Abstract
We have initiated a project to identify protein-protein interactions involved in the pathogenicity of the bacterial plant pathogen Xanthomonas axonopodis pv. citri. Using a yeast two-hybrid system based on Gal4 DNA-binding and activation domains, we have focused on identifying interactions involving subunits, regulators, and substrates of the type III secretion system coded by the hrp (for hypersensitive response and pathogenicity), hrc (for hrp conserved), and hpa (for hrp associated) genes. We have identified several previously uncharacterized interactions involving (i) HrpG, a two-component system response regulator responsible for the expression of X. axonopodis pv. citri hrp operons, and XAC0095, a previously uncharacterized protein encountered only in Xanthomonas spp.; (ii) HpaA, a protein secreted by the type III secretion system, HpaB, and the C-terminal domain of HrcV; (iii) HrpB1, HrpD6, and HrpW; and (iv) HrpB2 and HrcU. Homotropic interactions were also identified for the ATPase HrcN. These newly identified protein-protein interactions increase our understanding of the functional integration of phytopathogen-specific type III secretion system components and suggest new hypotheses regarding the molecular mechanisms underlying Xanthomonas pathogenicity.
A number of species of gram-negative bacteria are able to infect and cause disease in, or establish symbiotic relationships with, specific plant hosts (2). These phytopathogenic bacteria use a full barrage of molecular strategies by which to enter and colonize host tissues. This invasion eventually modifies, and in many cases compromises, plant homeostasis at the tissue level or at the level of the entire plant. Biochemical, genetic, and cellular studies of phytopathogenic bacteria have revealed that these mechanisms involve a variety of factors such as adhesins, pili, bacterial signaling factors, receptors of external and plant-derived factors, proteins involved in signal transduction, specialized transcription factors, alternate sigma factors, and proteins which generate, assemble, and regulate specific macromolecular secretion systems that transport bacterial macromolecular pathogenicity factors (1, 2, 8, 15, 25, 32, 53, 67). Only a few of these processes are understood fully at the molecular level and, in the few cases where they have been elucidated for more than one system, significant differences have been observed (32, 39)—differences that must eventually be attributed to the specific biologies of the interacting bacterium-plant pair.
During the infective process, a large variety of pathogenic gram-negative bacteria inject macromolecular pathogenic factors into their animal or plant host cells. Bacteria may use one of two systems to accomplish this: the type III (6, 12, 20, 25, 26, 32, 46) and type IV (3, 10, 11, 14, 18, 19, 60, 65, 69) secretion systems. Recently, the genomes of a number of bacterial phytopathogens have been sequenced, including Xylella spp. (52, 58), Ralstonia solanacearum (49), Xanthomonas spp. (21), Agrobacterium tumefaciens (66), and Pseudomonas syringae (9). These sequences have revealed the existence of several gene clusters that code for putative macromolecule secretion systems that are demonstrably or possibly involved in pathogenesis. The type III secretion system (TTSS) is coded for by a group of ca. 25 genes, most of which are localized to a single chromosomal locus (hrp). A subgroup of products encoded by these genes consists of homologues of the core flagellar secretory components, which has led to the conclusion that the TTSS and flagellar machines are evolutionarily related (6, 42, 57). In contrast, the type IV secretion systems (TFSS) responsible for secreting pathogenicity factors are related to the machines responsible for transfer of nucleic acid-protein complexes during bacterial conjugation (19).
We identify here new protein-protein interactions involving components, substrates, and regulators of the TTSS of the phytopathogen Xanthomonas axonopodis pv. citri, the causal agent of citrus canker. In the plant pathogens Xanthomonas, Ralstonia, Erwinia, and Pseudomonas spp., a subset of hrp genes are induced upon contact with the plant in response to a variety of diffusible or nondiffusible plant-derived factors (8, 25, 32, 51). Beyond their requirement for pathogenicity, the mechanisms of action of most of the components of this secretion machine are not well understood at the molecular level. This is especially true for phytopathogen-specific components with no homologs in animal TTSS or in flagella. Interactions among some components of TTSS from several bacterial pathogens have been elucidated, and in many cases these interactions have revealed possible functional roles of specific components (34). However, the fact that several of the gene products of these clusters are only distantly related, if at all, to other proteins of known function places limits on the homology-based approach for deduction of function. This limitation is further highlighted by the fact that the X. axonopodis pv. citri hrp cluster (Fig. 1) possesses open reading frames (ORFs) that code for previously uncharacterized proteins, in several cases specific to xanthomonads and in some cases specific to X. axonopodis pv. citri. To understand the specific mechanisms by which X. axonopodis pv. citri uses its TTSS to interact with and modify the metabolism of its host, it is of utmost importance to delimit the protein-protein interactions involving X. axonopodis pv. citri TTSS components and associated proteins. To this end, we have used a yeast two-hybrid system (17) to perform genome-scale protein-protein interaction screens by using specific X. axonopodis pv. citri hrp, hrc, and hpa proteins as baits. Our results reveal a number of previously uncharacterized interactions involving these proteins that may be of importance to the processes of hrp gene expression and TTSS function.
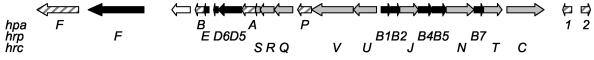
FIG. 1.
hrp locus of X. axonopodis pv. citri (21). Arrows: ▪, hrp (for hypersensitive response and pathogenicity) genes; ░⃞, hrc (for hrp conserved) genes; ▨, hpa (for hrp-associated) genes; □, genes coding for hypothetical proteins.
MATERIALS AND METHODS
X. axonopodis pv. citri (strain 306) genomic DNA library.
X. axonopodis pv. citri strain 306 genomic DNA was nebulized under nitrogen, repaired, and size separated in agarose gels. Fragments of 500 to 1,500 bp and 1,500 to 3,000 bp were purified and cloned into the plasmid vector pOAD (56) previously linearized with PvuII and dephosphorylated with calf intestine alkaline phosphatase. Then, 1-μl aliquots of the ligation reactions were used to transform competent Escherichia coli DH10B cells by electroporation. Cells from each set of 10 transformations were pooled and, after the addition of 1 volume of 50% glycerol, stored at −70°C. Before freezing, 20-μl aliquots were separated, frozen at −70°C, thawed, and plated to determine the total number of independent clones in that pool, which varied from 2,000 to 20,000. Transformations were performed until each library (500 to 1,500 bp and 1,500 to 3,000 bp) contained >106 independent clones.
Transformants were thawed and pooled into groups containing between 100,000 and 200,000 independent clones. These were diluted into 16 2-liter flasks containing a total of ca. 10 liters of 2xTY medium supplemented with 200 μg of carbenicillin/ml. After growth at 37°C for 8 h, 10-ml aliquots were removed from each flask, combined, diluted with 1 volume of 50% glycerol, and stored at −70°C. The rest of the culture was incubated at 37° until it reached an optical density at 600 nm of 1.2, at which point the cells were collection by centrifugation, and the plasmid DNA was purified (50). HindIII digests of each preparation were analyzed by agarose gel electrophoresis. DNA concentrations were determined, and preparations from all of the libraries were pooled in amounts proportional to the number of independent clones per unit mass of DNA. This pOAD-library mixture was used to transform yeast cells in two-hybrid screens.
Cloning of baits for two-hybrid screens.
X. axonopodis pv. citri DNA sequences coding proteins for use as baits were amplified by PCR by using X. axonopodis pv. citri genomic DNA and primers designed based on the X. axonopodis pv. citri genome sequence (21). The primers also contained unique restriction sites (usually NcoI and XhoI) to facilitate cloning into the NcoI and SalI sites of the pOBD vector (56) downstream of and in frame with the Gal4 DNA-binding domain. After transformation into DH10B E. coli cells, individual colonies were picked for plasmid isolation and confirmation by DNA sequencing. Most baits corresponded to the full-length proteins as annotated in the X. axonopodis pv. citri genome sequence (21). In some cases, analysis of the protein sequence by using the PSORT algorithm (43) indicated the presence of domains containing one or more putative transmembrane helices and protein fragments lacking the transmembrane helices were used. These cases are described in the text and in Table 1. In the case of XAC0095, the bait used contained a 12-amino-acid N-terminal extension encoded by the 36 nucleotides upstream of the annotated start codon. This 85-amino-acid bait is therefore coded for by nucleotides 111185 to 111442 of the X. axonopodis pv. citri chromosome.
TABLE 1.
Summary of protein-protein interactions involving X TTSS components observed in this studya
| Bait | Bait gene no. | Total no. of positive preys sequencedb | Specific prey(s) (name, gene no., no. of preys derived from this protein)c |
|---|---|---|---|
| TTSS and related proteins | |||
| HrpG | XAC1265 | 90 | HrpG, XAC1265, 48 (48); XAC0095, XAC0095, 36; two-component system histidine kinase, XAC3683, 1; XAC1568, XAC1568, 5 |
| XAC0095d | XAC0095 | 72 | XAC0095, XAC0095, 62 (61); XAC0095b, XAC0095b, 1; HrpG, XAC1265, 3; helix-turn-helix, XAC0524, 6 |
| HrpB2 | XAC0408 | 10 | HrcU, XAC0406, 4; 4-hydroxyphenylpyruvate dioxygenase, XAC0452, 5 |
| HrpB1 | XAC0407 | 29 | HrpD6, XAC0398, 10; HrpW, XAC2922, 2; PHA synthase subunit, XAC2047, 6; M13 metallopeptidasee, XAC3712, 3 |
| HrpB4 | XAC0410 | 10 | Two-component system composite sensor/His kinase/RR, XAC2054, 6 |
| HrpD5 | XAC0399 | 17 | DNA polymerase III β-subunit,e XAC0002, 9 |
| HrcN | XAC0412 | 6 | HrcN, XAC0412, 4 |
| HpaA | XAC0400 | 40 | HpaB, XAC0396, 40 |
| HpaB (full-length) | XAC0396 | 49 | HpaA, XAC0040, 37; HrcV, XAC0405, 12 |
| HrcV (residues 325 to 646) | XAC0405 | 18 | HrcV, XAC0405 (14); HpaB, XAC0396, 1 |
| Positive controls | |||
| FtsZ | XAC0784 | 13 | FtsZ, XAC0784, 11 (3) |
| DNA polymerase III δ′ subunit | XAC1132 | 4 | DNA polymerase δ′ subunit, XAC1132, 1 |
| GroES | XAC0541 | 24 | GroES, XAC0541, 22 (21) |
Only relevant interactions mentioned in the text are shown. Therefore, the number of listed preys may not equal the total number of preys sequenced. Gene number is as defined in the annotated X. axonopodis pv. citri genome (21). PHA, polyhydroxyalkanoic acid.
Not including false-positive preys mentioned in the text.
When applicable, the number of preys derived from recombination with the bait vector is indicated in parentheses.
Bait with 12-amino-acid N-terminal extension (see the text).
Not mentioned in the text.
Growth of yeast strains and transformation.
Saccharomyces cerevisae strain PJ694-a (MATa trp1-901 leu2-3 112 ura3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ) (35) was grown at 30°C in YAPD medium (1% yeast extract, 2% peptone, 2% glucose, 0.008% adenine) or in SC medium (0.66% nitrogen base [without amino acids], 2% glucose, 0.008% adenine, 0.8% amino acid mixture adjusted to pH 5.6) as described previously (45). Where indicated, SC medium was prepared lacking one or more specific components: adenine (−Ade), histidine (−His), tryptophan (−Trp), and leucine (−Leu). In the case of growth on solid media, 1.6% Bacto Agar and 3-aminotriazole (3AT) (see below) was added. Rapid transformations with pOBD-bait plasmids were carried out by using the PEG3350-lithium acetate protocol described by Gietz et al. (28) and selected on SC−Trp plates at 30°C for 2 to 4 days. These cells were then used in high-efficiency transformations with the pOAD-library using 30 μg of plasmid DNA and the 30× scale-up procedure described by Gietz and Woods (27), which resulted in, on average, 0.5 × 107 to 1 × 107 transformants on SC−Trp−Leu plates.
Tests for autoactivation and false-positive results.
To determine the amount of 3AT to be used for each bait, ca. 1,000 yeast cells transformed with the pOBD-bait plasmid were plated on SC−Trp−His medium containing 0, 1, 5, 10, 25, or 50 mM 3AT and incubated for 5 days at 30°C. Similarly, the small number of baits that autoactivate the GAL2-ADE2 reporter were identified by growth on SC−Trp−Ade plates. The pOAD-library was shown to lack any clones that autoactivate the His or Ade gene reporters on their own or simultaneously. On the other hand, when transformed into PJ694-a cells previously transformed with the pOBD vector (lacking a bait insert), ca. 1,000 colonies were found to grow in the presence of up to 3 mM 3AT. Sequencing of the preys in 66 of these colonies subsequently showed them to be derived from only a few specific X. axonopodis pv. citri genes (see Results).
Yeast two-hybrid assays and DNA sequencing.
After transformation with the pOAD-library, cells were resuspended in 5 ml of sterile water and spread on 10 plates with SC−Trp−Leu−His−Ade plus 3AT (150 by 15 mm). The amount of 3AT used varied between 5 and 50 mM, depending on the bait: 5 mM for all baits except for HpaA (10 mM) and HrcN (50 mM). Plates were incubated at 30°C for up to 14 days. Colonies that grew in the absence of His and Ade were transferred to fresh plates with SC−Trp−Leu−His−Ade plus 3AT. Plasmid DNA was isolated from yeast colonies that were able to grow under the selection of both reporters by using a method (50) scaled up and modified for execution in 96-well plates. Purified plasmid DNA mixtures were used to transform DH10B E. coli cells. After overnight growth at 37°C, plasmid DNA (a mixture of pOAD- and pOBD-derived vectors) was purified. The prey or bait DNA sequences were sequenced by using pOAD- or pOBD-specific primers, respectively. Sequences were analyzed by comparison with the X. axonopodis pv. citri genome database (21).
RESULTS AND DISCUSSION
X. axonopodis pv. citri two-hybrid library construction.
We constructed a X. axonopodis pv. citri total genomic DNA library (X. axonopodis pv. citri chromosome plus plasmids) containing fragments of 500 to 3,000 bp cloned into the PvuII site of the vector pOAD (56). The quality of the library was tested in a number of independent ways. (i) HindIII digests of the sublibraries after amplification but before pooling revealed a broad population of inserts of the expected sizes (500 to 1,500 bp or 1,500 to 3,000 bp). (ii) Initial sequencing of ca. 1,000 independently chosen clones did not reveal any obvious bias in the library and that, of the 3.0 million independent clones in the combined library, 78% (2.35 million) contained X. axonopodis pv. citri genomic DNA inserts. In the final pooled library, fragments in the size ranges of 500 to 1,500 bp and 1,500 to 3,000 bp were present in about equal proportions (1.2 and 1.15 million, respectively). If we assume a random distribution of fragmentation sites in the genome, the library contains fragments initiating every 2.2 bp or every 13 bp in a correct reading frame. Although some in-frame clones code for full-length proteins with N-terminal extensions, most fragments code a protein with an N-terminal deletion. (iii) Subsequent analysis of two-hybrid results revealed that, except for some special cases due to yeast-based recombination events (see below), clones of preys derived from a specific gene initiated at many unique sites within that gene. (iv) The X. axonopodis pv. citri genomic library in pOAD does not contain any clones that on their own activate the HIS3 and ADE2 reporter genes individually. However, in the presence of the empty pOBD vector, some library clones did lead to activation. In an experiment of a total of 2 × 107 transformations, ca. 1,000 colonies were observed to grow on plates with SC−Trp−Leu−His−Ade plus 3AT. Sequencing of the pOAD-derived vector from 62 of these colonies revealed that all contained DNA inserts derived from only a small number of X. axonopodis pv. citri genes: 89% of the preys coded for fragments of the TolC protein, 6.5% of the preys coded for fragments of the RibA protein, and single clones were found that were derived from the phoQ, glnA, and piuB genes. TolC-derived fragments seem to be promiscuous interactors in two-hybrid assays and should be considered false positives, as shown below. RibA preys have thus far only been detected when empty pOBD is used as bait.
Identification of false positives.
In using this X. axonopodis pv. citri genomic DNA library, we identified two classes of prey that we consider to be false positives. One class is composed of specific preys that appear to interact with a great number of apparently unrelated baits with a frequency that does not appear to be of physiological relevance, at least in the context of the physiological role of the baits. Of more than 100 baits tested thus far with this library (data not shown), ∼40% interacted with at least one prey derived from the TolC protein and, for several baits, the major fraction of interactors was derived from TolC. Due to the ubiquity of these sequences in the two-hybrid screens, these sequences were considered to be false positives and therefore disregarded. TolC tends to interact with the Gal4 DNA-binding domain (see above), and this tendency may be inhibited or enhanced by bait fusions. In addition to TolC, several other proteins were detected as preys at a frequency to warrant suspicion of their physiological significance. These proteins included (i) members of the large TonB-dependent receptor family, (ii) products of the two wapA genes (XAC1866 and XAC1305), (iii) the XAC3515 protein, and (iv) the plasmid-encoded PthA and KfrA proteins. For this reason, these preys were also considered to be false positives and are not considered further.
The majority of baits interacted with a large number of preys, most of which were derived from a small number of X. axonopodis pv. citri proteins (Table 1). In the case of some baits, however (for example, HrpF, Hpa2, and the N-terminal fragment of HpaB), no clear preference for any specific X. axonopodis pv. citri protein was observed, and almost all of the preys sequenced were derived from a different protein (data not shown). The apparent lack of specificity of these baits suggests that the majority of their preys do not reflect physiologically relevant interactions. In such a case, even if a small number of the preys of such a bait do in fact represent a true interactor, this fact is not immediately apparent from these data alone and must be confirmed by other experimental approaches.
Positive controls.
Three proteins known to participate in well-characterized protein-protein interactions were initially used as positive controls for the two-hybrid screen itself, as well as to further test the quality of the library. All three baits tested—GroES, the δ′ subunit of DNA polymerase III, and FtsZ—were found to interact with previously identified interacting partners: GroES with itself in 22 of 24 clones sequenced, the δ′ subunit of DNA polymerase III (36) with the product of the gene coding for the γ and τ subunits of the polymerase in one of four clones sequenced, and FtsZ (29) with itself in 11 of 13 preys sequenced (Table 1).
Prey-bait vector recombination events.
Detailed sequencing analysis of the Gal4 activation domain-prey junction in the prey vector could demonstrate that for some baits, including FtsZ and GroES controls, a fraction of the preys were derived from recombination events in which the bait gene was transferred into the pOAD vector (3 of 11 for FtsZ and 21 of 22 for GroES). Recombination is facilitated by the fact that pOBD and pOAD vectors (56) have identical sequences before (∼60 bp) and after (∼2,840 bp) their multiple cloning sites. To test whether these in vivo recombinations necessitate a specific library clone or could occur even with the empty pOAD vector, we transformed PJ694-a cells with both the empty pOAD plasmid and the pOBD-XAC0095 plasmid. XAC0095 is an X. axonopodis pv. citri conserved hypothetical protein that was observed to suffer the same type of recombination event (see below). Of a total of 5 × 106 transformants, 25 colonies were observed to grow on plates with SC−Trp−Leu−Hi−Ade plus 3AT. Sequencing of the pOAD vector in 16 of these clones revealed that they all now contained the full-length XACb0095 bait in the specific site in which it was cloned in the pOBD vector (distinct from the PvuII site in which the genomic DNA library was cloned in pOAD). Therefore, full-length copies of the bait in both vectors can be obtained via recombination of the full-length bait with, in principle, any library clone, including the empty pOAD vector if the bait protein can interact with itself forming dimers or higher-order complexes (as is in fact known to be the case for FtsZ and GroES). This phenomenon was observed for a few other TTSS-related proteins (see below).
Two-hybrid assays of proteins involved in the type III secretory pathways.
Two-hybrid assays were performed by using simultaneous screening of two reporter genes under the control of different inducible promoters (GAL1-HIS3 and GAL2-ADE2). This simultaneous screening in combination with the high stringency of the GAL2-ADE2 reporter significantly reduced the number of false positives (35). Only baits that did not simultaneously autoactivate these two reporters were tested in screens against the prey library. Table 1 lists the TTSS-related proteins analyzed and summarizes the results obtained in two-hybrid assays in which they were used as baits. The table presents the total number of positive interactions in which each prey was identified. False positives, as defined above, are not indicated. For some baits capable of homotropic interactions, the number of preys derived from recombination of the bait gene into the pOAD vector is indicated. In assays necessitating the simultaneous activation of the GAL1-HIS3 and GAL2-ADE2 gene reporters, interactions observed at least two times with preys derived from the same X. axonopodis pv. citri protein were considered to be of potential significance, especially if the physiological significance of the interaction was apparent. What follows is a detailed description of these results and their significance.
Interactions involving HrpG.
HrpG is a DNA-binding transcriptional regulatory protein that functions at the top of the hrp gene cluster regulatory cascade, controlling the expression of the hrpA gene in X. campestris pv. vesicatoria (hrcC in X. axonopodis pv. citri), as well as the gene for the downstream transcription regulator HrpX (62, 63). HrpX in turn activates expression of the hrpB-F loci, as well as of a number of Xanthomonas outer protein (xop) genes in X. campestris pv. vesicatoria (61). The 263-residue HrpG protein of X. axonopodis pv. citri, which belongs to the OmpR family of two-component system response regulators, contains an N-terminal response regulator receiver (RR) domain and a C-terminal DNA-binding motif (62). Although the means by which the Xanthomonas HrpG protein is regulated remains unknown, its homolog in R. solanacearum has been shown to be regulated by a cell-contact signal transduction cascade involving PrhA (a probable receptor for insoluble host factors), PrhR (a transmembrane protein that receives the signal from PrhA), PrhI (an ECF sigma factor), and PrhJ (a LuxR/UhpA family transcription regulator) (1, 7, 8). PrhA, -R, and -I are homologues of the FecA, -R, and -I proteins, which interact in the signaling pathway that controls genes involved in ferric citrate transport in E. coli (22, 54). In spite of this, no direct protein-protein interactions involving HrpG in Xanthomonas or Ralstonia spp. have as yet been identified. Furthermore, no candidate two-component system histidine kinase has been identified which could in principle regulate HrpG via phosphorylation of its RR domain.
When the full-length HrpG protein was used as bait, all preys were derived from one of four proteins (Table 1 and Fig. 2). Just over half of the preys (48 of 90) were derived from identical clones that code for the full-length HrpG protein. Sequencing analysis of the sequence coding for the junction between the Gal4 activation domain and the HrpG coding sequence demonstrated that all of these clones were derived from recombination events between the pOBD-HrpG bait plasmid and pOAD in which the full-length HrpG gene plus 25 nucleotides of the pOBD polylinker was transferred to the pOAD vector. HrpG-HrpG interactions have not been observed before. However, OmpR family proteins are known to bind to DNA as dimers, although dimerization has not been observed in solution (31, 37).
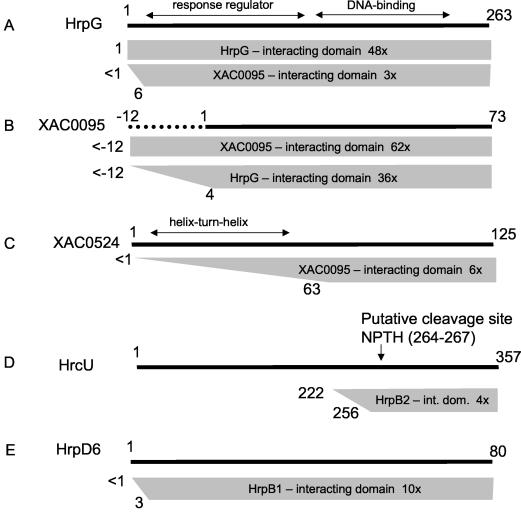
FIG. 2.
Summary of interactions observed in yeast two-hybrid assays involving HrpG, XAC0095, XAC0524, HrcU, HrpD6, HrpB2, and HrpB1. (A) Preys derived from HrpG that interacted with baits derived from HrpG and XAC0095; (B) preys derived from XAC0095 that interacted with baits derived from XAC0095 and HrpG (the XAC0095 bait used had a 12-amino-acid N terminus extension as described in the text); (C) preys derived from XAC0524 that interacted with the XAC0095 bait; (D) preys derived from HrcU that interacted with the bait derived from HrpB2; (E) preys derived from HrpD6 that interacted with the HrpB1 bait. Shaded trapezoids or rectangles indicate the maximum and minimum fragments within the set of preys that interacted with a specific bait. Numbers to the left of the trapezoid indicate the maximum and minimum positions of the N-terminal boundaries of the preys. The specific bait used is indicated within the trapezoid. Also indicated is the number of clones sequenced derived from that particular prey. The full-length prey is represented as a solid black line above.
The second largest set of preys (36 of 90) obtained using HrpG as bait were derived from the hypothetical protein (XAC0095), whose only known homologs are coded by three ORFs in the X. campestris pv. campestris genome (Fig. 3) (21). XAC0095 is a 73-residue protein (Fig. 3), and all of these clones coded for at least residues 4 to 73 (Fig. 2B). The HrpG-XAC0095 interaction was confirmed when we used the XAC0095 protein with a 12-residue N-terminal extension as bait: 3 of 71 prey clones coded for a fragment of either the full-length HrpG protein or a near full-length fragment beginning at amino acid residue 6 (Fig. 2A). Two factors may contribute to the relatively low fraction of preys mapping to HrpG: (i) the HrpG RR domain is located in the N-terminal portion of the protein (residues 11 to 130) and may be required for interaction with XAC0095, and (ii) the presence of an in-frame stop codon immediately upstream from the start codon (TAAATG) eliminates all clones containing inserts that begin upstream from the start codon. If HrpG-HrpG interactions are also mediated through N-terminal domain contacts, this second factor could also explain the predominance of recombination-derived full-length HrpG preys when HrpG is used as bait (above).
FIG. 3.
Primary structure alignment of XAC0095 with its homologs in X. axonopodis pv. citri (XAC0095b and XAC0095c) and in X. campestris pv. campestris (XCC0067, XCC1070, and XCC2900). Amino acid positions with identical (*), highly conserved (:), and less-well-conserved (.) residues are indicated. Two blocks of particularly well conserved residues are indicated by thin horizontal lines. Both blocks contain conserved heptad pseudorepeats (abcdefg) in which each a and d position is almost invariably a hydrophobic residue. The two heptad repeats are delimited with thick black horizontal lines. The C-terminal block has two hydrophobic heptad repeats superimposed on one another.
Five HrpG preys were derived from XAC1568, a 157-residue conserved hypothetical protein (Table 1). All five preys began at either residue 64 or 65, indicating that the C-terminal domain of XAC1568 mediates its interactions with HrpG. Interestingly, the N-terminal domain contains several well-conserved cysteine residues found in all homologs from a large number of bacteria, whereas the C-terminal domain is present in only a much smaller group of proteins found in X. campestris pv. campestris, A. tumefaciens, Mesorhizobium spp., Sinorhizobium spp., Caulobacter crescentus, Bradyrhizobium japonicum, Rhodopseudomonas palustris, Rhodospirillum rubrum, Rhodobacter sphaeroides, and P. aeruginosa. None of these homologs, however, have known functions.
Finally, HrpG was also found to interact with a single prey derived from the XAC3683 gene that encodes a two-component system composite sensor-histidine kinase/response regulator (Table 1). This result is consistent with a probable interaction between HrpG and a two-component system histidine kinase phosphoacceptor domain. The domain structure of XAC3683 includes a N-terminal PAS and PAC motifs frequently associated with signal sensor domains (47), central histidine kinase and phosphoacceptor domains, and a C-terminal RR receiver domain (44, 55). The fragment of XAC3683 detected as a positive prey begins at residue 96 located just before the first PAS domain (residues 110 to 172). This is the first time that HrpG has been shown to interact with a specific sensor protein and opens up the way to our understanding of the complete transmission pathway from external effector to hrp gene expression. Since only 1 of 90 HrpG preys was derived from XAC3683, the significance of this interaction will have to be confirmed in other biochemical tests.
Since 40% of HrpG preys were derived from XAC0095, we decided to use this protein as a bait in two-hybrid screens against the X. axonopodis pv. citri genomic prey library. The bait we used contains a 12-amino-acid N-terminal extension (Fig. 2B). The vast majority (62 of 72) of XAC0095 preys were mapped to the XAC0095 gene (Table 1 and Fig. 2). Of these, all clones coded for the whole XAC0095 ORF. Sequencing analysis could demonstrate that all but one of these preys are derived from recombination events in which the XAC0095 bait gene was transferred into the pOAD vector (Table 1 and see above) and that a single prey was derived from the library. This library-derived clone codes for the full-length XAC0095 protein plus a 23-residue N-terminus extension.
The XAC0095 bait also interacted with a single prey that is coded for by a previously unannotated ORF that has 45% identity with XAC0095. This ORF, which we name XAC0095b, is located between nucleotides 1331630 and 1331896 of the X. axonopodis pv. citri chromosome and codes for an 88-amino-acid protein (Fig. 3), residues 8 to 88 of which are coded by the XAC0095 prey. Interestingly, further analysis of the X. axonopodis pv. citri genome sequence allowed us to identify a third, also previously unidentified, X. axonopodis pv. citri ORF (which we name XAC0095c) between nucleotides 5030863 and 5031120 that codes for an 85-residue homolog with 35% identity with XAC0095 and 40% identity with XAC0095b (Fig. 3). Thus, both X. axonopodis pv. citri and X. campestris pv. campestris (see above) seem to have three XAC0095 homologs. An alignment of these three putative X. axonopodis pv. citri proteins and their corresponding homologs in X. campestris pv. campestris is shown in Fig. 3. Some specific features of the alignment are of interest. (i) There appear to be two blocks of particularly well conserved residues: one 29-residue block in the N-terminal half of the protein and one 23-residue block in the C-terminal half (indicated by thin horizontal lines in Fig. 3). Within these two blocks, 19 residues are absolutely conserved in all six homologs. (ii) Each N-terminal block is flanked by proline residues on both sides. The C-terminal blocks in all six proteins are also flanked by prolines on their N-terminal sides, and three are flanked by prolines on their C-terminal sides as well (Fig. 3). These conserved prolines may correspond to turns in specific structural features in these homologs. (iii) All six homologs have a conserved cysteine residue in their C-terminal block. (iv) Both blocks contain conserved heptad pseudo-repeats (abcdefg) in which each a and d position is almost invariably a hydrophobic residue (Fig. 3). In fact, the C-terminal block has two hydrophobic heptad repeats superimposed on one another (Fig. 3). Such heptad repeats mediate side-by-side hydrophobic interactions between amphipathic alpha-helices in coiled-coil proteins (41).
Finally, XAC0095 baits interacted with six preys derived from the XAC0524 protein (Table 1 and Fig. 2C). This uncharacterized 125-residue protein contains a helix-turn-helix DNA-binding domain in its N-terminal (residues 8 to 60) that is found in a number of transcription regulators, including the bacteriophage repressor proteins Cro, C1, and C2. The C-terminal half of the protein, however, has only a few homologs in the public databases. Of these, only the E. coli DicA protein (30% identity with XAC0524), derived from a cryptic prophage, has been characterized (5). The minimal XAC0524 domain observed to interact with XAC0095 is delimited by residues 63 to 125, the C-terminal half of the protein that follows the putative DNA-binding domain (Fig. 2C).
The function of the XAC0095 protein and its homologs is not known at the moment, but its interaction with HrpG and XAC0524 strongly suggest a role in the control of gene expression, particularly hrp gene expression. Since HrpG is a bifunctional protein with both a receiver/response regulator and DNA-binding domains, its interaction with XAC0095 could serve to modulate its interactions with downstream factors associated with the hrpX and hrcC/hrpA gene promoters, as observed in X. campestris pv. vesicatoria (62), or with upstream effectors such as a two-component system sensor or histidine kinase domains. XAC0095 could stabilize HrpG or stabilize the phosphoryated or dephosphorylated state of specific aspartic acid residues in its RR receiver domain (Asp60 in X. axonopodis pv. citri HrpG). Phosphorylation of RR receiver domain Asp residues is known to modulate the functions of neighboring effector domains in two-component signal transduction pathways (44, 55).
HrpB2 interacts with a C-terminal domain of HrcU that is proteolytically cleaved in HrcU homologs.
HrpB2 is a substrate of the type III secretion system: in X. campestris pv. vesicatoria, it is secreted to the exterior of the bacterial cell and is necessary for secretion of other proteins via the TTSS (48). HrpB2 homologs are found only in Xanthomonas spp. and R. solanacearum and are essential for pathogenesis in X. campestris pv. vesicatoria (48). When HrpB2 was used as bait in the two-hybrid assay, 4 of 10 clones sequenced were found to map to HrcU, a conserved member of the TTSS superfamily associated with the bacterial inner membrane (Table 1). All four clones code for HrcU fragments beginning between residues 222 and 256, and all terminate downstream of the termination codon (Fig. 2D). Interestingly, the HrcU paralog FlhB has been shown to have a direct role (along with FliK) in the switch that determines which substrates (hook versus filament subunits) are secreted by the type III secretion systems responsible for flagellar assembly (64). Another HrcU paralog, YscU is necessary for the secretion of Yersinia anti-host factors (Yops) (40). Both FlhB and YscU undergo site-specific proteolytic cleavage at a conserved Asn-Pro-Thr-His sequence, also found in X. axonopodis pv. citri HrcU (residues 264 to 267). In fact, three of four of the preys found to interact with HrpB2 map almost perfectly to this putative C-terminal cleavage fragment in HrcU (Fig. 2D). In Yersinia, overexpression of a full-length uncleavable form of YscU inhibits growth, whereas overexpression of the C-terminal cleavage fragment results in increased Yop secretion (40). It is therefore intriguing that a HrcU fragment possibly involved in the control of type III secretion system substrate specificity interacts physically with a known substrate (HrpB2) that, in turn, is required for the secretion of other proteins by this pathway (48). This direct physical interaction has not been demonstrated previously.
Interactions between HrpB1, HrpD6, and HrpW.
Preys obtained when HrpB1 is used as bait mapped to a number of different X. axonopodis pv. citri proteins (Table 1 and data not shown). Of 29 sequenced preys, 10 were derived from the HrpD6 protein, also coded for by the hrp locus. Both HrpB1 and HrpD6 are small (130 and 80 residues, respectively) cytoplasmic proteins whose homologs are essential for pathogenicity in X. campestris pv. vesicatoria (48). However, neither of the proteins has homologs in the flagellar apparatus or in the TTSSs of animal pathogens, and no functional analyses of these two proteins are available in the literature. In fact, HrpD6 homologs have been identified thus far only in Xanthomonas spp., whereas HrpB1 homologs are only found in Xanthomonas spp., as well as in R. solanacearum and Burkholderia pseudomallei. The smallest HrpD6 fragment found to interact with HrpB1 corresponded to residues 3 to 80 (Fig. 2E). The interactions between these two proteins, identified here for the first time, may indicate that they act together in a common function. Unfortunately, when HrpD6 was used as a bait in two-hybrid screens, all eight preys were derived from the false-positive TolC protein (data not shown).
Two other HrpB1 preys were derived from clones coding for a full-length X. axonopodis pv. citri HrpW protein (XAC2922) (Table 1). In spite of its name, the X. axonopodis pv. citri hrpW gene is not situated in the hrp locus. However, in X. campestris pv. campestris, the hrpW gene is located within the hrp locus, adjacent to the hpaB gene (21). In P. syringae and Erwinia amylovora, the >41-kDa HrpW protein binds to pectate, and its TTSS-dependent secretion can elicit the plant hypersensitive response (16, 38). The harpin domain in HrpW of P. syringae has seven glycine-rich repeats between residues 119 and 188. However, HrpW from X. axonopodis pv. citri (33 kDa) and X. campestris pv. campestris (36 kDa), which share 46% identity and 59% similarity, do not possess complete harpin domains and contain fewer glycine-rich repeats. The pectate lyase domain in X. axonopodis pv. citri and X. campestris pv. campestris HrpW seem complete however and shows >30% identity with the corresponding domain in the Pseudomonas and Erwinia HrpW proteins, as well as with pectate lyase of Bacillus spp. The interaction between HrpB1, a cytosolic protein, and HrpW, a homolog of known TTSS substrates, may indicate that HrpB1 (and perhaps HrpD6) are involved in directing HrpW and other substrates to the TTSS apparatus (see Fig. 5, below). Finally, we note that six of the HrpB1 preys were derived from a protein (XAC2047, 407 amino acids) with only very little identity (23% identity, 40% similarity) with a putative polyhydroxyalkanoate synthase subunit PhaE from Ectothiorhodospira shaposhnikovii (unpublished, gi|11096253) (Table 1 and see below).
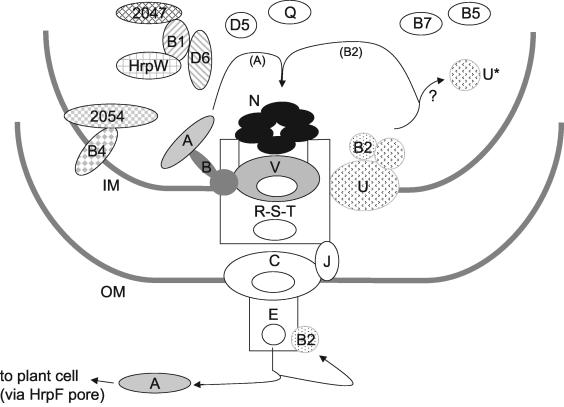
FIG. 5.
Summary of the interactions observed in the present study and their integration into a functional model of the Xanthomonas spp. TTSS derived from the work from several laboratories (2, 6, 12, 13, 32, 33, 48). Proteins involved in interactions identified in the present study are represented by shaded objects, whereas white objects represent proteins not studied here. HpaA secretion may be mediated via interactions with HpaB, which in turn interacts with the pore-forming component HrcV. HrpB2 secretion may be mediated via interactions with the C-terminal domain of HrcU. HrcU homologs in flagella and Yersinia TTSSs suffer proteolysis in which the C-terminal domain is released. Specific interactions between HrpB1, HrpD6, the pectate lysase harpin homolog HrpW, and XAC2047 are shown. Also shown are interactions with between HrpB4 and XAC2054, a two-component system sensor/histidine kinase/response regulator. Note that XAC2047 and XAC2054 are near neighbors on the X. axonopodis pv. citri chromosome. See the text for details. Legend: A, HpaA; B, HpaB; B1, HrpB1; B2, HrpB2; B4, HrpB4; B5, HrpB5; B7, HrpB7; D5, HrpD5; D6, HrpD6; E, HrpE; J, HrcJ; N, HrcN; Q, HrcQ; R, HrcR; S, HrcS; T, HrcT; U, full-length HrcU; U*, C-terminal proteolytic fragment of HrcU; V, HrcV; IM, inner membrane; OM, outer membrane.
HrpB4.
HrpB4 is a 209-residue protein highly conserved (>90% identity) in Xanthomonas spp. Its only other known homolog is the HrpH protein of R. solanacearum. HrpB4 has been shown to fractionate mostly to the soluble fraction of X. campestris pv. vesicatoria lysates (48), although primary structure analysis has detected putative transmembrane helices (13). When used as bait, HrpB4 was found to interact six times (out of a total of ten) with XAC2054. XAC2054 is a two-component system composite sensor/histidine kinase/response regulator. The domain architecture of XAC2054 includes multiple N-terminal PAS, PAC, and GAF domains frequently associated with signal sensors, central histidine kinase, and phosphoacceptor domains and a C-terminal RR receiver domain. The smallest HrpB4-interacting fragment begins at residue 313 of this 1,127-amino-acid protein. Interestingly, the XAC2054 gene is located 3.6 kb from the XAC2047 gene whose product was observed to interact with HrpB1 (see above). These observations point to the involvement of HrpB4 and HrpB1 in interactions that may be integrating the TTSS with other X. axonopodis pv. citri proteins.
HrcN.
HrcN is a conserved component of all type III secretion systems, localized to the bacterial cytoplasm, and possibly associated with inner membrane components of the TTSS (46). HrcN is highly similar to FliI, which is essential for bacterial flagellar assembly (64). HrcN is also similar to InvC, Spa47 (MxiB), HrpB6, and YscN, all components of the type III protein secretion systems in Salmonella spp., Shigella flexneri, X. campestris, and Yersinia, respectively (6, 46). All of these proteins are homologs of the catalytic beta (and alpha) subunit of the F0F1-ATPase. When HrcN is used as a bait, four of the six preys sequenced were derived from the HrcN protein itself, the minimal prey fragment corresponding to residues 30 to 442. (Interestingly, in this case, no preys were derived from recombination events from the bait plasmid.) This may reflect a hexameric ring state for the HrcN protein in vivo similar to the a3b3 ring structure in the F0F1-ATPase, as proposed recently for the HrcN homolog Spa47 in Shigella sp. by Blocker et al. (6).
HpaA-HpaB-HrcV interactions.
HpaA and HpaB are two proteins coded by genes located within the Xanthomonas hrp locus (Fig. 1). HpaA homologs are found only in Xanthomonas spp. and R. solanacearum, and HpaA has been shown to be secreted by X. campestris pv. vesicatoria (33). HpaA may function as an effector molecule in X. campestris pv. vesicatoria as disruptions of the hpaA gene eliminate disease symptoms in tomato and pepper plants without affecting the ability to elicit hypersensitive response (33). Furthermore, the HpaA primary sequence contains two nuclear localization signals, one in the N-terminal and one in the C-terminal half of the protein, and HpaA protein, transiently expressed in onion cells, has been shown to localize to the nucleus (33). Less is known about the putative function of HpaB, except that it may be localized to the bacterial inner membrane due to a single putative transmembrane helix (43). Tn3-gus insertion mutagenesis of the hpaB gene of X. axonopodis pv. glycines reduced bacterial pathogenicity (39). HpaB homologs have been found only in Xanthomonas spp., R. solanacearum, and B. pseudomallei.
Both HpaA and HpaB were used as baits in two-hybrid screens. When HpaA (271 amino acids) was used as a bait, 100% of the 40 preys sequenced were found to be HpaB (Table 1). Interestingly, all of these clones coded for polypeptides containing at least all except the first six amino acids of HpaB, suggesting that the HpaA-HpaB interaction requires an almost complete N-terminal region of the HpaB polypeptide chain (Fig. 4). We also tested two baits derived from the hpaB gene: one coding for the full-length sequence of 156 residues and a second coding for residues 1 to 92. This N-terminal fragment was used since residues 92 to 108 are predicted to form a transmembrane helix by the PSORT algorithm (43). Each of the 16 preys sequenced when the HpaB N-terminal fragment was used was derived from a different gene in the X. axonopodis pv. citri genome. This result indicates that this fragment, on its own, does not make specific interactions with other peptides in the X. axonopodis pv. citri proteome. On the other hand, the results obtained with the full-length HpaB sequence were much more specific. Of 49 clones sequenced, 37 were derived from the hpaA gene (Fig. 4). The smallest HpaA fragment found to interact with HpaB corresponded to residues 126 to 271, the C-terminal half of the protein. Other than the presence of one of the nuclear localization signals, no functional information is available regarding this or any other region of HpaA.
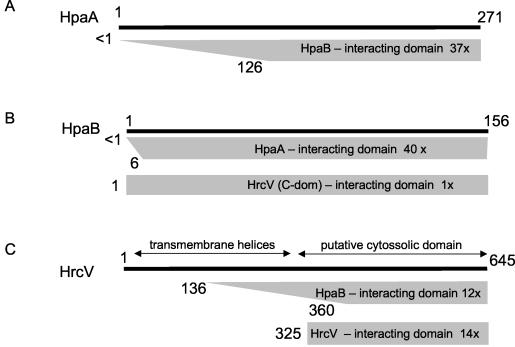
FIG. 4.
Summary of interactions observed in yeast two-hybrid assays involving HpaA, HpaB, and HrcV. (A) Preys derived from HpaA that interacted with the full-length HpaB bait; (B) preys derived from HpaB that interacted with baits derived from full-length HpaA and from the C-terminal domain of HrcV (residues 325 to 645); (C) preys derived from HrcV that interacted with baits derived from full-length HpaB and from the C-terminal domain of HrcV (residues 325 to 645). Shaded trapezoids or rectangles indicate the maximum and minimum fragments within the set of preys that interacted with a specific bait. Numbers to the left of the trapezoid indicate the maximum and minimum positions of the N-terminal boundaries of the preys. The specific bait used is indicated within the trapezoid. Also indicated is the number of clones sequenced derived from that particular prey. The full-length prey is represented as a solid black line above.
The remaining 12 preys obtained when the full-length HpaB protein was used as bait were derived from the HrcV protein, a component of the type-III secretion machinery. The minimal fragment of HrcV found to interact with full-length HpaB corresponded to residues 360 to 645 (Fig. 4). This observation is consistent with the prediction that the N-terminal domain of HrcV (up to residue 316) codes for at least six transmembrane segments (43), whereas the C-terminal half of the protein forms a soluble cytosolic domain. When the HrcV C-terminal domain (residues 325 to 646) was used as a bait, 14 of 18 preys were derived from the HrcV protein. All of these preys were derived from recombination events with the bait vector; therefore, the minimal domain necessary for this interaction could not be determined. Furthermore, one prey was derived from the full-length HpaB protein, confirming the HpaB-HrcV interaction observed when HpaB was used as bait. A summary of the interactions observed between HpaA, HpaB, and HrcV is shown in Fig. 4. Since HrcV (and possibly HpaB) is localized to the inner membrane, whereas HpaA is a soluble protein with domains indicative of activity in the host cell nucleus (see above), a functional role for these interactions becomes immediately apparent: HpaA, HpaB, and the C-terminal cytosolic domain of HrcV may interact in a manner that results in the targeting of HpaA to the type III secretion machinery and its subsequent translocation into the host cell (Fig. 5). The above results seem to point to HpaB possibly functioning as a chaperone or protein usher for HpaA, perhaps facilitating its interaction with HrcV at the cytoplasmic entrance of the TTSS channel (Fig. 5).
Concluding remarks.
The yeast two-hybrid system has been used to identify protein-protein interactions in protein complexes such as the yeast pheromone-response pathway complex (23) and yeast RNA polymerase III (24) and between Drosophila cyclin-dependent protein kinase interactors and cyclin-dependent kinases involved in cell cycle regulation (30). It has also been used to elucidate an interaction map of proteins involved in Caenorhabditis elegans vulval development (59), to screen an oligopeptide expression library (68), and to identify protein-protein interactions on the proteome-scale (4, 56). Ward et al. (60) recently used this methodology to delimit interactions between components of the type IV secretion system of A. tumefaciens. The two-hybrid system has only been used in one other study to investigate interactions between TTSS components in Yersinia pestis (34), responsible for the export of 12 Yersinia outer proteins. In that study, specific interactions were observed for YscQ with YscK and YscL and for YscL with YscQ and YscN. Those authors suggested that YscKQLN may form a complex peripherally associated with the inner membrane in a manner similar to the F1 and V1 multiprotein complexes of the F0F1 and V0V1 proton-translocating ATPases. YscQ and YscN are homologs of X. axonopodis pv. citri proteins HrcQ and HrcN, whereas YscK and YscL have no X. axonopodis pv. citri homologs. The interactors and interactions observed in the Yersinia study (34) were therefore different from those observed in the present study.
In the present study, we have identified a number of potentially physiologically relevant interactions between subunits, regulators, and substrates of the X. axonopodis pv. citri type III secretion system. We have identified interactions involving proteins previously known or suspected to be involved in X. axonopodis pv. citri pathogenicity, including HrpG, HpaA, HpaB, HrcV, HrpB1, HrpD6, HrpB2, HrcU, HrpW, HrpB4, and HrcN. The fact that our prey library consists of whole genomic X. axonopodis pv. citri DNA significantly increased the possibility of observing so-called “false-positive” interactions. In spite of this, relatively few and in many cases easily identifiable false-positives were observed, and it is highly significant that the majority of the interactions observed in these assays “make sense” physiologically. In fact, the multiple interactions observed between known Hrp proteins when a whole genomic DNA prey library was used is a strong confirmation of the physiological relevance of these interactions. A similar high degree of internal consistency has been observed by us in two-hybrid assays in which the baits were derived from X. axonopodis pv. citri type IV secretion system components (M. C. Alegria et al., unpublished data).
The protein-protein interactions identified here have clear implications for our understanding of the molecular mechanisms underlying Xanthomonas pathogenicity in general and the workings and regulation of the TTSS in particular. Figure 5 presents a summary of the interactions observed in the present study and their integration into a functional model of the Xanthomonas spp. TTSS that has emerged from the important contributions of many other laboratories. What is particularly interesting is that we now have subsets of protein-protein interactions that point to a relationship between specific TTSS substrates (HpaA and HrpB2) and specific conserved components of the TTSS machinery (HrcV and HrcU, respectively). In the case of HpaA and HrcV, their association appears to be mediated via HpaB. These results point to more specific studies that should be carried out in the near future, including investigations into (i) the molecular interactions important for HpaA secretion, including those involving HpaB and HrcV; (ii) the possible occurrence and role of HrcU proteolytic cleavage in TTSS assembly and regulation of TTSS substrate specificity, in particular the secretion of HrpB2; and (iii) specific interactions between HrpB1, HrpD6, and the pectate lysase harpin homolog HrpW and the possibility of their functional integration with HrpB4 and the conserved hypothetical proteins XAC2047 and XAC2054. Finally, of special interest for future studies are the specific molecular interactions between HrpG and its upstream and downstream regulators, including members of the newly identified XAC0095 family of proteins, transcription factors, and two-component system sensor histidine kinases.
Acknowledgments
We are deeply grateful for the excellent technical assistance of Izaura Nabuko Toma, Ilda de Souza Costa, and Elizabeth S.N. Mandetta. We thank Stanley Fields for kindly providing the pOBD and pOAD plasmids. We also thank Phillip James for kindly providing the PJ694-a yeast cells.
This study was supported by the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) and the Conselho Nacional de Pesquisa of Brazil. M.C.A., C.D., and L.K. are graduate fellows of FAPESP.
REFERENCES
- 1.Aldon, D., B. Brito, C. Boucher, and S. Genin. 2000. A bacterial sensor of plant cell contact controls the transcriptional induction of Ralstonia solanacearum pathogenicity genes. EMBO J. 19:2304-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alfano, J. R., and A. Collmer. 2001. Mechanisms of bacterial pathogenesis in plants: familiar foes in a foreign kingdom, p. 179-226. In E. A. Groisman (ed.), Principles of bacterial pathogenesis. Academic Press, Inc., New York, N.Y.
- 3.Baron, C., D. O'Callaghan, and E. Lanka. 2002. Bacterial secrets of secretion: EuroConference on the biology of type IV secretion processes. Mol. Microbiol. 43:1359-1365. [DOI] [PubMed] [Google Scholar]
- 4.Bartel, P. L., J. A. Roecklein, D. SenGupta, and S. Fields. 1996. A protein linkage map of Escherichia coli bacteriophage T7. Nat. Genet. 12:72-77. [DOI] [PubMed] [Google Scholar]
- 5.Bejar, S., K. Cam, and J. P. Bouche. 1986. Control of cell division in Escherichia coli: DNA sequence of dicA and of a second gene complementing mutation dicA1, dicC. Nucleic Acids Res. 14:6821-6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blocker, A., K. Komoriya, and S. Aizawa. 2003. Type III secretion systems and bacterial flagella: insights into their function from structural similarities. Proc. Natl. Acad. Sci. USA 100:3027-3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brito, B., M. Marenda, P. Barberis, C. Boucher, and S. Genin. 1999. prhJ and hrpG, two new components of the plant signal-dependent regulatory cascade controlled by PrhA in Ralstonia solanacearum. Mol. Microbiol. 31:237-251. [DOI] [PubMed] [Google Scholar]
- 8.Brito, B., D. Aldon, P. Barberis, C. Boucher, and S. Genin. 2002. A signal transfer system through three compartments transduces the plant cell contact-dependent signal controlling Ralstonia solanacearum hrp genes. Mol. Plant-Microbe Interact. 15:109-119. [DOI] [PubMed] [Google Scholar]
- 9.Buell, C. R., V. Joardar, M. Lindeberg, J. Selengut, I. T. Paulsen, M. L. Gwinn, R. J. Dodson, R. T. Deboy, A. S. Durkin, J. F. Kolonay, R. Madupu, S. Daugherty, L. Brinkac, M. J. Beanan, D. H. Haft, W. C. Nelson, T. Davidsen, N. Zafar, L. Zhou, J. Liu, Q. Yuan, H. Khouri, N. Fedorova, B. Tran, D. Russell, K. Berry, T. Utterback, S. E. Van Aken, T. V. Feldblyum, M. D'Ascenzo, W. L. Deng, A. R. Ramos, J. R. Alfano, S. Cartinhour, A. K. Chatterjee, T. P. Delaney, S. G. Lazarowitz, G. B. Martin, D. J. Schneider, X. Tang, C. L. Bender, O. White, C. M. Fraser, and A. Collmer. 2003. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 100:10181-10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burns, D. L. 1999. Biochemistry of type IV secretion. Curr. Opin. Microbiol. 2:25-29. [DOI] [PubMed] [Google Scholar]
- 11.Burns, D. L. 2003. Type IV transporters of pathogenic bacteria. Curr. Opin. Microbiol. 6:29-34. [DOI] [PubMed] [Google Scholar]
- 12.Büttner, D., and U. Bonas. 2002. Getting across-bacterial type III effector proteins on their way to the plant cell. EMBO J. 21:5313-5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Büttner, D., and U. Bonas. 2002. Port of entry: the type III secretion translocon. Trends Microbiol. 10:186-192. [DOI] [PubMed] [Google Scholar]
- 14.Cascales, E., and P. J. Christie. 2003. The versatile bacterial type IV secretion systems. Nature Rev. 1:137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan, J. W., and P. H. Goodwin. 1999. The molecular genetics of virulence of Xanthomonas campestris. Biotechnol. Adv. 17:489-508. [DOI] [PubMed] [Google Scholar]
- 16.Charkowski, A. O., J. R. Alfano, G. Preston, J. Yuan, S. Y. He, and A. Collmer. 1998. The Pseudomonas syringae pv. tomato HrpW protein has domains similar to harpins and pectate lyases and can elicit the plant hypersensitive response and bind to pectate. J. Bacteriol. 180:5211-5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chien, C. T., P. L. Bartel, R. Sternglanz, and S. Fields. 1991. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc. Natl. Acad. Sci. USA 88:9578-9582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Christie, P. J. 2001. Type IV secretion: intercellular transfer of macromolecules by systems ancestrally related to conjugation machines. Mol. Microbiol. 40:294-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christie, P. J., and J. P. Vogel. 2000. Bacterial type IV secretion: conjugation systems adapted to deliver effector molecules to host cells. Trends Microbiol. 8:354-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cornelis, G. R., and F. Van Gijsegem. 2000. Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 54:735-774. [DOI] [PubMed] [Google Scholar]
- 21.da Silva, A. C., J. A. Ferro, F. C. Reinach, C. S. Farah, L. R. Furlan, R. B. Quaggio, C. B. Monteiro-Vitorello, M. A. Van Sluys, N. F. Almeida, L. M. Alves, A. M. do Amaral, M. C. Bertolini, L. E. Camargo, G. Camarotte, F. Cannavan, J. Cardozo, F. Chambergo, L. P. Ciapina, R. M. Cicarelli, L. L. Coutinho, J. R. Cursino-Santos, H. El-Dorry, J. B. Faria, A. J. Ferreira, R. C. Ferreira, M. I. Ferro, E. F. Formighieri, M. C. Franco, C. C. Greggio, A. Gruber, A. M. Katsuyama, L. T. Kishi, R. P. Leite, E. G. Lemos, M. V. Lemos, E. C. Locali, M. A. Machado, A. M. Madeira, N. M. Martinez-Rossi, E. C. Martins, J. Meidanis, C. F. Menck, C. V. Miyaki, D. H. Moon, L. M. Moreira, M. T. Novo, V. K. Okura, M. C. Oliveira, V. R. Oliveira, H. A. Pereira, A. Rossi, J. A. Sena, C. Silva, R. F. de Souza, L. A. Spinola, M. A. Takita, R. E. Tamura, E. C. Teixeira, R. I. Tezza, M. Trindade dos Santos, D. Truffi, S. M. Tsai, F. F. White, J. C. Setubal, and J. P. Kitajima. 2002. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417:459-463. [DOI] [PubMed] [Google Scholar]
- 22.Enz, S., S. Mahren, U. H. Stroeher, and V. Braun. 2000. Surface signaling in ferric citrate transport gene induction: interaction of the FecA, FecR, and FecI regulatory proteins. J. Bacteriol. 182:637-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Evangelista, C., D. Lockshon, and S. Fields. 1996. The yeast two-hybrid system: prospects for protein linkage maps. Trends Cell Biol. 6:196-201. [DOI] [PubMed] [Google Scholar]
- 24.Flores, A., J. F. Briand, O. Gadal, J. C. Andrau, L. Rubbi, V. Van Mullem, C. Boschiero, M. Goussot, C. Marck, C. Carles, P. Thuriaux, A. Sentenac, and M. Werner. 1999. A protein-protein interaction map of yeast RNA polymerase III. Proc. Natl. Acad. Sci. USA 96:7815-7820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Francis, M. S., H. Wolf-Watz, and A. Forsberg. 2002. Regulation of type III secretion systems. Curr. Opin. Microbiol. 5:166-172. [DOI] [PubMed] [Google Scholar]
- 26.Galan, J. E., and A. Collmer. 1999. Type III secretion machines: bacterial devices for protein delivery into host cells. Science 284:1322-1328. [DOI] [PubMed] [Google Scholar]
- 27.Gietz, R. D., and R. A. Woods. 2002. Transformation of yeast by the LiAc/SS carrier DNA/PEG method. Methods Enzymol. 350:87-96. [DOI] [PubMed] [Google Scholar]
- 28.Gietz, R. D., R. A. Woods, P. Manivasakam, and R. H. Schiestl. 1998. Growth and transformation of Saccharomyces cerevisiae. In D. Spector, R. Goldman, and L. Leinwand (ed.), Cells: a laboratory manual, vol. I. Culture and biochemical analysis of cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Hale, C. A., and P. A. de Bôer. 1999. Recruitment of ZipA to the septal ring of Escherichia coli is dependent on FtsZ and independent of FtsA. J. Bacteriol. 181:167-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harper, J. W., G. R. Adami, N. Wei, K. Keyomars, and S. J. Ellege. 1993. The p21 cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75:805-816. [DOI] [PubMed] [Google Scholar]
- 31.Harrison-McMonagle, P., N. Denissova, E. Martinez-Hackert, R. H. Ebright, and A. M. Stock. 1999. Orientation of OmpR monomers within an OmpR:DNA complex determined by DNA affinity cleaving. J. Mol. Biol. 285:555-566. [DOI] [PubMed] [Google Scholar]
- 32.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huguet, E., K. Hahn, K. Wengelnik, and U. Bonas. 1998. hpaA mutants of Xanthomonas campestris pv. vesicatoria are affected in pathogenicity but retain the ability to induce host-specific hypersensitive reaction. Mol. Microbiology 29:1379-1390. [DOI] [PubMed] [Google Scholar]
- 34.Jackson, M. W., and G. V. Plano. 2000. Interactions between type III secretion apparatus components from Yersinia pestis detected using the yeast two-hybrid system. FEMS Microbiol. Lett. 186:85-90. [DOI] [PubMed] [Google Scholar]
- 35.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jeruzalmi, D., M. O'Donnell, and J. Kuriyan. 2001. Crystal structure of the processivity clamp loader gamma (γ) complex of E. coli DNA polymerase III. Cell 106:429-441. [DOI] [PubMed] [Google Scholar]
- 37.Kenny, L. K. 2002. Structure/function relationships in OmpR and other winged-helix transcription factors. Curr. Opin. Microbiol. 5:135-141. [DOI] [PubMed] [Google Scholar]
- 38.Kim, J. F., and S. V. Beer. 1998. HrpW of Erwinia amylovora, a new harpin that contains a domain homologous to pectate lyases of a distinct class. J. Bacteriol. 180:5203-5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim, J.-G., B. K. Park, C.-H. Yoo, E., Jeon, J. Oh, and I. Hwang. 2003. Characterization of the Xanthomonas axonopodis pv. glycines Hrp pathogenicity island. J. Bacteriol. 185:3155-3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lavander, M., L. Sundberg, P. J. Edqvist, S. A. Lloyd, H. Wolf-Watz, and A. Forsberg. 2002. Proteolytic cleavage of the FlhB homologue YscU of Yersinia pseudotuberculosis is essential for bacterial survival but not for type III secretion. J. Bacteriol. 184:4500-4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lupas, A. 1996. Coiled coils: new structures and new functions. Trends Biochem. Sci. 21:375-382. [PubMed] [Google Scholar]
- 42.Macnab, R. M. 1996. Flagella and motility, p. 123-145. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 43.Nakai, K., and M. Kanehisa. 1991. Expert system for predicting protein localization sites in gram-negative bacteria proteins. Struct. Funct. Genet. 11:95-110. [DOI] [PubMed] [Google Scholar]
- 44.Pao, G. M., and M. H. Saier. 1995. Response regulators of bacterial signal transduction systems: selective domain shuffling during evolution. J. Mol. Evol. 40:136-154. [DOI] [PubMed] [Google Scholar]
- 45.Parchaliuk, D. L., R. D. Kirkpatrick, R. Agatep, S. L. Simon, and R. D. Gietz. 1999. Yeast two-hybrid system. C. Characterizing positives. Technical Tips Online 1:69:P01714 [Online.] http://tto.trends.com. [Google Scholar]
- 46.Plano, G. V., J. B. Day, and F. Ferracci. 2001. Type III export: new uses for an old pathway. Mol. Microbiol. 40:284-293. [DOI] [PubMed] [Google Scholar]
- 47.Ponting, C. P., and L. Aravind. 1997. PAS: a multifunctional domain family comes to light. Curr. Biol. 7:674-677. [DOI] [PubMed] [Google Scholar]
- 48.Rossier, O., G. Van den Ackerveken, and U. Bonas. 2000. HrpB2 and HrpF from Xanthomonas are type III-secreted proteins and essential for pathogenicity and recognition by the host plant. Mol. Microbiol. 38:828-838. [DOI] [PubMed] [Google Scholar]
- 49.Salanoubat, M, S. Genin, F. Artiguenave, J. Gouzy, S. Mangenot, M. Arlat, A. Billault, P. Brottier, J. C. Camus, L. Cattolico, M. Chandler, N. Choisne, C. Claudel-Renard, S. Cunnac, N. Demange, C. Gaspin, M. Lavie, A. Moisan, C. Robert, W. Saurin, T. Schiex, P. Siguier, P. Thebault, M. Whalen, P. Wincker, M. Levy, J. Weissenbach, and C. A. Boucher. 2002. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415:497-502. [DOI] [PubMed] [Google Scholar]
- 50.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 51.Schulte, R., and U. Bonas. 1992. Expression of the Xanthomonas campestris pv. vesicatoria hrp gene cluster, which determines pathogenicity and hypersensitivity on pepper and tomato, is plant inducible. J. Bacteriol. 174:815-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simpson, A. J., F. C. Reinach, P. Arruda, F. A. Abreu, M. Acencio, R. Alvarenga, L. M. Alves, J. E. Araya, G. S. Baia, C. S. Baptista, M. H. Barros, E. D. Bonaccorsi, S. Bordin, J. M. Bove, M. R. Briones, M. R. Bueno, A. A. Camargo, L. E. Camargo, D. M. Carraro, H. Carrer, N. B. Colauto, C. Colombo, F. F. Costa, M. C. Costa, C. M. Costa-Neto, L. L. Coutinho, M. Cristofani, E. Dias-Neto, C. Docena, H. El-Dorry, A. P. Facincani, A. J. Ferreira, V. C. Ferreira, J. A. Ferro, J. S. Fraga, S. C. Franca, M. C. Franco, M. Frohme, L. R. Furlan, M. Garnier, G. H. Goldman, M. H. Goldman, S. L. Gomes, A. Gruber, P. L. Ho, J. D. Hoheisel, M. L. Junqueira, E. L. Kemper, J. P. Kitajima, J. E. Krieger, E. E. Kuramae, F. Laigret, M. R. Lambais, L. C. Leite, E. G. Lemos, M. V. Lemos, S. A. Lopes, C. R. Lopes, J. A. Machado, M. A. Machado, A. M. Madeira, H. M. Madeira, C. L. Marino, M. V. Marques, E. A. Martins, E. M. Martins, A. Y. Matsukuma, C. F. Menck, E. C. Miracca, C. Y. Miyaki, C. B. Monteriro-Vitorello, D. H. Moon, M. A. Nagai, A. L. Nascimento, L. E. Netto, A. J. Nhani, F. G. Nobrega, L. R. Nunes, M. A. Oliveira, M. C. de Oliveira, R. C. de Oliveira, D. A. Palmieri, A. Paris, B. R. Peixoto, G. A. Pereira, H. A. Pereira, Jr., J. B. Pesquero, R. B. Quaggio, P. G. Roberto, V. Rodrigues, A. J. de M Rosa, V. E. de Rosa, Jr., R. G. de Sa, R. V. Santelli, H. E. Sawasaki, A. C. R. da Silva, A. M. da Silva, F. R. da Silva, W. A. da Silva, Jr., J. F. da Silveira, M. L. Silvestri, W. J. Siqueira, A. A. de Souza, A. P. de Souza, M. F. Terenzi, D. Truffi, S. M. Tsai, M. H. Tsuhako, H. Vallada, M. A. Van Sluys, S. Verjovski-Almeida, A. L. Vettore, M. A. Zago, M. Zatz, J. Meidanis, and J. C. Setubal. 2000. The genome sequence of the plant pathogen Xylella fastidiosa. Nature 406:151-157. [DOI] [PubMed] [Google Scholar]
- 53.Slater, H., A. Alvarez-Morales, C. E. Barber, M. J. Daniels, and J. M. Dow. 2000. A two-component system involving an HD-GYP domain protein links cell-cell signalling to pathogenicity gene expression in Xanthomonas campestris. Mol. Microbiol. 38:986-1003. [DOI] [PubMed] [Google Scholar]
- 54.Stiefel, A., S. Mahren, M. Ochs, P. T. Schindler, S. Enz, and V. Braun. 2001. Control of the ferric citrate transport system of Escherichia coli: mutations in region 2.1 of the FecI extracytoplasmic-function sigma factor suppress mutations in the FecR transmembrane regulatory protein. J. Bacteriol. 183:162-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 56.Uetz, P., L. Giot, G. Cagney, T. A. Mansfield, R. S. Judson, J. R. Knight, D. Lockshon, V. Narayan, M. Srinivasan, P. Pochart, A. Qureshi-Emili, Y. Li, B. Godwin, D. Conover, T. Kalbfleisch, G. Vijayadamodar, M. Yang, M. Johnston, S. Fields, and J. M. Rothberg. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403:623-627. [DOI] [PubMed] [Google Scholar]
- 57.Van Gijsegem, F., C. Gough, C. Zischek, E. Niqueux, M. Arlat, S. Genin, P. Barberis, S. German, P. Castello, and C. Boucher. 1995. The hrp gene locus of Pseudomonas solanacearum, which controls the production of a type III secretion system, encodes eight proteins related to components of the bacterial flagellar biogenesis complex. Mol. Microbiol. 15:1095-1114. [DOI] [PubMed] [Google Scholar]
- 58.Van Sluys, M. A., M. C. de Oliveira, C. B. Monteiro-Vitorello, C. Y. Miyaki, L. R. Furlan, L. E. Camargo, A. C. da Silva, D. H. Moon, M. A. Takita, E. G. Lemos, M. A. Machado, M. I. Ferro, F. R. da Silva, M. H. Goldman, G. H. Goldman, M. V. Lemos, H. El-Dorry, S. M. Tsai, H. Carrer, D. M. Carraro, R. C. de Oliveira, L. R. Nunes, W. J. Siqueira, L. L. Coutinho, E. T. Kimura, E. S. Ferro, R. Harakava, E. E. Kuramae, C. L. Marino, E. Giglioti, I. L. Abreu, L. M. Alves, A. M. do Amaral, G. S. Baia, S. R. Blanco, M. S. Brito, F. S. Cannavan, A. V. Celestino, A. F. da Cunha, R. C. Fenille, J. A. Ferro, E. F. Formighieri, L. T. Kishi, S. G. Leoni, A. R. Oliveira, V. E. Rosa, Jr., F. T. Sassaki, J. A. Sena, A. A. de Souza, D. Truffi, F. Tsukumo, G. M. Yanai, L. G. Zaros, E. L. Civerolo, A. J. Simpson, N. F. Almeida, Jr., J. C. Setubal, and J. P. Kitajima. 2003. Comparative analyses of the complete genome sequences of Pierce's disease and citrus variegated chlorosis strains of Xylella fastidiosa. J. Bacteriol. 185:1018-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walhout, A. J. M., R. Sordella, X. Lu, J. L. Hartley, G. F. Temple, M. A. Brasch, N. Thierry-Mieg, and M. Vidal. 2000. Protein interaction mapping in Caenorhabditis elegans using proteins involved in vulval development. Science 287:116-122. [DOI] [PubMed] [Google Scholar]
- 60.Ward, D. V., O. Draper, J. R. Zupan, and P. C. Zambryski. 2002. Peptide linkage mapping of the Agrobacterium tumefaciens vir-encoded type IV secretion system reveals protein subassemblies. Proc. Natl. Acad. Sci. USA 99:11493-11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wengelnik, K., and U. Bonas. 1996. HrpXv, an AraC-type regulator, activates expression of five of the six loci in the hrp cluster of Xanthomonas campestris pv. vesicatoria. J. Bacteriol. 178:3462-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wengelnik, K., M. M. Russel, and U. Bonas. 1996. Expression and localization of HrpA1, a protein of Xanthomonas campestris pv. vesicatoria essential for pathogenicity and induction of the hypersensitive reaction. J. Bacteriol. 178:1061-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wengelnik, K., O. Rossier, and U. Bonas. 1999. Mutations in the regulatory gene hrpG of Xanthomonas campestris pv. vesicatoria result in constitutive expression of all hrp genes. J. Bacteriol. 181:6828-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Williams, A. W., S. Yamaguchi, F. Togashi, S. Aizawa, I. Kawagishi, and R. M. Macnab. 1996. Mutations in fliK and flhB affecting flagellar hook and filament assembly in Salmonella typhimurium. J. Bacteriol. 178:2960-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Winther-Larsen, H. C., and M. Koomey. 2002. Transcriptional, chemosensory, and cell-contact-dependent regulation of type IV pilus expression. Curr. Opin. Microbiol. 5:173-178. [DOI] [PubMed] [Google Scholar]
- 66.Wood, D. W., J. C. Setubal, R. Kaul, D. E. Monks, J. P. Kitajima, V. K. Okura, Y. Zhou, L. Chen, G. E. Wood, N. F. Almeida, Jr., L. Woo, Y. Chen, I. T. Paulsen, J. A. Eisen, P. D. Karp, D. Bovee, Sr, P. Chapman, J. Clendenning, G. Deatherage, W. Gillet, C. Grant, T. Kutyavin, R. Levy, M. J. Li, E. McClelland, A. Palmieri, C. Raymond, G. Rouse, C. Saenphimmachak, Z. Wu, P. Romero, D. Gordon, S. Zhang, H. Yoo, Y. Tao, P. Biddle, M. Jung, W. Krespan, M. Perry, B. Gordon-Kamm, L. Liao, S. Kim, C. Hendrick, Z. Y. Zhao, M. Dolan, F. Chumley, S. V. Tingey, J. F. Tomb, M. P. Gordon, M. V. Olson, and E. W. Nester. 2001. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science 294:2317-2323. [DOI] [PubMed] [Google Scholar]
- 67.Wösten, M. M. S. M. 1998. Eubacterial sigma-factors. FEMS Microbiol. Rev. 22:127-150. [DOI] [PubMed] [Google Scholar]
- 68.Yang, M., Z. Wu, and S. Fields. 1995. Protein-protein interactions analyzed with the yeast two-hybrid system. Nucleic Acids Res. 23:1152-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhu, J., P. M. Oger, B. Schrammeijer, P. J. Hooykaas, S. K. Farrand, and S. C. Winans. 2000. The basis of crown gall tumorigenesis J. Bacteriol. 182:3885-3895. [DOI] [PMC free article] [PubMed] [Google Scholar]