under Vmax conditions: d-fructose, 100 mM; NADP+, 2 mM.
Concentrations under Vmax conditions: d-arabinose, 100 mM; NADP+, 2 mM.
ND, not determined.
FIG. 4.
Rate dependence of xylose dehydrogenase purified from H. marismortui on the concentrations of xylose (A) and glucose (B). The inserts show double-reciprocal plots of the rates versus the corresponding substrate concentrations. The assay mixture contained 20 mM Tris-HCl (pH 8.3), 1.5 M KCl, 1 mM NADP+, various concentrations of xylose or glucose, and enzyme.
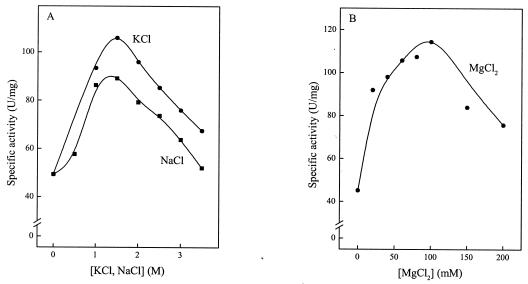
Effects of salt, pH, and temperature on xylose dehydrogenase activity.
The activity of xylose dehydrogenase from H. marismortui was strongly stimulated by high concentrations of NaCl or KCl and by moderate concentrations of MgCl2. Maximal activities were obtained at about 1.5 M both KCl and NaCl and at 100 mM MgCl2 (Fig. 5). The rate dependence of the enzyme on pH and temperature was tested in the presence of 1.5 M KCl. The pH optimum was about pH 8.3, and 50% activity was found at pHs 7 and 9. Xylose dehydrogenase activity increased exponentially with temperature between 25 and 45°C; from the corresponding linear part of an Arrhenius plot, an activation energy of 64 kJ/mol was calculated. The highest catalytic activity of xylose dehydrogenase was found at 50°C.
FIG. 5.
Effects of NaCl and KCl (A) and MgCl2 (B) on xylose dehydrogenase purified from H. marismortui. The assay mixture contained 20 mM Tris-HCl (pH 8.3), 1 mM NADP+, 10 mM xylose, enzyme, and the concentrations of NaCl, KCl, and MgCl2 indicated.
Identification, sequence analysis, and cloning of the gene encoding xylose dehydrogenase from H. marismortui and its functional overexpression in E. coli.
On the basis of the N-terminal amino acid sequence determined from the subunit of xylose dehydrogenase, MNVDALTGGFDRRDWQEQTATDNPVRFAA, an ORF that exactly matches the 29 N-terminal amino acid residues was identified by BLASTP search in contig 97 of the partially sequenced genome of H. marismortui (Zhang et al., personal communication). The ORF contains 1,083 bp coding for a polypeptide of 360 amino acids with a calculated molecular mass of 39.9 kDa; the protein contains large amounts of negatively charged amino acids, 10% Asp and 11% Glu, which is typical for halophilic enzymes (10, 27), and had a predicted pI of 4.2. The G+C content of the ORF is 62 mol%. The coding sequence starts with ATG and stops with TGA. A putative archaeal box A (TATA box) promoter signal (5′-TAATAT-3′) was identified between positions −23 and −28 upstream from the ATG start codon (25, 37). Immediately upstream of the initiation codon, a putative ribosome binding site with the sequence 5′-GTGGT-3′ is present (32). Downstream of the gene, a pyrimidine-rich sequence beginning at position 1069 and a short inverted repeat located between positions 1112 and 1121 were identified, indicating a transcription termination site (38).
The ORF was characterized as the xdh gene encoding xylose dehydrogenase by its functional overexpression in E. coli. The xdh gene was amplified by PCR and cloned into the vector pET17b. The recombinant plasmid was used to transform E. coli BL21 codon plus(DE3)-RIL. After induction with IPTG, a polypeptide of about 59 kDa was overexpressed, which was recovered almost completely in inclusion bodies. The protein purified from inclusion bodies was a catalytically active, extremely halophilic xylose dehydrogenase.
Solubilization, refolding, and purification of recombinant xylose dehydrogenase.
Recombinant xylose dehydrogenase was purified from inclusion bodies by dissolving with 8 M urea in the presence of dithioerythritol, followed by refolding with a buffer containing a high salt concentration (3 M KCl), substrates, and glutathione (see Materials and Methods). Maximal xylose dehydrogenase activity was obtained after 6 days of incubation. The refolded activated xylose dehydrogenase was purified by chromatography on Superdex and Phenyl Resource. The recombinant xylose dehydrogenase showed molecular and kinetic properties almost identical to those of the enzyme purified from H. marismortui (Table 2). The molecular masses of the native enzyme and subunits were 180 ± 10 and 59 ± 3 kDa (Fig. 3), respectively, and the apparent Km values were very similar for xylose, ribose, glucose, and NADP+ (data not shown); however, the apparent Vmax of the recombinant enzyme was significantly (about 40%) lower.
DISCUSSION
In the present communication, we describe the purification and characterization of the first archaeal d-xylose dehydrogenase and the gene that encodes it from the halophilic archaeon H. marismortui. The enzyme was induced during growth on xylose, suggesting that xylose dehydrogenase represents the initial enzyme of the xylose degradation pathway in this archaeon. The enzyme constitutes a novel type of xylose dehydrogenase related to GFOR from Z. mobilis.
Molecular and kinetic properties.
Xylose dehydrogenase was characterized as a homotetrameric enzyme of about 175 kDa; the calculated subunit molecular mass is 39.9 kDa. The apparent molecular mass of subunits on SDS-PAGE of about 57 kDa obtained for xylose dehydrogenase was overestimated as reported for several halophilic proteins, probably because of the presence of large amounts of negatively charged amino acids. The same degree of overestimation as described for xylose dehydrogenase was observed with glucose dehydrogenase from Haloferax mediterranei. The enzyme has a calculated molecular mass of 39.3 kDa (27) and showed an apparent molecular mass on SDS-PAGE of 53 ± 3 kDa (6).
Xylose dehydrogenase showed dual cofactor specificity for pyridine nucleotides with a high preference for NADP+ over NAD+, indicating that NADP+ is the physiological electron acceptor. The enzyme catalyzed the oxidation of xylose, ribose, and glucose; however, the catalytic activity for the latter was about 70-fold lower. The archaeal xylose dehydrogenase can be discriminated from archaeal glucose dehydrogenases characterized from various organisms including Haloferax, Sulfolobus, Thermoplasma, and Thermoproteus spp., which all show xylose dehydrogenase activity. These archaeal glucose dehydrogenases are tetrameric or dimeric enzymes composed of 40-kDa subunits, show dual cofactor specificity for NADP+ and NAD+ with a high preference for NADP+, and utilize various aldoses (hexose and pentoses) including xylose in addition to glucose. However, in contrast to xylose dehydrogenase from H. marismortui, all archaeal glucose dehydrogenases showed significantly higher catalytic efficiencies for NADP+-dependent oxidation of glucose compared to that of xylose (6, 22, 33, 34).
Few reports of purified xylose dehydrogenases from Eucarya and Bacteria are available. An NADP+-specific xylose dehydrogenase from pig liver was characterized (46). The enzyme is a homodimer composed of 32-kDA subunits showing the highest catalytic activity with xylose but also accepts ribose and glucose at about 7- or 30-fold lower catalytic efficiency. The enzyme was specific for NADP+ and did not reduce NAD+. Recently, Aoki et al. (1) demonstrated that the NADP+-dependent d-xylose dehydrogenase of pig liver is identical to dimeric dihydrodiol dehydrogenase (DD). Copurification of DD activity and xylose dehydrogenase activity from pig liver, molecular mass and kinetic analyses, and inhibitor studies showed that the two enzymes are identical (1). Dimeric DDs catalyze the NADP+-dependent oxidation of various aromatic hydrocarbons, e.g., naphthalene dihydrodiol, to the corresponding catechols and also the oxidation of various sugars, with xylose as the most effective sugar substrate. The oxidation rate of naphthalene dihydrodiol was about twofold higher than that of xylose (1), suggesting that dihydrodiols are the preferred substrates of the dimeric DD-xylose dehydrogenase. The genes coding for various mammalian dimeric DD-xylose dehydrogenases, including pig liver, rabbit lens, human intestine, and monkey kidney, were sequenced (2), and thus, sequences of eucaryal xylose dehydrogenases are known.
In bacteria, oxidation of xylose to xylonate has been reported for several species (9); an NAD+-reducing xylose dehydrogenase activity was reported for two Caulobacter species (28); however—to our knowledge—the only xylose dehydrogenase from bacteria characterized to some detail is the enzyme from Arthrobacter sp. The enzyme was induced by xylose and showed a high specificity for xylose (apparent Km, 17.4 mM) and for NAD+ as an electron acceptor. Other pentoses and hexoses, as well as NADP+ as a cofactor, were not accepted as substrates (44). The amino acid sequence of the enzyme has not been reported.
Thus, both characterized eucaryal and bacterial d-xylose dehydrogenases showed significant differences in molecular and kinetic properties compared to the archaeal xylose dehydrogenase from H. marismortui.
Sequence comparison and phylogenetic analysis of archaeal xylose dehydrogenase.
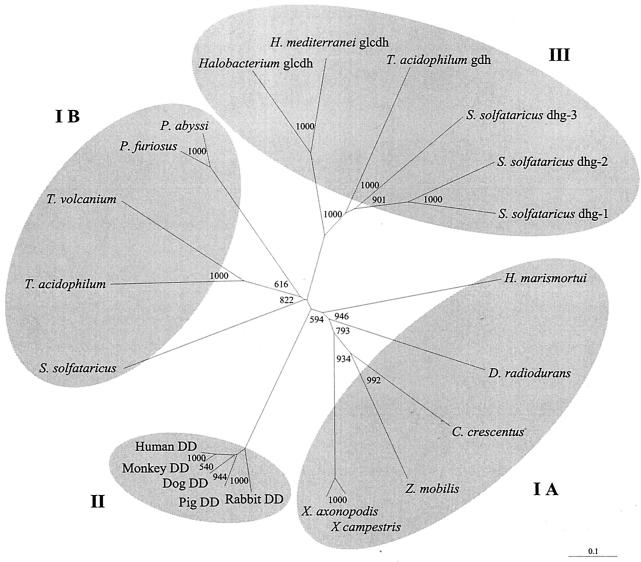
On the basis of the N-terminal amino acid sequence of the subunit, the xdh gene encoding the xylose dehydrogenase from H. marismortui was identified in contig 97 of the partially sequenced genome of the organism by functional overexpression in E. coli. BLASTP searches of nonredundant databases with the deduced amino acid sequence of the xdh gene from H. marismortui revealed various hits. Almost all of them were putative oxidoreductases or dehydrogenases. The highest degrees of similarity were found with bacterial GFOR from Z. mobilis (41%) and hypothetical GFORs from the bacteria Deinococcus radiodurans, Caulobacter crescentus, Streptococcus pneumoniae, and Bacillus halodurans (35 to 40%). Similarities of 31 to 34% with the putative archaeal oxidoreductases or dehydrogenases from Pyrococcus sp., Sulfolobus solfataricus, and Thermoplasma sp. were found, the best scores being obtained for the putative dehydrogenases from Pyrococcus furiosus (PF1919) and S. solfataricus (SSO3015). Lower degrees of similarity were found with characterized eucaryal xylose dehydrogenases-dimeric DDs (28 to 29%) and with archaeal glucose dehydrogenases (17 to 21%) from halophiles (Haloferax and Halobacterium spp.) and thermoacidophiles (Sulfolobus and Thermoplasma spp.). For accession numbers, see the legend to Fig. 7.
FIG. 7.
Phylogenetic relationships of the archaeal xylose dehydrogenase from H. marismortui, oxidoreductases and dehydrogenases from bacteria (IA) and archaea (IB), xylose dehydrogenases-dimeric DDs from eucarya (II), and glucose dehydrogenases from archaea (III). The numbers at the nodes are bootstrapping values according to neighbor joining (generated by using the neighbor-joining options of ClustalX). National Center for Biotechnology Information accession numbers: C. crescentus, AAK23207; D. radiodurans, B75475; dog DD, BAA83487; Halobacterium sp. NRC-1 glcdh, AAG18991; H. mediterranei glcdh, CAC4250; human DD, BAA83490; monkey DD, BAA83488; pig DD, BAA83486; Pyrococcus abyssi ORF PAB1139, B75025; P. furiosus ORF PF1919, AAL82043; rabbit DD, BAA83485; S. solfataricus ORF SSO3015, AAK43117; S. solfataricus dhg-1 (ORF SSO3003), AAK43106; S. solfataricus dhg-2 (ORF SSO3042), AAK43143; S. solfataricus dhg-3 (ORF SSO3204), AAK43301; Thermoplasma acidophilum ORF TA1182, CAC12307; T. acidophilum gdh, CAA42450; T. volcanium TVG1418453, BAB60511; X. campestris, AAM40130; X. axonopodis, AAM35776; Z. mobilis GFOR, 1H6DK.
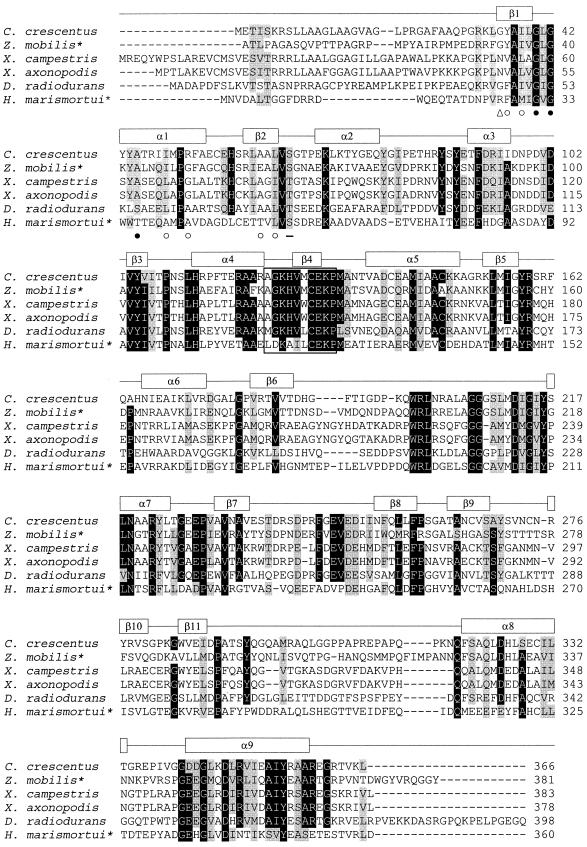
The most similar enzyme of xylose dehydrogenase from H. marismortui, i.e., GFOR from Z. mobilis, is a homotetrameric enzyme composed of 40-kDa subunits and containing tightly bound cofactor NADP+. The enzyme catalyzes the coupled intermolecular oxidation-reduction of glucose and fructose to form gluconolactone and sorbitol. The periplasmic enzyme is synthesized as a precursor with an N-terminal signal peptide of 52 amino acid residues (14). A multiple-sequence alignment of xylose dehydrogenase of H. marismortui, mature GFOR from Z. mobilis, and other putative bacterial oxidoreductases is given in Fig. 6. The alignment includes a prediction of secondary structure, which is in accordance with the crystal structure of GFOR from Zymomonas oxidoreductase (20). Sequence comparisons indicate a variety of conserved regions including a typical βαβ dinucleotide binding pocket (Rossman fold, amino acids 26 to 57) (16, 43) and—with few deviations—the recently postulated consensus sequence for a novel class of dehydrogenases, including the highly conserved EKP motif (42) (amino acids 113 to 122). Although xylose dehydrogenase shows a high degree of similarity to GFOR from Z. mobilis, the two enzymes catalyze different reactions; xylose dehydrogenase is a dehydrogenase with dual cofactor specificity for NADP+ and NAD+, whereas GFOR catalyzes the coupled oxidation-reduction of glucose and fructose with tightly bound NADP+, i.e., in the absence of added cofactors. However, it has been shown that substitution of a single amino acid alters GFOR from Z. mobilis to a glucose dehydrogenase with dual cofactor specificity for NADP+ and NAD+ (42). Thus, one might speculate that H. marismortui xylose dehydrogenase represents a natural mutant of an aldose-ketose oxidoreductase.
FIG. 6.
Multiple-sequence alignment of deduced amino acid sequences of the xylose dehydrogenase from H. marismortui, of GFOR from Z. mobilis, and of putative bacterial oxidoreductases. Characterized proteins are marked by asterisks. The alignment was generated by ClustalX with the Gonnet matrix (39). The predicted secondary structure of the xylose dehydrogenase from H. marismortui is shown above the sequences. Symbols denote residues of the Rossman fingerprint motif (16, 43): ▵, basic or hydrophilic; ○, small and hydrophobic; •, glycine; —, acid. National Center for Biotechnology Information accession numbers: C. crescentus, AAK23207; D. radiodurans, B75475; Xanthomonas axonopodis, AAM35776; X. campestris, AAM40130; Z. mobilis GFOR, 1H6DK.
The phylogenetic relationship of the xylose dehydrogenase of H. marismortui with oxidoreductase and dehydrogenase sequences, showing significant similarity according to BLASTP searches (see above), is given in the phylogram shown in Fig. 7. They include the characterized GFOR from Z. mobilis and putative bacterial GFORs (cluster IA), putative archaeal oxidoreductases and dehydrogenases (IB), eucaryal xylose dehydrogenases-dimeric DDs (II), and archaeal glucose dehydrogenases (III), each forming a separate cluster. In accordance with the highest degree of similarity, the H. marismortui sequence clusters within the bacterial oxidoreductases for which only the GFOR from Z. mobilis has been functionally characterized. Cluster IB contains only putative archaeal oxidoreductases and dehydrogenases, including dehydrogenases from Sulfolobus and Pyrococcus spp. Determination of whether the putative bacterial or archaeal sequences of clusters IA and IB represent functional oxidoreductases or (xylose) dehydrogenases must await their biochemical characterization following expression of the genes that encode them. Eukaryotic xylose dehydrogenases-dimeric DDs, (cluster II), which probably represent a novel family of dehydrogenases (1, 2), and archaeal glucose dehydrogenases (cluster III), which belong to the medium-chain dehydrogenase-reductase superfamily (26), form distinct phylogenetic clusters separate from the xylose dehydrogenase from H. marismortui, which is in accordance with differences in their molecular and catalytic properties.
Is xylose dehydrogenase the first enzyme of archaeal xylose catabolism?
The xylose dehydrogenase of H. marismortui showed specific induction during growth on xylose; together with the findings that xylose isomerase and xylulose kinase, as well as the genes that encode them, were absent from H. marismortui, we suggest that this novel type of xylose dehydrogenase represents the first step in xylose degradation by this archaeon. Since xylose isomerase and xylulose kinase and the genes that encode them, xylA and xylB, have not been reported for any archaeal species, one might speculate that the initial reaction of xylose metabolism in archaea in general might involve a xylose dehydrogenase rather than xylose isomerase and xylulose kinase, the typical reactions in bacterial xylose catabolism. Further steps in the xylose degradation pathway in H. marismortui following the fate of xylonate remain to be elucidated. Experiments using a proteomic approach to identify xylose-inducible proteins as analyzed by two-dimensional gel electrophoresis are in progress.
Acknowledgments
We thank R. Schmid (Mikrobiologie, Universität Osnabrück, Osnabrück, Germany) for performing N-terminal amino acid sequencing. We thank Shiladitya DasSarma for getting access to available contigs of the genome of H. marismortui (National Science Foundation grant reference MCB-0135595; University of Maryland Biotechnology Institute website [http://zdna2.umbi.umd.edu]). The expert technical assistance of A. Brandenburger is gratefully acknowledged.
This work was supported by the EU grant Extremophiles as Cell Factories and by the Fonds der Chemischen Industrie.
Footnotes
Dedicated to Rolf Thauer on the occasion of his 65th birthday.
REFERENCES
- 1.Aoki, S., S. Ishikura, Y. Asada, N. Usami, and A. Hara. 2001. Identity of dimeric dihydrodiol dehydrogenase as NADP+-dependent d-xylose dehydrogenase in pig liver. Chemico-Biol. Interact. 130-132:775-784. [DOI] [PubMed] [Google Scholar]
- 2.Arimitsu, E., S. Aoki, S. Ishikura, K. Nakanishi, K. Matsuura, and A. Hara. 1999. Cloning and sequencing of the cDNA species for mammalian dimeric dihydrodiol dehydrogenases. Biochem. J. 342(Pt. 3):721-728. [PMC free article] [PubMed] [Google Scholar]
- 3.Bhosale, S. H., M. B. Rao, and V. V. Deshpande. 1996. Molecular and industrial aspects of glucose isomerase. Microbiol. Rev. 60:280-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biesterveld, S., M. D. Kok, C. Dijkema, A. J. B. Zehnder, and A. J. M. Stams. 1994. d-Xylose catabolism in Bacteriodes xylanolyticus X5-1. Arch. Microbiol. 161:521-527. [DOI] [PubMed] [Google Scholar]
- 5.Bode, C., H. Goebell, and E. Stahler. 1968. Elimination of errors caused by turbidity in the determination of protein by the biuret method. Z. Klin. Chem. Klin. Biochem. 6:418-422. [PubMed] [Google Scholar]
- 6.Bonete, M. J., C. Pire, F. I. LLorca, and M. L. Camacho. 1996. Glucose dehydrogenase from the halophilic archaeon Haloferax mediterranei: enzyme purification, characterisation and N-terminal sequence. FEBS Lett. 383:227-229. [DOI] [PubMed] [Google Scholar]
- 7.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 8.Brückner, J. 1955. Estimation of monosaccharides by the orcinol-sulphuric acid reaction. Biochem. J. 60:200-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buchert, J., L. Viikari, M. Linko, and P. Markkanen. 1986. Production of xylonic acid by Pseudomonas fragi. Biotechnol. Lett. 8:541-546. [Google Scholar]
- 10.Cendrin, F., J. Chroboczek, G. Zaccai, H. Eisenberg, and M. Mevarech. 1993. Cloning, sequencing, and expression in Escherichia coli of the gene coding for malate dehydrogenase of the extremely halophilic archaebacterium Haloarcula marismortui. Biochemistry 32:4308-4313. [DOI] [PubMed] [Google Scholar]
- 11.Connaris, H., J. B. Chaudhuri, M. J. Danson, and D. W. Hough. 1999. Expression, reactivation, and purification of enzymes from Haloferax volcanii in Escherichia coli. Biotechnol. Bioeng. 64:38-45. [PubMed] [Google Scholar]
- 12.David, J. D., and H. Wiesemeyer. 1970. Control of xylose metabolism in Escherichia coli. Biochim. Biophys. Acta 201:497-499. [DOI] [PubMed] [Google Scholar]
- 13.Erlandson, K. A., J. H. Park, Wissam, K. El, H. H. Kao, P. Basaran, S. Brydges, and C. A. Batt. 2000. Dissolution of xylose metabolism in Lactococcus lactis. Appl. Environ. Microbiol. 66:3974-3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Halbig, D., T. Wiegert, N. Blaudeck, R. Freudl, and G. A. Sprenger. 1999. The efficient export of NADP-containing glucose-fructose oxidoreductase to the periplasm of Zymomonas mobilis depends both on an intact twin-arginine motif in the signal peptide and on the generation of a structural export signal induced by cofactor binding. Eur. J. Biochem. 263:543-551. [DOI] [PubMed] [Google Scholar]
- 15.Hansen, T., D. Wendorff, and P. Schönheit. 2004. Bifunctional phosphoglucose/phosphomannose isomerases from the Archaea Aeropyrum pernix and Thermoplasma acidophilum constitute a novel enzyme family within the phosphoglucose isomerase superfamily. J. Biol. Chem. 279:2262-2272. [DOI] [PubMed] [Google Scholar]
- 16.Hanukoglu, I., and T. Gutfinger. 1989. cDNA sequence of adrenodoxin reductase. Identification of NADP-binding sites in oxidoreductases. Eur. J. Biochem. 180:479-484. [DOI] [PubMed] [Google Scholar]
- 17.Horecker, B. L. 1988. d-Xylulose and d-xylose, p. 465-473. In H.-U. Bergmeyer (ed.), Methods of enzymatic analysis. VCH Verlagsgesellschaft mbH, Weinheim, Germany.
- 18.Johnsen, U., T. Hansen, and P. Schönheit. 2003. Comparative analysis of pyruvate kinases from the hyperthermophilic archaea Archaeoglobus fulgidus, Aeropyrum pernix, and Pyrobaculum aerophilum and the hyperthermophilic bacterium Thermotoga maritima: unusual regulatory properties in hyperthermophilic archaea. J. Biol. Chem. 278:25417-25427. [DOI] [PubMed] [Google Scholar]
- 19.Johnsen, U., M. Selig, K. B. Xavier, H. Santos, and P. Schönheit. 2001. Different glycolytic pathways for glucose and fructose in the halophilic archaeon Halococcus saccharolyticus. Arch. Microbiol. 175:52-61. (Erratum, 180:503, 2003.) [DOI] [PubMed] [Google Scholar]
- 20.Kingston, R. L., R. K. Scopes, and E. N. Baker. 1996. The structure of glucose-fructose oxidoreductase from Zymomonas mobilis: an osmoprotective periplasmic enzyme containing non-dissociable NADP. Structure 4:1413-1428. [DOI] [PubMed] [Google Scholar]
- 21.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 22.Lamble, H. J., N. I. Heyer, S. D. Bull, D. W. Hough, and M. J. Danson. 2003. Metabolic pathway promiscuity in the archaeon Sulfolobus solfataricus revealed by studies on glucose dehydrogenase and 2-keto-3-deoxygluconate aldolase. J. Biol. Chem. 278:34066-34072. [DOI] [PubMed] [Google Scholar]
- 23.Lin, E. C. C. 1996. Dissimilatory pathways of sugars, polyols, and carboxylates, p. 307-342. In F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger. (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, D.C.
- 24.Oren, A., M. Ginzburg, B. Z. Ginzburg, L. I. Hochstein, and B. E. Volcani. 1990. Haloarcula marismortui (Volcani) sp. nov., nom. rev., an extremely halophilic bacterium from the Dead Sea. Int. J. Syst. Bacteriol. 40:209-210. [DOI] [PubMed] [Google Scholar]
- 25.Palmer, J. R., and C. J. Daniels. 1995. In vivo definition of an archaeal promoter. J. Bacteriol. 177:1844-1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Persson, B., J. S. J. Zigler, and H. Jornvall. 1994. A super-family of medium-chain dehydrogenases/reductases (MDR). Sub-lines including zeta-crystallin, alcohol and polyol dehydrogenases, quinone oxidoreductase enoyl reductases, VAT-1 and other proteins. Eur. J. Biochem. 226:15-22. [DOI] [PubMed] [Google Scholar]
- 27.Pire, C., J. Esclapez, J. Ferrer, and M. J. Bonete. 2001. Heterologous overexpression of glucose dehydrogenase from the halophilic archaeon Haloferax mediterranei, an enzyme of the medium chain dehydrogenase/reductase family. FEMS Microbiol. Lett. 200:221-227. [DOI] [PubMed] [Google Scholar]
- 28.Poindexter, J. 1964. Biological properties and classification of the Caulobacter group. Bacteriol. Rev. 28:231-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodionov, D. A., A. A. Mironov, and M. S. Gelfand. 2001. Transcriptional regulation of pentose utilisation systems in the Bacillus/Clostridium group of bacteria. FEMS Microbiol. Lett. 205:305-314. [DOI] [PubMed] [Google Scholar]
- 30.Schönheit, P., and T. Schäfer. 1995. Metabolism of hyperthermophiles. World J. Microbiol. Biotechnol. 11:26-57. [DOI] [PubMed] [Google Scholar]
- 31.Selig, M., K. B. Xavier, H. Santos, and P. Schönheit. 1997. Comparative analysis of Embden-Meyerhof and Entner-Doudoroff glycolytic pathways in hyperthermophilic archaea and the bacterium Thermotoga. Arch. Microbiol. 167:217-232. [DOI] [PubMed] [Google Scholar]
- 32.Shine, J., and L. Dalgarno. 1975. Determinant of cistron specificity in bacterial ribosomes. Nature 254:34-38. [DOI] [PubMed] [Google Scholar]
- 33.Siebers, B., V. F. Wendisch, and R. Hensel. 1997. Carbohydrate metabolism in Thermoproteus tenax: in vivo utilization of the non-phosphorylative Entner-Doudoroff pathway and characterization of its first enzyme, glucose dehydrogenase. Arch. Microbiol. 168:120-127. [DOI] [PubMed] [Google Scholar]
- 34.Smith, L. D., N. Budgen, S. J. Bungard, M. J. Danson, and D. W. Hough. 1989. Purification and characterization of glucose dehydrogenase from the thermoacidophilic archaebacterium Thermoplasma acidophilum. Biochem. J. 261:973-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Staley, J. T. 1968. Prosthecomicrobium and Ancalomicrobium: new prosthecate freshwater bacteria. J. Bacteriol. 95:1921-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takeda, Y., K. Takase, I. Yamato, and K. Abe. 1998. Sequencing and characterization of the xyl operon of a gram-positive bacterium, Tetragenococcus halophila. Appl. Environ. Microbiol. 64:2513-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomm, M. 1996. Archaeal transcription factors and their role in transcription initiation. FEMS Microbiol. Rev. 18:159-171. [DOI] [PubMed] [Google Scholar]
- 38.Thomm, M., W. Hausner, and C. Hethke. 1994. Transcription factors and termination of transcription in Methanococcus. Syst. Appl. Microbiol. 16:648-655. [Google Scholar]
- 39.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tindall, B. J. 1992. The family Halobacteriaceae, p. 768-808. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K. H. Schleifer (ed.), The prokaryotes. A handbook of bacteria: ecophysiology, isolation, identification, applications. Springer-Verlag, New York, N.Y.
- 41.Verhees, C. H., S. W. Kengen, J. E. Tuininga, G. J. Schut, M. W. Adams, W. M. De Vos, and J. Van der Oost. 2003. The unique features of glycolytic pathways in Archaea. Biochem. J. 375:231-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wiegert, T., H. Sahm, and G. A. Sprenger. 1997. The substitution of a single amino acid residue (Ser-116 → Asp) alters NADP-containing glucose-fructose oxidoreductase of Zymomonas mobilis into a glucose dehydrogenase with dual coenzyme specificity. J. Biol. Chem. 272:13126-13133. [DOI] [PubMed] [Google Scholar]
- 43.Wierenga, R. K., M. De Maeyer, and W. G. Hol. 1985. Interaction of pyrophosphate moieties with α-helixes in dinucleotide binding proteins. Biochemistry 24:1346-1357. [Google Scholar]
- 44.Yamanaka, K., M. Gino, and R. Kaneda. 1977. A specific NAD-d-xylose dehydrogenase from Arthrobacter sp. Agric. Biol. Chem. 41:1493-1499. [Google Scholar]
- 45.Zaigler, A., S. C. Schuster, and J. Soppa. 2003. Construction and usage of a onefold-coverage shotgun DNA microarray to characterize the metabolism of the archaeon Haloferax volcanii. Mol. Microbiol. 48:1089-1105. [DOI] [PubMed] [Google Scholar]
- 46.Zepeda, S., O. Monasterio, and T. Ureta. 1990. NADP(+)-dependent d-xylose dehydrogenase from pig liver. Purification and properties. Biochem. J. 266:637-644. [DOI] [PMC free article] [PubMed] [Google Scholar]