Abstract
Bacillus subtilis is an endospore-forming bacterium. There are indications that protein disulfide linkages occur in spores, but the role of thiol-disulfide chemistry in spore synthesis is not understood. Thiol-disulfide oxidoreductases catalyze formation or breakage of disulfide bonds in proteins. CcdA is the only B. subtilis thiol-disulfide oxidoreductase that has previously been shown to play some role in endospore biogenesis. In this work we show that lack of the StoA (YkvV) protein results in spores sensitive to heat, lysozyme, and chloroform. Compared to CcdA deficiency, StoA deficiency results in a 100-fold-stronger negative effect on sporulation efficiency. StoA is a membrane-bound protein with a predicted thioredoxin-like domain probably localized in the intermembrane space of the forespore. Electron microscopy of spores of CcdA- and StoA-deficient strains showed that the spore cortex is absent in both cases. The BdbD protein catalyzes formation of disulfide bonds in proteins on the outer side of the cytoplasmic membrane but is not required for sporulation. Inactivation of bdbD was found to suppress the sporulation defect of a strain deficient in StoA. Our results indicate that StoA is a thiol-disulfide oxidoreductase that is involved in breaking disulfide bonds in cortex components or in proteins important for cortex synthesis.
Bacteria of the genera Bacillus and Clostridium can differentiate into endospores in response to unfavorable growth conditions. This dormant state is more resistant to heat, desiccation, UV radiation, hydrolytic enzymes, and toxic chemicals than the vegetative cell. The outermost protective layers of Bacillus subtilis endospores are the coat and the cortex (7). The spore coat is a proteinaceous barrier against bactericidal enzymes and destructive chemicals. The cortex is composed of a thick peptidoglycan layer that helps to maintain the dehydrated state of the spore core and is required for the extreme heat resistance of spores. There are indications that proteins in the coat are cross-linked by disulfide bonds (1, 26). These bonds may contribute to the overall resistance of the spore. Some proteins, e.g., YkvU and SpmB, encoded by σE-dependent genes (8, 11) and possibly involved in cortex assembly are also rich in cysteine residues. However, the importance of disulfides and free thiol groups for the function of sporulation proteins is not understood.
Thiol-disulfide oxidoreductases catalyze the formation or breakage of disulfide bonds in other proteins. These enzymes have in their active site a pair of cysteine residues that participate in the reaction. These cysteine residues are often arranged in a Cys-X-X-Cys motif. Stable disulfide bonds in proteins in Bacteria are normally only found in extracytoplasmic compartments and in secreted proteins. Thioredoxin and other reductants break disulfide bonds formed in cytoplasmic proteins. In both gram-positive and -negative bacteria, several thiol-disulfide oxidoreductases have been identified that are involved in forming or breaking disulfide bonds in proteins on the outer side of the cytoplasmic membrane (for a review, see reference 19).
Six membrane-bound thiol-disulfide oxidoreductases that function on the outer side of the cytoplasmic membrane have so far been identified in B. subtilis. Four of these proteins function as pairs, i.e., BdbA/BdbB and BdbD/BdbC, and are similar to Escherichia coli DsbA/DsbB. These protein pairs catalyze formation of disulfide bonds in proteins (4, 6, 10). B. subtilis CcdA most likely transfers reducing equivalents from thioredoxin in the cytoplasm across the cytoplasmic membrane to ResA on the outer side of the membrane (32). ResA has a thioredoxin-like domain (5) and functions to break a disulfide bond in the heme binding site of apo-cytochrome c (9). CcdA is also required for efficient spore synthesis (31). The exact role of CcdA in sporulation has not been determined, but we have proposed that it transfers reducing equivalents to one or more not-yet-identified thiol-disulfide oxidoreductases that function in spore synthesis. As deduced from the B. subtilis genome sequence, YkvV and YneN are predicted membrane-bound proteins with a thioredoxin-like domain and are overall similar to ResA. These proteins could therefore possibly interact with CcdA. The ykvV gene is in a dicistronic operon together with ykvU located at 123° on the B. subtilis chromosome (Fig. 1). The ykvUV operon is transcribed from a σE-dependent promoter (8, 11). YkvU is of unknown function but has sequence similarity to SpoVB, which is important for cortex synthesis. The yneN gene is monocistronic and located at 164° on the chromosomal map.
FIG. 1.
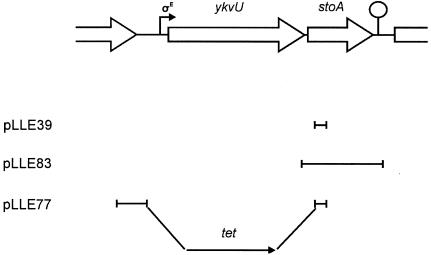
Map of the ykvU-stoA operon in the B. subtilis chromosome. Shown also are DNA segments cloned in plasmids used for disruption of stoA (pLLE39), for complementation analysis (pLLE83), and for deletion of ykvU-stoA (pLLE77). The hooked arrow and the loop indicate a σE-dependent promoter and a transcription terminator, respectively.
We have analyzed the role of YkvV, YkvU, and YneN in B. subtilis and show that YkvV is involved in spore formation. The ykvV gene is therefore renamed stoA (sporulation thiol-disulfide oxidoreductase A). We show that StoA is a membrane-bound thiol-disulfide oxidoreductase important for spore cortex synthesis.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The B. subtilis strains and plasmids used in this work are listed in Table 1. Oligonucleotides used as primers are listed in Table 2. E. coli strain JM109 {recA1 supE44 endA1 hsdR17 gyrA96 relA1 thi Δ(lac-proAB) F′ [traD36 proAB+ lacIq lacZΔM15]} and TOP10 [mcrA Δ(mrr-hsdRMS-mcrBC) φ80ΔlacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL endA1 nupG] were used for the propagation of plasmids. E. coli strains BL21(DE3) [F− dcm ompT hsdS(rB− mB−) gal λ(DE3)] and TOP10 were used for recombinant protein production.
TABLE 1.
Bacterial strains and plasmids used in this work
| Strains and plasmids | Descriptiona | Source or referenceb |
|---|---|---|
| B. subtilis strains | ||
| 1A1 | trpC2 | BGSCc |
| LU60A1 | trpC2 ΔccdA::ble; Pmr | 32 |
| LUA13 | trpC2 ΩcotC-lacZ; Cmr | 31 |
| LUA14 | trpC2 ΔspoIIIG::neo; Nmr | 31 |
| LUL3 | trpC2 bdbCΩpLLE21; Emr | 10 |
| LUL9 | trpC2 resAΩpLLE36; Emr | 9 |
| LUL10 | trpC2 bdbDΩTn10; Spr | 10 |
| LUL20 | trpC2 stoAΩpLLE39; Cmr | This work; pLLE39 → 1A1 |
| LUL21 | trpC2 stoAΩpLLE39 ΔccdA::ble; Cmr Pmr | This work; LUL20 → LU60A1 |
| LUL23 | trpC2 stoAΩpLLE39 bdbDΩTn10; Cmr Spr | This work; LUL10 → LUL20 |
| LUL24 | trpC2 stoAΩpLLE39 Δ(bdbA-yolJ-bdbB)::kan; Cmr Kmr | This work; LUL110 → LUL20 |
| LUL25 | trpC2 stoAΩpLLE39 ΔyneN::erm; Cmr Emr | This work; LUL121 → LUL20 |
| LUL27 | trpC2 bdbDΩTn10 Δ(bdbA-yolJ-bdbB)::kan; Spr Kmr | This work; LUL10 → LUL110 |
| LUL30 | trpC2 Δ(ykvU-stoA)::tet; Tcr | This work; pLLE77 → 1A1 |
| LUL35 | trpC2 ΩcotC-lacZ; Spr | This work; pCm::Sp → LUA13 |
| LUL36 | trpC2 stoAΩpLLE39 ΩcotC-lacZ; Spr | This work; LUL35 → LUL20 |
| LUL110 | trpC2 Δ(bdbA-yolJ-bdbB)::kan; Kmr | This work; pLLE9 → 1A1 |
| LUL121 | trpC2 ΔyneN::erm; Emr | This work; pLLE16 → 1A1 |
| Plasmids | ||
| pBAD-HisA/B | Expression vectors for His-tagged proteins; Amr | In vitrogen |
| pDG148 | Expression vector; Emr Kmr | 35 |
| pDG647 | Carries antibiotic resistance cassette; Emr Amr | 13 |
| pDG780 | Carries antibiotic resistance cassette; Kmr Amr | 13 |
| pDG1515 | Carries antibiotic resistance cassette; Tcr Amr | 13 |
| pHV32 | Integration vector for B. subtilis; Cmr Tcr Amr | 25 |
| pPHO1 | pET21-(+) with a 1.4-kb SacI-HindIII fragment containing truncated E. coli phoA from pPhoA; Amr | Mimmi Throne-Holst and reference 28 |
| pRAN1 | pHPSK with a 0.73-kb fragment containing B. subtilis resA; Cmr Emr | 9 |
| pCm::Sp | Plasmid for changing antibiotic resistance; Spr | 34 |
| pLLE9 | pDG780 with B. subtilis chromosomal sequences flanking the kanamycin resistance gene; Kmr Amr | This work |
| pLLE16 | pDG647 with B. subtilis chromosomal sequences flanking the erythromycin resistance gene; Emr Amr | This work |
| pLLE34 | pBAD-HisA with a 609-bp fragment containing a truncated bdbD gene; Amr | This work |
| pLLE39 | 310-bp internal fragment from stoA in pHV32; Cmr Tcr Amr | This work |
| pLLE64 | pPHO1 with a 271-bp fragment containing the 5′ part of stoA; Amr | This work |
| pLLE65 | pBAD-HisB with a 443-bp fragment containing a truncated stoA gene; Amr | This work |
| pLLE77 | pDG1515 with B. subtilis chromosomal sequences flanking the tetracycline gene; Tcr Amr | This work |
| pLLE82 | pDG148 with a 0.73-kb HindIII-ScaI fragment containing B. subtilis resA from pRAN1; Emr Kmr | This work |
| pLLE83 | pDG148 with a 537-bp fragment containing the stoA gene; Emr Kmr | This work |
Amr, Cmr, Emr, Kmr, Nmr, Spr, Pmr, and Tcr indicate resistance to ampicillin, chloramphenicol, erythromycin, kanamycin, neomycin, spectinomycin, phleomycin, and tetracycline, respectively.
An arrow indicates transformation of the indicated strain with chromosomal or plasmid DNA.
Bacillus Genetic Stock Center, Columbus, Ohio.
TABLE 2.
Oligonucleotides used as primers in this work
| Name | Sequencea | Restriction site |
|---|---|---|
| LE001 | 5′-GCTCTAGAGCACAATTGTTAGGACTC-3′ | XbaI |
| LE002 | 5′-CGGGATCCAATAAGAAGTAACCCGCC-3′ | BamHI |
| LE003 | 5′-CCATCGATAATACTAATGGCTGCTGC-3′ | ClaI |
| LE004 | 5′-GGGGTACCCTTGGACAAGCAGTACAG-3′ | KpnI |
| LE009 | 5′-GCGAATTCTCGAGGTAGATGTTGATG-3′ | EcoRI |
| LE010 | 5′-TCCCCCGGGTTCAGCATATGCCACCTC-3′ | SmaI |
| LE011 | 5′-GCTCTAGAGGATCTTGATTAGATTCAG-3′ | XbaI |
| LE012 | 5′-AACTGCAGGAACATATTGAGGCTGAC-3′ | PstI |
| LE030 | 5′-CGGGGTACCTAGCAGCCATTGTCATC-3′ | KpnI |
| LE031 | 5′-GTGCAAGCTTGTTACTTCCCTTTCAGCTC-3′ | HindIII |
| LE034 | 5′-AAAACTGCAGGATTGCATGGTTTCCAGGTG-3′ | PstI |
| LE035 | 5′-GTGCAAGCTTGGACAATCGGAAACGTCAG-3′ | HindIII |
| LE047 | 5′-GTGCAAGCTTGGCAAGCTAATTGAAAAGC-3′ | HindIII |
| LE048 | 5′-GTGCTCTAGACTCAGCTATTCTTCCGTC-3′ | XbaI |
| LE049 | 5′-CGGGATCCCTGCAAAGCATTGAAGG-3′ | BamHI |
| LE050 | 5′-GGGGTACCTTATTCGGAATCGAGATGTC-3′ | KpnI |
| LE051 | 5′-CGGGGTACCGGTGCGGCACAAGCTGAG-3′ | KpnI |
| LE052 | 5′-GTGCAAGCTTGCTCTCAGCTATTCTTCC-3′ | HindIII |
| LE053 | 5′-CTGGTCTAGAGTCTCGTTCACGCAGAGG-3′ | XbaI |
| LE054 | 5′-CGGGATCCTTCACAAATCGATTCATGATG-3′ | BamHI |
| LE055 | 5′-CACAGTCGACGAATAGCTGAGAGCATAGAC-3′ | SalI |
| LE056 | 5′-CTGGCTCGAGCAGCGTTTTGGATTCGAG-3′ | XhoI |
| HV32P01 | 5′-CGGCATAAATGCGTGGTC-3′ |
The restriction site is underlined.
Media and growth conditions.
E. coli cells were grown at 37°C in Luria-Bertani (LB) medium or on LB agar plates (29). B. subtilis strains were cultivated at 30 or 37°C in LB medium or nutrient sporulation medium with phosphate (NSMP) (12) or on tryptose blood agar base (TBAB) plates (Difco). Antibiotics were used at the following concentrations when appropriate: for B. subtilis, chloramphenicol at 3 mg/liter, erythromycin at 1 mg/liter, kanamycin at 10 mg/liter, neomycin at 5 mg/liter, spectinomycin at 150 mg/liter, and tetracycline at 15 mg/liter; for E. coli, ampicillin at 50 mg/liter and chloramphenicol at 12.5 mg/liter.
DNA techniques.
Standard DNA techniques were used (29). Plasmid DNA was isolated by using a Quantum prep plasmid miniprep kit (Bio-Rad) or by CsCl density gradient centrifugation. Chromosomal DNA from B. subtilis was isolated according to the method of Marmur (22). E. coli was transformed by electroporation, and B. subtilis was grown to natural competence essentially as described by Hanahan et al. and by Hoch, respectively (15, 17).
Construction of plasmids.
Plasmid pLLE9 was constructed by amplifying a region upstream of bdbA using primers LE001 and LE002. The PCR product was cloned into pDG780 at restriction sites XbaI and BamHI. A region downstream of bdbB was amplified using primers LE003 and LE004 and cloned into the plasmid at restriction sites ClaI and KpnI. Plasmid pLLE16 was constructed by amplifying regions up- and downstream of yneN using primers LE009 and LE010 (fragment I) and LE011 and LE012 (fragment II), respectively. The PCR products were cloned into pDG647 at restriction sites EcoRI and SmaI (for fragment I) and XbaI and PstI (for fragment II). Plasmid pLLE34 was constructed using primers LE030 and LE031. The amplified DNA fragment was cut with KpnI and HindIII and cloned into the vector pBAD-HisA that had been digested with the same restriction enzymes. Plasmid pLLE39 was constructed by amplifying an internal fragment of stoA using primers LE034 and LE035. The PCR product was cut with PstI and HindIII and cloned into pHV32 that had been cut with the same restriction enzymes. Plasmid pLLE65 was constructed using primers LE051 and LE052. The amplified DNA fragment was cut with KpnI and HindIII and cloned into the vector pBAD-HisB that had been digested with the same restriction enzymes. Plasmid pLLE77 was constructed by amplifying a region upstream of ykvU using primers LE053 and LE054. The PCR product was cloned into pDG1515 at restriction sites XbaI and BamHI. A region downstream of stoA was amplified using primers LE055 and LE056 and cloned into the plasmid at restriction sites SalI and XhoI. Plasmid pLLE83 was constructed using primers LE047 and LE048. The amplified DNA fragment was cut with HindIII and XbaI and cloned into the vector pDG148 that had been cut with the same restriction enzymes. The template used for all the PCRs described above was chromosomal DNA isolated from B. subtilis strain 1A1. Plasmid pLLE82 was constructed by moving a HindIII and ScaI fragment containing resA from pRAN1 to pDG148.
Construction of B. subtilis strains.
Strain LUL20 was obtained by transforming 1A1 with pLLE39 and selecting transformants on TBAB plates containing chloramphenicol. The disruption of the stoA gene by the integrated plasmid was confirmed by PCR amplification of a DNA fragment using primers LE034, which hybridizes just upstream of stoA in the chromosome, and HV32P01, which hybridizes to a sequence in the vector part of pLLE39. Strain LUL30 was constructed by transforming 1A1 with SalI-cut pLLE77 and selecting transformants on plates containing tetracycline. Strain LUL110 was obtained by transforming 1A1 with SalI-cut pLLE9 and selection on plates containing kanamycin. Strain LUL121 was isolated after transforming 1A1 with SalI-cut pLLE16 and selection on plates containing erythromycin.
Production and affinity purification of water soluble His-tagged variants of StoA and BdbD.
Soluble His-tagged StoA was produced in E. coli TOP10 containing pLLE65. Cells were grown in 100 ml of LB medium at 37°C. At an optical density at 600 nm of 0.6, the expression of stoA in the plasmid was induced with 0.2% (wt/vol) arabinose. Three hours after induction, the cells were collected by centrifugation and washed in buffer A (50 mM NaH2PO4, 300 mM NaCl, pH 8.0) containing 10 mM imidazole. The cells were then broken using a French press. The lysate was centrifuged at 31,000 × g for 30 min, and the supernatant was mixed with 1 ml of Ni-nitrilotriacetic acid resin (QIAGEN). After mixing for 1 h at 4°C, the resin was washed with 20 mM imidazole in buffer A and the protein was eluted from the resin by 250 mM imidazole in buffer A. Soluble His-tagged BdbD was produced in E. coli TOP10 containing pLLE34. Expression and purification of the protein was done in the same way as for soluble His-tagged StoA except that gene expression was induced with 0.02% (wt/vol) arabinose. The purity of the isolated soluble His-tagged StoA and BdbD was assessed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis (30).
Analysis of StoA transmembrane topology.
DNA corresponding to a truncated variant of the stoA reading frame was amplified by PCR using B. subtilis 1A1 chromosomal DNA as template and the primer pair LE049/LE050. The PCR product was cut with BamHI and KpnI and cloned into pPHO1 digested with the same enzymes, resulting in pLLE64. Alkaline phosphatase activity of cell lysates of E. coli BL21(DE3) containing pLLE64 or pPHO1 grown in LB medium with 50 mM phosphate was measured using p-nitrophenyl phosphate as substrate (36).
Spore assay.
Cultures were grown in 25 ml of NSMP at 30°C in 500-ml baffled Erlenmeyer flasks for 2 days. Sporulation efficiency of strains was analyzed by heating 5 ml of culture at 80°C for 15 min or by adding 0.6 ml of chloroform to a 5-ml culture followed by vigorous mixing for 10 s. Lysozyme sensitivity of cells was analyzed by diluting the culture 100-fold in minimal salts solution [80.4 mM K2HPO4, 44.1 mM KH2PO4, 15.1 mM (NH4)2SO4, 3 mM sodium-citrate] followed by incubation with lysozyme (0.5 g/liter) at 30°C for 30 min. Serial dilutions of treated and untreated samples were spread on TBAB plates. After overnight incubation of the plates at 37°C, colonies were counted.
Electron microscopy.
Preparation of B. subtilis cells for analysis by electron microscopy was performed essentially as described by Asai et al. (2). Cells were grown in NSMP medium at 37°C for 24 h after entry into stationary phase. After fixation in 3% glutaraldehyde in 50 mM phosphate buffer (pH 6.5), the cells were postfixed in 1% osmium tetroxide for 1 h, dehydrated, and embedded in Epon. Sections were stained in 2% uranyl acetate and in lead citrate, according to the method of Reynolds (27). Sections were examined using a JEOL 1230 transmission electron microscope.
Microarray analysis.
B. subtilis strains 1A1 and LUL20 were grown in 300 ml of NSMP medium at 37°C. Samples (20 ml) were harvested at the point of entry into stationary phase (T0) and two hours into stationary phase (T2). RNA extraction, cDNA synthesis, hybridization of cDNA to glass microarrays (Eurogentec), and acquisition and analysis of data were performed essentially as described by Hambraeus et al. (14).
Other methods.
Membranes were isolated from strains grown in NSMP at 37°C essentially as described previously (16). Protein concentrations were determined using the bicinchoninic acid protein assay (Pierce Chemical Co.) with bovine serum albumin as standard. N,N,N′,N′-Tetramethyl-p-phenylenediamine (TMPD) oxidation assay of colonies on NSMP plates and cytochrome c oxidation activity measurements were performed as described previously (10, 21). Insulin reduction assay was performed essentially as described by Holmgren (18). Insulin and E. coli thioredoxin were purchased from Sigma Chemical Co. β-Galactosidase activity measurements using 4-methylumbelliferyl-β-d-galactoside as substrate were done as described previously (31).
RESULTS AND DISCUSSION
StoA and YneN are predicted membrane-bound thiol-disulfide oxidoreductases.
StoA and YneN both have a dicysteine motif (Cys-X-Pro-Cys) that is characteristic for proteins in the thioredoxin family. Alignment of StoA and YneN with known thiol-disulfide oxidoreductases shows sequence similarity, particularly around the dicysteine motif. Secondary structure prediction (using PSIPRED [23]) based on comparison to E. coli thioredoxin (TrxA) indicates that StoA and YneN have a thioredoxin-like fold. The program TMHMM (20) predicts that StoA and YneN each have one transmembrane segment, constituted by the N-terminal part of the protein and with the thioredoxin-like domain exposed on the outer side of the cytoplasmic membrane.
StoA is involved in sporulation.
To investigate the physiological functions of B. subtilis StoA and YneN, the stoA gene in the chromosome was disrupted by a Campbell-type integration of plasmid pLLE39 and the yneN gene was deleted and replaced by an erythromycin resistance gene. The StoA- and YneN-deficient strains were named LUL20 and LUL121, respectively. Survival after heat treatment of LUL20 cells, grown for sporulation, was only 0.05% compared to that of untreated cells, indicating that StoA has a role in sporulation (Table 3). In a recent study, Eichenberger et al. (8) found a sporulation efficiency of 0.001% with an independent B. subtilis StoA (YkvV)-deficient strain. StoA deficiency does not completely block formation of heat-resistant spores, as can be seen by comparing the sporulation efficiency of LUL20 with that of LUA14, a strain blocked in sporulation (Table 4). As presented in Table 4, the survival of LUL20 cells, grown for sporulation, was also low after lysozyme and chloroform treatment compared to that of wild-type cells. The sporulation defect of LUL20 cells was complemented by stoA expressed in trans from plasmid pLLE83 which contains stoA under control of the IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible spac promoter (Table 5 and Fig. 1). This confirmed that the stoA gene is important for sporulation.
TABLE 3.
Sporulation efficiency of B. subtilis strainsa
| Strain | Relevant genotype | Total cell titerb | Spore titerc | Sporulation efficiency (%)d |
|---|---|---|---|---|
| 1A1 | Wild type | 6.5 × 108 | 6.4 × 108 | 98 |
| LU60A1 | ΔccdA::ble | 8.7 × 107 | 3.9 × 106 | 4 |
| LUL20 | stoAΩpLLE39 | 7.0 × 107 | 3.3 × 104 | 0.05 |
| LUL121 | ΔyneN::erm | 5.0 × 108 | 5.0 × 108 | 100 |
| LUL21 | stoAΩpLLE39 ΔccdA::ble | 9.7 × 107 | 5.6 × 103 | 0.006 |
| LUL25 | stoAΩpLLE39 ΔyneN::erm | 1.1 × 108 | 4.9 × 104 | 0.04 |
| LUL30 | Δ(ykvU-stoA)::tet | 3.7 × 108 | 3.2 × 102 | 0.0001 |
| LUL3 | bdbCΩpLLE21 | 5.1 × 108 | 4.9 × 108 | 96 |
| LUL10 | bdbDΩTn10 | 5.2 × 108 | 4.9 × 108 | 94 |
| LUL110 | Δ(bdbA-yolJ-bdbB)::kan | 5.3 × 108 | 4.8 × 108 | 90 |
| LUL23 | stoAΩpLLE39 bdbDΩTn10 | 6.0 × 108 | 5.9 × 108 | 98 |
| LUL24 | stoAΩpLLE39 Δ(bdbA-yolJ-bdbB)::kan | 8.8 × 107 | 5.4 × 104 | 0.06 |
| LUL27 | bdbDΩTn10 Δ(bdbA-yolJ-bdbB)::kan | 5.7 × 108 | 4.8 × 108 | 84 |
In all sporulation assays 1A1 and LU60A1 were included as controls. Experimental results were considered reliable if the numbers for 1A1 and LU60A1 did not deviate more than 20% from previously obtained (10, 31) mean values.
Titer is CFU per milliliter.
Titer after heat treatment for 15 min at 80°C.
Sporulation efficiency is calculated as the spore titer divided by the total cell titer.
TABLE 4.
Effect of heat, lysozyme, and chloroform treatment on spore survival of different B. subtilis strains
| Strain | Relevant genotype | Sporulation efficiency (%)
|
||
|---|---|---|---|---|
| Heat | Lysozyme | Chloroform | ||
| 1A1 | Wild type | 97 | 81 | 86 |
| LUL20 | stoAΩpLLE39 | 0.04 | 2.6 | 0.05 |
| LUA14 | ΔspoIIIG::neo | <0.0001 | 2.4 | <0.0001 |
TABLE 5.
Complementation of StoA deficiency in strains LUL20 and LUL30 by IPTG-dependent expression of StoA from plasmid pLLE83 (a derivative of vector pDG148)
| Strain | [IPTG]a | Total cell titer | Spore titerb | Sporulation efficiency (%) |
|---|---|---|---|---|
| 1A1/pDG148 | 0 | 8.0 × 108 | 6.6 × 108 | 84 |
| 1A1/pDG148 | 1 | 7.0 × 108 | 5.9 × 108 | 84 |
| 1A1/pLLE83 | 1 | 7.0 × 108 | 7.1 × 108 | 100 |
| LUL20/pDG148 | 0 | 1.6 × 108 | 1 × 105 | 0.06 |
| LUL20/pDG148 | 1 | 1.0 × 108 | 5.9 × 104 | 0.06 |
| LUL20/pLLE83 | 0 | 1.7 × 108 | 2.7 × 106 | 2 |
| LUL20/pLLE83 | 1 | 2.1 × 108 | 1.7 × 108 | 79 |
| LUL30/pDG148 | 1 | 1.6 × 108 | 1.2 × 104 | 0.008 |
| LUL30/pLLE83 | 1 | 2.4 × 108 | 5.0 × 107 | 20 |
IPTG concentration (mM) in the growth medium.
Assessed by heat treatment (see Table 3).
Strain LUL121, with the yneN gene deleted, was found to have a normal sporulation phenotype. A double mutant strain, LUL25, deficient in both YneN and StoA, showed the same low sporulation efficiency as LUL20 (Table 3). These results demonstrate that YneN is not important for sporulation and indicate that StoA and YneN do not functionally overlap, i.e., YneN does not contribute to sporulation in a strain deficient in StoA. Strain LU60A1, which is deficient in CcdA, has 2 to 5% of wild-type sporulation efficiency (31). Strain LUL21, deficient in both StoA and CcdA, showed approximately 10% sporulation efficiency compared to LUL20 (Table 3). The additive effect of the two defects suggest that StoA and CcdA act independently of each other, i.e., probably do not function in the same pathway in spore formation.
Morphology of strains lacking StoA.
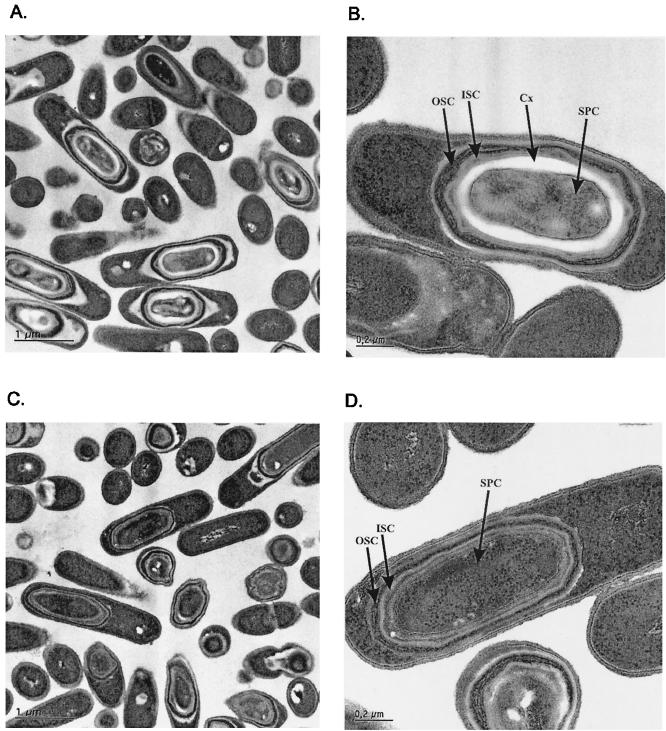
The effect of StoA deficiency on the morphology of sporulating cells was examined by light microscopy. LUL20 cells did not show the characteristic bright light refraction seen for cells of the parental strain 1A1 grown under the same conditions. Electron microscopic examination of 1A1 cells showed a clear spore cortex 24 h after the initiation of sporulation (Fig. 2A and B). LUL20 cells formed spores but without a visible cortex (Fig. 2C and D). The electron-dense outer coat and the lamellar inner coat were seen in spores of both the mutant and the wild type. The heat sensitivity of LUL20 spores can be explained by the lack of the cortex layer, which is essential for heat resistance. The spore coat is the major protection barrier against lysozyme and chloroform (7, 33). A defect in the spore coat as indicated by the lysozyme and chloroform sensitivity of LUL20 spores was not revealed by electron microscopy. The defect might originate from the lack of cortex which serves as a support base for the synthesis of the spore coat. Strains defective in cortex synthesis but with spore coat visible in electron microscopy images have been reported to be sensitive to lysozyme and chloroform (2, 3)
FIG. 2.
Electron microscopy analysis of B. subtilis endospores of strains 1A1 (parental strain) (A and B) and LUL20 (StoA-deficient strain) (C and D) grown for 24 h after onset of sporulation. Abbreviations: OSC, outer spore coat; ISC, inner spore coat; Cx, cortex; SPC, spore core.
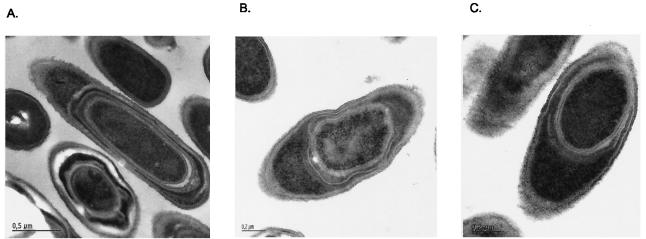
Electron microscopy showed that CcdA deficiency also affects spore cortex synthesis (Fig. 3A). Endospores of strain LUL21, which lacks both CcdA and StoA, contained inner and outer coat layers but appeared deficient in cortex (Fig. 3B). Thus, both StoA and CcdA deficiency affect cortex synthesis, and endospores of strains LUL20, LU60A1, and LUL21 looked the same as judged from electron microscopy analysis. From the available data we cannot exclude the possibility that the lack of a visible cortex in the mutants is due to hydrolysis of cortex material. However, such hydrolysis seems unlikely since at 12 h after the onset of sporulation spores of strain LUL20 lacked a visible cortex whereas the wild-type control contained cortex.
FIG. 3.
Electron microscopy analysis of B. subtilis endospores of strains LU60A1 (CcdA-deficient strain) (A), LUL21 (StoA- and CcdA-deficient strain) (B), and LUL30 (YkvU- and StoA-deficient strain) (C) grown for 24 h after onset of sporulation.
The sporulation sigma factor cascade is normal in StoA-deficient strains.
In two recent studies it has been shown that transcription of the ykvU-stoA operon is dependent on σE (8, 11). Genome-wide analysis of mRNA extracted from cells harvested from cultures in early stationary phase (i.e., T0 and T2) by using DNA microarrays showed no apparent difference between strains 1A1 and LUL20 in upregulation of genes known to be under sigma factor σF, σE, or σG control (array data not shown). Genes dependent on σK were not assayed in the microarray experiment. To determine if StoA deficiency affects σK-dependent gene transcription, expression of a cotC-lacZ gene fusion integrated in single copy into the amyE locus was analyzed as before (31). β-Galactosidase activity measurements with this strain, LUL36, and the parental strain LUL35 showed that both the time point for induction during sporulation and activity levels were the same for both strains (data not shown). The results showed that the sporulation sigma factor cascade is normal in StoA-deficient cells.
StoA is a membrane protein.
The transmembrane topology of StoA was analyzed by using the N-terminal segment of StoA (residues 1 to 45) fused to E. coli alkaline phosphatase (PhoA) lacking its native signal sequence. Alkaline phosphatase requires two disulfide bonds and is therefore only active in E. coli if it is transported to the outer side of the cytoplasmic membrane. Lysates of E. coli BL21(DE3) cells harboring plasmid pLLE64 (containing the stoA-phoA fusion) showed alkaline phosphatase activity {0.15 μmol/[min × (mg of protein)]} when expression was induced with IPTG, whereas lysates of E. coli BL21(DE3) cells containing the vector pPHO1 showed no detectable activity {<0.01 μmol/[min × (mg of protein)]}. Furthermore, the alkaline phosphatase activity of BL21(DE3)/pLLE64 was found to be associated with the particulate subfraction of cell lysates. From these results, and the predicted topology, we conclude that the N-terminal part of StoA functions as a membrane anchor and the C-terminal thioredoxin-like domain of StoA in B. subtilis is most likely exposed on the outer side of the cytoplasmic membrane.
Known oxidizing thiol-disulfide oxidoreductases are not required for efficient sporulation.
BdbD/BdbC and the paralogous BdbA/BdbB system catalyze disulfide bond formation in proteins on the outer side of the membrane in B. subtilis (4, 6, 10). The BdbA/BdbB system seems specifically involved in the maturation of the lanthionine sublancin 168. The BdbC/BdbD system is more generally involved in disulfide bond formation in extracytoplasmic proteins. The BdbD/BdbC and BdbA/BdbB systems are not important for sporulation, i.e., strains LUL3, LUL10, LUL110, and LUL27 showed normal sporulation efficiency (Table 3). Thus, known oxidizing thiol-disulfide oxidoreductase systems are not important for spore formation.
Effects of StoA deficiency are suppressed by BdbD deficiency.
The sporulation defect caused by StoA deficiency was found to be suppressed by inactivation of bdbD (strain LUL23; Table 3). This indicated that StoA is a thiol-disulfide oxidoreductase with a reductive function. Addition of the reducing thiol reagent dithiothreitol (DTT) to the growth medium can overcome cytochrome c deficiency in strains lacking CcdA or ResA (9, 10). However, inclusion of DTT in the growth medium (15 mM DTT in NSMP medium added at different time points during vegetative growth and sporulation) did not complement the sporulation defect of strain LUL20 and only slightly complemented the sporulation defect of strain LU60A1 (data not shown). One reason for these negative results might be that DTT is unable to penetrate the cytoplasmic membrane and thus will not reach the relevant location, i.e., the space surrounding the engulfed forespore. Colonies lacking CcdA or StoA lyse on plates after a few days of incubation at room temperature. Microcolonies growing among lysed cells are observed after prolonged incubation at room temperature. Bacteria isolated from such microcolonies behave as wild-type cells on agar plates and show normal sporulation efficiency (31) (data not shown). Mutations that suppress CcdA deficiency are located in the bdbC or bdbD gene (10). The StoA deficiency suppressor mutations have not been identified but are most likely in bdbC or bdbD.
StoA cannot complement ResA deficiency and StoA is not involved in cytochrome c synthesis.
CcdA and ResA function in cytochrome c biogenesis by reducing the heme binding site of apo-cytochrome c prior to covalent attachment of the heme cofactor (9, 10). StoA and ResA show 51% sequence similarity and 28% identity. Both proteins have the thioredoxin-like domain anchored to the membrane by one transmembrane segment constituted by the N-terminal part of the polypeptide. To investigate if StoA has any role in cytochrome c synthesis, TMPD oxidation of colonies and cytochrome c oxidation activity assay of isolated membranes were preformed on strains 1A1 and LUL20. Both assays test for the presence of functional cytochrome caa3 oxidase (9). LUL20 showed the same cytochrome c oxidase phenotype as 1A1 (data not shown). Therefore, StoA is not involved in cytochrome c synthesis or assembly of the oxidase.
A strain lacking ResA (LUL9) remained TMPD oxidation negative when stoA was expressed in trans from the plasmid pLLE83. StoA therefore does not complement ResA deficiency. Also, plasmid pLLE82 (containing resA) in LUL20 was unable to rescue the sporulation defect. This indicated that ResA cannot function as a substitute for StoA. Thus, ResA and StoA are either targeted to different subcellular locations and/or they have very different substrate specificity.
StoA has thioredoxin-like activity.
The thioredoxin-like domain of StoA and BdbD were produced in E. coli TOP10 containing plasmid pLLE65 and pLLE34, respectively. The water-soluble His-tagged StoA and BdbD proteins were purified as described in Materials and Methods. In the in vitro insulin reduction assay where the rate of precipitation of reduced insulin is measured using a spectrophotometer, the water-soluble StoA showed an activity of 7.5 × 103 A650/(min × mol). Purified E. coli thioredoxin, used as a reference, showed an activity of 30.8 × 103 A650/(min × mol). Water-soluble BdbD showed no detectable activity in this assay, as could be expected from the oxidative function of the protein. The results demonstrated that the water-soluble domain of StoA has thioredoxin-like activity.
YkvU is not an electron donor to StoA.
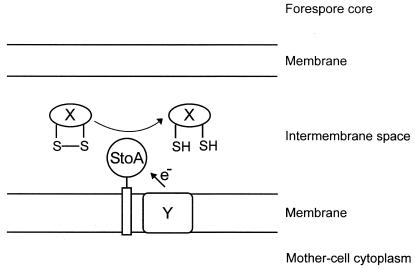
The stoA gene is transcribed from a σE-dependent promoter (8), which is only active in the mother cell, and StoA is seemingly involved in cortex synthesis. Therefore, the thioredoxin-like domain of StoA is probably localized in the intermembrane compartment in the forespore during spore maturation. At this location the protein is apparently involved in breaking disulfide bonds (Fig. 4). Reducing equivalents need to be transported to StoA from the mother cell or forespore cytoplasm to recycle StoA after each of its catalytic steps. In cytochrome c synthesis, ResA seems recycled by CcdA which transfers the reducing equivalents required for disulfide bond breakage from the thioredoxin system in the cytoplasm. As was mentioned, CcdA is unlikely to be an electron donor to StoA because StoA deficiency results in a ∼100-fold-stronger negative effect on sporulation efficiency than CcdA deficiency (Table 3). The ykvU gene upstream of stoA (Fig. 1) encodes a protein of unknown function but with sequence similarity to SpoVB, which is involved in cortex synthesis. YkvU has 12 predicted transmembrane segments and four cysteine residues. The protein seems selectively localized in the forespore outer membrane during spore synthesis as determined by using a fusion to green fluorescent protein and microscopy of cells (8). YkvU could be a reductase or a substrate protein for StoA. The sporulation efficiency of strain LUL30, with both ykvU and stoA deleted, was found to be lower than that for LUL20 (Table 3). This indicated that YkvU might play some role in sporulation. Electron microscopy examination of endospores of strain LUL30 showed that they were similar to LUL20 endospores, i.e., contained inner and outer spore coat layers but were deficient in cortex (Fig. 3C). Strains LUL20 and LUL30 containing pLLE83 (which carries the stoA gene) showed approximately 1,000-fold-higher sporulation efficiency than LUL20 and LUL30 containing only the plasmid vector, pDG148 (Table 5). Thus, stoA on a plasmid can complement the sporulation defect of strain LUL30 to the same relative level as it complements that of LUL20. YkvU is therefore probably not an electron donor to StoA. However, it cannot be excluded that YkvU is a cortex synthesis protein and a substrate for StoA because some other thiol-disulfide oxidoreductase might partially complement for StoA deficiency.
FIG. 4.
Proposed subcellular localization, topology, and function of StoA in the intermembrane space of a forespore engulfed by the mother cell. The sporulation protein(s) (X) that requires disulfide bond reduction catalyzed by StoA and the electron donor (Y) to StoA remain to be identified. Thioredoxin (TrxA) in the mother-cell cytoplasm might be the donor of reducing equivalents to the electron donor and indirectly to StoA.
Conclusion.
B. subtilis StoA is a membrane-bound thiol-disulfide oxidoreductase important for spore maturation. Spores of StoA-deficient strains are heat, lysozyme, and chloroform sensitive and seemingly lack the cortex layer. The thioredoxin-like domain of StoA is probably present in the intermembrane space of the forespore and catalyzes disulfide bond breakage in cortex components or in proteins that are important for cortex synthesis (Fig. 4). StoA is not required for efficient spore synthesis if BdbD is absent. BdbD together with BdbC constitute a membrane-bound system that catalyzes disulfide bond formation in proteins on the outer side of the cytoplasmic membrane. The BdbD/BdbC system is required for competence by catalyzing disulfide bond formation in two Com proteins (10, 24) but is not needed for spore synthesis. It therefore remains uncertain whether disulfide bond formation catalyzed by protein factors is important for endospore synthesis. Our findings suggest that BdbD catalyzes formation of disulfide bonds in some presently unknown substrate molecules resulting in spore cortex deficiency. StoA counteracts (or corrects) this effect of BdbD activity by breaking disulfide bonds. Such functional counteraction would be analogous to the situation in cytochrome c synthesis where BdbD catalyzes the formation of a disulfide bond in apo-cytochrome c and ResA specifically breaks this bond (9).
Acknowledgments
We are grateful to Ingrid Stål for technical assistance and Rita Wallén for the expert help with electron microscopy. We thank Fredrik Johansson for contributions regarding the construction and analysis of strain LUL30.
This work was supported by a grant from The Swedish Research Council (621-2001-3125) to L.H.
REFERENCES
- 1.Aronson, A. I., and P. Fitz-James. 1976. Structure and morphogenesis of the bacterial spore coat. Bacteriol. Rev. 40:360-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asai, K., H. Takamatsu, M. Iwano, T. Kodama, K. Watabe, and N. Ogasawara. 2001. The Bacillus subtilis yabQ gene is essential for formation of the spore cortex. Microbiology 147:919-927. [DOI] [PubMed] [Google Scholar]
- 3.Beall, B., and C. P. Moran, Jr. 1994. Cloning and characterization of spoVR, a gene from Bacillus subtilis involved in spore cortex formation. J. Bacteriol. 176:2003-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolhuis, A., G. Venema, W. J. Quax, S. Bron, and J. M. van Dijl. 1999. Functional analysis of paralogous thiol-disulfide oxidoreductases in Bacillus subtilis. J. Biol. Chem. 274:24531-24538. [DOI] [PubMed] [Google Scholar]
- 5.Crow, A., R. M. Acheson, N. E. Le Brun, and A. Oubrie. 2004. Structural basis of redox-coupled protein substrate selection by the cytochrome c biosynthesis protein ResA. J. Biol. Chem. 279:23654-23660. [DOI] [PubMed] [Google Scholar]
- 6.Dorenbos, R., T. Stein, J. Kabel, C. Bruand, A. Bolhuis, S. Bron, W. J. Quax, and J. M. Van Dijl. 2002. Thiol-disulfide oxidoreductases are essential for the production of the lantibiotic sublancin 168. J. Biol. Chem. 277:16682-16688. [DOI] [PubMed] [Google Scholar]
- 7.Driks, A. 1999. Bacillus subtilis spore coat. Microbiol. Mol. Biol. Rev. 63:1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eichenberger, P., S. T. Jensen, E. M. Conlon, C. van Ooij, J. Silvaggi, J. E. Gonzalez-Pastor, M. Fujita, S. Ben-Yehuda, P. Stragier, J. S. Liu, and R. Losick. 2003. The σE regulon and the identification of additional sporulation genes in Bacillus subtilis. J. Mol. Biol. 327:945-972. [DOI] [PubMed] [Google Scholar]
- 9.Erlendsson, L. S., R. M. Acheson, L. Hederstedt, and N. E. Le Brun. 2003. Bacillus subtilis ResA is a thiol-disulfide oxidoreductase involved in cytochrome c synthesis. J. Biol. Chem. 278:17852-17858. [DOI] [PubMed] [Google Scholar]
- 10.Erlendsson, L. S., and L. Hederstedt. 2002. Mutations in the thiol-disulfide oxidoreductases BdbC and BdbD can suppress cytochrome c deficiency of CcdA-defective Bacillus subtilis cells. J. Bacteriol. 184:1423-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feucht, A., L. Evans, and J. Errington. 2003. Identification of sporulation genes by genome-wide analysis of the σE regulon of Bacillus subtilis. Microbiology 149:3023-3034. [DOI] [PubMed] [Google Scholar]
- 12.Fortnagel, P., and E. Freese. 1968. Analysis of sporulation mutants. II. Mutants blocked in the citric acid cycle. J. Bacteriol. 95:1431-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guérout-Fleury, A. M., K. Shazand, N. Frandsen, and P. Stragier. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167:335-336. [DOI] [PubMed] [Google Scholar]
- 14.Hambraeus, G., C. Von Wachenfeldt, and L. Hederstedt. 2003. Genome-wide survey of mRNA half-lives in Bacillus subtilis identifies extremely stable mRNAs. Mol. Genet. Genomics 269:706-714. [DOI] [PubMed] [Google Scholar]
- 15.Hanahan, D., J. Jessee, and F. R. Bloom. 1991. Plasmid transformation of Escherichia coli and other bacteria. Methods Enzymol. 204:63-113. [DOI] [PubMed] [Google Scholar]
- 16.Hederstedt, L. 1986. Molecular properties, genetics, and biosynthesis of Bacillus subtilis succinate dehydrogenase complex. Methods Enzymol. 126:399-414. [DOI] [PubMed] [Google Scholar]
- 17.Hoch, J. A. 1991. Genetic analysis in Bacillus subtilis. Methods Enzymol. 204:305-320. [DOI] [PubMed] [Google Scholar]
- 18.Holmgren, A. 1979. Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J. Biol. Chem. 254:9627-9632. [PubMed] [Google Scholar]
- 19.Kadokura, H., F. Katzen, and J. Beckwith. 2003. Protein disulfide bond formation in prokaryotes. Annu. Rev. Biochem. 72:111-135. [DOI] [PubMed] [Google Scholar]
- 20.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 21.Le Brun, N. E., J. Bengtsson, and L. Hederstedt. 2000. Genes required for cytochrome c synthesis in Bacillus subtilis. Mol. Microbiol. 36:638-650. [DOI] [PubMed] [Google Scholar]
- 22.Marmur, J. 1961. A procedure for the isolation of deoxyribonucleic acid from microorganisms. J. Mol. Biol. 3:208-218. [Google Scholar]
- 23.McGuffin, L. J., K. Bryson, and D. T. Jones. 2000. The PSIPRED protein structure prediction server. Bioinformatics 16:404-405. [DOI] [PubMed] [Google Scholar]
- 24.Meima, R., C. Eschevins, S. Fillinger, A. Bolhuis, L. W. Hamoen, R. Dorenbos, W. J. Quax, J. M. van Dijl, R. Provvedi, I. Chen, D. Dubnau, and S. Bron. 2002. The bdbDC operon of Bacillus subtilis encodes thiol-disulfide oxidoreductases required for competence development. J. Biol. Chem. 277:6994-7001. [DOI] [PubMed] [Google Scholar]
- 25.Niaudet, B., A. Goze, and S. D. Ehrlich. 1982. Insertional mutagenesis in Bacillus subtilis: mechanism and use in gene cloning. Gene 19:277-284. [DOI] [PubMed] [Google Scholar]
- 26.Pandey, N. K., and A. I. Aronson. 1979. Properties of the Bacillus subtilis spore coat. J. Bacteriol. 137:1208-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reynolds, E. S. 1963. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 17:208-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roth, R., and C. Hägerhäll. 2001. Transmembrane orientation and topology of the NADH:quinone oxidoreductase putative quinone binding subunit NuoH. Biochim. Biophys. Acta 1504:352-362. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
- 30.Schägger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 31.Schiött, T., and L. Hederstedt. 2000. Efficient spore synthesis in Bacillus subtilis depends on the CcdA protein. J. Bacteriol. 182:2845-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schiött, T., M. Throne-Holst, and L. Hederstedt. 1997. Bacillus subtilis CcdA-defective mutants are blocked in a late step of cytochrome c biogenesis. J. Bacteriol. 179:4523-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Setlow, P. 1995. Mechanisms for the prevention of damage to DNA in spores of Bacillus subtilis. Annu. Rev. Microbiol. 49:29-54. [DOI] [PubMed] [Google Scholar]
- 34.Steinmetz, M., and R. Richter. 1994. Plasmids designed to alter the antibiotic resistance expressed by insertion mutations in Bacillus subtilis, through in vivo recombination. Gene 142:79-83. [DOI] [PubMed] [Google Scholar]
- 35.Stragier, P., C. Bonamy, and C. Karmazyn-Campelli. 1988. Processing of a sporulation sigma factor in Bacillus subtilis: how morphological structure could control gene expression. Cell 52:697-704. [DOI] [PubMed] [Google Scholar]
- 36.von Wachenfeldt, C., and L. Hederstedt. 1990. Bacillus subtilis holo-cytochrome c-550 can be synthesised in aerobic Escherichia coli. FEBS Lett. 270:147-151. [DOI] [PubMed] [Google Scholar]