Abstract
General stress proteins protect Bacillus subtilis cells against a variety of environmental insults. This adaptive response is particularly important for nongrowing cells, to which it confers a multiple, nonspecific, and preemptive stress resistance. Induction of the general stress response relies on the alternative transcription factor, SigB, whose activity is controlled by a partner switching mechanism that also involves the anti-sigma factor, RsbW, and the antagonist protein, RsbV. Recently, the SigB regulon has been shown to be continuously induced and functionally important in cells actively growing at low temperature. With the exception of this chill induction, all SigB-activating stimuli identified so far trigger a transient expression of the SigB regulon that depends on RsbV. Through a proteome analysis and Northern blot and gene fusion experiments, we now show that the SigB regulon is continuously induced in cells growing actively at 51°C, close to the upper growth limit of B. subtilis. This heat induction of SigB-dependent genes requires the environmental stress-responsive phosphatase RsbU, but not the metabolic stress-responsive phosphatase RsbP. RsbU dependence of SigB activation by heat is overcome in mutants that lack RsbV. In addition, loss of RsbV alone or in combination with RsbU triggers a hyperactivation of the general stress regulon exclusively at high temperatures detrimental for cell growth. These new facets of heat induction of the SigB regulon indicate that the current view of the complex genetic and biochemical regulation of SigB activity is still incomplete and that SigB perceives signals independent of the RsbV-mediated signal transduction pathways under heat stress conditions.
In its natural habitats, Bacillus subtilis often faces hostile conditions that limit or prevent cell growth. Prevalence and proliferation of the bacterial cells require the constant monitoring of the environmental conditions and the mounting of appropriate genetic and cellular defense reactions (33, 51). B. subtilis is well known for its ability to form highly resistant endospores as a measure of last resort, thereby allowing survival of part of the bacterial population for extended time periods even under extreme conditions (44). The synthesis of stress proteins constitutes another route for cellular adaptation to unfavorable conditions. While stress-specific proteins defend the cell against particular environmental insults such as osmotic (12) or oxidative (26) stress, there is increasing evidence that general stress proteins provide cellular protection against a variety of environmental insults (24, 39). In B. subtilis, synthesis of these general stress proteins is controlled by the alternative transcription factor SigB (8, 11, 22, 47). The SigB-dependent general stress regulon comprises approximately 150 members (3, 25, 38, 40), and its induction affords cells with a multiple, nonspecific, and preemptive stress resistance (4, 20, 21, 48).
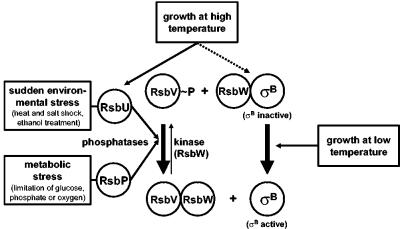
The activity of the sigma factor SigB is subjected to a tight biochemical regulation to (i) suppress the expression of the SigB regulon under nonstress conditions and (ii) allow rapid induction of the whole regulon following the imposition of diverse stresses (24, 39). During exponential growth at 37°C, where increased levels of general stress proteins are not required, the activity of SigB is inhibited by binding to its primary regulatory protein, the anti-sigma factor RsbW (Fig. 1), thereby preventing SigB's association with core RNA polymerase (5). Release and thus activation of SigB from this inhibitory RsbW/SigB complex require the activity of the antagonist protein RsbV, which is capable of forming an alternative complex with RsbW (18). This partner switching of RsbW critically depends on the phosphorylation state of RsbV (1, 18). During growth, RsbV is rapidly phosphorylated and thereby inactivated by the kinase activity of the anti-sigma factor RsbW. After exposure to stress, RsbV is dephosphorylated by one of two specific PP2C-type phosphatases (RsbU and RsbP) (46, 53), allowing binding of RsbV to RsbW and thereby disruption of the inhibitory RsbW/SigB complex (Fig. 1).
FIG. 1.
Model for the regulation of SigB activity in B. subtilis. The environmental- and metabolic-stress-sensing branches of the signal transduction cascade convey their output via two PP2C-type phosphatases, RsbU and RsbP, to the antagonist protein RsbV (24, 39). Low-temperature growth (13) triggers SigB activation via a thus unresolved RsbV-independent pathway, which also functions in an rsbVUP triple mutant. Continuous growth at high temperature (51°C) prompts a sustained induction that strictly requires RsbU in a wild-type background (solid black line). However, the presence of the rsbV312 frameshift allele relieves this requirement for RsbU and allows SigB activation by growth at 51°C even in a triple mutant lacking RsbP, RsbU, and RsbV (dashed line).
SigB-activating stimuli can be allocated to two different groups based on the utilization of either the RsbU or the RsbP phosphatase. Sudden environmental stresses such as osmotic or thermal upshift and exposure to ethanol activate the RsbU phosphatase (Fig. 1) via the action of additional regulatory proteins (e.g., RsbR, RsbS, RsbT, and RsbX) (53). Metabolic stresses such as limitation of glucose, phosphate, or oxygen in turn are thought to activate the RsbP phosphatase (46). Each of these signals triggers a transient dephosphorylation of RsbV∼P (46, 49, 53) and the concomitant transient induction of the SigB-dependent general stress regulon (11, 50). This already complex picture of the control of SigB activity was further complicated by the recent finding that continued growth of B. subtilis in the cold triggers a long-lasting induction of the SigB regulon independently of the antagonist protein RsbV and the phosphatase RsbU or RsbP (Fig. 1) (13).
One of the environmental cues that transiently induces the SigB regulon is a heat shock from 37 to 48°C (8, 11, 47). Preadaptation through a moderate heat shock (48°C) provides wild-type cells with resistance against a strong heat challenge (54°C), a resistance that is largely lost in a sigB mutant (48). However, sigB mutants still display a considerable degree of inducible heat resistance that depends on the induction of other classes of heat shock genes (43). The heat shock stimulon of B. subtilis (25) comprises, besides the SigB regulon, at least four other groups of heat shock genes: (i) the HrcA regulon encoding the GroESL and DnaK-GrpE-DnaJ chaperones (42); (ii) the CtsR regulon coding for the proteases and/or chaperones of the Clp family (ClpP, ClpC, and ClpE) (17, 31); (iii) the CssRS-controlled htrA and htrB genes (16); and (iv) a diverse group of heat shock genes including htpG, lonA, ftsH, and clpX, for which no genetic control mechanism has been elucidated yet (43). In addition to the changes in gene expression that occur subsequent to a heat shock (25, 38), uptake of compatible solutes such as glycine betaine can also provide thermoprotection to B. subtilis cells propagated close to their upper growth limit of 52°C (29).
In contrast to all other SigB-activating stimuli observed so far, growth of B. subtilis in the cold (15°C) triggered a delayed but long-lasting induction of the entire SigB regulon that does not depend on the antagonist protein RsbV (13). These observations raise the question whether a similar atypical induction of the SigB-dependent general stress response also occurs when cells are propagated close to the maximal growth temperature of B. subtilis. In this report, we show that the SigB regulon is induced in cells that are exponentially growing at 51°C. Like the previously observed chill induction (13), heat induction of the SigB regulon does not depend on the key regulatory protein RsbV. In contrast, a hyperinduction of the entire general stress regulon is observed in mutants lacking RsbV.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
The experiments conducted in this study were performed with the B. subtilis strain 168 (33) or isogenic mutant derivatives of this wild-type strain (Table 1). The chromosomal gsiB-bgaB reporter gene fusion was transferred into the B. subtilis strain 168 background or its derivatives by transformation (15). The parental gene fusion strain BSM52 and the resulting derivatives are listed in Table 1. Bacteria were routinely grown under vigorous agitation (220 rpm) in Spizizen's Minimal Medium (SMM) with 0.5% (wt/vol) glucose as the carbon source, l-tryptophan (20 mg/liter), and a solution of trace elements (23). Precultures of B. subtilis strains were inoculated from exponentially growing overnight cultures propagated in SMM to a final optical density at 578 nm (OD578) of 0.1. These precultures were allowed to grow to an OD578 of 0.5, diluted to an OD578 of 0.1, and subsequently transferred to the higher growth temperatures indicated in the individual experiments. Ethanol stress was imposed onto the cells during exponential growth by the addition of ethanol to a final concentration of 4% (vol/vol). For drug resistance selection in B. subtilis, the following antibiotics were used (final concentrations are given in parentheses): chloramphenicol (5 μg/ml), spectinomycin (200 μg/ml), kanamycin (20 μg/ml), and erythromycin (1 μg/ml).
TABLE 1.
B. subtilis strains used in this study
| Strain | Relevant genotype | Construction or referenced |
|---|---|---|
| 168 | trpC2 | 33 |
| BSA73 | trpC2 rsbU::kan rsbV312 SPβ ctc::lacZ | 6e |
| BSM21 | trpC2 rsbU::kan | 13 |
| BSM24 | trpC2 rsbV312 rsbX::pWH25-spca | 13 |
| BSM29 | trpC2 sigBΔ2::spc | 13 |
| BSM30 | trpC2 rsbP::spc | 13 |
| BSM52 | trpC2 amyE::pGK11gsiB::bgaB cat86 erm | G. Kuhnke and U. Völkere |
| BSM201 | trpC2 rsbP::spc | 13e |
| BSM269 | trpC2 amyE::pGK30cgsiB::gfp cat86 | 13 |
| BSM275 | trpC2 rsbV312 rsbX::pWH25-spca amyE::pGK30c-gsiB::gfp cat86 | 13 |
| BSM276 | trpC2 sigBΔ2::spc amyE::pGK30c-gsiB::gfp cat86 | 13 |
| BSM277 | trpC2 rsbU::kan amyE::pGK30c-gsiB::gfp cat86 | 13 |
| BSM278 | trpC2 rsbP::spc amyE::pGK30c-gsiB::gfp cat86 | 13 |
| BSM279 | trpC2 rsbP::spc rsbU::kan rsbV312 | 13 |
| BSM280 | trpC2 rsbP::spc rsbU::kan rsbV312 amyE::pGK30c-gsiB::gfp cat86 | 13 |
| BSG1 | trpC2 rsbU::kan rsbV312 amyE::pGK30c-gsiB::gfp cat86 | BSA73 ⇒ BSM269 |
| BSG13 | trpC2 rsbP::spc rsbU::kan amyE::pGK30c-gsiB::gfp cat86 | BSM201 ⇒ BSM277 |
| 168F | trpC2 amyE::pGK11b-gsiB::bgaB cat86 erm | BSM52 ⇒ 168 |
| BSM21F | trpC2 rsbU::kan amyE::pGK11b-gsiB::bgaB cat86 erm | BSM52 ⇒ BSM21 |
| BSM24F | trpC2 rsbV312 rsbX::pWH25-spca-amyE::pGK11b-gsiB::bgaB cat86 erm | BSM52 ⇒ BSM24 |
| BSM29F | trpC2 sigBΔ2::spc amyE::pGK11b-gsiB::bgaB cat86 erm | BSM52 ⇒ BSM29 |
| BSM30F | trpC2 rsbP::spc amyE::pGK11b-gsiB::bgaB cat86 erm | BSM52 ⇒ BSM30 |
| BSM279F | trpC2 rsbP::spc rsbU::kan rsbV312 amyE::pGK11b-gsiB::bgaB cat86 erm | BSM52 ⇒ BSM279 |
The integrative plasmid pWH25 contains a 2-kb EcoRI-SphI fragment, including the 3′ end of rsbX and 1.9 kb downstream of rsbX.
The strain contains part of plasmid pGK11 inserted into the amyE locus of the chromosome via a double-crossover event, thus linking the transcriptional gsiB-bgaB fusion to the chloramphenicol (cat86) and erythromycin (erm) resistance genes.
The strain contains part of plasmid pGK30 inserted into the amyE locus of the chromosome via a double-crossover event, thus linking the transcriptional gsiB-gfp fusion to the chloramphenicol resistance gene (cat86).
The arrow indicates the construction of the strain by transformation.
BSA73, BSM52, and BSM201 are derivatives of the B. subtilis wild-type strain PY22 (P. Youngman, University of Georgia, Athens, Ga.)
RNA isolation and Northern blot analysis.
Total RNA was isolated from B. subtilis cells growing exponentially (OD578, 0.5 to 1.0) at 37 or 51°C by the acid-phenol method (2, 34) with the modifications introduced by Völker et al. (47). Aliquots (4 μg) of the total RNA were used for the Northern blot analysis of the expression profile of gsiB. Digoxigenin-labeled antisense RNA probes were generated by in vitro transcription using a StripEZ kit (Ambion, Inc., Woodward, Tex.) and a gene-specific PCR product as the template. In the PCR with chromosomal DNA prepared from the B. subtilis strain 168, one of the DNA primers carried the sequence of the T7 promoter (gsiB-for, 5′-GAGACCCGGGTTTTGTTTGTTTAAAAGAATTG-3′, gsiB-rev, 5′-TAATACGACTCACTATAGGGAGGTCGTTGTTGCGGGCGT-3′). The PCR fragment was subsequently used for in vitro RNA synthesis with commercially available T7 RNA polymerase (Ambion, Inc.), yielding a hybridization probe internal to the gsiB gene of 469 nt. Denaturing RNA electrophoresis on agarose gels, RNA transfer by diffusion onto a nylon membrane (NY13N; Schleicher & Schuell, Dassel, Germany), hybridization to gene-specific probes, and signal detection were performed as described by Holtmann et al. (28).
2-DE.
Crude protein extracts for the separation by two-dimensional protein gel electrophoresis (2-DE) were prepared from 200-ml cultures grown in 2-liter flasks to an OD578 of 0.6. High-resolution 2-DE with immobilized pH gradients (pH 4 to 7) in the first dimension was performed as previously described (27). Analytical gels were stained with silver nitrate according to the method of Blum et al. (10). After scanning, analysis and quantification of the 2-DE gel images were performed with the Delta 2D software package (Decodon GmbH, Greifswald, Germany). Subsequent to the matching of the gel sets, the background intensity of all spots was subtracted and relative spot volumes (background corrected fraction of total spot intensity on the gel contained in the individual spot) of cultures grown at 37°C were compared with those of cultures grown at 51°C. Proteins were considered to be repressed or induced when the relative spot volume differences between cultures grown at 37°C and those grown at 51°C were observed to be at least fourfold. Two separate gels of each condition were analyzed, and only changes in the protein pattern passing the cutoff criterion on both parallels were considered significant. Proteins were identified by peptide mass fingerprinting as previously described (27) or by comparison with the master gel of B. subtilis 168 (http://microbio1.biologie.uni-greifswald.de/sub2d.htm) (14).
Western blot analysis.
The Western blot analysis was carried out as described previously (7, 50). The monoclonal antibodies raised against RsbS, RsbV, RsbW, SigB, and RsbX have been described (7, 18, 19).
Determination of BgaB activity.
In this set of experiments, a β-galactosidase gene (bgaB) from Bacillus stearothermophilus whose product (BgaB) is stable at high temperature was used (41). For the determination of β-galactosidase activity of gsiB-bgaB fusion strains (Table 1), cultures were propagated as described above. At appropriate time points, 1-ml aliquots were harvested by centrifugation in a Heraeus tabletop centrifuge at 4°C. Cells were resuspended in 800 μl of modified Z-buffer (41) and permeabilized by addition of 10 μl of toluol; β-galactosidase enzyme activity was then assayed at 52°C as previously described (41). BgaB activity is given in units according to Miller (37).
RESULTS
High-temperature induction of the SigB-dependent general stress regulon.
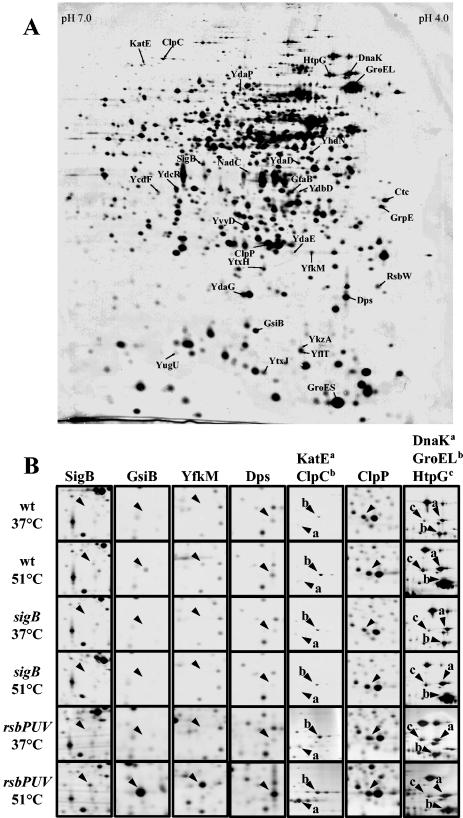
Brigulla et al. (13) recently reported that continued growth of B. subtilis in the cold (15°C) triggers a long-lasting induction of the SigB-controlled general stress regulon. This study challenged the belief that induction of the general stress regulon is only a transient response to various environmental stresses (24, 39), and it raised the question whether long-term induction also occurred as a response to heat stress. A sudden temperature upshift from 37 to 48°C is known to transiently induce the SigB regulon (8, 11, 47), but how the SigB regulon responds to cultivation of B. subtilis close to its maximal growth temperature of 52°C has not yet been addressed (29). High-resolution 2-DE is a convenient tool to monitor changes in the protein synthesis profile of the cells on a global scale (9). We therefore carried out a comparative proteome analysis of cultures of B. subtilis 168 propagated at 37 or 51°C, and these experiments revealed extensive temperature-dependent differences in the cytosolic protein profile with more than 128 repressed and 72 induced proteins at 51°C (data not shown). These changes included increased levels of well-known SigB-dependent general stress proteins such as GsiB, Dps, Ctc, SigB, KatE, YfkM, and YkzA at 51°C (Fig. 2). Hence, continued growth at 51°C prompts the induction of the SigB regulon, and, as expected, this phenomenon was not observed in a sigB mutant (Fig. 2).
FIG. 2.
Influence of growth at high temperature on the protein profile of B. subtilis. After staining with silver nitrate, the gels were scanned with an imaging system and analyzed with the DELTA-2D software package from DECODON GmbH (Greifswald, Germany). (A) Cytosolic protein pattern of B. subtilis 168 grown in SMM at 51°C. Proteins induced at high temperature are labeled with arrows and their corresponding names. (B) Enlarged sections of gels prepared from crude protein extracts of cells of the B. subtilis wild-type strain 168 or its mutant derivatives BSM29 (sigB) and BSM279 (rsbP rsbU rsbV) grown at either 37 or 51°C. Representative examples of the different classes of heat shock proteins are displayed as follows: for the SigB regulon, SigB, GsiB, YfkM, Dps, and KatE; for the CtsR regulon, ClpC and ClpP; for the HrcA regulon, DnaK and GroEL; for Class IV, HtpG.
As noted above, induction of proteins at high growth temperature was not confined to members of the SigB regulon but also included increased production of other typical heat shock proteins such as DnaK, GrpE, GroEL, GroES, and HtpG (Fig. 2). Consistent with their previously published induction pattern (43), these proteins remained induced in a sigB mutant (Fig. 2B). Notably, the amount of the heat shock proteins ClpP and ClpC, which are under the dual control of the CtsR repressor and SigB (17, 31), increased only in the high-temperature-grown cells of the wild-type and not in those of the sigB mutant (Fig. 2).
High-temperature regulation of the sigB operon.
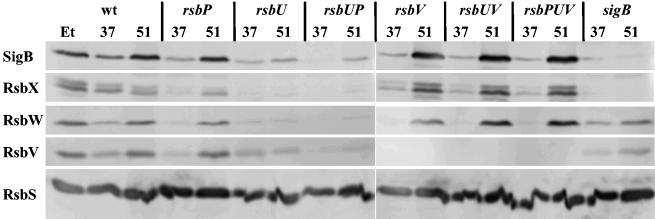
SigB, its main regulatory proteins RsbW and RsbV, the phosphatase RsbU, and four additional regulatory proteins (RsbR, RsbS, RsbT, and RsbX) are encoded within the sigB operon (rsbR-rsbS-rsbT-rsbU-rsbV-rsbW-sigB-rsbX). Basal levels of the sigB operon products are mainly provided by transcription initiating at a vegetative promoter positioned upstream of the rsbR gene (30, 52). A second SigB-dependent promoter located between rsbU and rsbV is induced by all SigB-activating environmental and metabolic stimuli described thus far (13, 24, 39), thereby accounting for an amplification of the SigB-mediated general stress response (8, 11, 13). The proteome experiments described above (Fig. 2) demonstrate that the SigB regulon is induced in cells cultured at 51°C. Therefore, one would predict that the RsbV, RsbW, SigB, and RsbX proteins are synthesized in greater amounts in cells growing exponentially at 51°C than in cells cultivated at 37°C. To test this prediction experimentally, we used a set of monoclonal antibodies directed against these proteins in Western blot analyses of crude protein extracts prepared from B. subtilis 168 cultures grown either at 37 or at 51°C. As a control, we used a monoclonal antibody directed against RsbS, which is also encoded by the sigB operon but whose gene is located upstream of the SigB-dependent promoter. Thus, the cellular levels of RsbS do not respond to SigB activity and should not increase during heat stress. As documented in Fig. 3, growth of the cells at 51°C triggered an increase in the levels of RsbV, RsbW, and SigB similar to that observed after a 60-min exposure to 4% (vol/vol) ethanol, a well-known strong inducer of the SigB regulon (11, 50). This increase was almost completely abolished in a sigB mutant. Residual low-level SigB-independent increases in the levels of RsbV and RsbW by heat shock have been observed before (50). Neither ethanol nor heat stress significantly influenced the level of the control protein RsbS (Fig. 3). Thus, this Western blot analysis supports the initial observation of the proteome study of a high-temperature-mediated increase in the level of SigB, the master regulator of the general stress response, and its main regulatory proteins, RsbW and RsbV.
FIG. 3.
Influence of high temperature on the levels of the products of the sigB operon in B. subtilis 168 and its isogenic SigB regulatory mutants. Crude protein extracts were prepared from cultures grown at either 37°C (37) or 51°C (51). As a control, we treated the wild-type strain with 4% (vol/vol) ethanol (Et) during exponential growth at 37°C for 60 min to induce the SigB regulon. After separation of the crude protein extracts by sodium dodecyl sulfate-polyacrylamide protein gel electrophoresis and transfer of the proteins to a nitrocellulose membrane, the proteins were allowed to react with a set of monoclonal antibodies. Specific antibody binding was detected by an alkaline phosphatase-conjugated goat anti-mouse secondary antibody. Proteins are indicated with their designations, and the following strains were included in the experiment: BSM269 (wild type), BSM275 (rsbV), BSM276 (sigB), BSM277 (rsbU), BSM278 (rsbP), BSM280 (rsbPUV), BSG01 (rsbUV), and BSG13 (rsbUP).
High-temperature regulation of the sigB operon products in mutants lacking SigB regulatory proteins.
Heat shock has been shown to induce the SigB regulon via the environmental branch of the signal transduction pathway that is dependent on a functional RsbU protein (50). Disruption of the rsbP gene did not prevent heat induction of SigB and its main regulators (RsbV and RsbW), whereas insertional inactivation of rsbU alone or in combination with an rsbP mutation almost completely abolished the accumulation of SigB, RsbV, and RsbW in cells growing actively at 51°C (Fig. 3). With the exception of the chill induction of the SigB regulon (13), all SigB-activating stimuli depend on an intact RsbV protein to elicit the induction of the general stress regulon (24, 39). We now observed that RsbV was not required for heat induction of either RsbW, SigB, or RsbX (Fig. 3). Surprisingly, heat induction of these proteins occurred in an rsbUV double mutant, and their increased levels were even manifested in an rsbPUV triple mutant (Fig. 3). Thus, RsbU seems to be essential for heat induction of sigB operon products in the presence of RsbV but is not required in cells carrying the rsbV312 frameshift allele (thus, lacking RsbV).
Hyperinduction of the SigB-dependent gsiB gene in an rsbV312 mutant.
The Western blot analysis already provided hints for a hyperinduction of sigB operon products in strains lacking RsbV (Fig. 3). To investigate this phenomenon further, we carried out a Northern blot analysis of the gsiB gene, whose transcription is exclusively driven by a SigB-dependent promoter and is therefore a suitable reporter for SigB activity in the cell (35). The gsiB gene was induced in cells cultured at 51°C, and this induction was prevented in a sigB mutant (Fig. 4). A much stronger accumulation of the gsiB transcript was detected in rsbV312 mutant cells grown at 51°C than in the wild type (Fig. 4). This strong transcription of the gsiB gene, however, was strictly confined to the high growth temperature, because there was no gsiB transcript detectable when the cells were grown at 37°C. The Northern blot analysis of gsiB thus provides strong evidence that RsbV is not necessary for high-temperature induction of SigB-dependent genes. On the contrary, its loss leads to hyperactivation of SigB. Hyperinduction of the whole SigB regulon in an rsbPUV triple mutant at 51°C was also observed in the proteome analysis (Fig. 2B).
FIG. 4.
Heat induction of the SigB-dependent gsiB gene. Total RNA was isolated from cultures of the wild-type strain 168 and its isogenic mutant derivatives grown at 37 and 51°C, electrophoretically separated on an agarose gel, blotted onto a nylon membrane, and hybridized with a single-stranded RNA probe specific for gsiB. Lanes 1 and 2, strain BSM24 (rsbV); lanes 3 and 4, strain BSM29 (sigB); lanes 5 and 6, wild-type strain 168. Lanes 1, 3, and 5, total RNA isolated from cultures grown at 37°C; lanes 2, 4, and 6, total RNA isolated from cultures grown at 51°C.
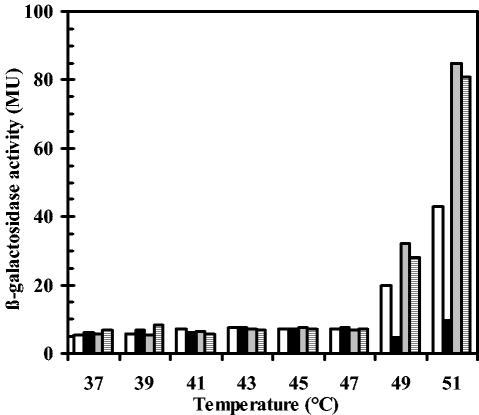
High-temperature induction of the gsiB gene is restricted to a narrow temperature range.
A strain carrying a chromosomal gsiB-bgaB reporter gene fusion was precultured at 37°C. Subsequent propagation of the cells was continued in a range of temperatures between 37 and 51°C. This experiment was carried out to determine the growth temperature above which SigB is permanently released from its negative regulation by the anti-sigma factor RsbW. Expression of the gsiB-bgaB fusion remained repressed in cells that were cultivated between 37 and 47°C (Fig. 5). At 49°C, transcription of the reporter gene fusion was induced in the wild-type strain, and the level of induction was further increased when the cells were grown at 51°C (Fig. 5). Fully consistent with the Northern blot analysis of the gsiB transcript in temperature-challenged cells (Fig. 4), a hyperinduction of the gsiB-bgaB transcription in strains that lack RsbV alone or that are simultaneously defective in the PP2C-type phosphatases RsbU and RsbP was observed (Fig. 5).
FIG. 5.
Temperature-dependent induction of a chromosomal gsiB-bgaB reporter gene fusion. The B. subtilis gsiB-bgaB fusion strains were grown in SMM at the indicated temperatures, and samples were withdrawn at an OD578 of 1.0 and assayed for β-galactosidase activity. β-Galactosidase activities are expressed in Miller units (MU). Open bars, wild-type strain 168; black bars, strain BSM29 (sigB); gray bars, strain BSM24 (rsbV); hatched bars, strain BSM279 (rsbPUV).
Time-resolved high-temperature induction of the gsiB-bgaB reporter gene fusion.
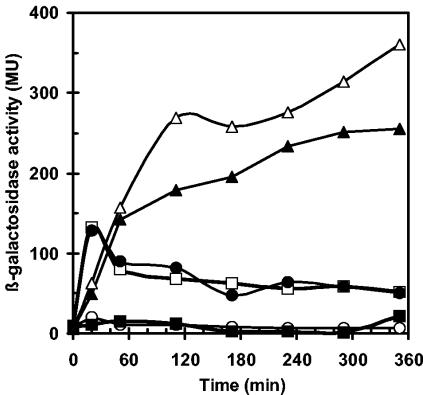
The data reported in Fig. 5 argue for a permanent expression of SigB-dependent genes in B. subtilis cells growing at 51°C, close to their maximal growth temperature of 52°C (29). By analyzing the β-galactosidase activity of the gsiB-bgaB fusion during the first 6 h after the temperature shift from 37 to 51°C, we investigated the kinetics of high-temperature-mediated induction of SigB-dependent genes. In accordance with previous data, a sudden temperature upshift triggered an immediate activation of the gsiB-bgaB reporter gene in a wild-type strain that peaked at 20 min after the shift from 37 to 51°C (Fig. 6). Thereafter, β-galactosidase activity slowly declined but remained significantly elevated above the preinduction basal level (37°C) throughout the experiment (Fig. 6). Thermoinduction of the gsiB-bgaB fusion was entirely prevented by the presence of a sigB mutation.
FIG. 6.
Time-resolved heat induction of a chromosomal gsiB-bgaB reporter gene fusion in the wild-type strain 168 and its isogenic mutant derivatives. A set of B. subtilis gsiB-bgaB reporter gene fusion strains was cultivated to mid-exponential phase (OD578, 0.5) at 37°C and utilized for the inoculation of cultures (OD578, 0.1) that were subsequently propagated at 51°C. Samples were removed for β-galactosidase assays at the indicated time points. The following strains were included in the experiment: 168F (wild type) (open squares); BSM29F (sigB) (filled squares); BSM21F (rsbU) (open circles); BSM30F (rsbP) (filled circles); BSM24F (rsbV) (open triangles); BSM279F (rsbPUV) (filled triangles).
A pattern of β-galactosidase activity like that of the wild type was also observed in a gsiB-bgaB fusion strain carrying a mutation in the gene (rsbP) encoding the metabolic stress responsive phosphatase RsbP (Fig. 6). This observation indicates that this branch of the SigB signal transduction cascade (Fig. 1) is not involved in thermosensing. In contrast, the introduction of an rsbU mutation into the reporter strain completely abolished the induction of the gsiB-bgaB gene fusion, thus demonstrating that the PP2C-phosphatase RsbU is critically involved in sensing high temperature. Inactivation of the antagonist protein RsbV by the frameshift mutation rsbV312 caused a short delay in the induction of the gsiB-bgaB fusion, but then the β-galactosidase activity of this fusion continued to rise throughout the experiment, reaching sevenfold-higher levels than in the wild-type strain approximately 6 h after the temperature upshift (Fig. 6). Furthermore, delayed hyperinduction of the SigB-dependent reporter gene construct also occurred in an rsbPUV triple mutant, lacking the antagonist protein RsbV as well as both RsbV∼P phosphatases (Fig. 6).
Growth at high temperature.
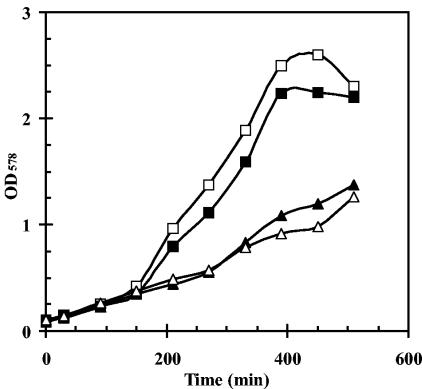
Growth experiments of B. subtilis under chill stress conditions (15°C) in a chemically defined minimal medium have shown that a sigB mutant has a distinct growth disadvantage in comparison to the wild-type strain (13). Furthermore, such a sigB mutant is highly sensitive to severe heat shock (54°C) (48). Even after preadaptation by a mild heat shock (48°C), a sigB mutant displayed extreme sensitivity to the strong heat challenge (54°C), whereas the same pretreatment conferred complete resistance to the wild-type strain (48). We therefore investigated whether a growth phenotype is associated with a loss of SigB activity in cells cultured at 51°C. We found that this was not the case (Fig. 7). However, the growth experiments revealed a strong reduction in growth rate, which was associated with the inactivation of rsbV (Fig. 7) that triggered hyperactivation of SigB (Fig. 3, 4, and 5). This reduced growth rate (at 51°C) was also observed at high temperature in a strain combining mutations in rsbV, rsbU, and rsbP (Fig. 7).
FIG. 7.
Growth of a B. subtilis wild-type strain and isogenic SigB regulatory mutants at high temperature. Strains were precultured to mid-exponential growth phase (OD578, 0.5) and used for the inoculation of cultures (OD578, 0.1) that were then propagated at 51°C. Strains: 168 (wild type) (open squares); BSM29 (sigB) (filled squares); BSM24 (rsbV) (open triangles); BSM279 (rsbPUV) (filled triangles).
DISCUSSION
The SigB-dependent general stress regulon of the soil bacterium B. subtilis is engaged when cells experience a wide range of environmental or metabolic stresses (24, 39). The physiological role of general stress proteins for cellular protection is emphasized by the finding that the disruption of the structural gene for its master regulator (SigB) causes sensitivity of the cell to a variety of stress factors, such as severe heat and salt shocks, ethanol treatment, low or high pH, and free oxygen radicals (4, 20, 21, 48).
Thus far, increased synthesis of the general stress proteins was believed to be particularly important under harsh stress conditions under which the bacterial cells are no longer able to grow (24, 39). A new facet of the physiological function of the SigB regulon was recently discovered by the finding that this regulon is induced to high levels in cells that actively grow under chill stress conditions (15°C) (13) or that are subjected to a sudden temperature downshift from 37 to 20°C (36). A heat shock at temperatures from 37 to 48°C has long been known to be one of the environmental cues that induce the SigB regulon (8, 11, 47). The data reported here now demonstrate a continuous high-level expression of SigB-dependent general stress proteins in cells that actively grow at 51°C (Fig. 2, 5, and 6), close to the maximal growth temperature (52°C) of B. subtilis (29). Growth at very high and low temperatures is likely to reflect more closely the situation in natural habitats of B. subtilis than the sudden harsh temperature up- or downshifts used in the laboratory to induce the SigB response. A sigB mutant cannot cope effectively with low-temperature (15°C) growth conditions (13), but such a mutant is not at a significant growth disadvantage when it is cultured at 51°C (Fig. 7). Lack of a specific growth phenotype of a sigB mutant strain at high temperature probably reflects the presence of redundant heat stress adaptation mechanisms in B. subtilis (43). Indeed, we observed strong induction of heat shock proteins (GroEL, GroES, DnaK, and GrpE) controlled by the HrcA repressor and of the heat shock protein HtpG in cells that were exponentially growing at 51°C (Fig. 2). It is well known that increased levels of chaperones are needed when cells have to cope with high growth temperatures (43, 54).
It is firmly established that environmental stimuli such as salt shock, ethanol treatment, and heat shock transiently induce the SigB response (8, 11, 47, 50). In contrast, continued growth close to the upper (Fig. 2, 5, and 6) or lower (13) temperature limits of B. subtilis leads to a sustained high-level expression of the general stress response to provide cells with a sufficient level of general stress proteins. At these temperature boundaries, permanent high-level expression of the SigB regulon occurs only in narrow temperature ranges (Fig. 5) (13).
Heat and salt shock and ethanol treatment activate SigB via the environmental branch of the signal transduction cascade that relies on the activity of the PP2C-type phosphatase RsbU (Fig. 1) (49, 50, 53). Whereas the requirement for RsbV in the sensing of salt shock and ethanol stress is firmly established, the role of RsbV in heat shock activation of SigB has been a matter of debate. Initially, SigB accumulation following heat shock had been observed to persist in cells lacking RsbV (8, 11). Due to the autoregulation of the sigB gene, this observation was interpreted as indicating an RsbV-independent SigB activation. Subsequently, the majority of the heat shock induction of the SigB regulon was shown to proceed via RsbU and RsbV and residual SigB accumulation in rsbV or rsbU mutants seemed to be largely independent of SigB activity (50). In a wild-type background, such a dependence on RsbU for thermoactivation of SigB was also observed for cells that were continuously cultured at 51°C (Fig. 6). In contrast, the metabolic stress-sensing PP2C-type phosphatase RsbP (Fig. 1) was not required for thermoinduction of SigB-dependent genes (Fig. 6). The RsbU dependence of thermoactivation of SigB both after a heat shock and during continued growth at 51°C suggests that in a wild-type background both processes are mediated by the same signal transduction pathway relying on a functional RsbV protein (Fig. 1).
However, a distinctively different regulation seems to occur in mutants that lack RsbV due to the frameshift mutation rsbV312. Thermoactivation of SigB during continuous growth at 51°C was also observed with strains carrying this rsbV312 mutation (Fig. 3 and 6), indicating that the requirement of RsbV can be bypassed (Fig. 1). To our great surprise, introduction of the rsbV312 allele into an rsbU mutant background overrides the RsbU dependence and restores thermoactivation of SigB (Fig. 3 and 6).
An RsbV-independent induction of general stress genes was also observed in chill-stressed B. subtilis cells (13). However, under heat stress, one observes a hyperinduction of the SigB regulon in an rsbV312 mutant (Fig. 4 and 6) that is not seen in chill-stressed rsbV312 mutant cells (13). This hyperinduction of SigB-dependent general stress genes is conserved both in an rsbUV double mutant and in an rsbPUV triple mutant (Fig. 2, 3, and 6) and is detrimental to the B. subtilis cell, since it results in a reduced growth rate at 51°C (Fig. 7).
Even if one cannot completely exclude that secondary effects such as a slightly modified RsbW/SigB ratio might contribute to the observed high-temperature activation of SigB in strains lacking RsbV, our data indicate that the current model for the genetic and biochemical control of SigB activity (24, 39) after exposure to heat seems to be incomplete (Fig. 1).
Heat stress is also a strong inducer of the SigB regulon in other gram-positive bacteria such as Staphylococcus aureus (32) and Bacillus cereus (45). Neither microorganism encodes orthologues of RsbT, RsbR, RsbS, and RsbX that are critically involved in signaling of environmental stress in B. subtilis (30, 46, 52, 53), indicating that the perception of heat stress in these organisms might also involve signal transduction components that have not yet been detected.
Acknowledgments
We are grateful to W. G. Haldenwang for providing the monoclonal antibodies directed against SigB and its regulatory proteins and F. Spiegelhalter and G. Kuhnke for the construction of plasmids. We thank V. Koogle for her help in editing the manuscript.
Financial support for this study was provided by the Deutsche Forschungsgemeinschaft through the SFB-395 and the Graduiertenkolleg “Proteinfunktion auf atomarer Ebene,” the Bundesministerium für Bildung und Forschung through the “Genomnetzwerk Göttingen,” the Max-Planck-Society, and the Fonds der Chemischen Industrie.
REFERENCES
- 1.Alper, S., A. Dufour, D. A. Garsin, L. Duncan, and R. Losick. 1996. Role of adenosine nucleotides in the regulation of a stress-response transcription factor in Bacillus subtilis. J. Mol. Biol. 260:165-177. [DOI] [PubMed] [Google Scholar]
- 2.Ambulos, N. P., Jr., E. J. Duvall, and P. S. Lovett. 1987. Method for blot-hybridization analysis of mRNA molecules from Bacillus subtilis. Gene 51:281-286. [DOI] [PubMed] [Google Scholar]
- 3.Antelmann, H., J. Bernhardt, R. Schmid, H. Mach, U. Völker, and M. Hecker. 1997. First steps from a two-dimensional protein index towards a response-regulation map for Bacillus subtilis. Electrophoresis 18:1451-1463. [DOI] [PubMed] [Google Scholar]
- 4.Antelmann, H., S. Engelmann, R. Schmid, and M. Hecker. 1996. General and oxidative stress responses in Bacillus subtilis—cloning, expression, and mutation of the alkyl hydroperoxide reductase operon. J. Bacteriol. 178:6571-6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benson, A. K., and W. G. Haldenwang. 1993. Bacillus subtilis σB is regulated by a binding protein (RsbW) that blocks its association with core RNA polymerase. Proc. Natl. Acad. Sci. USA 90:2330-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benson, A. K., and W. G. Haldenwang. 1992. Characterization of a regulatory network that controls σB expression in Bacillus subtilis. J. Bacteriol. 174:749-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benson, A. K., and W. G. Haldenwang. 1993. Regulation of σB levels and activity in Bacillus subtilis. J. Bacteriol. 175:2347-2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benson, A. K., and W. G. Haldenwang. 1993. The σB dependent promoter of the Bacillus subtilis sigB operon is induced by heat shock. J. Bacteriol. 175:1929-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernhardt, J., J. Weibezahn, C. Scharf, and M. Hecker. 2003. Bacillus subtilis during feast and famine: visualization of the overall regulation of protein synthesis during glucose starvation by proteome analysis. Genome Res. 13:224-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blum, H., H. Beier, and H. Gross. 1987. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis 8:93-99. [Google Scholar]
- 11.Boylan, S. A., A. R. Redfield, M. S. Brody, and C. W. Price. 1993. Stress-induced activation of the σB transcription factor of Bacillus subtilis. J. Bacteriol. 175:7931-7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bremer, E. 2002. Adaptation to changing osmolarity, p. 385-391. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives. From genes to cells. ASM Press, Washington, D.C.
- 13.Brigulla, M., T. Hoffmann, A. Krisp, A. Völker, E. Bremer, and U. Völker. 2003. Chill induction of the SigB-dependent general stress response in Bacillus subtilis and its contribution to low-temperature adaptation. J. Bacteriol. 185:4305-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Büttner, K., J. Bernhardt, C. Scharf, R. Schmid, U. Mader, C. Eymann, H. Antelmann, A. Völker, U. Völker, and M. Hecker. 2001. A comprehensive two-dimensional map of cytosolic proteins of Bacillus subtilis. Electrophoresis 22:2908-2935. [DOI] [PubMed] [Google Scholar]
- 15.Cutting, S. M., and P. B. Vander Horn. 1990. Genetic analysis, p. 27-74. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley & Sons, Chichester, United Kingdom.
- 16.Darmon, E., D. Noone, A. Masson, S. Bron, O. P. Kuipers, K. M. Devine, and J. M. van Dijl. 2002. A novel class of heat and secretion stress-responsive genes is controlled by the autoregulated CssRS two-component system of Bacillus subtilis. J. Bacteriol. 184:5661-5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Derre, I., G. Rapoport, and T. Msadek. 1999. CtsR, a novel regulator of stress and heat shock response, controls clp and molecular chaperone gene expression in Gram-positive bacteria. Mol. Microbiol. 31:117-132. [DOI] [PubMed] [Google Scholar]
- 18.Dufour, A., and W. G. Haldenwang. 1994. Interactions between a Bacillus subtilis anti-sigma factor (RsbW) and its antagonist (RsbV). J. Bacteriol. 176:1813-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dufour, A., U. Völker, A. Völker, and W. G. Haldenwang. 1996. Relative levels and fractionation properties of Bacillus subtilis σB and its regulators during balanced growth and stress. J. Bacteriol. 178:3701-3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Engelmann, S., and M. Hecker. 1996. Impaired oxidative stress resistance of Bacillus subtilis sigB mutants and the role of katA and katE. FEMS Microbiol. Lett. 145:63-69. [DOI] [PubMed] [Google Scholar]
- 21.Gaidenko, T. A., and C. W. Price. 1998. General stress transcription factor σB and sporulation transcription factor σH each contribute to survival of Bacillus subtilis under extreme growth conditions. J. Bacteriol. 180:3730-3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haldenwang, W. G., and R. Losick. 1980. Novel RNA polymerase sigma factor from Bacillus subtilis. Proc. Natl. Acad. Sci. USA 77:7000-7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harwood, C. R., and A. R. Archibald. 1990. Growth, maintenance and general techniques, p. 1-26. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley & Sons, Chichester, United Kingdom.
- 24.Hecker, M., and U. Völker. 2001. General stress response of Bacillus subtilis and other bacteria. Adv. Microb. Physiol. 44:35-91. [DOI] [PubMed] [Google Scholar]
- 25.Helmann, J. D., M. F. Wu, P. A. Kobel, F. J. Gamo, M. Wilson, M. M. Morshedi, M. Navre, and C. Paddon. 2001. Global transcriptional response of Bacillus subtilis to heat shock. J. Bacteriol. 183:7318-7328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herbig, A. F., and J. D. Helmann. 2002. Metal ion uptake and oxidative stress, p. 405-414. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives. From genes to cells. ASM Press, Washington, D.C.
- 27.Hoffmann, T., A. Schütz, M. Brosius, A. Völker, U. Völker, and E. Bremer. 2002. High-salinity-induced iron limitation in Bacillus subtilis. J. Bacteriol. 184:718-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holtmann, G., E. P. Bakker, N. Uozumi, and E. Bremer. 2003. KtrAB and KtrCD: two K+ uptake systems in Bacillus subtilis and their role in the adaptation to hypertonicity. J. Bacteriol. 185:1289-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holtmann, G., and E. Bremer. 2004. Thermoprotection of Bacillus subtilis by exogenously provided glycine betaine and structurally related compatible solutes: involvement of the Opu transporters. J. Bacteriol. 186:1683-1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kalman, S., M. L. Duncan, S. M. Thomas, and C. W. Price. 1990. Similar organization of the sigB and spoIIA operons encoding alternate sigma factors of Bacillus subtilis RNA polymerase. J. Bacteriol. 172:5575-5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krüger, E., and M. Hecker. 1998. The first gene of the Bacillus subtilis clpC operon, ctsR, encodes a negative regulator of its own operon and other class III heat shock genes. J. Bacteriol. 180:6681-6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kullik, I., and P. Giachino. 1997. The alternative sigma factor σB in Staphylococcus aureus: regulation of the sigB operon in response to growth phase and heat shock. Arch. Microbiol. 167:151-159. [DOI] [PubMed] [Google Scholar]
- 33.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, and A. Danchin. 1997. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 34.Majumdar, D., Y. J. Avissar, and J. H. Wyche. 1991. Simultaneous and rapid isolation of bacterial and eukaryotic DNA and RNA: a new approach for isolating DNA. BioTechniques 11:94-101. [PubMed] [Google Scholar]
- 35.Maul, B., U. Völker, S. Riethdorf, S. Engelmann, and M. Hecker. 1995. σB-dependent regulation of gsiB in response to multiple stimuli in Bacillus subtilis. Mol. Gen. Genet. 248:114-120. [DOI] [PubMed] [Google Scholar]
- 36.Mendez, M. B., L. M. Orsaria, V. Philippe, M. E. Pedrido, and R. R. Grau. 2004. Novel roles of the master transcription factors Spo0A and σB for survival and sporulation of Bacillus subtilis at low growth temperature. J. Bacteriol. 186:989-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 38.Petersohn, A., M. Brigulla, S. Haas, J. D. Hoheisel, U. Völker, and M. Hecker. 2001. Global analysis of the general stress response of Bacillus subtilis. J. Bacteriol. 183:5617-5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Price, C. W. 2000. Protective function and regulation of the general stress response in Bacillus subtilis and related gram-positive bacteria, p. 179-197. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 40.Price, C. W., P. Fawcett, H. Ceremonie, N. Su, C. K. Murphy, and P. Youngman. 2001. Genome-wide analysis of the general stress response in Bacillus subtilis. Mol. Microbiol. 41:757-774. [DOI] [PubMed] [Google Scholar]
- 41.Schrögel, O., and R. Allmansberger. 1997. Optimisation of the BgaB reporter system: determination of transcriptional regulation of stress responsive genes in Bacillus subtilis. FEMS Microbiol. Lett. 153:237-243. [DOI] [PubMed] [Google Scholar]
- 42.Schulz, A., and W. Schumann. 1996. hrcA, the first gene of the Bacillus subtilis dnaK operon, encodes a negative regulator of class I heat shock genes. J. Bacteriol. 178:1088-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schumann, W., M. Hecker, and T. Msadek. 2002. Regulation and function of heat-inducible genes in Bacillus subtilis, p. 359-368. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives. From genes to cells. ASM Press, Washington, D.C.
- 44.Sonenshein, A. L. 2000. Bacterial sporulation: a response to environmental signals, p. 199-215. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 45.van Schaik, W., M. H. Tempelaars, J. A. Wouters, W. M. de Vos, and T. Abee. 2004. The alternative sigma factor σB of Bacillus cereus: response to stress and role in heat adaptation. J. Bacteriol. 186:316-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vijay, K., M. S. Brody, E. Fredlund, and C. W. Price. 2000. A PP2C phosphatase containing a PAS domain is required to convey signals of energy stress to the σB transcription factor of Bacillus subtilis. Mol. Microbiol. 35:180-188. [DOI] [PubMed] [Google Scholar]
- 47.Völker, U., S. Engelmann, B. Maul, S. Riethdorf, A. Völker, R. Schmid, H. Mach, and M. Hecker. 1994. Analysis of the induction of general stress proteins of Bacillus subtilis. Microbiology 140:741-752. [DOI] [PubMed] [Google Scholar]
- 48.Völker, U., B. Maul, and M. Hecker. 1999. Expression of the σB-dependent general stress regulon confers multiple stress resistance in Bacillus subtilis. J. Bacteriol. 181:3942-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Völker, U., A. Völker, and W. G. Haldenwang. 1996. Reactivation of the Bacillus subtilis anti-σB antagonist, RsbV, by stress- or starvation-induced phosphatase activities. J. Bacteriol. 178:5456-5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Völker, U., A. Völker, B. Maul, M. Hecker, A. Dufour, and W. G. Haldenwang. 1995. Separate mechanisms activate σB of Bacillus subtilis in response to environmental and metabolic stresses. J. Bacteriol. 177:3771-3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wipat, A., and C. R. Harwood. 1999. The Bacillus subtilis genome sequence: the molecular blueprint of a soil bacterium. FEMS Microbiol. Ecol. 28:1-9. [Google Scholar]
- 52.Wise, A. A., and C. W. Price. 1995. Four additional genes in the sigB operon of Bacillus subtilis that control activity of the general stress factor σB in response to environmental signals. J. Bacteriol. 177:123-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang, X. F., C. M. Kang, M. S. Brody, and C. W. Price. 1996. Opposing pairs of serine protein kinases and phosphatases transmit signals of environmental stress to activate a bacterial transcription factor. Genes Dev. 10:2265-2275. [DOI] [PubMed] [Google Scholar]
- 54.Yura, T., M. Kanemori, and M. T. Morita. 2000. The heat shock response: regulation and function, p. 3-18. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.