Abstract
Expression of ica operon-mediated biofilm formation in Staphylococcus epidermidis RP62A is subject to phase variable regulation. Reversible transposition of IS256 into icaADBC or downregulation of icaADBC expression are two important mechanisms of biofilm phenotypic variation. Interestingly, the presence of IS256 was generally associated with a more rapid rate of phenotypic variation, suggesting that IS256 insertions outside the ica locus may affect ica transcription. Consistent with this, we identified variants with diminished ica expression, which were associated with IS256 insertions in the σB activator rsbU or sarA. Biofilm development and ica expression were activated only by ethanol and not NaCl in rsbU::IS256 insertion variants, which were present in ∼11% of all variants. σB activity was impaired in rsbU::IS256 variants, as evidenced by reduced expression of the σB-regulated genes asp23, csb9, and rsbV. Moreover, expression of sarA, which is σB regulated, and SarA-regulated RNAIII were also suppressed. A biofilm-forming phenotype was restored to rsbU::IS256 variants only after repeated passage and was not associated with IS256 excision from rsbU. Only one sarA::IS256 insertion mutant was identified among 43 biofilm-negative variants. Both NaCl and ethanol-activated ica expression in this sarA::IS256 variant, but only ethanol increased biofilm development. Unlike rsbU::IS256 variants, reversion of the sarA::IS256 variant to a biofilm-positive phenotype was accompanied by precise excision of IS256 from sarA and restoration of normal ica expression. These data identify new roles for IS256 in ica and biofilm phenotypic variation and demonstrate the capacity of this element to influence the global regulation of transcription in S. epidermidis.
Biofilm formation by staphylococci on implanted biomaterials is now recognized as an important virulence factor contributing to the development of a significant proportion of all device-related infections. Enzymes encoded by the icaADBC operon (20, 23, 27) are required for synthesis of an extracellular polysaccharide termed polysaccharide intercellular adhesin (PIA) in Staphylococcus epidermidis (37, 58) and poly-N-acetylglucosamine (PNAG) in S. aureus (12, 31, 41, 43, 44), which plays an important role in both the initial attachment and cellular proliferation processes characteristic of staphylococcal biofilm development (27, 38, 38, 40).
Much effort has been focused on understanding the regulation of ica operon expression, and it is now known that increased transcription of the ica operon can be observed under anaerobic growth conditions (13), in the presence of subinhibitory concentrations of certain antibiotics, and in response to osmotic stress (32, 52). We have recently reported that the icaR gene encodes a transcriptional repressor with a central role in the environmental regulation of ica operon expression in S. epidermidis (10, 11). Jefferson et al. (29) demonstrated that purified IcaR protein from S. aureus bound the ica operon promoter region close to the icaA start codon. Consistent with this, deletion of the icaR gene in S. aureus is also associated with activation of ica operon expression (30; C. A. Kennedy and J. P. O'Gara, unpublished data). A new negative regulator of ica operon expression, the teicoplanin-associated locus regulator (TcaR), has also been recently described in S. aureus (30).
The S. aureus global stress response regulator, σB, and the S. epidermidis rsbU gene (a positive regulator of σB) have also been implicated in the regulation of biofilm development in S. aureus and S. epidermidis (32, 51). Interestingly, induction of biofilm by NaCl is affected in S. epidermidis rsbU and S. aureus σB mutants (32, 51). However, the role of σB in the regulation of ica operon expression remains unclear given the absence of an identifiable consensus binding site for this sigma factor upstream of the icaA or icaR start codons. In addition, Valle et al. (54) recently reported that the staphylococcal accessory regulator, sarA, which controls the expression of over 100 genes (16), and not σB was required for ica operon expression, PIA/PNAG synthesis, and biofilm development in S. aureus, although σB was found to influence the regulation of ica operon transcription. In contrast, a transposon mutation in the rsbU gene in S. epidermidis was associated with both a reduction in PIA/PNAG levels and the loss of biofilm-forming capacity (32).
Coagulase-negative staphylococci and S. aureus are capable of rapid phenotypic switching involving properties such as colony morphology, growth rate, antibiotic susceptibility, and biofilm-forming capacity (9, 11, 14, 25, 45, 46, 58, 59). In terms of pathogenesis, staphylococcal phenotypic variation may contribute to dissemination, invasive disease, and sepsis. Ziebuhr et al. (59) demonstrated that reversible inactivation of the ica operon by the insertion sequence element IS256 can result in the production of 25 to 33% of phenotypic variants. More recently, we characterized multiple S. epidermidis isolates and demonstrated that downregulation of ica operon expression and mutation are the primary mechanisms responsible for biofilm phenotypic variation (25). It therefore appears that more than one mechanism contributes to the production of phenotypic variants in S. epidermidis. In this context, it is interesting to note the reported association between the levels of resistance to methicillin, oxacillin, and penicillin and biofilm-forming capacity in phenotypic variants of S. epidermidis (45, 46), suggesting that the phenotypic switch associated with impaired biofilm-forming capacity may also impact on other properties. In addition, the global regulators Agr and SarA, which influence methicillin resistance in S. aureus (50), have also been demonstrated to regulate staphylococcal biofilm formation (2, 54, 56, 57).
In the present study we characterized the contribution of IS256 to the production of biofilm-negative variants with diminished ica operon expression. Two genetic switches were identified that involved IS256 insertions at the rsbU and sarA genes. The impact of IS256 insertions in both genes on biofilm, σB regulon, and agr and sarA expression was examined. The data presented here provide new insights into biofilm phenotypic variation and the impact of IS256-mediated dynamic genetic events on the activity of at least three global regulators and the regulation of staphylococcal biofilm formation.
MATERIALS AND METHODS
Bacterial strains, media, growth conditions, and isolation of biofilm-negative phenotypic variants.
The bacterial strains used in the present study are shown in Table 1. Bacteria were routinely grown at 37°C on brain heart infusion (BHI) medium (Oxoid) supplemented with 4% ethanol, 4% NaCl, or 10% NaCl. Semiquantitative determinations of biofilm formation in 96-well tissue culture plates (Nunc) were performed as described previously (11). To screen for biofilm-negative phenotypic variants bacteria were grown on Congo red agar (CRA) plates as described previously (10, 11, 59).
TABLE 1.
Rate of phenotypic switching in IS256-positive and IS256-negative clinical isolates of S. epidermidis
| S. epidermidis strain | Relevant characteristic(s) | IS256 | Avg ON-to-OFF switching rate (SD)a | Source or reference |
|---|---|---|---|---|
| RP62A | Biofilm-positive, blood culture isolate | + | 5.5 (1.4) × 10−5 | ATCC 35984b |
| CSF24047 | Biofilm-positive, blood culture isolate (Beaumont Hospital, Dublin, Ireland) | + | 5.3 (2.5) × 10−5 | This study |
| 13652 | Biofilm-positive, blood culture isolate (Duke University, Durham, NC.) | + | 4.1 (1.6) × 10−5 | Vance Fowler, Duke University |
| 17174 | Biofilm-positive, blood culture isolate (Duke University, Durham, NC.) | + | 4.4 (2.3) × 10−5 | Vance Fowler, Duke University |
| 8621 | Biofilm-positive, blood culture isolate (Duke University, Durham, NC.) | + | 3.0 (0.5) × 10−5 | Vance Fowler, Duke University |
| SE56 | Biofilm-positive, blood culture isolate (Medical College of Virginia, Richmond, Va.) | + | 3.2 (1.0) × 10−5 | Paul Fey, University of Nebraska |
| CSF41498 | Biofilm-positive, cerebrospinal fluid isolate | − | 0.17 (0.02) × 10−5 | 11 |
| SE5 | Biofilm-positive, blood culture isolate (Medical College of Virginia, Richmond, Va.) | − | 0.15 (0.06) × 10−5 | Paul Fey, University of Nebraska (53) |
| 1457 | Biofilm-positive, blood culture isolate (Hamburg, Germany) | − | 1.3 (0.6) × 10−5 | D. Mack, Hamburg, Germany |
| BC78032 | Biofilm-positive, blood culture isolate (Beaumont Hospital, Dublin, Ireland) | − | 0.87 (0.55) × 10−5 | This study |
| BC70837 | Biofilm-positive, blood culture isolate (Beaumont Hospital, Dublin, Ireland) | − | 0.76 (0.23) × 10−5 | This study |
| 14765 | Biofilm-positive, blood culture isolate (Duke University, Durham, NC.) | − | 1.0 (0.7) × 10−5 | Vance Fowler, Duke University |
That is, the rate of biofilm-positive to biofilm-negative switching per CFU per generation. Assays were performed on at least three independent occasions for each strain. Average values and standard deviations are shown.
ATCC, American Type Culture Collection.
Measurement of biofilm phase variation (switching) rates.
To examine the rates of phenotypic switching, we adapted the method of Eisenstein (17) as described previously (48) to measure the rate at which variant cells were produced during the growth of a colony from a single biofilm-forming CFU. Black colonies from CRA were restreaked onto BHI agar and incubated overnight at 37°C. Individual colonies from these plates, which are almost exclusively derived from an initially biofilm-positive CFU, were resuspended and serially diluted in sterile H2O, plated onto CRA plates, and incubated at 37°C for 24 h. Plates were inspected by using a colony microscope to identify and count the number of variant colonies as a proportion of the total number of parental, biofilm-positive, black colonies, which in turn reflects the proportion of variant and parental cells in the original colony. Since each biofilm-positive colony analyzed in this way originates from a biofilm-positive bacterial CFU, the proportion of bacterial cells that have switched to the variant phenotype in the resulting colony is a reflection of the phenotypic switching rate. The number of generations required for an individual biofilm-positive CFU to give rise to a colony is calculated by dividing the log of the total viable count by the log of 2 [i.e., log(total viable count)/log(2)]. The data were recorded as phenotypic switch frequencies per CFU per generation.
Isolation of biofilm-positive revertants.
A single colony of a biofilm-negative phenotypic variant was grown in BHI medium at 37°C in a tissue culture flask. After 24 h the medium was replaced. This procedure was repeated until a biofilm of adhering bacteria became visible on the bottom of the tissue culture flask or at the liquid-air interface (minimum of 4 days). After a wash with phosphate-buffered saline (PBS), the adhering bacterial cells were scratched from the bottom and streaked on CRA. After incubation at 37°C overnight and an additional 24 h at room temperature, single, black colonies were isolated.
Genetic techniques.
Genomic and plasmid DNA purification and manipulations were performed as described previously (10, 11). The oligonucleotide primers used in the present study were supplied by MWG Biotech (Germany) and are listed in Table 2.
TABLE 2.
Oligonucleotide primers used in this study
| Target gene(s) | Primers | Primers sequence (5′-3′) |
|---|---|---|
| rsbU, rsbV, rsbW, sigB | SESigFor | TCACCAGTTCAAGGGTCTGA |
| SESigRev | TCTTTGGAGCTTCGTCTGTG | |
| icaR, icaADBC | ICAR1 | CTCGAATTTGTTACATACTAG |
| ICAC1 | CCATAGCTTGAATAAGGGAC | |
| icaR, ica operon promoter | PV1 | TTTGAAATCTCGAATTTGTTACAT |
| PV2 | CAATGATCGATTAAAGGGTTTTTC | |
| rsbUVFor | GGAAGTAAGGAGGCGCATTT | |
| rsbUVRev | GCGCCAGCTCTTGAAAATAC | |
| rsbU, rsbV | rsbUFor | GGAAGTAAGGAGGCGCATTT |
| rsbU2 | CGACTTCTGGTAACACATCGAG | |
| rsbU4 | AAAATTTGGCATGGATGCTT | |
| rsbU5 | CGTCCAATTCTCCACCAACT | |
| asp23 | SEasp23For | CATGAAAGGTGGCTTCACAG |
| SEasp23Rev | CATTACGTCGTCAACTTGCAT | |
| csb9 | SEcsb9For | ATTCGATGCGAGTGGAGATT |
| SEcsb9Rev | CGTGGAATTGATCCTGCTTT | |
| rsbV | SErsbVFor | TTGGTGGAGAATTGGACGTA |
| SErsbVRev | CCTATCCGCTCTGAAACACC | |
| RNAIII | SERNAIIIFor | TGAAGTTATGATGGCAGCAGAT |
| SERNAIIIRev | GTTGGGATGGCTCAACAACT | |
| SEsarAFor | TCAGCTTTGAAGAATTTGCAG | |
| SEsarARev | TCTTTCATCGTGTTCATTACGTTT | |
| sarA | SEsarA1 | TCCCTTCAAAACCAAACGAA |
| SEsarA2 | AATTCAGGACATGCACCACA | |
| SEsarA3 | AAAAGATGGGTTTTAAGATTTATGGA | |
| SEsarA4 | CTGTCAGCATAAGTGACCATAGC | |
| icaA | KCA1 | AACAAGTTGAAGGCATCTCC |
| KCA2 | GATGCTTGTTTGATTCCCT | |
| icaR | KCR1 | GGTAAAGTCCGTCAATGGAA |
| KCR2 | CGCAATAACCTTATTTTCCG | |
| gyrB | GYR1 | TTATGGTGCTGGACAGATACA |
| GYR2 | CACCGTGAAGACCGCCAGATA |
The primers ICAR1 and ICAC1 were used to amplify a 4,204-bp fragment comprising the entire ica operon under the following conditions: 35 cycles of 94°C for 30 s, 47°C for 30 s, and 72°C for 5 min. The primers PV1 and PV2 were used to amplify an 871-bp product comprising the icaR gene and the ica operon promoter region under the following conditions: 35 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 1 min.
Amplification of a 3,910-bp fragment encompassing the rsbU, rsbV, rsbW, and sigB genes was achieved by using the primers SEsigFor and SEsigRev under the following conditions: 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 5 min. The primers rsbUFor and rsbU2 were used to amplify a 240-bp product comprising an internal portion at the 5′ end of the rsbU gene under the following conditions: 35 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 1 min. The primers rsbUVFor and rsbUVRev were used to amplify a 1,552-bp product comprising the rsbU and rsbV genes under the following conditions: 35 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 1 min.
The sarA locus was amplified on a 1,498-bp fragment by using the primers SEsarA1 and SEsarA2 under the following conditions: 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 2 min.
S. epidermidis strains were screened for the presence of IS256 by using a PCR assay as described previously (11).
Nucleotide sequencing was performed commercially by MWG Biotech (Germany) with the following primers: rsbUFor, rsbU2, rsbU4, rsbU5, SEsarAFor, SEsarARev, PV1, and PV2.
RNA purification and RT-PCR.
RNA purification, reverse transcription-PCR (RT-PCR) and analysis of RT-PCR data was performed as described previously (10, 11), with the following oligonucleotide primer pairs: GYR1 and GYR2 (gyrB), KCA1 and KCA2 (icaA), KCR1 and KCR2 (icaR), SEasp23For and SEasp23Rev (asp23), SEcsb9For and SEcsb9Rev (csb9), SErsbVFor and SersbVRev (rsbV), SEsarAFor and SEsarARev or SEsarA3 and SEsarA4 (sarA), and SERNAIIIFor and SERNAIIIRev (RNAIII). RT was performed at 55°C for 30 min, followed by 14 to 24 amplification cycles of 94°C 30 s, 50°C 30 s, and 72°C 30 sec.
Hemagglutination assays.
PIA/PNAG expressed by S. epidermidis strains has been shown to be responsible for their ability to agglutinate red blood cells (18, 31, 39). Thus, the hemagglutination assay can be used as an indirect assay for PIA/PNAG production. Briefly, S. epidermidis RP62A cultures were grown to early stationary phase (optical density at 600 nm [OD600] = 6.0) in BHI medium. A 1% sheep red blood (SRB) cell suspension was made by reconstituting lyophilized SRB cells (Sigma, St. Louis, Mo.) in PBS supplemented with 1% bovine serum albumin. The bacterial cultures were washed once in PBS and resuspended in PBS supplemented with an additional 2% NaCl. Then, 50 μl of the cell suspension was added to each well in the top row of a round-bottom 96-well plate, and subsequent twofold dilutions were made in PBS supplemented with an additional 2% NaCl. Subsequently, a 50-μl aliquot of 1% SRB was added to each well, and the plate was incubated without mixing at room temperature for 2 h before visual examination. A positive result was defined as the production of diffuse red blood cells with no red blood cells pelleting at the bottom of the well.
RESULTS
Contribution of IS256 to biofilm phenotypic switching.
A role for IS256 in phenotypic variation of biofilm formation in S. epidermidis was originally described by Ziebuhr et al. (58). Insertional inactivation of the ica operon by this mobile genetic element via a unique and reversible mechanism results in the production of between 25 and 33% of phenotypic variants in the reference strain RP62A (59). More recently, we conducted an analysis of multiple S. epidermidis isolates which suggested that mutation and downregulation of ica operon expression are the primary mechanisms governing biofilm phenotypic variation (25).
To further investigate biofilm phenotypic variation in S. epidermidis strains, we measured the rate of phenotypic switching in six strains harboring IS256 (RP62A, CSF24047, SE56, 13652, 8621, and 17174) and six strains lacking IS256 (CSF41498, SE5, 14765, 1457, BC78032, and BC70837) (Table 1). To facilitate this analysis, we adapted the method of Eisenstein (17) as described previously (48) to measure the rate at which variant cells were produced during the growth of a colony from a biofilm-forming CFU (Materials and Methods). The data are expressed as the rate of switching per CFU per generation. To assess the validity of this methodology, we confirmed that rate of phenotypic switching for S. epidermidis RP62A was ca. 5.5 × 10−5 per cell per generation, which was in good agreement with previous measurements for this strain (59).
Similar to RP62A, the biofilm-positive to biofilm-negative switching frequencies in the five clinical isolates that harbor the IS256 element (CSF24047, 13652, 8621, 17174, and SE56) were between 3 × 10−5 and 5.5 × 10−5 per CFU per generation (Table 1). In contrast, the switching frequencies in the six IS256-negative isolates were significantly reduced compared to the IS256-positive strains (P = 1.8 × 10−5) (Table 1). This low switching frequency in the IS256-negative strains also contrasted with the switching frequencies reported by others in both S. epidermidis (58) and S. aureus (1), all of which were ca. 10−5 per cell per generation. These data suggest that the presence of IS256 is generally associated with more rapid phenotypic variation of biofilm-forming capacity in S. epidermidis.
Regulation of ica operon expression and biofilm formation in a phenotypic variant of S. epidermidis RP62A.
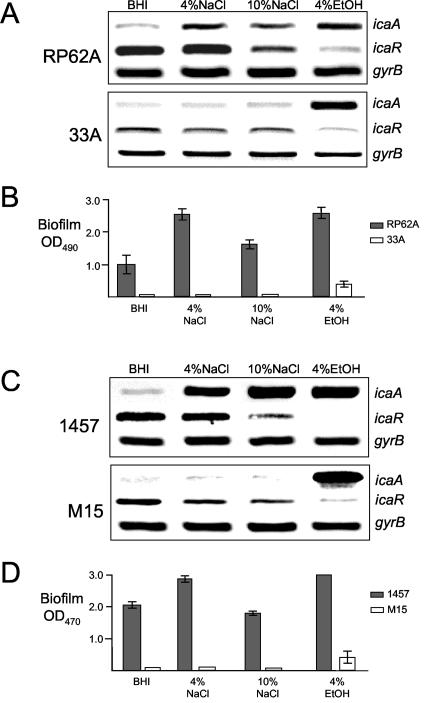
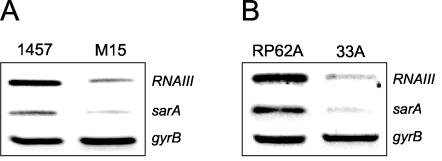
To investigate the possibility that IS256 insertions outside the ica operon may play a role in the more rapid production of biofilm-negative variants with reduced ica operon transcription, we characterized the environmental regulation of ica operon expression in biofilm-negative variants of RP62A. RT-PCR was used to measure icaA and icaR transcription in variants grown in BHI broth or BHI medium supplemented with 4% NaCl, 10% NaCl, or 4% ethanol. This analysis revealed that growth of one variant, designated 33A, in the presence of 4% ethanol but not in the presence of 4% or 10% NaCl resulted in ica operon activation (Fig. 1A). Consistent with this, at the phenotypic level, the capacity of 33A to form biofilm was partially restored (albeit not to wild-type levels) in the presence of ethanol but not NaCl (Fig. 1B). Interestingly, Knobloch et al. (32) also reported that a transposon mutation in the rsbU gene of S. epidermidis 1457 resulted in a biofilm-negative phenotype, which could be reversed by growth in ethanol but not NaCl. rsbU is the first gene of the sigB operon and encodes a phosphatase, which positively regulates σB (32). Using RT-PCR we demonstrated that, a finding consistent with the biofilm phenotypes, growth of the 1457 rsbU transposon mutant M15 only in ethanol and not NaCl was associated with activation of ica operon expression (Fig. 1C and D). In addition, because PIA/PNAG expressed by S. epidermidis strains mediates erythrocyte agglutination (18, 31, 39), the ability of 33A to agglutinate SRB cells was tested. Consistent with the reduced levels of ica operon transcription, hemagglutination assays revealed a significant reduction in PIA/PNAG levels in 33A compared to RP62A (data not shown). These data suggested that altered σB activity in the RP62A variant 33A may be responsible for diminished ica operon expression and the biofilm-negative phenotype.
FIG. 1.
Characterization of S. epidermidis RP62A, 33A, 1457, and M15 genotypes and phenotypes. (A) Comparative measurement of icaA, icaR, and gyrB (control) transcription in RP62A and 33A. RT-PCR analysis was performed on RNA prepared from cultures grown at 37°C to an OD600 of 4.0 in BHI medium or BHI medium supplemented with 4% NaCl, 10% NaCl, or 4% ethanol. (B) Biofilm formation in tissue culture treated 96-well plates by RP62A and 33A in BHI medium or BHI medium supplemented with 4% NaCl, 10% NaCl, or 4% ethanol. (C) Comparative measurement of icaA, icaR, and gyrB (control) transcription in 1457 and M15. RT-PCR analysis was performed on RNA prepared from cultures grown at 37°C to an OD600 of 4.0 in BHI medium or BHI medium supplemented with 4% NaCl, 10% NaCl, or 4% ethanol. (D) Biofilm formation in tissue culture treated 96-well plates by RP62A and 33A in BHI medium or BHI medium supplemented with 4% NaCl, 10% NaCl, or 4% ethanol. Biofilm values represent OD490 readings after staining with crystal violet and are the means of at least three independent assays. Standard deviations are indicated where applicable. EtOH, ethanol.
Analysis of σB activity in phenotypic variants.
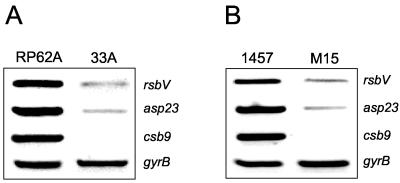
In order to assess the levels of σB activity in the RP62A variant 33A, we used RT-PCR to measure σB-dependent gene expression. Three σB-regulated genes were chosen for this analysis: asp23 (21, 34, 47, 51), csb9 (22), and rsbV (24, 32, 49).
This analysis revealed that transcription of the asp23, csb9, and rsbV genes in the RP62A variant 33A were substantially reduced compared to the wild-type parent (Fig. 2A). In addition, a similarly dramatic decrease in asp23, csb9, and rsbV expression was observed in the S. epidermidis rsbU transposon mutant M15 compared to its wild-type parent 1457 (Fig. 2B). These findings strongly suggest that σB activity is impaired in variant 33A and that this change in the activity of a global regulator is responsible for diminished ica operon expression and a biofilm-negative phenotype.
FIG. 2.
Comparative measurement of rsbV, asp23, csb9, and gyrB (control) transcription in RP62A and 33A (A) and 1457 and M15 (B). RT-PCR analysis was performed on RNA prepared from cultures grown at 37°C to an OD600 of 4.0 in BHI medium.
PCR amplification and nucleotide sequence analysis of the sigB locus from biofilm-negative variants.
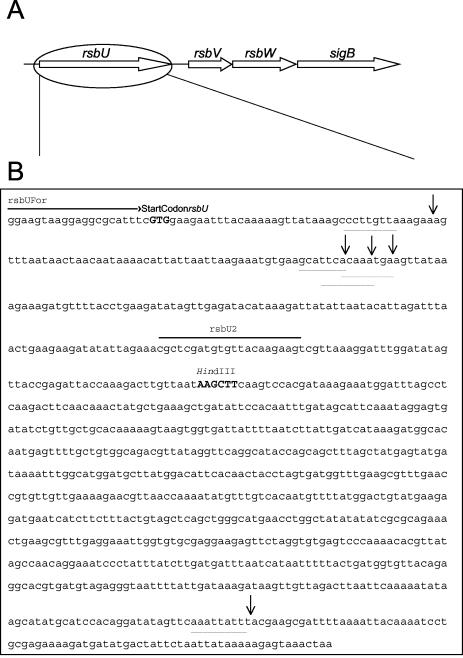
Analysis of ica operon environmental regulation and σB-dependent gene expression in the RP62A variant 33A suggested that impaired σB activity may be responsible for the biofilm-negative phenotype. We therefore decided to characterize the sigB operon of variant 33A. The sigB operon of S. epidermidis has previously been characterized and comprises the four genes rsbU, rsbV, rsbW, and sigB (32). Long-range PCR amplification of the sigB operon generated products of ca. 5,200-bp for variant 33A compared to the expected size of 3,910 bp for RP62A (data not shown). This observation suggested that a genetic rearrangement may have occurred at the sigB locus of variant 33A and was particularly interesting given that IS256 insertions in the ica operon can also generate biofilm-negative variants (59). Restriction enzyme and nucleotide sequence analysis subsequently revealed the presence of an IS256 insertion at the 5′ end of the rsbU gene in variant 33A and thus identified the genetic basis for the impaired σB activity in this strain (Fig. 3). Consistent with the findings of Ziebuhr et al. (59), the IS256 element, which was oriented in the opposite transcriptional orientation to the rsbU gene, was flanked by an 8-bp duplicated target sequence (Fig. 3).
FIG. 3.
Location of IS256 insertions in the rsbU gene. (A) Diagrammatic representation of the rsbU, rsbV, rsbW, sigB operon structure in S. epidermidis. (B) Exact locations of IS256 insertions detected in the rsbU gene of RP62A variants. The nucleotide binding sites for the PCR primers rsbUFor and rsbU2, the rsbU start codon, and the location of a unique HindIII site are all indicated. The duplicated 8-bp IS256 target sequences—TTAAAGAA for Red 1; AGCATTCA for 33A; CAAATGAA for Red 2, Red 3, and Red 4; and AATTATTT for Red 5—are underlined, and arrows indicate the exact insertion sites.
Frequency of rsbU::IS256 insertion variant production.
To determine the frequency at which rsbU::IS256 insertions occurred, we performed three independent experiments to isolate 43 biofilm-negative variants from RP62A. PCR analysis of all 43 variants revealed that 11 (25.5%) contained ica::IS256 insertions, whereas 5 (11.6%) contained IS256 insertions in the rsbU gene of the sigB locus. Nucleotide sequence analysis confirmed the presence of IS256 insertions in the rsbU gene and identified two new IS256 insertion sites at the 5′ end of rsbU in four of these variants, which were designated Red 1, Red 2, Red 3, and Red 4 (Fig. 3). The fifth variant, Red 5, contained an IS256 insertion at the 3′ end of the rsbU gene (Fig. 3). In contrast to the IS256 insertion in variant 33A, IS256 was oriented in the same transcriptional orientation to the rsbU gene in all five of these variants. Similar to 33A, we identified 8-bp duplicated IS256 target sequences in all five of these rsbU::IS256 insertion variants; however, no consensus target sequence could be identified. These data suggest that the rsbU gene, and in particular the 5′ end of this gene, may represent a chromosomal hot spot for IS256 insertions.
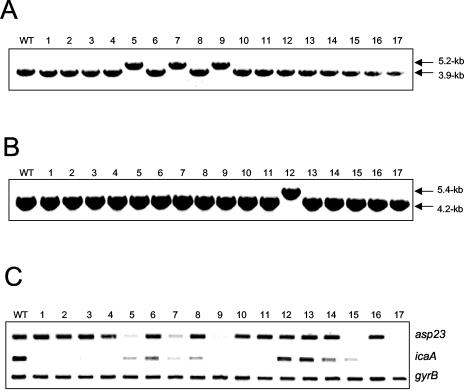
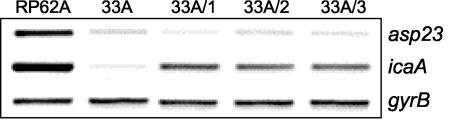
Seventeen phenotypic variants isolated from one experiment were characterized further by using PCR to amplify the ica and sigB operons and RT-PCR to measure ica operon and asp23 expression (Fig. 4). In this particular experiment, 3 of the 17 variants examined were found to contain IS256 insertions in the rsbU gene of the sigB operon (Fig. 4A). Only one variant contained an ica::IS256 insertion (Fig. 4B), indicating that considerable variation in the rates of IS256 insertions at specific loci exists between experiments. Interestingly, RT-PCR analysis revealed that ica operon expression was substantially reduced in 14 of the 17 variants, including the three variants with rsbU::IS256 insertions (Fig. 4C). Importantly, five variants including the three rsbU::IS256 insertion variants were found to have reduced levels of asp23 expression (Fig. 4C). The molecular basis for reduced asp23 expression in the two variants that do not contain IS256 insertions in the sigB operon is unknown.
FIG. 4.
Analysis of 17 phenotypic variants produced by S. epidermidis RP62A. (A and B) Long-range PCR analysis of the sigB operon (A) and ica operon (B) in the wild-type RP62A and in 17 phenotypic variants. (C) Comparative measurement of asp23, icaA, and gyrB (control) transcription in wild-type RP62A and the 17 variants. RT-PCR analysis was performed on RNA prepared from cultures grown at 37°C to an OD600 of 4.0 in BHI medium.
Selection and analysis of biofilm-positive revertant strains from an rsbU::IS256 insertion variant.
Ziebuhr et al. (59) reported previously that biofilm-negative icaC::IS256 insertion variants were capable of reverting, after repeated passage, to a biofilm-positive phenotype after the complete excision of the insertion sequence element. In order to determine whether rsbU::IS256 insertion variants could also revert to a biofilm-positive phenotype, we serially passaged variant 33A in a tissue culture flask for up to 15 days. Seven independent flasks were used and revertants, which were isolated from a weak biofilm of adhering bacteria formed after 5 to 9 days, were identified as displaying a stable, black colony morphology on CRA. Interestingly, PCR revealed that in all revertants tested, the rsbU::IS256 insertion remained intact, suggesting that a secondary or compensatory mutation was responsible for the reversion to a biofilm-positive phenotype. To further investigate this possibility, we examined the ica operon and asp23 gene expression in three revertants, 33A/1 to 33A/3. Consistent with the presence of an rsbU::IS256 allele, asp23 expression levels were substantially lower in 33A, and all three revertants compared to wild-type RP62A (Fig. 5). In contrast, ica operon expression levels were predictably higher in all revertants compared to 33A but were not comparable to the levels of ica expression in RP62A (Fig. 5). Semiquantitative biofilm assays revealed that the revertant strains were only weakly biofilm positive when grown in BHI broth but were strongly biofilm positive when grown in NaCl or ethanol (data not shown). Thus, the biofilm phenotype of these revertants was different from both 33A and wild-type RP62A. We have previously reported that the icaR gene encodes a negative regulator involved in the environmental regulation of ica operon expression (10, 11). In order to determine whether mutations in the ica operon regulatory regions were responsible for the increased levels of ica operon expression in the revertant strains, we determined the nucleotide sequence of the icaR gene and ica operon promoter region in all seven revertants. However, no sequence differences were identified in the icaR gene and ica operon promoter region of these revertants (data not shown).
FIG. 5.
Comparative measurement of asp23, icaA, and gyrB (control) transcription in wild-type RP62A, biofilm-negative variant 33A and three revertants from 33A. RT-PCR analysis was performed on RNA prepared from cultures grown at 37°C to an OD600 of 4.0 in BHI medium.
Impact of rsbU::IS256 insertions on the expression of sarA and agr.
The absence of a recognizable σB promoter upstream of the ica operon (32, 51) may suggest an indirect role for σB in the regulation of ica operon transcription. In addition, Valle et al. (54) and Beenken et al. (2) recently demonstrated an essential role for the staphylococcal accessory regulator SarA in S. aureus biofilm development. Valle et al. (54) also demonstrated that inactivation of sigB in S. aureus was only associated with slightly decreased ica operon expression. Because σB-dependent promoters have been identified upstream of the sarA gene in both S. epidermidis (19) and S. aureus (5, 15, 42), we decided to investigate the expression of sarA in the rsbU::IS256 insertion variant 33A. This analysis revealed that sarA transcription was reduced in variant 33A compared to wild-type RP62A and that a similar pattern of regulation was evident in the rsbU transposon mutant M15 compared to its parental strain 1457 (Fig. 6). Thus, these data suggest that decreased levels of sarA transcription in phenotypic variants harboring rsbU::IS256 insertions may also contribute to diminished levels of ica operon expression.
FIG. 6.
Comparative measurement of RNAIII, sarA, and gyrB (control) transcription in 1457 and M15 (A) and RP62A and 33A (B). RT-PCR analysis was performed on RNA prepared from cultures grown at 37°C to an OD600 of 4.0 in BHI medium.
A number of studies have revealed that SarA is required for maximal expression of the two staphylococcal accessory gene regulatory promoters agr P2 and agr P3 (3, 4, 6-8, 28, 35). Transcription of agrP3-driven RNAIII was also substantially reduced in the rsbU transposon mutant M15 and the rsbU::IS256 insertion variant 33A compared to 1457 and RP62A, respectively (Fig. 6), which was consistent with the decreased levels of sarA expression in these strains. Thus, taken together, these data suggest that IS256 insertions at rsbU directly affect σB activity and accordingly sarA expression, which in turn is associated with reduced levels of RNAIII transcription.
Isolation and characterization of a sarA::IS256 insertion variant.
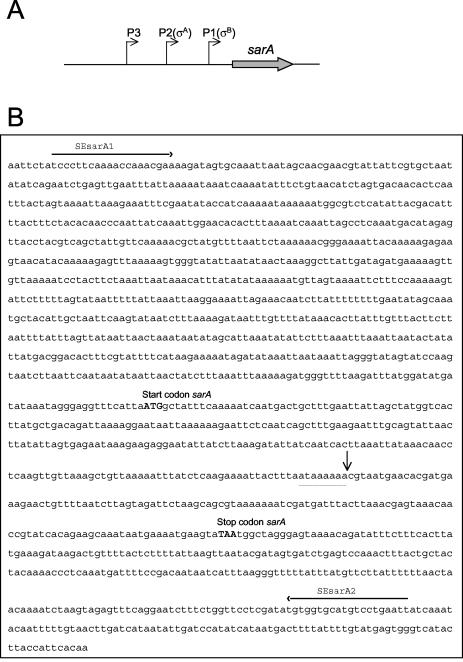
To investigate the possibility that IS256 insertions at other sites may also play an important role in the production of biofilm-negative variants with diminished ica operon expression, we used PCR to amplify the sarA locus in all 43 RP62A variants that we had previously characterized (see above). One variant, designated Red S1, was identified in which the amplified sarA fragment was ca. 2,800 bp compared to the expected 1,498-bp fragment produced by the wild-type RP62A. Restriction enzyme and nucleotide sequence analysis subsequently confirmed the presence of an IS256 insertion within the sarA gene in variant Red S1 (Fig. 7). The IS256 element was oriented in the same transcriptional orientation to the sarA gene and, as observed in the rsbU::IS256 insertion variants (Fig. 3), was flanked by an 8-bp duplicated target sequence (Fig. 7). Interestingly, the RP62A sarA::IS256 insertion mutant showed a bright red, smooth colony morphology phenotype compared to the darker color of the rsbU::IS256 variants on CRA.
FIG. 7.
Location of IS256 insertion in the sarA gene. (A) Diagrammatic representation of the sarA open reading frame and triple promoter regulatory region in S. epidermidis. The P1 promoter has homology to σB-dependent promoters, whereas P2 appears to be σA dependent (19). (B) Exact location of the IS256 insertion detected in the sarA gene of RP62A variant Red S1. The nucleotide binding sites for the PCR primers SEsarA1 and SEsarA2 and the sarA start and stop codons are indicated. The duplicated 8-bp IS256 target sequence, ATAAAAAA, for Red S1 is underlined, and an arrow indicates the exact insertion site.
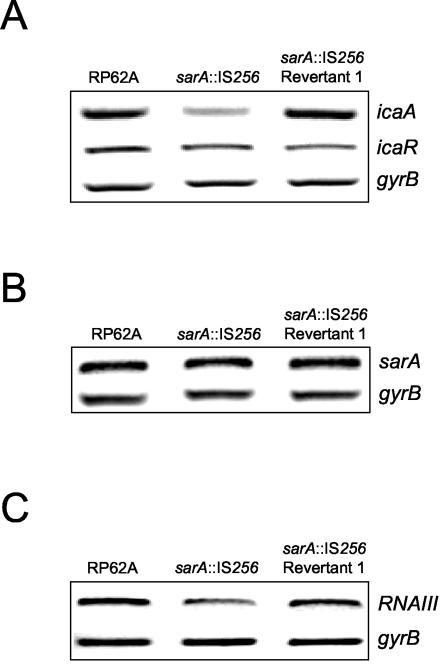
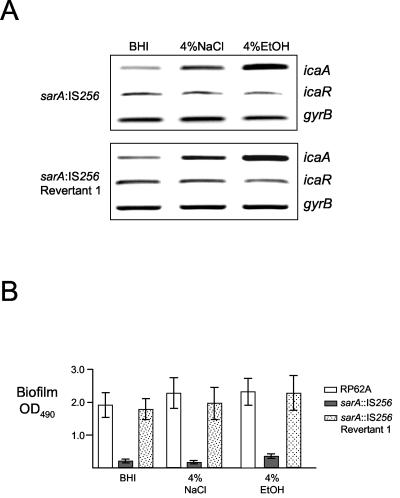
RT-PCR analysis revealed a reduction in ica operon transcription in the sarA::IS256 mutant compared to wild-type RP62A, particularly during early exponential growth (Fig. 8A). Consistent with the reduced levels of ica operon transcription, hemagglutination assays also revealed a significant reduction in PIA/PNAG levels in the sarA::IS256 mutant compared to RP62A (data not shown). These findings suggested that SarA is a positive regulator of ica operon expression in S. epidermidis and was consistent with the negative impact of a sarA mutation on ica operon expression in S. aureus (54). In contrast, decreased ica operon expression in the sarA mutant was not associated with altered regulation of icaR (Fig. 8A), suggesting that icaR expression is SarA-independent. Similarly, the IS256 insertion in sarA did not appear to result in altered expression of sarA itself, suggesting that SarA does not regulate its own transcription (Fig. 8B). Based on our earlier findings, which revealed that reduced sarA expression in rsbU::IS256 insertion variants was accompanied by decreased RNAIII expression, we also examined RNAIII transcription in the sarA::IS256 mutant. Surprisingly, this analysis only revealed no significant difference in RNAIII expression in the sarA mutant compared to wild-type RP62A during early exponential growth (data not shown) and only a small reduction in RNAIII expression during early stationary growth (Fig. 8C), suggesting that sarA may play a more important role in the regulation of agr expression in S. aureus than in S. epidermidis.
FIG. 8.
Analysis of ica operon, icaR, sarA, and RNAIII expression in a sarA::IS256 insertion mutant. (A) Comparative measurement of icaA, icaR, and gyrB (control) transcription in RP62A, Red S1 (sarA::IS256), and Red S1/R1 (revertant 1). RT-PCR analysis was performed on RNA prepared from cultures grown at 37°C to an OD600 of 2.0 in BHI medium. (B) Comparative measurement of sarA and gyrB (control) transcription in RP62A and Red S1 (sarA::IS256) and Red S1/R1 (revertant 1). RT-PCR analysis was performed on RNA prepared from cultures grown to an OD600 of 2.0 at 37°C in BHI medium. Primers SEsarA3 and SEsarA4, located between the sarA P1 promoter and the IS256 insertion site, were used to measure sarA transcription in the sarA mutant. (C) Comparative measurement of RNAIII and gyrB (control) transcription in RP62A, Red S1 (sarA::IS256), and Red S1/R1 (revertant 1). RT-PCR analysis was performed on RNA prepared from cultures grown to an OD600 of 8.0 at 37°C in BHI medium.
Interestingly, unlike the rsbU::IS256 mutants, a wild-type pattern of ica operon environmental regulation was observed in the sarA::IS256 variant grown in NaCl or ethanol (Fig. 9A), suggesting that SarA may not be required for ica operon environmental regulation. However, biofilm assays revealed that ethanol- and NaCl-induced ica operon transcription was not accompanied by wild-type levels of biofilm-forming capacity in the sarA mutant (Fig. 9B), suggesting that sarA may also be required for normal PIA/PNAG synthesis or an ica-independent mechanism of biofilm formation.
FIG. 9.
Characterization of biofilm and ica operon environmental regulation in S. epidermidis RP62A, Red S1 (sarA::IS256) and Red S1/R1 (revertant 1). (A) Comparative measurement of icaA, icaR, and gyrB (control) transcription in Red S1 (sarA::IS256) and Red S1/R1 (revertant 1). RT-PCR analysis was performed on RNA prepared from cultures grown at 37°C to an OD600 of 4.0 in BHI medium or in BHI medium supplemented with 4% NaCl or 4% ethanol. (B) Biofilm formation in tissue culture-treated 96-well plates by RP62A, Red S1 (sarA::IS256), and Red S1/R1 (revertant 1) in BHI medium or BHI medium supplemented with 4% NaCl or 4% ethanol. EtOH, ethanol.
Interestingly, PCR revealed a wild-type sarA allele (1,498-bp amplification product) in three biofilm-positive revertants isolated from the sarA::IS256 mutant (data not shown). Nucleotide sequencing of the sarA locus from one revertant, Red S1/R1, revealed that IS256 had been precisely excised, leaving behind a fully intact sarA gene (data not shown). RT-PCR analysis revealed that ica operon transcription was restored to RP62A levels in revertant Red S1/R1 (Fig. 8A). Moreover, as observed in wild-type RP62A and unlike the sarA::IS256 insertion variant, growth of the revertant Red S1/R1 in NaCl or ethanol was associated with both ica operon activation and increased biofilm formation (Fig. 8).
DISCUSSION
Staphylococcal device-related infections involving biofilms are not always localized to the site of the implanted biomaterial but are sometimes characterized by dissemination of the causative organisms and sepsis. In this context, the contribution of phase variation to the regulation of ica operon expression and PIA production might represent an important pathogenic mechanism facilitating the release of planktonic cells from the biofilm. In the present study we investigated the molecular basis for the production of biofilm-negative phenotypic variants with diminished ica operon transcription. Our data reveal that inactivation of the rsbU or sarA genes by the insertion sequence element IS256 results in diminished levels of ica operon transcription and is responsible for the production of ca. 11% of biofilm-negative variants in RP62A. Phenotypic variants harboring rsbU::IS256 insertions also have impaired σB activity, as well as reduced levels of ica, sarA, and agr expression. Importantly, IS256 insertion mutations in the rsbU and sarA genes were also identified in biofilm-negative variants of the clinical isolate CSF24047 (data not shown), suggesting that IS256 insertion mutants may be produced by all IS256-positive strains of S. epidermidis. These findings are consistent with a recent report which revealed that IS256 may play a more significant role in staphylococcal virulence than other IS elements. These study demonstrated that not only was IS256 present in multiple copies on the genomes of disease-associated S. epidermidis strains but also that IS256 was typically associated with biofilm-forming capacity, the presence of the ica operon, and antibiotic resistance (33).
A direct role for IS256 in the ON-to-OFF switching of biofilm-forming capacity was initially characterized by Ziebuhr et al. (59), who demonstrated that reversible transposition of IS256 into hotspots within the ica operon was responsible for the production of 25 to 33% of variants. Our finding that ca. 11% of variants harbor rsbU::IS256 insertions and that IS256 can also integrate into sarA, albeit at a lower frequency, reveals that this IS element is responsible for the production of up to 50% of biofilm-negative variants in S. epidermidis RP62A. The mechanism(s) responsible for the production of the remaining biofilm-negative variants remains unknown. However, consistent with our previous findings (25), we were able to demonstrate that up to 80% of RP62A variants have diminished levels of ica operon expression, indicating that this is the genetic basis for the biofilm-negative phenotype in the majority of variants produced by this strain.
In addition to the direct impact on σB activity, we also obtained evidence that the rsbU::IS256 insertion mutation indirectly affected the expression of two global regulators: sarA, which is σB dependent, and agr (RNAIII), which is SarA dependent. These data highlight the potential of IS256, through insertions at both rsbU and sarA, to alter the global regulation of transcription and reveal that the switch to biofilm-negative is likely to be one of multiple phenotypic changes in rsbU and sarA IS256-generated mutants. Interestingly, the absence of an identifiable σB-consensus binding site upstream of the ica operon (32, 51) and the observations that deletion of sigB does not affect biofilm development by S. aureus (2, 54) have led to the suggestion that σB may not directly regulate ica operon expression. Moreover, data which revealed that sarA is required for S. aureus ica operon expression and biofilm development (54) suggested that in S. epidermidis mutation of sarA or reductions in the levels of sarA transcription may also contribute to repression of ica operon expression (2). Consistent with this, analysis of the sarA::IS256 insertion mutant revealed that SarA is directly or indirectly involved in the transcriptional regulation of ica operon expression. Comparative analysis of RP62A, 33A (rsbU::IS256), and Red S1 (sarA::IS256) revealed that ica operon transcript levels were similar in the rsbU and sarA mutants (data not shown) and may further suggest that the impact of a σB mutation on ica operon expression is at least in part due to decreased expression of sarA.
A distinctive phenotype associated with S. epidermidis rsbU mutants is the ability of ethanol but not NaCl to activate ica operon transcription. In contrast, both medium supplements activated ica operon expression in the sarA::IS256 mutant. These findings may suggest that σB and SarA control ica operon expression through separate regulatory pathways. We have previously proposed that ica operon activation by ethanol is solely icaR dependent, whereas NaCl can activate ica expression via alterations in σB and icaR activity (10, 11). The data presented in the present study suggest that NaCl-induced ica operon activation is sarA independent. Interestingly, analysis of sarA and σB mutants in S. aureus revealed that PIA/PNAG levels did not reflect levels of ica transcription and were higher in a σB/sarA double mutant than in a sarA single mutant, suggesting that SarA and σB may compete to enhance or repress, respectively, the activity of an unknown regulatory factor involved in the synthesis or turnover of PIA/PNAG (54). Given that activation of ica operon expression by NaCl in the S. epidermidis sarA::IS256 variant does not result in any increase in biofilm development and that ethanol-induced ica expression in both the sarA and rsbU mutants was not accompanied by wild-type levels of biofilm formation, it is possible that both σB and SarA are also involved in the posttranscriptional regulation of PIA/PNAG synthesis in S. epidermidis.
In S. aureus, sarA plays an important role in the regulation of agr transcription (4). However, our findings only revealed a small reduction in RNAIII expression in the S. epidermidis sarA::IS256 mutant. Interestingly, although expression of the S. aureus and S. epidermidis sarA genes are both driven by three separate promoters, one of which is σB dependent, distinct differences possibly reflecting functional divergence between these two organisms also exist (19). For example, the sarA promoters are clustered much closer together in S. epidermidis than in S. aureus and as a result two small potential open reading frames (ORF3 and ORF4) that are present between the S. aureus sarA promoters are absent in S. epidermidis (19). Because ORF3, together with SarA protein, may play a role in regulating agr expression in S. aureus (4), it is tempting to speculate that structural differences in the sarA regulatory sequences may be reflected in the different effects of sarA mutations on RNAIII transcription in S. epidermidis and S. aureus.
A recent study by Vuong et al. (55) revealed that mutation of the agr locus in S. epidermidis was actually associated with increased biofilm-forming capacity and expression of the autolysin AtlE, which is involved in primary attachment (26), although the levels of PIA/PNAG product were not substantially affected. Similarly the recent studies of Valle et al. (54) and Beenken et al. (2) also demonstrated that mutation of agr did not influence biofilm forming capacity in S. aureus. Thus, it seems unlikely that reduced expression of RNAIII in the sarA and rsbU IS256 insertion variants is an important determinant in their biofilm-negative phenotypes.
In summary, these findings provide new insights into biofilm phenotypic variation and identify two molecular mechanisms of phenotypic switching involving insertion of a transposable element into two global regulatory genes, rsbU and sarA. These genetic switches lead not only to decreased ica operon transcription and impaired biofilm-forming capacity but also reduced expression of σB, sarA, and agr, which may in turn modulate the global regulation of transcription in this opportunistic bacterial pathogen. In addition, given that IS256 is highly active in multiresistant staphylococci and enterococci (36), our findings further highlight the potential of transposable elements to influence the genetic flexibility and perhaps virulence of many other important gram-positive pathogens.
Acknowledgments
This study was funded by grants from the Research Committee of the Royal College of Surgeons in Ireland and the Irish Research Council for Science, Engineering, and Technology to J. P. O'Gara.
We are grateful to Pfizer (Ireland) for generously supporting the establishment of the RCSI Microbiology Laboratory at the RCSI Education and Research Centre. S. epidermidis 1457 and M15 were kindly provided by Johannes Knobloch and Dietrich Mack. Luke D. Handke and Paul D. Fey, University of Nebraska Medical Center, and Vance Fowler, Duke University, generously provided clinical isolates. We thank Ciara Kennedy, Sinead O'Donnell, Fidelma Fitzpatrick, and Tracey Dillane for experimental advice and assistance throughout the study and Charles J. Dorman for critical reading of the manuscript.
REFERENCES
- 1.Baselga, R., I. Albizu, M. De La Cruz, E. Del Cacho, M. Barberan, and B. Amorena. 1993. Phase variation of slime production in Staphylococcus aureus: implications in colonization and virulence. Infect. Immun. 61:4857-4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beenken, K. E., J. S. Blevins, and M. S. Smeltzer. 2003. Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect. Immun. 71:4206-4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blevins, J. S., K. E. Beenken, M. O. Elasri, B. K. Hurlburt, and M. S. Smeltzer. 2002. Strain-dependent differences in the regulatory roles of sarA and agr in Staphylococcus aureus. Infect. Immun. 70:470-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheung, A. L., M. G. Bayer, and J. H. Heinrichs. 1997. sar genetic determinants necessary for transcription of RNAII and RNAIII in the agr locus of Staphylococcus aureus. J. Bacteriol. 179:3963-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheung, A. L., Y. T. Chien, and A. S. Bayer. 1999. Hyperproduction of alpha-hemolysin in a sigB mutant is associated with elevated SarA expression in Staphylococcus aureus. Infect. Immun. 67:1331-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung, A. L., and S. J. Projan. 1994. Cloning and sequencing of sarA of Staphylococcus aureus, a gene required for the expression of agr. J. Bacteriol. 176:4168-4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chien, Y., and A. L. Cheung. 1998. Molecular interactions between two global regulators, sar and agr, in Staphylococcus aureus. J. Biol. Chem. 273:2645-2652. [DOI] [PubMed] [Google Scholar]
- 8.Chien, Y., A. C. Manna, and A. L. Cheung. 1998. SarA level is a determinant of agr activation in Staphylococcus aureus. Mol. Microbiol. 30:991-1001. [DOI] [PubMed] [Google Scholar]
- 9.Christensen, G. D., L. M. Baddour, B. M. Madison, J. T. Parisi, S. N. Abraham, D. L. Hasty, J. H. Lowrance, J. A. Josephs, and W. A. Simpson. 1990. Colonial morphology of staphylococci on Memphis agar: phase variation of slime production, resistance to beta-lactam antibiotics, and virulence. J. Infect. Dis. 161:1153-1169. [DOI] [PubMed] [Google Scholar]
- 10.Conlon, K. M., H. Humphreys, and J. P. O'Gara. 2002. Regulation of icaR gene expression in Staphylococcus epidermidis. FEMS Microbiol. Lett. 216:173-179. [DOI] [PubMed] [Google Scholar]
- 11.Conlon, K. M., H. Humphreys, and J. P. O'Gara. 2002. icaR encodes a transcriptional repressor involved in environmental regulation of ica operon expression and biofilm formation in Staphylococcus epidermidis. J. Bacteriol. 184:4400-4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cramton, S. E., C. Gerke, N. F. Schnell, W. W. Nichols, and F. Gotz. 1999. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 67:5427-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cramton, S. E., M. Ulrich, F. Gotz, and G. Doring. 2001. Anaerobic conditions induce expression of polysaccharide intercellular adhesin in Staphylococcus aureus and Staphylococcus epidermidis. Infect. Immun. 69:4079-4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deighton, M., S. Pearson, J. Capstick, D. Spelman, and R. Borland. 1992. Phenotypic variation of Staphylococcus epidermidis isolated from a patient with native valve endocarditis. J. Clin. Microbiol. 30:2385-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deora, R., T. Tseng, and T. K. Misra. 1997. Alternative transcription factor σSB of Staphylococcus aureus: characterization and role in transcription of the global regulatory locus sar. J. Bacteriol. 179:6355-6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunman, P. M., E. Murphy, S. Haney, D. Palacios, G. Tucker-Kellogg, S. Wu, E. L. Brown, R. J. Zagursky, D. Shlaes, and S. J. Projan. 2001. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J. Bacteriol. 183:7341-7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eisenstein, B. I. 1981. Phase variation of type 1 fimbriae in Escherichia coli is under transcriptional control. Science 214:337-339. [DOI] [PubMed] [Google Scholar]
- 18.Fey, P. D., J. S. Ulphani, F. Gotz, C. Heilmann, D. Mack, and M. E. Rupp. 1999. Characterization of the relationship between polysaccharide intercellular adhesin and hemagglutination in Staphylococcus epidermidis. J. Infect. Dis. 179:1561-1564. [DOI] [PubMed] [Google Scholar]
- 19.Fluckiger, U., C. Wolz, and A. L. Cheung. 1998. Characterization of a sar homolog of Staphylococcus epidermidis. Infect. Immun. 66:2871-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerke, C., A. Kraft, R. Sussmuth, O. Schweitzer, and F. Gotz. 1998. Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J. Biol. Chem. 273:18586-18593. [DOI] [PubMed] [Google Scholar]
- 21.Gertz, S., S. Engelmann, R. Schmid, K. Ohlsen, J. Hacker, and M. Hecker. 1999. Regulation of σB-dependent transcription of sigB and asp23 in two different Staphylococcus aureus strains. Mol. Gen. Genet. 261:558-566. [DOI] [PubMed] [Google Scholar]
- 22.Gertz, S., S. Engelmann, R. Schmid, A. K. Ziebandt, K. Tischer, C. Scharf, J. Hacker, and M. Hecker. 2000. Characterization of the σB regulon in Staphylococcus aureus. J. Bacteriol. 182:6983-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gotz, F. 2002. Staphylococcus and biofilms. Mol. Microbiol. 43:1367-1378. [DOI] [PubMed] [Google Scholar]
- 24.Haldenwang, W. G. 1995. The sigma factors of Bacillus subtilis. Microbiol. Rev. 59:1-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Handke, L. D., K. M. Conlon, S. R. Slater, S. Elbaruni, F. Fitzpatrick, H. Humphreys, W. P. Giles, M. E. Rupp, P. D. Fey, and J. P. O'Gara. 2004. Genotypic and phenotypic analysis of biofilm phenotypic variation in multiple Staphylococcus epidermidis isolates. J. Med. Microbiol. 53:367-374. [DOI] [PubMed] [Google Scholar]
- 26.Heilmann, C., M. Hussain, G. Peters, and F. Gotz. 1997. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol. Microbiol. 24:1013-1024. [DOI] [PubMed] [Google Scholar]
- 27.Heilmann, C., O. Schweitzer, C. Gerke, N. Vanittanakom, D. Mack, and F. Gotz. 1996. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol. Microbiol. 20:1083-1091. [DOI] [PubMed] [Google Scholar]
- 28.Heinrichs, J. H., M. G. Bayer, and A. L. Cheung. 1996. Characterization of the sar locus and its interaction with agr in Staphylococcus aureus. J. Bacteriol. 178:418-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jefferson, K. K., S. E. Cramton, F. Gotz, and G. B. Pier. 2003. Identification of a 5-nucleotide sequence that controls expression of the ica locus in Staphylococcus aureus and characterization of the DNA-binding properties of IcaR. Mol. Microbiol. 48:889-899. [DOI] [PubMed] [Google Scholar]
- 30.Jefferson, K. K., D. B. Pier, D. A. Goldmann, and G. B. Pier. 2004. The teicoplanin-associated locus regulator (TcaR) and the intercellular adhesin locus regulator (IcaR) are transcriptional inhibitors of the ica locus in Staphylococcus aureus. J. Bacteriol. 186:2449-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Joyce, J. G., C. Abeygunawardana, Q. Xu, J. C. Cook, R. Hepler, C. T. Przysiecki, K. M. Grimm, K. Roper, C. C. Ip, L. Cope, D. Montgomery, M. Chang, S. Campie, M. Brown, T. B. McNeely, J. Zorman, T. Maira-Litran, G. B. Pier, P. M. Keller, K. U. Jansen, and G. E. Mark. 2003. Isolation, structural characterization, and immunological evaluation of a high-molecular-weight exopolysaccharide from Staphylococcus aureus. Carbohydr. Res. 338:903-922. [DOI] [PubMed] [Google Scholar]
- 32.Knobloch, J. K., K. Bartscht, A. Sabottke, H. Rohde, H. H. Feucht, and D. Mack. 2001. Biofilm formation by Staphylococcus epidermidis depends on functional RsbU, an activator of the sigB operon: differential activation mechanisms due to ethanol and salt stress. J. Bacteriol. 183:2624-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kozitskaya, S., S. H. Cho, K. Dietrich, R. Marre, K. Naber, and W. Ziebuhr. 2004. The bacterial insertion sequence element IS256 occurs preferentially in nosocomial Staphylococcus epidermidis isolates: association with biofilm formation and resistance to aminoglycosides. Infect. Immun. 72:1210-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kullik, I., P. Giachino, and T. Fuchs. 1998. Deletion of the alternative sigma factor σB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J. Bacteriol. 180:4814-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kupferwasser, L. I., M. R. Yeaman, C. C. Nast, D. Kupferwasser, Y. Q. Xiong, M. Palma, A. L. Cheung, and A. S. Bayer. 2003. Salicylic acid attenuates virulence in endovascular infections by targeting global regulatory pathways in Staphylococcus aureus. J. Clin. Investig. 112:222-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loessner, I., K. Dietrich, D. Dittrich, J. Hacker, and W. Ziebuhr. 2002. Transposase-dependent formation of circular IS256 derivatives in Staphylococcus epidermidis and Staphylococcus aureus. J. Bacteriol. 184:4709-4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mack, D., W. Fischer, A. Krokotsch, K. Leopold, R. Hartmann, H. Egge, and R. Laufs. 1996. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear β-1,6-linked glucosaminoglycan: purification and structural analysis. J. Bacteriol. 178:175-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mack, D., M. Nedelmann, A. Krokotsch, A. Schwarzkopf, J. Heesemann, and R. Laufs. 1994. Characterization of transposon mutants of biofilm-producing Staphylococcus epidermidis impaired in the accumulative phase of biofilm production: genetic identification of a hexosamine-containing polysaccharide intercellular adhesin. Infect. Immun. 62:3244-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mack, D., J. Riedewald, H. Rohde, T. Magnus, H. H. Feucht, H. A. Elsner, R. Laufs, and M. E. Rupp. 1999. Essential functional role of the polysaccharide intercellular adhesin of Staphylococcus epidermidis in hemagglutination. Infect. Immun. 67:1004-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mack, D., N. Siemssen, and R. Laufs. 1992. Parallel induction by glucose of adherence and a polysaccharide antigen specific for plastic-adherent Staphylococcus epidermidis: evidence for functional relation to intercellular adhesion. Infect. Immun. 60:2048-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maira-Litran, T., A. Kropec, C. Abeygunawardana, J. Joyce, G. Mark III, D. A. Goldmann, and G. B. Pier. 2002. Immunochemical properties of the staphylococcal poly-N-acetylglucosamine surface polysaccharide. Infect. Immun. 70:4433-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Manna, A. C., M. G. Bayer, and A. L. Cheung. 1998. Transcriptional analysis of different promoters in the sar locus in Staphylococcus aureus. J. Bacteriol. 180:3828-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McKenney, D., J. Hubner, E. Muller, Y. Wang, D. A. Goldmann, and G. B. Pier. 1998. The ica locus of Staphylococcus epidermidis encodes production of the capsular polysaccharide/adhesin. Infect. Immun. 66:4711-4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McKenney, D., K. Pouliot, Y. Wang, V. Murthy, M. Ulrich, G. Doring, J. C. Lee, D. A. Goldmann, and G. B. Pier. 2000. Vaccine potential of poly-1-6 β-d-N-succinylglucosamine, an immunoprotective surface polysaccharide of Staphylococcus aureus and Staphylococcus epidermidis. J. Biotechnol. 83:37-44. [DOI] [PubMed] [Google Scholar]
- 45.Mempel, M., H. Feucht, W. Ziebuhr, M. Endres, R. Laufs, and L. Gruter. 1994. Lack of mecA transcription in slime-negative phase variants of methicillin-resistant Staphylococcus epidermidis. Antimicrob. Agents Chemother. 38:1251-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mempel, M., E. Muller, R. Hoffmann, H. Feucht, R. Laufs, and L. Gruter. 1995. Variable degree of slime production is linked to different levels of beta-lactam susceptibility in Staphylococcus epidermidis phase variants. Med. Microbiol. Immunol. 184:109-113. [DOI] [PubMed] [Google Scholar]
- 47.Miyazaki, E., J. M. Chen, C. Ko, and W. R. Bishai. 1999. The Staphylococcus aureus rsbW (orf159) gene encodes an anti-sigma factor of SigB. J. Bacteriol. 181:2846-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O'Gara, J. P., and C. J. Dorman. 2000. Effects of local transcription and H-NS on inversion of the fim switch of Escherichia coli. Mol. Microbiol. 36:457-466. [DOI] [PubMed] [Google Scholar]
- 49.Petersohn, A., J. Bernhardt, U. Gerth, D. Hoper, T. Koburger, U. Volker, and M. Hecker. 1999. Identification of σB-dependent genes in Bacillus subtilis using a promoter consensus-directed search and oligonucleotide hybridization. J. Bacteriol. 181:5718-5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Piriz Duran, S., F. H. Kayser, and B. Berger-Bachi. 1996. Impact of sar and agr on methicillin resistance in Staphylococcus aureus. FEMS Microbiol. Lett. 141:255-260. [DOI] [PubMed] [Google Scholar]
- 51.Rachid, S., K. Ohlsen, U. Wallner, J. Hacker, M. Hecker, and W. Ziebuhr. 2000. Alternative transcription factor σB is involved in regulation of biofilm expression in a Staphylococcus aureus mucosal isolate. J. Bacteriol. 182:6824-6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rachid, S., K. Ohlsen, W. Witte, J. Hacker, and W. Ziebuhr. 2000. Effect of subinhibitory antibiotic concentrations on polysaccharide intercellular adhesin expression in biofilm-forming Staphylococcus epidermidis. Antimicrob. Agents Chemother. 44:3357-3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rupp, M. E., N. Sloot, H. G. Meyer, J. Han, and S. Gatermann. 1995. Characterization of the hemagglutinin of Staphylococcus epidermidis. J. Infect. Dis. 172:1509-1518. [DOI] [PubMed] [Google Scholar]
- 54.Valle, J., A. Toledo-Arana, C. Berasain, J. M. Ghigo, B. Amorena, J. R. Penades, and I. Lasa. 2003. SarA and not σB is essential for biofilm development by Staphylococcus aureus. Mol. Microbiol. 48:1075-1087. [DOI] [PubMed] [Google Scholar]
- 55.Vuong, C., C. Gerke, G. A. Somerville, E. R. Fischer, and M. Otto. 2003. Quorum-sensing control of biofilm factors in Staphylococcus epidermidis. J. Infect. Dis. 188:706-718. [DOI] [PubMed] [Google Scholar]
- 56.Vuong, C., F. Gotz, and M. Otto. 2000. Construction and characterization of an agr deletion mutant of Staphylococcus epidermidis. Infect. Immun. 68:1048-1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vuong, C., H. L. Saenz, F. Gotz, and M. Otto. 2000. Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J. Infect. Dis. 182:1688-1693. [DOI] [PubMed] [Google Scholar]
- 58.Ziebuhr, W., C. Heilmann, F. Gotz, P. Meyer, K. Wilms, E. Straube, and J. Hacker. 1997. Detection of the intercellular adhesion gene cluster (ica) and phase variation in Staphylococcus epidermidis blood culture strains and mucosal isolates. Infect. Immun. 65:890-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ziebuhr, W., V. Krimmer, S. Rachid, I. Lossner, F. Gotz, and J. Hacker. 1999. A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol. Microbiol. 32:345-356. [DOI] [PubMed] [Google Scholar]