Abstract
Bacillus subtilis implements several adaptive strategies to cope with nutrient limitation experienced at the end of exponential growth. The DegS-DegU two-component system is part of the network involved in the regulation of postexponential responses, such as competence development, the production of exoenzymes, and motility. The degU32(Hy) mutation extends the half-life of the phosphorylated form of DegU (DegU-P); this in turn increases the production of alkaline protease, levan-sucrase, and other exoenzymes and inhibits motility and the production of flagella. The expression of the flagellum-specific sigma factor SigD, of the flagellin gene hag, and of the fla-che operon is strongly reduced in a degU32(Hy) genetic background. To investigate the mechanism of action of DegU-P on motility, we isolated mutants of degU32(Hy) that completely suppressed the motility deficiency. The mutations were genetically mapped and characterized by PCR and sequencing. Most of the mutations were found to delete a transcriptional termination signal upstream of the main flagellar operon, fla-che, thus allowing transcriptional readthrough from the cod operon. Two additional mutations improved the σA-dependent promoter sequence of the fla-che operon. Using an electrophoretic mobility shift assay, we have demonstrated that purified DegU binds specifically to the PA promoter region of the fla-che operon. The data suggest that DegU represses transcription of the fla-che operon, and they indicate a central role of the operon in regulating the synthesis and assembly of flagella.
Swimming motility in bacteria depends upon the presence on the cell surface of the flagellar organelle, composed of the basal body, the hook, and the filament. The production of flagella is of such adaptive value that most bacterial species are endowed with flagella, despite the high energy requirement for the synthesis of the numerous flagellin monomers that are necessary to build and maintain the flagellar filament.
In enterobacteria, the genes involved in flagellar formation are organized into regulons which are arranged into three hierarchical classes. The first class is constituted by the flhDC master operon, whose expression is necessary to turn on class II genes, coding for components of the export machinery and for the hook and basal body. The class II gene fliA encodes σ28, the transcription factor for the class III genes, which include flagellar filament structural genes and the chemotaxis signal transduction system (7, 19). In addition, many global regulators, such as CAP, H-NS, H-HU, Lrp, etc., have been reported to affect flagellar synthesis and assembly (5, 13, 24, 34).
In Bacillus subtilis, a bona fide master operon is missing and all genes corresponding to the enteric class II are clustered in a single fla-che operon. The expression of the operon depends upon a σA-recognized promoter (fla-che PA), with an additional σD-dependent promoter (PD-3) playing a minor role (1, 9). Deletion of the fla-che PA promoter renders the cells completely nonmotile, or Mot− (40). The sigD gene, encoding σD, is at the end of the fla-che operon, and its expression is required for the transcription of class III genes for flagellin, motor components, receptors, autolysins, and chemotaxis (27). One of the late flagellar gene products, FlgM, is an anti-sigma factor involved in the control of expression of σD-dependent promoters (3).
The regulation of motility in B. subtilis is integrated into a complex net of regulatory circuits controlling different adaptive responses, such as competence development and degradative enzyme synthesis and secretion (22). The response regulator DegU controls the production of exoenzymes such as proteases, levan-sucrase, and α-amylase and is involved in competence development and motility. DegU is the second element of a two-component signaling system that is phosphorylated by the first component, DegS. Missense mutations within the degS and degU genes, designated degS(Hy) and degU(Hy), that increase the half-life of the phosphorylated form of DegU (DegU-P) result in the overexpression of secreted enzymes and a lack of flagella (2, 8, 17, 23, 37). One of the best-characterized such mutations is degU32(Hy) (15). This mutation causes a strong reduction in the expression of sigD, as seen with a sigD-lacZ translational fusion, and prevents transcription from the cwlB σD-dependent promoter (38). The mechanism through which the phosphorylated form of DegU negatively regulates the expression of sigD is completely obscure.
To investigate the mechanism of action of DegU-P, we isolated and characterized mutations that suppress the effect of degU32(Hy) on motility. Our results indicate that DegU-P represses the transcription of the fla-che operon, thereby preventing the expression of genes coding for the hook and basal body components of the flagellum and for the σD transcription factor. Furthermore, we show that purified DegU binds to the regulatory region of the fla-che operon.
MATERIALS AND METHODS
Bacterial strains and media.
Table 1 lists the bacterial strains used for this study. B. subtilis strains were grown in Schaeffer sporulation medium (SM), PY broth (Pennassay antibiotic medium 3; Difco), or the minimal medium described by Kunst and Rapoport (18). Motility was tested by spotting samples obtained from overnight colonies by use of toothpicks. The motility medium had the following composition:1% Bacto peptone, 8% Bacto gelatin, 1% Bacto agar, 0.5% NaCl, and 25 μg of the appropriate growth requirement/ml. Escherichia coli cells were grown in Luria-Bertani broth. E. coli DH5α supE44 lacU169 (φ80 lacΔZM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 was used as a host for the construction of recombinant plasmids. E. coli C600 thi-1 thr-1 leuB6 lacY1 tonA21 supE44 was used as an intermediate host for plasmids prior to the transformation of B. subtilis. For protein expression and purification, we used E. coli BL21 λDΞ3 (hsdS gal [λ cIts857 ind1 Sam7 nin5 lacUV5-T7] gene1). Antibiotics were used at the following concentrations: ampicillin, 100 μg/ml; erythromycin, 1 μg/ml; kanamycin, 2.5 μg/ml; chloramphenicol, 5 μg/ml; phleomycin, 5 μg/ml.
TABLE 1.
Bacterial strains used in this study
| Strain | Relevant phenotype | Source or referencea |
|---|---|---|
| PB168 | trpC2 | Lab strain |
| PB1894 | hisA1 thr5 trpC2 amyE::pJM115 (Kanr) | Lab strain |
| PB5094 | trpC2 ΔdegSU (Kanr) | QB4238 (23) |
| PB5128 | trpC2 pks::P458hag-lagZ (Catr) | 11 |
| PB5213 | trpC2 degU32(Hy) | BG156 (E. Ferrari) |
| PB5246 | trpC2 leuA8 degS200(Hy) | BG105 (E. Ferrari) |
| PB5248 | trpC2 degU32(Hy)ΔdhsA245 (1690149-1690393)b | This work |
| PB5267 | trpC2 degU32(Hy) pks::P458hag-lacZ (Catr) | PB5213 (tf) PB5128 |
| PB5279 | trpC2 degU32(Hy) ylxF::pMutin4 (Eryr) | PB5213 (tf) BFA2666 |
| PB5280 | trpC2 degU32(Hy) ΔdhsA245 pks::P458hag-lacZ (Catr) | PB5248 (tf) PB5128 |
| PB5284 | trpC2 degU32(Hy) ΔdhsA245 ylxF::pMutin4 (Eryr) | PB5248 (tf) BFA2666 |
| PB5290 | trpC2 ΔcodY::pMutin4 (Eryr) | This work |
| PB5291 | trpC2 degU32(Hy) codY::pMutin4 (Eryr) | PB5213 (tf) PB5290 |
| PB5295 | trpC2 unkU::spcΔcodY (Spcr) | PS37 (33) |
| PB5297 | trpC2 degU32(Hy) unkU::spcΔcodY (Spcr) | PB5213 (tf) PB5295 |
| PB5303 | trpC2 degU32(Hy) unkU::spcΔcodY P458hag-lacZ (Catr) | PB5297 (tf) PB5128 |
| PB5306 | trpC2 ΔPD-3 (Kanr) | This work |
| PB5307 | trpC2 degU32(Hy) ΔPD-3 (Kanr) (1690203-1690393)b | PB5213 (tf) PB5306 |
| PB5314 | trpC2 degU32(Hy) ΔdhsA1 (1690199-1690323)b | This work |
| PB5315 | trpC2 degU32(Hy) ΔdhsA2 (1690201-1690285)b | This work |
| PB5316 | trpC2 degU32(Hy) ΔdhsA3 (1690149-1690393)b | This work |
| PB5317 | trpC2 degU32(Hy) ΔdhsA4 (1689893-1690359)b | This work |
| PB5318 | trpC2 degU32(Hy) ΔdhsA5 (1689172-1690236)b | This work |
| PB5319 | trpC2 degU32(Hy) dhsA6 | This work |
| PB5320 | trpC2 degU32(Hy) ΔdhsA8 (1690142-1690235)b | This work |
| PB5321 | trpC2 degU32(Hy) dhsA10 | This work |
| PB5322 | trpC2 degU32(Hy) ΔdhsA11 (1690169-1690241)b | This work |
| PB5323 | trpC2 degU32(Hy) ΔdhsA4 codY::pB01 | PB5317 (tf) pB01 |
| PB5324 | trpC2 ΔPD-3 (Kanr) pks::P458hag-lacZ (Catr) | PB5306 (tf) PB5128 |
| PB5325 | trpC2 degU32(Hy) ΔPD-3 (Kanr) pks::P458hag-lacZ (Catr) | PB5307 (tf) PB5128 |
| PB5326 | trpC2leuA8 degS200(Hy) pks::P458hag-lacZ (Catr) | PB5246 (tf) PB5128 |
| PB5327 | trpC2 degU32(Hy) ΔdhsA245 ΔdegSU (Kanr) | PB5248 (tf) PB5094 |
| PB5328 | trpC2 ΔPD-3 (Kanr) ylxF::pMutin4 (Eryr) | PB5306 (tf) BFA2666 |
| PB5329 | trpC2 degU32(Hy) ΔPD-3 (Kanr) ylxF::pMutin4 (Eryr) | PB5307 (tf) BFA2666 |
| PB5368 | trpC2 degU32(Hy)ΔdhsA245 codY::pMutin4 (Eryr) | This work |
| BFA2666 | trpC2 ylxF::pMutin4 (Eryr) | This work |
For strains constructed by transformation, the recipient strain number is reported first and the donor strain is reported after “tf.”
The numbers refer to the first and the last nucleotide involved in the deletion (Subtilist coordinates).
Genetic techniques.
B. subtilis strains were transformed with chromosomal or plasmid DNA by the procedure of Kunst and Rapoport (18). Transduction mapping with the PBS1 phage was performed according to Hoch et al. (16). For genetic landmarks, we used a set of mutants constructed by several laboratories (32). E. coli transformation was performed according to standard protocols (31).
Construction of mutants.
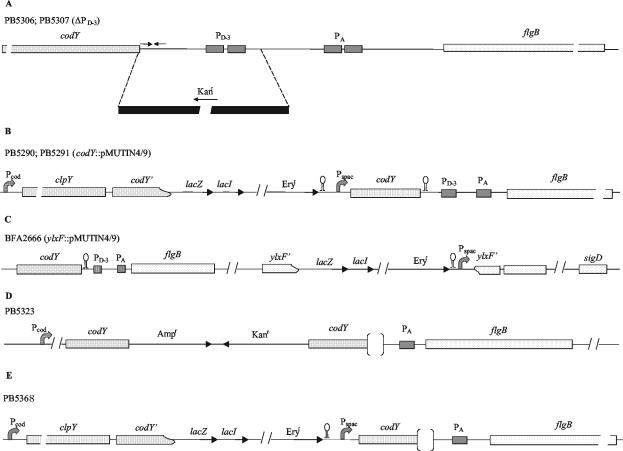
For the construction of strains with a deletion of the promoter PD-3, a sequence of 191 bp overlapping the promoter was replaced with a kanamycin resistance determinant (Fig. 1A). First a 485-bp DNA fragment corresponding to the intercistronic region downstream of PD-3 and extending into the coding sequence of flgB was amplified by a PCR with primers E8601 and B9070, which contained an EcoRI and a BamHI site, respectively. The template was chromosomal DNA from B. subtilis 168. The amplified fragment was purified with a Qiaquick PCR purification kit (Qiagen) and restricted with EcoRI and BamHI. After digestion, the DNA fragment was purified with the same kit and ligated to EcoRI- and BamHI-digested pJM114 (28). The ligated plasmid was transformed into E. coli DH5α and called pGA10. A second DNA fragment of 474 bp extending from the middle of the codY coding sequence to four nucleotides after the codY stop codon was obtained by a PCR with primers K7936 and S8409, which contained a KpnI and a SalI recognition sequence, respectively. The amplified DNA was treated as described above and cloned into pGA10 that had been digested with KpnI and SalI. The final plasmid (pGA11) contained two fragments of DNA derived from B. subtilis strain 168 flanking the kanamycin resistance determinant. The plasmid was verified by sequencing. The plasmid DNA was linearized by ScaI digestion and used to transform competent cells of B. subtilis 168, selecting for Kanr (2.5 μg/ml). One of the transformants (PB5306) was characterized by PCR and sequencing and its DNA was used to transfer the deletion mutation into strain PB5213, generating strain PB5307.
FIG. 1.
Schematic presentation of construction of B. subtilis mutant strains. (A) Deletion of PD-3 promoter sequence upstream of fla-che operon. flgB is the first gene of the operon. The region between the end of the codY gene and the PA promoter was replaced with the kanamycin resistance determinant of plasmid pJM114 by a double-crossover event. (B) Placement of codY gene under control of the IPTG-inducible Pspac promoter. A chromosomal fragment corresponding to the end of the clpY gene and to one-third of the codY gene was amplified by PCR and cloned into pMutin4. The pMutin4 derivative was inserted into the chromosome by a single crossover event (Campbell-type integration), thus placing the codY gene under the control of Pspac. (C) Construction of ylxF-lacZ transcriptional fusion. The ylxF gene of the fla-che operon was disrupted with pMutin4 by a Campbell-type integration, placing the lacZ reporter gene under the control of the Pspac promoter. (D) Insertion of the pJM114 plasmid sequence upstream of the dhsA4 deletion mutation. The codY coding sequence was amplified and cloned into plasmid pJM114. The plasmid derivative was inserted into the chromosome of strain PB5317 (dhsA4) by a Campbell-type event. (E) Placement of fla-che operon gene under control of Pspac promoter. The codY gene under the control of Pspac was combined with a deletion of the transcription terminator dhsA245. The deletion is represented by an empty space between the two brackets. The densely dotted boxes refer to the clpY and codY genes, which are part of the cod operon. The fla-che operon genes (flgB, ylxF, and sigD) are shown as sparsely dotted boxes. The stem-loops indicate transcription termination signals. lacI, E. coli lacI gene; Ampr, ampicillin resistance marker; Kanr, kanamycin resistance marker; Eryr, erythromycin resistance marker.
To place gene codY under the control of the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible Pspac promoter, we proceeded in the following way (Fig. 1B). A 451-bp DNA fragment corresponding to the end of gene clpY (upstream of codY) and part of codY was obtained by amplification of the chromosomal DNA with primers 7541 and 7971 (Table 2). The purified DNA was cloned into pUC18 by use of the SureClone ligation kit (Amersham Pharmacia Biotec). After sequencing, one plasmid was chosen and digested with EcoRI and BamHI, and a DNA band of approximately 450 bp was purified and cloned into plasmid pMutin4 (39), which had previously been digested with EcoRI and BamHI and dephosphorylated. The new plasmid, pMutin4/9, was verified by sequencing, transformed into E. coli strain C600, extracted, and used to transform B. subtilis 168 competent cells. Selection was done on tryptose blood agar base (Difco) plates with erythromycin at 1 μg/ml. One transformant was retained (PB5290), and after PCR verification, its DNA was used to transform strain PB5213 to obtain strain PB5291. Strains PB5290 and PB5291 are thus derivatives of PB168 and PB5213, respectively, with the codY gene under the control of the IPTG-inducible Pspac promoter.
TABLE 2.
Oligonucleotide primers
| Primer | Sequence (5′-3′) |
|---|---|
| 7541 | CGGAACGATAGCCAAAAACAA |
| 7971 | TCTTTCCCCGCCTCCGATGAT |
| 8390 | TCTAAAATCTCATTAATCAC |
| 8601 | CGAGAAATGTAGTTCTAACAATC |
| 8630r | GTCCTAGATTGTTAGAACTAC |
| 8740 | TCTCGACAAGGATATTGAGG |
| 8756 | CACCCTCAATATCCTTGTCGAG |
| 9090 | TGCGCCAATTCTGTCATCTC |
| B9070 | CGGGATCCAATTCTGTCATCTCTTTG |
| codY2 | CGGAATTCCTAGGAGGATTATTTATCATGGCT |
| codYB | CGGGATCCTTAATGAGATTTTAGATTTTCTAATTCAATTAGGA |
| degU-Nt | GGAATTCCATATGACTAAAGTAAACATTG |
| degU-Ct | GGAATTCCATATGTCACTATCTCATTTCTACCCAGC |
| E8601 | CGGAATTCGAGAAATGTAGTTCTAAC |
| FLGb1 | GGATTCTTGATCTAATAAATTTTGGA |
| K7936 | GCGGTACCTGTTTCAAGCTGGTTTAAC |
| PA | TCTCGACAAGGATATTGAGG |
| RTFLGb2 | GCTTAATATCCGCTCTGCTCAAGGCA |
| S8409 | ACGCGTCGACGTGATTAATGAGATTTTAG |
| ylxF2 | GGGATCCTCCTCAGCCGTCTTTTCA |
| ylxF3 | GGAATTCAGTTCCGTTCGGTTTTGC |
Plasmid pMutin4/9 was also used to transform strain PB5248 to obtain strain PB5368, in which the codY gene under the control of the Pspac promoter was combined with the dhsA245 deletion (Fig. 1E). For a tighter repression of transcription from Pspac in the absence of the IPTG inducer, plasmid pMAP65, which overproduces LacI, was introduced by transformation and selection for Phleor (30).
For the construction of the ylxF-lacZ transcriptional fusion, an internal fragment of the ylxF sequence was amplified by a PCR with the primers ylxF3 and ylxF2 (Table 2). The amplified fragment (331 nucleotides) was restricted with EcoRI and BamHI and cloned into EcoRI- and BamHI-digested pMutin4 to generate pMutin331EB. The B. subtilis ylxF::pMutin4 (BFA2666) strain was obtained by Campbell-type integration of pMutin331EB into the chromosome of B. subtilis 168. The mutant construction was verified by PCRs with combinations of the ylxF3 and ylxF2 primers and oligonucleotides corresponding to sequences of the vector.
To construct strain PB5323, in which the dhsA4 mutation was separated from the cod operon, we cloned the complete coding sequence of gene codY into the integrative plasmid pJM114 (28). The codY gene was generated from the chromosomal DNA by a PCR with primers codY2 and codYB, which contained an EcoRI and a BamHI site, respectively (Table 2). The amplified fragment was purified, digested, and ligated into pJM114, which had been digested with EcoRI and BamHI as reported above. The new plasmid (pPB01) was expanded in E. coli strain C600 and used to transform competent cells of strain PB5317 (dhsA4), selecting for Kanr (Fig. 1D).
β-Galactosidase activity assay.
To assay β-galactosidase activity, we diluted overnight cultures in SM in fresh medium and took samples at 30-min intervals for optical density readings at 525 nm and β-galactosidase activity determinations. The calculation of β-galactosidase units was done in modified Miller units (29).
Preparation of RNA.
The following protocol, adapted from the work of Caldwell et al. (6), was used to prepare B. subtilis RNA samples for reverse transcription-PCR (RT-PCR). Samples (5 ml) of cultures in SM were collected and rapidly frozen by dripping into approximately 40 ml of liquid nitrogen in a 50-ml Falcon tube. The tubes were stored at −80°C overnight. The frozen droplets were transferred to a commercial hand-held coffee grinder that was prechilled and contained crushed dry ice. The sample was ground for 2 min in a cold room and the powder was transferred to a new Falcon tube and stored at −20°C overnight to allow the dry ice to sublime. The frozen sample was treated with 5 ml of phenol saturated with Tris-EDTA buffer (pH 7.8), mixed, and incubated at 65°C for 3 min. After cooling for 3 min on ice, the mixture was centrifuged at 12,000 × g for 10 min at 4°C. The upper aqueous phase was extracted with an equal volume of phenol, and the procedure was repeated four additional times. The last aqueous phase was transferred to a 1.5-ml Eppendorf tube, and 5 volumes of Trizol reagent (GIBCO BRL) was added. After mixing, the sample was incubated at room temperature for 5 min. Chloroform was added (1 volume of the aqueous phase), mixed for 15 s, and incubated for 3 min at room temperature. After centrifugation at 12,000 × g for 15 min at 4°C, the aqueous phase was carefully removed and transferred to a new tube, and the RNA was precipitated with a one-half volume of isopropanol at room temperature for 10 min. After centrifugation at 12,000 × g for 10 min at 4°C, the supernatant was removed and the pellet was washed with 70% ethanol and centrifuged at 7,500 × g for 5 min at room temperature. The pellet was air dried for 15 min and dissolved in diethyl pyrocarbonate-treated water. The yield was determined by UV spectroscopy.
RT-PCR.
We used RT-PCR to evaluate the presence of readthrough transcription from the cod operon into the fla-che operon. Wild-type, degU32, and degU32 dhs mutant cells grown in SM were harvested at T−1 (1 h before the transition point), T0 (the transition point between the exponential and postexponential growth phases), and T2 (2 h after the transition point), and RNAs were isolated as described above.
A cDNA were synthesized by RT, with purified RNA as a template and with primer flgB1 (Table 2; also see Fig. 8), which was complementary to the flgB gene mRNA. For these experiments, 3 μg of RNA template was mixed in a final volume of 12 μl with 20 pmol of the oligonucleotide flgB1. The sample was denatured for 5 min at 70°C and cooled for 1 min on ice. The following reagents were added to this mixture: 4 μl of 5× first-strand buffer (GIBCO BRL), 2 μl of 0.1 M dithiothreitol, and 1 μl of 10 mM (each) deoxynucleoside triphosphates. After incubation at 42°C for 2 min, 1 μl (200 U) of SuperScript II reverse transcriptase (GIBCO BRL) was added, and the incubation was prolonged for 1 h at 42°C. The sample was treated with 1 μl of RNase H (2 U/μl; GIBCO BRL) for 10 min at 55°C and then allowed to cool. The resulting cDNA was used as a template to synthesize and amplify DNA fragments with Taq DNA polymerase (5 U/μl; GIBCO BRL). The downstream primer was flgB2, which was complementary to the noncoding strand. Two different upstream primers, 7901 and PA, were used (Table 2; also see Fig. 8). Primer 7901, which was complementary to the codY transcript, was used to detect readthrough cod-flgB transcripts, whereas primer PA was used to detect both readthrough transcripts and transcripts initiating at the PA promoter. The second reaction contained 1.5 μl of the product of the first reaction and 15 pmol each of primer flgB2 and primer PA (or primer 7901) in a final volume of 20 μl. The conditions for PCR were as follows: 94°C for 1 min, 54°C for 1 min, and 72°C for 1 min for 40 cycles. The resulting RT-PCR products were analyzed by agarose gel electrophoresis (see Fig. 8). RT-PCRs in the absence of reverse transcriptase were performed in parallel to check for DNA contamination. PCR products from chromosomal DNAs from the parental and mutant strains obtained with the same primers (flgB2 and 7901) were run in parallel with the samples.
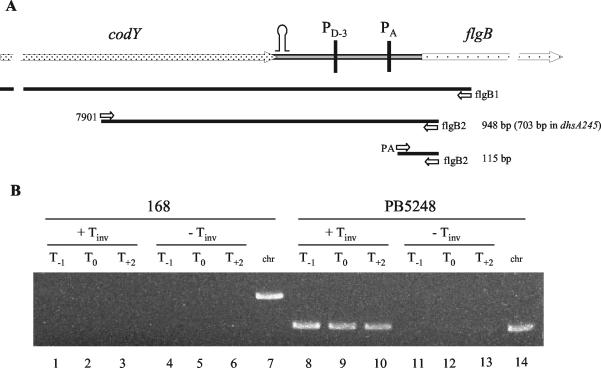
FIG. 8.
RT-PCR analysis of transcriptional readthrough. (A) Diagram of codY-flgB region. PD-3 and PA indicate the σD-dependent and σA-dependent promoters, respectively. The stem-loop indicates the transcription terminator at the end of the cod operon. The open arrows indicate the oligonucleotides used to prime RT (flgB1) and to amplify the cDNA products (7901 and flgB2; PA and flgB2). The calculated sizes of the PCR products are reported to the right. (B) Ethidium bromide-stained 1% agarose gel of RT-PCR and PCR products. RT reactions with the RNAs obtained from strains PB168 and PB5248 (as indicated) were performed with the flgB1 primer. The cDNAs were amplified with primers 7901 and flgB1. T0 refers to the time of transition between the exponential growth and stationary phases; T−1 and T2 refer to 1 h earlier and 2 h later, respectively. Lanes 1 to 6, the template for RT-PCRs was RNA from PB168 (wild type); lanes 1 to 3, RT-PCRs with reverse transcriptase; lanes 4 to 6, reactions in the absence of reverse transcriptase. Lane 7, PCR with chromosomal DNA from strain PB168 as a template and with primers 7901 and flgB2. Lanes 8 to 13, RT-PCRs with RNA from strain PB5248 (ΔdhsA245) as a template; lanes 8 to 10, reactions with reverse transcriptase; lanes 11 to 13, reactions in the absence of reverse transcriptase. Lane 14, PCR with chromosomal DNA from strain PB5248 as a template and with primers 7901 and flgB2.
Purification of DegU.
The degU gene was PCR amplified from wild-type (PB168) and degU32(Hy) mutant (PB5213) chromosomal DNAs with primers degU-Nt and degU-Ct (Table 2). Both primers contained a restriction site for NdeI, which was used to clone the genes into the expression plasmids pET12a and pET16B (Novagen). Four plasmids were thus obtained: two were derivatives of pET12a (pET12Uwt and pET12Uhy) coding for the wild-type and mutant proteins, respectively, and two were derivatives of pET16b (pET16Uwt and pET16Uhy) coding for the two proteins with an N-terminal His6-tag fusion (named H6-DegU and H6-DegU32, respectively). For expression of the recombinant proteins, E. coli ΒL21 λDE3(pLys) competent cells were transformed with the plasmids, inoculated in Luria-Bertani medium containing 100 μg of ampicillin/ml, grown at 37°C to an optical density at 600 nm of 0.6, and induced with 1 mM IPTG, and growth was then continued for 2.5 additional hours. Cells from 5-ml cultures were collected by centrifugation and washed twice in phosphate-buffered saline (140 mM NaCl, 27 mM KCl, 101 mM Na2HPO4, 18 mM KH2PO4, pH 7.3). The pellets were resuspended in buffer A (20 mM Tris-HCl [pH 8.0], 200 mM NaCl) supplemented with 0.5 mM phenylmethylsulfonyl fluoride and 1 mg of lysozyme/ml. After the cells were incubated on ice for 30 min, Triton X-100 (1% final concentration), 5 μg of DNase/ml, and 5 μg of RNase/ml were added. The samples were further incubated on ice for 15 min, and the cells were then broken by sonication (five pulses of 15 s each with 15-s intervals, on ice) and centrifuged for 10 min at 12,000 × g. The pellet was dissolved in 400 μl of buffer B (20 mM Tris-HCl [pH 8.0], 200 mM NaCl, 5 M urea) and gently stirred on ice for 1 h. The samples were centrifuged in a microcentrifuge for 10 min at 12,000 rpm, and the supernatants were recovered. For purification of the His6-tagged proteins, the supernatants were each mixed with 20 μl of Ni-nitriloacetate agarose (Qiagen) and stirred on ice for 45 min. The resin was recovered by centrifugation and washed three times with buffer A. The fusion proteins were eluted with 100 mM imidazole (for H6-DegU) or 200 mM imidazole (for H6-DegU32) in buffer A. Prior to their utilization for gel retardation analysis, the proteins were dialyzed against water.
Electrophoretic mobility shift assays.
Electrophoretic mobility shift assays were performed as described by Hamoen et al. (12). DNA fragments from the codY-flgB intercistronic region were obtained by PCR amplification with the primers described in Fig. 10A and Table 2. Chromosomal DNA from strain PB168 was used as a template. After purification with a PCR purification kit (Qiagen), the fragments were end labeled with T4 polynucleotide kinase and [γ-32P]ATP (2,500 Ci/mmol, 10 mCi/ml; Amersham). The probes were purified with a PCR purification kit (Qiagen), and approximately 1 ng of probe was mixed with the protein in binding buffer (20 mM Tris-HCl [pH 8.0], 100 mM KCl, 5 mM MgCl2, 0.5 mM dithiothreitol, 10% [vol/vol] glycerol, 0.05% Nonidet P-40, 0.05 mg of bovine serum albumin/ml) containing 0.05 μg of sonicated salmon sperm DNA/ml as a nonspecific competitor. After incubation at 37°C for 20 min, the samples were loaded into a nondenaturing 6.5% polyacrylamide gel. Gels were run in TAE buffer (40 mM Tris-acetate [pH 8.0], 2 mM EDTA) at 10 mA (65 V) for about 90 min and then were autoradiographed (Hyperfilm MP). B. subtilis specific competitor DNA was obtained by PCR and used at a 100-fold excess over the amount of labeled probe.
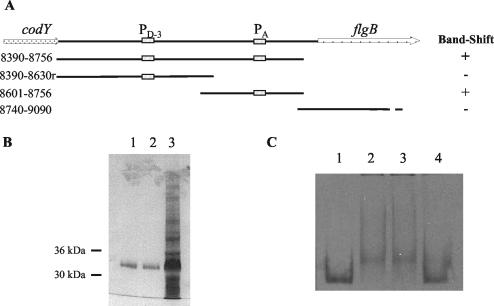
FIG. 10.
Gel mobility shifts of a 32P-labeled fla-che operon promoter fragment. (A) Scheme of codY-flgB intercistronic region. The PD-3 and PA boxes represent the σD-dependent and σA-dependent promoters, respectively. DNA fragments tested in gel mobility shift assays in the presence of DegU are indicated. Numbers to the left refer to the oligonucleotide primers used to amplify the fragments (Table 2). (B) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of H6-DegU preparation used in this study. Lanes 1 and 2, H6-DegU eluted from Ni-nitrilotriacetic acid-agarose with 100 and 150 mM imidazole, respectively; lane 3, crude extract after 2.5 h of induction with 1 mM IPTG. (C) Gel retardation by DegU. Lane 1, probe 8601-8756 alone; lane 2, probe 8601-8756 incubated with H6-DegU32 (0.2 μM); lane 3, probe 8601-8756 incubated with H6-DegU32 (0.2 μM) in the presence of cold competitor 8740-9090 DNA (100 ng); lane 4, probe 8601-8756 incubated with H6-DegU32 (0.2 μM) in the presence of cold competitor 8601-8756 (100 ng).
RESULTS
DegU-P represses motility.
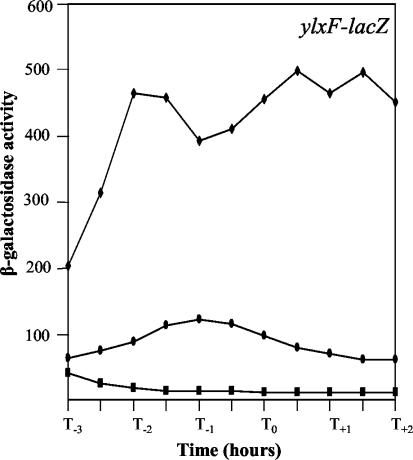
B. subtilis strains with the degU32(Hy) mutation are nonmotile (2, 17) (see Fig. 3). Using the lacZ reporter gene transcriptionally fused to the hag flagellin promoter (Phag), we confirmed that transcription is completely abolished in the degU32(Hy) background (Table 3). It has been reported that the degU32(Hy) mutation causes a strong reduction in the expression of a sigD-lacZ translational fusion (38). Using anti-SigD antibodies for an immunoblot analysis, we confirmed that in the mutant strain the SigD protein is barely detectable (data not shown). In addition, we measured the expression of the fla-che operon, in which sigD is present, by using a lacZ transcriptional fusion. For this purpose, we used the pMutin4 vector (39), in which a fragment derived from the ylxF open reading frame was cloned. ylxF is the ninth open reading frame of the fla-che operon. As reported in Fig. 2, in the wild-type background β-galactosidase activity reached the maximal value at approximately the end of exponential growth and eventually decreased. In the presence of the degU32(Hy) mutation, no β-galactosidase activity could be detected (Fig. 2 and Table 3). The expression of hag-lacZ could be rescued by the production of SigD from a plasmid (Table 3). This was obtained by the transformation of strain PB5267 (degU32) with plasmid pSigD, which contains sigD under the control of the IPTG-inducible Pspac promoter (3). Nevertheless, the transformants were nonmotile. In accordance with this phenotype, the expression of the fla-che operon, measured as the β-galactosidase activity of the ylxF-lacZ fusion, was not affected by the induction of SigD and remained at undetectable levels (Table 3). This suggests that the lack of flagella and motility cannot be ascribed only to the absence of an active SigD factor.
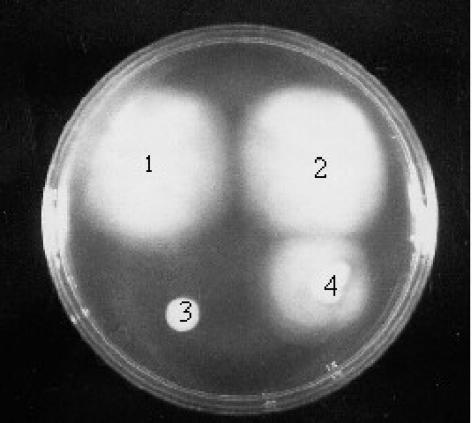
FIG. 3.
Motility of B. subtilis strains. Overnight colonies were transferred to a motility plate and incubated at 37°C overnight. 1, strain PB5248 [degU32(Hy) ΔdhsA245]; 2, strain PB5327 (ΔdhsA245 ΔdegSU); 3, strain PB5213 [degU32(Hy)]; 4, strain PB168 (wild type).
TABLE 3.
Expression of hag and fla-che operon in various mutant strains
| Relevant mutation | Level of expression (Miller units)
|
|
|---|---|---|
| hag-lacZ | fla-che (ylxF::pMutin4) | |
| Wild type | 2,388 | 121 |
| degU32(Hy) | 11 | 6 |
| degS200 | 13 | NDa |
| degU32(Hy) pSigD | 764 | 5 |
| degU32(Hy) dhsA245 | 1,536 | 498 |
| degU32(Hy) ΔcodY | 7 | NDa |
ND, not determined.
FIG. 2.
Analysis of fla-che operon expression in different genetic backgrounds. The ylxF-lacZ fusion schematically shown in Fig. 1C was used to monitor the expression of the fla-che operon in different B. subtilis strains. ylxF is the ninth open reading frame of the operon. The strains were BFA 2666 (wild type; solid circles) PB5279 [dehU32(Hy); solid squares], and PB5284 [degU32(Hy) dhsA245; solid diamonds]. The β-galactosidase specific activity is expressed in Miller units per milligram of protein. Zero time (T0) indicates the transition point between the exponential and stationary phases of growth.
We observed the same drastic reduction in Phag-dependent transcription in the presence of the degS200(Hy) mutation (Table 3). Since both mutations cause an increase in the half-life of the phosphorylated form of DegU (DegU-P) (8, 37), we propose that the observed transcriptional repression is dependent upon the presence of DegU-P. This interpretation is indirectly supported by the lack of any effect on hag-lacZ expression of a deletion involving degS and degU (data not shown). In addition, we transferred the deletion of degSU by transformation into the degU32(Hy) strain. For this purpose, we used a mutant in which the degSU operon is replaced with a kanamycin resistance determinant (23). The Kanr transformants recovered motility completely (data not shown). The same deletion introduced into the Mot+ suppressed strain PB5248 (see below) did not alter the Mot+ phenotype. These results support an active role for DegU-P in repressing motility.
Isolation and mapping of a suppressor mutant.
To understand the mechanism of action of DegU-P, we searched for a spontaneous suppressor mutant of the Mot− phenotype in a degU32(Hy) background. Strain PB5213 (degU32 Mot−) was spotted onto a motility plate. After incubation, cells from the peripheral boundaries of the growth zone were isolate and characterized. One motile strain, PB5248, which retained the original degU32(Hy) mutation, as assessed by sequencing and the protease hyperproduction phenotype, was chosen for characterization. The suppressing mutation was called dhsA245, for degU high suppressor. The Mot+ phenotype was stable, even after several passages through a single colony. On motility plates, the suppressed strain moved to an extent that was comparable to that of the parental strain PB168 (Fig. 3). It also largely recovered expression from the hag promoter, as shown by the level of β-galactosidase activity (Table 3).
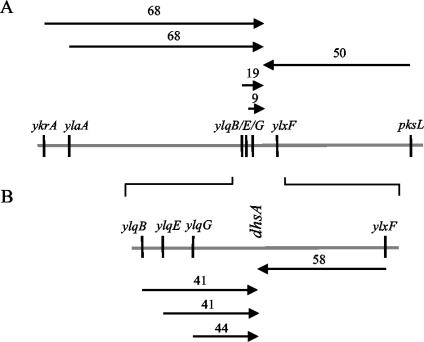
The suppressor was genetically mapped by PBS1-mediated transduction, with lysates obtained from a collection of derivatives of strain PB168 with the erythromycin resistance marker present in defined genes around the chromosome used as donors (32). Transductants selected for Eryr were screened for motility; the replacement of the dhsA245 mutant allele with the wild-type gene was expected to confer a Mot− phenotype. Of 18 markers tested, 5 consistently showed significant linkage to dhsA245, indicating that it mapped at approximately 144° (Fig. 4). It should be pointed out that this position is far from the location of gene degU at 311°. Transformation crosses allowed us to restrict the region to the portion of the chromosome comprising the area between the ylqB and ylxF genes (Fig. 4). Further refinement of the genetic map was not attempted. To pinpoint the dhsA245 mutation, we determined the nucleotide sequence of approximately 20 kb in the region. The sequence derived from strain PB5248 [degU32(Hy) dhsA245] differed from the sequence of the reference strain 168 by only a 245-bp deletion (see Fig. 7). The deletion encompassed the end of the codY gene and part of the intercistronic region between codY and flgB. As a result of the mutation, the deduced CodY protein is 251 residues long, instead of 259, and the last 8 residues differ from the wild-type protein. The span of the deletion should be sufficient to abolish the activity of the protein, as reported for an insertion mutation, cod-37, that causes a frame shift and results in a polypeptide of 251 amino acids (35). We monitored the expression of a hut-lacZ and a dpp-lacZ fusion, which were previously reported to be negatively controlled by CodY (10). The expression of the two fusions was significantly increased in strains with the dhsA245 mutation, in agreement with the inactivation of gene codY in the suppressed mutant (data not shown). Downstream of gene codY, the deletion eliminates the σD-dependent promoter PD-3 of the fla-che operon, which contributes marginally to the expression of the operon (9, 40).
FIG. 4.
Genetic map of dhsA245 mutation. (A) Transduction crosses; (B) transformation crosses. The arrows are based on the selected marker and point to the nonselected marker. Donor strains had an erythromycin resistance determinant present in the indicated gene that was used as a selective marker. After selection, the transductants (transformants) were screened for motility. Distances are expressed as percentages of cotransduction (cotransformation). The gray line in panel A represents the region between 130° and 155° on the genetic map.
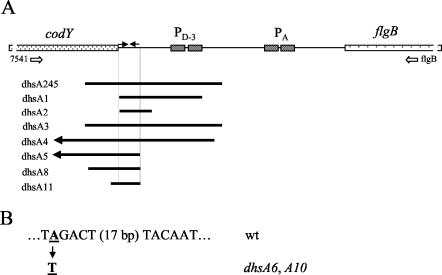
FIG. 7.
Mutations of dhs locus. (A) Schematic physical map of codY-flgB region of B. subtilis chromosome. The open arrows labeled 7541 and flgB indicate the positions of the oligonucleotide primers used to amplify the codY-flgB region (Fig. 6). The positions and extension of eight dhs deletion mutations are indicated by the solid bars below the physical map. Deletions Δdhs4 and Δdhs5 extend further into the cod operon and are represented by arrows pointing to the left. (B) Sequence of the σA-dependent promoter PA and of the dhsA6 and dhsA10 point mutations.
To evaluate the role of the deletion as a suppressor of degU32(Hy), we transduced the recipient strain PB5248 [degU32(Hy) dhsA245] with a PBS1 lysate derived from the BFA 2626 strain, which has an Eryr determinant linked to the pksL gene (Fig. 4). As expected from the distance between pksL and dhsA245, approximately 50% of the Eryr transductants were Mot−. We assayed for the presence or absence of the deletion by PCR; all 11 Mot− transductants tested had the donor (wild-type) configuration and all 10 Mot+ isolates retained the deletion of the recipient. We concluded that the deletion is responsible for the suppression of the degU32(Hy) Mot− phenotype.
Gene codY and the promoter PD-3 are not responsible for suppression.
Two explanations for the mechanism of suppression by the dhsA245 deletion mutation are possible: it occurs either by the inactivation of gene codY or by the elimination of the PD-3 promoter region. CodY has been implicated in the nutritional repression of flagellin expression observed during early stages of exponential growth in complex media (4, 21). Even though DegU phosphorylation is predominant late in exponential growth and at the beginning of stationary phase, a possible involvement of CodY in the repression exerted by DegU-P could not be ignored. Transcription from the PD-3 promoter does not contribute significantly to the expression of the fla-che operon (40), but the effects of a deletion mutation affecting only PD-3 have not been investigated. To address these questions, we analyzed the effects of single deletions of codY and PD-3.
First, we constructed a strain with one copy of the codY gene disrupted and with a second copy under the control of the IPTG-inducible Pspac promoter (Fig. 1B). To this end, we used the integrative plasmid pMutin4 (39), in which we cloned a fragment of 450 bp, extending from the middle part of clpY (the gene immediately upstream of codY) to the middle of codY. We transformed the parental PB168 strain with the pMutin4 derivative. The transformants were Mot+ in both the presence and the absence of the inducer IPTG (data not shown). We concluded that the inactivation of codY (absence of inducer) or (over)expression of CodY (in the presence of 1 and 3 mM IPTG) did not interfere with motility in an otherwise wild-type background. The chromosomal DNA of one of the transformants (PB5290) was used to transfer the construction into PB5213 [degU32(Hy)]. The new transformants were still Mot−, in both the presence and the absence of the IPTG inducer (data not shown). The same result, a lack of suppression of the degU32(Hy) Mot− phenotype, was observed by the transfer into PB5213 of the codY deletion mutation described by Serror and Sonenshein (34). All transformants with the deletion were Mot−, and the level of expression of hag-lacZ fusions was very low, comparable to that obtained in the parental PB5213 strain (Table 3). We concluded that the codY gene is not involved in the repression exerted by DegU-P.
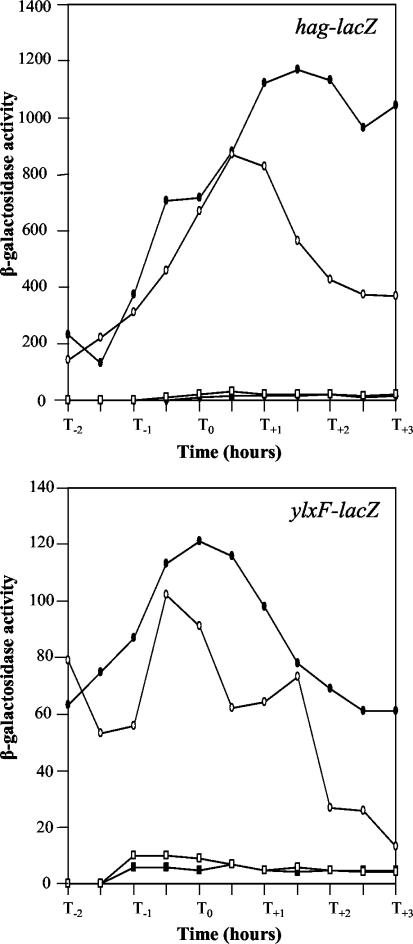
The possible involvement of the σD-dependent promoter PD-3 was evaluated through the construction of a 191-bp deletion extending from immediately downstream of the codY stop codon to approximately half of the codY-flgB intercistronic region. The PD-3 sequence was completely deleted, whereas the PA and surrounding sequences were unaltered. The deleted sequence was replaced with the kanamycin resistance determinant, with the direction of transcription being opposite that of the fla-che operon (Fig. 1A). In an otherwise wild-type background, the PD-3 deletion did not interfere with motility, with the expression of hag-lacZ fusions, or with transcription of the fla-che operon (Fig. 5 and data not shown). When it was transferred into the degU32(Hy) strain, the deletion did not restore the Mot+ phenotype and did not allow expression of the hag-lacZ transcriptional fusion and the fla-che operon (Fig. 5). We thus conclude that, under these experimental conditions, the PD-3 promoter does not affect the expression of the fla-che operon and is not involved in the DegU-P-dependent repression of motility.
FIG. 5.
Deletion of the PD-3 promoter does not affect hag gene (A) and fla-che operon (B) expression. hag-lacZ (9) and ylxF-lacZ (Fig. 1C) transcriptional fusions were used to monitor the expression of the flagellin hag gene and the fla-che operon, respectively. The strains were PB5128 and BFA2666 (wild type; filled circles), PB5324 and PB5328 (ΔPD-3; open circles), PB5267 and PB5279 [degU32(Hy); open squares], and PB5325 and PB5329 [degU32(Hy) ΔPD-3; filled squares].
Isolation of additional degU32(Hy) suppressor mutants.
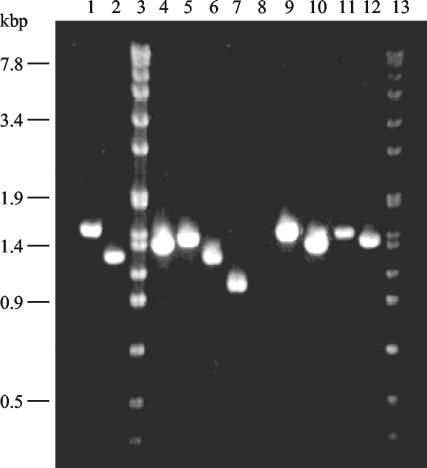
From the above results, it appears that suppression of the Mot− phenotype of the degU32(Hy) mutants depends on the presence of a deletion extending from the end of codY to upstream of the PA promoter of the fla-che operon. It is also evident that deletion of the codY gene or of the sequence of the PD-3 promoter does not restore the Mot+ phenotype. To obtain independent indications of the nature of the suppression and the mechanism of action of DegU(-P), we isolated additional suppressor mutants. Nine independent mutants were isolated and characterized at the genetic and molecular level, and they were designated dhsA1 to -6, -8, -10, and -11. All of the mutants maintained the original high level of exoprotease production and the degU32 mutation (data not shown). All of the suppressor mutations were genetically linked to the erythromycin resistance determinant present in ylq. The cotransduction frequencies were high, ranging from 81% for the dhsA6 mutant to 98% for the dhsA11 mutant. The mutations were characterized by PCR amplification (Fig. 6) and DNA sequencing. Seven mutations were deletions in the codY-flgB region and extending from bp 73 of dhsA11 to bp 1065 of dhsA5. This last deletion extended into gene clpY, upstream of codY, and did not give a PCR product with the pair of oligonucleotides used (Fig. 6, lane 8). The dhsA3 mutation was identical to the deletion in strain PB5248. The only region missing from all of the deletions started at the nucleotide immediately following the codY stop codon and ended 37 nucleotides further downstream. The consequence of the deletion was the complete removal of the putative intrinsic termination site of transcription that is normally present immediately downstream of the codY gene (Fig. 7). The simplest interpretation of this result is that in the absence of the termination signal, transcription from the cod operon can proceed through the intercistronic region and bypass the control exerted by DegU(-P). Two of the dhs mutations (dhsA6 and dhsA10) were identical point mutations, changing one A residue to a T residue (on the noncoding strand). The mutation altered the second position of the −35 hexamer of the PA promoter in front of the fla-che operon. The change, from TAGACT to TTGACT, increases the match of the −35 hexamer to the consensus sequence (TTGACA) and should increase the affinity of RNA polymerase for the promoter (14).
FIG. 6.
Molecular analysis of codY-flgB chromosomal region. Chromosomal DNAs from B. subtilis strains were PCR amplified with primers 7541 and flgB (Table 2 and Fig. 7) and were analyzed by agarose gel (0.6%) electrophoresis. Lanes 3 and 13, molecular weight standard (SPP1 phage DNA digested with EcoRI). The DNA samples were as follows: lane 1, PB5213 (wild type); lane 2, PB5248 (Δdhs245); lane 4, PB5314 (ΔdhsA1); lane 5, PB5315 (ΔdhsA2); lane 6, PB5316 (ΔdhsA3); lane 7, PB5317 (ΔdhsA4); lane 8, PB5318 (ΔdhsA5); lane 9, PB5319 (dhsA6); lane 10, PB5320 (ΔdhsA8); lane 11, PB5321 (dhsA10); lane 12, PB5322 (ΔdhsA11).
Transcriptional readthrough.
To evaluate the presence of readthrough transcripts in the suppressed strain PB5248, we used RT-PCR analysis. RNAs were purified from both parental (PB168) and dhsA245 (PB5248) cells at three time points during the growth curve, i.e., at the transition from exponential to stationary phase (T0), 1 h earlier (T−1), and 2 h into stationary phase (T2). The in vivo transcription products were analyzed by a two-step procedure (Fig. 8). First, a single filament cDNA was obtained by RT primed with oligonucleotide flgB1, which corresponded to a sequence within the flgB open reading frame (Fig. 8A). Subsequently, a small aliquot of the product of the first reaction was diluted 13 times, and two different PCRs were performed. The primer flgB2 was used in both PCRs; this primer was derived from within the flgB sequence, upstream of the flgB1 primer used for RT. In one set of reactions, amplification was carried out with primer PA, corresponding to the PA promoter, to obtain a product of 115 bp derived from the transcript synthesized from the PA promoter and from the hypothetical readthrough product. The second set of reactions used oligonucleotide 7901, which hybridizes to the codY gene, in addition to the flgB2 primer (Fig. 8A). The amplification product derived from the codY-flgB readthrough transcript was expected to be 948 bp long. In the presence of the dhsA245 deletion, the amplification product was expected to be 703 bp long. Control reactions without reverse transcriptase were included in each experiment. As expected, a 115-bp product was obtained from reactions derived from extracts of both the parental (PB168) and the suppressed PB5248 Mot+ strain; the degU32(Hy) RNA did not give any product (data not shown). For the parental strain, the 115-bp band was present at T−1 and T0 and absent from the T2 sample. For the suppressed mutant, the band was present at T2 as well. The 703-bp PCR product corresponding to a readthrough from the cod operon was detected at all time points examined for the strain with the dhsA245 deletion (Fig. 8B). The product was absent from the RT-PCR performed with RNA from the parental strain PB168 (Fig. 8B). These results confirm the interpretation that the Mot+ phenotype shown by the degU32(Hy) suppressed strains is due to expression of the fla-che operon by readthrough transcription driven from the cod operon promoter. For some experiments, a faint band of 948 bp was detected in the reaction mixture from the parental strain RNA, suggesting that some readthrough may occur in wild-type cells.
The significance of transcriptional readthrough in the suppression of the degU32 Mot− phenotype was also supported by the behavior of strain PB5323, in which the dhsA4 mutation was moved away from the cod operon. The strain was derived from the suppressed Mot+ dhsA4 mutant by the insertion of plasmid pPB01, a derivative of pJM114 carrying the complete codY coding sequence, into the chromosome. As a result of a Campbell-type insertion (Fig. 1D), an intact codY gene was present at the original position in the cod operon, whereas the fla-che operon and its regulatory region were separated from the cod operon by plasmid sequences. As a result of this manipulation, the strain derived from the suppressed Mot+ dhsA4 mutant became Mot− (Fig. 3). This result can be interpreted as due to a lack of readthrough and thus a lack of expression of the fla-che operon, whose transcription is repressed by DegU(-P).
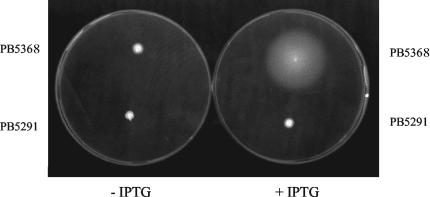
Further support for the interpretation that transcriptional readthrough can account for the suppression of the degU32 Mot− phenotype was obtained by combining the dhsA245 deletion with a codY gene under the control of the Pspac promoter (Fig. 1E). In this genetic background, the suppression (Mot+) was strictly dependent on IPTG induction (Fig. 1E and 9). In the control strain PB5291, with the putative transcription terminator downstream of codY, IPTG induction did not affect the Mot− phenotype (Fig. 1B and 9).
FIG. 9.
The deletion of ΔdhsA245 in combination with Pspac-dependent codY renders the strain IPTG dependent for motility. The plates were prepared as described in the legend to Fig. 3. The IPTG inducer was present at 2 mM.
DegU specifically binds the PA promoter of the fla-che operon.
The results reported above strongly suggest that DegU(-P) acts as a repressor of transcription of the fla-che operon. DegU has been shown to bind to the comK and aprE promoter regions (12, 26). Electrophoretic mobility shift assays were used to monitor the ability of DegU to bind to the fla-che promoter region. Probes of various lengths derived from the intercistronic region between codY and flgB (Fig. 10A) were radiolabeled and incubated with the DegU protein. We used purified DegU and DegU32 as N-terminal His6-tag fusion proteins (Fig. 10B). The same results were obtained with both proteins. In addition, we also tested partially purified DegU and DegU32 proteins without the His tag, again with the same type of results (data not shown).
As shown in Fig. 10C, DegU binds specifically to the DNA region containing the PA promoter of the fla-che operon. The DegU protein failed to bind to probes from upstream or downstream of PA (Fig. 10A); moreover, these DNA fragments failed to compete with the specific probe for DegU binding (Fig. 10C). The binding specificity was further demonstrated by the complete competition of the reaction obtained with an excess of unlabeled DNA probe (Fig. 10C). Thus, we conclude that the PA promoter region of the fla-che operon is a target for DegU binding.
DISCUSSION
The isolation and characterization of suppressor mutations of the Mot− phenotype due to the presence of the degU32(Hy) allele have shed some light on the mechanism of action of DegU on motility. All of the independent mutants isolated were altered in the codY-flgB region of the chromosome.
No additional locus was found to be involved in suppression, strongly suggesting that DegU(-P) is the only trans-acting factor and that it has only one target, the codY-flgB intercistronic region.
The observed mutations fell into two groups, namely, deletions and point mutations. The deletions had different lengths, but all had in common the removal of the transcription termination signal of the cod operon. Our results indicate that as an effect of the deletion, transcription from the cod promoter can continue into the fla-che operon. Since this readthrough is sufficient to allow the production of flagella and the acquisition of the Mot+ phenotype, it means that DegU(-P) acts by preventing transcription of the fla-che operon. This is a simple, straightforward model: DegU(-P) binds to the sequence in the proximity of PA, thus preventing binding of the RNA polymerase and transcription initiation. In fact, by performing a gel retardation analysis, we demonstrated that DegU binds to the PA promoter region of the fla-che operon. DegU has been shown to bind to the comK promoter (12), and more recently, to the aprE promoter (26). In our model, deletion of the transcription terminator allows RNA polymerase to proceed beyond the cod operon and to overcome the block represented by the DegU(-P) repressor bound to the fla-che operator site.
The repressor-operator model was also supported by the effect of two point mutations that changed the −35 hexamer of the PA promoter to a nearly perfect consensus sequence (14). Such a modification should allow a more efficient interaction between the RNA polymerase and the PA promoter and should increase the stability of the closed complex. If the operator sequence were to interdigitate with the promoter sequence, which is possible, a point mutation increasing the affinity of RNA polymerase for the promoter would reduce the effect of steric hindrance of a negative regulator, favoring transcription against repression.
A repressive role of DegU(-P) on the fla-che operon is supported by the results of a whole-genome transcriptional analysis, in which the overexpression of DegU mimicked the phosphorylation state of the response regulator (25). The fla-che operon genes were among those that were down-regulated by the overproduction of DegU (20, 25). Furthermore, the expression of degSU is induced by growth in a high-salinity medium, and under these conditions, transcription of the fla-che operon is strongly repressed (36).
Upon entering stationary phase, several different options are available to the starving bacterial cell. Among these for B. subtilis is the production of extracellular enzymes that scavenge the environment and free new resources. We may assume that the synthesis and secretion of degradative enzymes require a fair amount of energy, which is especially valuable in cells confronted with the depletion of nutrients. A different option consists of motility and chemotactic behavior, enabling the cells to move to new territories. This reaction also requires a large amount of energy, especially to assemble and maintain the flagellar filaments. Hence, it is not unexpected that the two responses are alternative. In fact, motility and enzyme secretion appear to be sequentially activated, with motility reaching its maximal activity at approximately T−1 and the secretion of exoenzymes peaking only after the beginning of the stationary phase. The DegS-DegU two-component system is involved in the regulation of the two alternative adaptive pathways. We suggest that the level of phosphorylated DegU negatively controls motility and activates exoenzyme production. An increase in DegU phosphorylation at the end of the exponential growth phase could be responsible for the observed decrease in transcription of the flagellum-related genes (3). The role of DegU-P on degradative enzyme synthesis is well documented (22). The temporal sequence of events occurring around the transition phase may be dictated by the state of the DegU response regulator, which in turn affects the stability of the protein.
The fla-che operon comprises the majority of the genes whose products are structural components of the flagellum, in addition to several genes involved in chemotaxis. The sigD cistron, coding for σD, is at the end of the 26-kbp operon, and its expression is essential for transcription of the late genes. In addition to being a target of DegU, the fla-che operon promoter region is the target of the CodY nutritional repressor that controls the expression of the operon in response to nutrient availability (4). The high expression level of the fla-che operon observed in strains with the dhsA245 deletion (Fig. 2) is probably a consequence of a lack of control exerted by both CodY and DegU upon the operon. The deletion inactivates the codY gene and removes the transcription terminator, allowing readthrough. The operon is released from the combined effects of CodY and DegU and is transcribed at an unusual level.
The on-off switch of the fla-che operon appears to be the first step in the cascade of events leading to the synthesis of the flagellar components and to the assembly and functioning of the organelle. Thus, the fla-che operon is in a pivotal position in the regulation of motility and chemotaxis in B. subtilis.
Acknowledgments
We are grateful to Eugenio Ferrari for suggestions during the early stages of this work, to Cinzia Calvio for critical readings of the manuscript, and to Elisabetta Andreoli for technical help. We thank the anonymous reviewer who suggested the experiment reported in Fig. 9.
This work was supported by Università degli Studi di Pavia (Fondo d'Ateneo per la Ricerca), the “C.N.R. Target Project on Biotechnology,” and grants from MIUR (Ministero dell'Istruzione, Università e Ricerca, Italy).
REFERENCES
- 1.Allmansberger, R. 1997. Temporal regulation of sigD from Bacillus subtilis depends on a minor promoter in front of the gene. J. Bacteriol. 179:6531-6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ayusawa, D., Y. Yoneda, K. Yamane, and B. Maruo. 1975. Pleiotropic phenomena in autolytic enzyme content, flagellation, and simultaneous hyperproduction of extracellular α-amylase and protease in a Bacillus subtilis mutant. J. Bacteriol. 124:459-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barillà, D., T. Caramori, and A. Galizzi. 1994. Coupling of flagellin gene transcription to flagellar assembly in Bacillus subtilis. J. Bacteriol. 176:4558-4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergara, F., C. Ibarra, J. Iwamasa, J. C. Patarroyo, R. Aguilera, and L. M. Márquez-Magaña. 2003. CodY is a nutritional repressor of flagellar gene expression in Bacillus subtilis. J. Bacteriol. 185:3118-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertin, P., E. Terrao, E. H. Lee, P. Lejoune, C. Colson, A. Danchin, and E. Collatz. 1994. The H-NS protein is involved in the biogenesis of flagella in Escherichia coli. J. Bacteriol. 176:5537-5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caldwell, R., R. Sapolsky, W. Weyler, R. R. Maile, S. C. Causey, and E. Ferrari. 2001. Correlation between Bacillus subtilis scoC phenotype and gene expression determined using microarrays for transcriptome analysis. J. Bacteriol. 183:7329-7340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chilcott, G. S., and K. T. Hughes. 2000. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 64:694-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dahl, M. K., T. Msadek, F. Kunst, and G. Rapoport. 1992. The phosphorylation state of the DegU response regulator acts as a molecular switch allowing either degradative enzyme synthesis or expression of genetic competence in Bacillus subtilis. J. Biol. Chem. 267:14509-14514. [PubMed] [Google Scholar]
- 9.Estacio, W., S. Santa Anna-Arriola, M. Adedipe, and L. M. Márquez-Magaña. 1998. Dual promoters are responsible for transcription initiation of the fla/che operon in Bacillus subtilis. J. Bacteriol. 180:3548-3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fisher, S. H., K. Rohrer, and A. E. Ferson. 1996. Role of CodY in regulation of the Bacillus subtilis hut operon. J. Bacteriol. 178:3779-3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fredrick, K., T. Caramori, Y.-F. Chen, A. Galizzi, and J. D. Helmann. 1995. Promoter architecture in the flagellar regulon of Bacillus subtilis: high-level expression of flagellin by the σD RNA polymerase requires an upstream promoter element. Proc. Natl. Acad. Sci. USA 92:2582-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamoen, L. W., A. F. Van Werkhoven, G. Venema, and D. Dubnau. 2000. The pleiotropic response regulator DegU functions as a priming protein in competence development in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 97:9246-9251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hay, N. A., D. J. Tipper, D. Gygi, and C. Hughes. 1997. A nonswarming mutant of Proteus mirabilis lacks the Lrp global transcriptional regulator. J. Bacteriol. 179:4741-4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helmann, J. D. 1995. Compilation and analysis of Bacillus subtilis σA-dependent promoter sequences: evidence for extended contact between RNA polymerase and upstream promoter DNA. Nucleic Acids Res. 23:2351-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henner, D. J., E. Ferrari, M. Perego, and J. A. Hoch. 1988. Location of the targets of the hpr-97, sacU32 (Hy), and sacQ36 (Hy) mutations in upstream regions of the subtilisin promoter. J. Bacteriol. 170:296-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoch, J. A., M. Barat, and C. Anagnostopoulos. 1967. Transformation and transduction in recombination-defective mutants of Bacillus subtilis. J. Bacteriol. 93:1925-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kunst, F., M. Pascal, J. Lepesant-Kejzlarová, J.-A. Lepesant, A. Billault, and R. Dedonder. 1974. Pleiotropic mutations affecting sporulation conditions and the synthesis of extracellular enzymes in Bacillus subtilis 168. Biochimie 56:1481-1489. [DOI] [PubMed] [Google Scholar]
- 18.Kunst, F., and G. Rapoport. 1995. Salt stress is an environmental signal affecting degradative enzyme synthesis in Bacillus subtilis. J. Bacteriol. 177:2403-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macnab, R. M. 1996. Flagella and motility, p. 123-145. In F. C. Neidhart et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 20.Mäder, U., H. Antelmann, T. Buder, M. K. Dahl, M. Hecker, and G. Homuth. 2002. Bacillus subtilis functional genomics: genome-wide analysis of the DegS-DegU regulon by transcriptomics and proteomics. Mol. Genet. Genomics 268:455-467. [DOI] [PubMed] [Google Scholar]
- 21.Mirel, D. B., W. F. Estacio, M. Mathieu, E. Olmsted, J. Ramirez, and L. M. Márquez-Magaña. 2000. Environmental regulation of Bacillus subtilis σD-dependent gene expression. J. Bacteriol. 182:3055-3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Msadek, T. 1999. When the going gets tough: survival strategies and environmental signalling networks in Bacillus subtilis. Trends Microbiol. 7:201-207. [DOI] [PubMed] [Google Scholar]
- 23.Msadek, T., F. Kunst, D. Henner, A. Klier, G. Rapoport, and R. Dedonder. 1990. Signal transduction pathway controlling synthesis of a class of degradative enzymes in Bacillus subtilis: expression of the regulatory genes and analysis of mutations in degS and degU. J. Bacteriol. 172:824-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishida, S., T. Mitzushima, T. Miki, and K. Sekimizu. 1997. Immotile phenotype of an Escherichia coli mutant lacking the histone-like protein HU. FEMS Microbiol. Lett. 150:297-301. [DOI] [PubMed] [Google Scholar]
- 25.Ogura, M., H. Yamaguchi, K. Yoshida, Y. Fujita, and T. Tanaka. 2001. DNA microarray analysis of Bacillus subtilis DegU, ComA and PhoP regulons: an approach to comprehensive analysis of B. subtilis two-component regulatory systems. Nucleic Acids Res. 29:3804-3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogura, M., K. Shimane, K. Asai, N. Ogasawara, and T. Tanaka. 2003. Binding of response regulator DegU to the aprE promoter is inhibited by RapG, which is counteracted by extracellular PhrG in Bacillus subtilis. Mol. Microbiol. 49:1685-1697. [DOI] [PubMed] [Google Scholar]
- 27.Ordal, G. W., L. Márquez-Magaña, and M. J. Chamberlin. 1993. Motility and chemotaxis, p. 765-784. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, D.C.
- 28.Perego, M. 1993. Integrational vectors for genetic manipulation in Bacillus subtilis, p. 615-624. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, D.C.
- 29.Perego, M., and J. A. Hoch. 1988. Sequence analysis and regulation of the hpr locus, a regulatory gene for protease production and sporulation in Bacillus subtilis. J. Bacteriol. 170:2560-2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petit, M., E. Dervyn, M. Rose, K. Entian, S. McGovern, S. D. Ehrlich, and C. Bruand. 1998. PcrA is an essential DNA helicase of Bacillus subtilis fulfilling functions both in repair and rolling-circle replication. Mol. Microbiol. 29:261-273. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Schumann, W., S. D. Ehrlich, and N. Ogasawara (ed.). 2001. Functional analysis of bacterial genes. A practical manual. John Wiley and Sons, Ltd. Chichester, United Kingdom.
- 33.Serror, P., and A. L. Sonenshein. 1996. CodY is required for nutritional repression of Bacillus subtilis genetic competence. J. Bacteriol. 178:5910-5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Silverman, M., and M. Simon. 1974. Characterization of Escherichia coli flagellar mutants that are insensitive to catabolite repression. J. Bacteriol. 120:1196-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Slack, F. J., P. Serror, E. Joyce, and A. L. Sonenshein. 1995. A gene required for nutritional repression of the Bacillus subtilis dipeptide permease operon. Mol. Microbiol. 15:689-702. [DOI] [PubMed] [Google Scholar]
- 36.Steil, L., T. Hoffmann, I. Budde, U. Völker, and E. Bremer. 2003. Genome-wide transcriptional profiling analysis of adaptation of Bacillus subtilis to high salinity. J. Bacteriol. 185:6358-6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanaka, T., M. Kawata, and K. Mukai. 1991. Altered phosphorylation of Bacillus subtilis DegU caused by single amino acid changes in DegS. J. Bacteriol. 173:5507-5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tokunaga, T., M. H. Rashid, A. Kuroda, and J. Sekiguchi. 1994. Effect of degS-degU mutations on the expression of sigD, encoding an alternative sigma factor, and autolysin operon of Bacillus subtilis. J. Bacteriol. 176:5177-5180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vagner, V., E. Dervyn, and S. D. Ehrlich. 1998. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 144:3097-3104. [DOI] [PubMed] [Google Scholar]
- 40.West, J. T., W. Estacio, and L. M. Márquez-Magaña. 2000. Relative roles of the fla/che PA, PD-3, and PsigD promoters in regulating motility and sigD expression in Bacillus subtilis. J. Bacteriol. 182:4841-4848. [DOI] [PMC free article] [PubMed] [Google Scholar]