Abstract
Brucella is the causative agent of the zoonotic disease brucellosis, which is endemic in many parts of the world. Genome sequencing of B. suis and B. melitensis revealed that both are complete denitrifiers. To learn more about the role of denitrification in these animal pathogens, a study of the role of denitrification in the closely related B. neotomae was undertaken. In contrast to B. suis and B. melitensis, it was found that B. neotomae is a partial denitrifier that can reduce nitrate to nitrite but no further. Examination of the B. neotomae genome showed that a deletion in the denitrification gene cluster resulted in complete loss of nirV and the partial deletion of nirK and nnrA. Even though the nor operon is intact, a norC-lacZ promoter fusion was not expressed in B. neotomae. However, the norC-lacZ fusion was expressed in the related denitrifier Agrobacterium tumefaciens, suggesting that the lack of expression in B. neotomae is due to inactivation of NnrA. A narK-lacZ promoter fusion was found to exhibit nitrate-dependent expression consistent with the partial denitrifier phenotype. Complementation of the deleted region in B. neotomae by using nirK, nirV, and nnrA from B. melitensis restored the ability of B. neotomae to reduce nitrite. There was a significant difference in the death of IRF-1−/− mice when infected with B. neotomae containing nirK, nirV, and nnrA and those infected with wild-type B. neotomae. The wild-type strain killed all the infected mice, whereas most of the mice infected with B. neotomae containing nirK, nirV, and nnrA survived.
Denitrification, the reduction of nitrate to gaseous nitrogen oxides, is most commonly found in eubacteria, but it has been observed in several archaeal species as well (37). Among the eubacteria, denitrification is most frequently observed in members of the α-proteobacteria. Denitrification has been actively studied in several free-living members of this group, such as Paracoccus denitrificans and Rhodobacter sphaeroides. Denitrification occurs in a number of symbiotic or pathogenic members of this group as well, but this process has not been as heavily studied in these bacteria. Denitrifying members of the α-proteobacteria that are symbionts or pathogens include the genera Rhizobium, Sinorhizobium, Bradyrhizobium, and Agrobacterium. Recently, genomic analysis has revealed that members of the genus Brucella, which are α-proteobacteria closely related to Agrobacterium and rhizobia, are denitrifiers (6, 24). Species in this genus are unique among this group of denitrifiers, since they are animal pathogens. Brucella is the causative agent of the zoonotic disease brucellosis, which is endemic in many parts of the world (15).
Even though only a few pathogens have been shown to be denitrifiers, recent work suggests genes encoding nitrogen oxide reductases may be important determinants of a pathogenic lifestyle. In Neisseria, a member of the β-proteobacterial group, nitrite and nitric oxide (NO) reductase (Nir and Nor, respectively) have been shown to be required for anaerobic growth and have been suggested to play a role in mitigating NO toxicity in the macrophage (1). In Brucella suis, transposon inactivation of the gene encoding nitrate reductase affected growth inside the macrophage (16). The utility of denitrification genes in pathogenesis is easy to rationalize, given the importance of NO in the host cell's defense against infection. Pathogens able to respire NO using Nor can decrease NO levels in their surroundings and gain an additional advantage by coupling this reaction to energy conservation. NO production can increase survival of Brucella in macrophages, consistent with this compound serving as a terminal respiratory oxidant (34).
This study was undertaken to examine the role of denitrification in the growth of Brucella in more detail. The strain chosen for the study was Brucella neotomae, isolated from desert wood rats but nonpathogenic for humans and domestic animals (23). This bacterium's genome has not been sequenced, but initial studies suggest its genome is very similar to the genomes of other Brucella species (8). Brucellae are facultative intracellular pathogens, and this study was initially designed to examine regulation and activity of the various nitrogen oxide reductases under free-living conditions. Unexpectedly, it was found that B. neotomae failed to grow under denitrifying conditions and accumulated nitrite when grown under oxygen-limiting conditions in nitrate-supplemented medium. Sequence analysis revealed differences between the denitrification gene cluster of B. neotomae and that of other Brucella genomes. The extent of these differences and their effect on denitrification and in vivo growth of B. neotomae provide insight into the regulation of denitrification and its physiological role in Brucella.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The B. neotomae strain used in this study, 5K33, was lyophilized in 1966 (5) (Table 1). The dried material was rehydrated with Brucella broth (Difco, Detroit, Mich.). One of the colonies arising from the rehydrated stock was saved and used for all further studies. Extraction of chromosomal DNA was done using the Wizard genomic DNA kit (Promega). A fragment of the 16S rRNA gene was amplified and sequenced as previously described and found to have >99% identity with the 16S rRNA gene of other Brucella strains (7, 11). Sequence analysis revealed it was identical to B. neotomae ATCC 23459, which is also derived from the 5K33 strain. Sequencing was carried out at the Cornell BioResource Center DNA Sequencing facility using an Applied Biosystems automated 3700 DNA analyzer.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Reference or source |
|---|---|---|
| Strains | ||
| DH5αF′ | Host for E. coli cloning; F′ φ80lacZΔM15 recA1 endA1 gyrA96 thi-1 hsdR17 (rK−mB−) supE44 relA1 deoR (ΔlacZYA-argF)U169 | 35 |
| S17-1 | For conjugational transfer of plasmids; recA thi pro hasdRM+ RP4:2-Tc:Mu:Km:TnZ | 28 |
| 5K33 | Wild-type strain of B. neotomae | 5 |
| 16M | B. melitensis strain used to amplify nirK-nnrA region for complementation | ATCC 23456 |
| C58 | Wild-type strain of A. tumefaciens | ATCC 33970 |
| A011 | nnrR strain of A. tumefaciens with nnrR being replaced by Ω Spr Smr cassette; Smr Spr | Baek and Shapleigh |
| Plasmids | ||
| pUC19 | Used for cloning in E. coli (Apr) | 36 |
| pRK415 | Broad-host-range plasmid (Tcr) | 12 |
| pBBR1MCS-5 | Broad-host-range plasmid (Gmr) | 18 |
| pKOK6 | Source of lacZ-Km cassette (Tcr Kmr Apr) | 17 |
| pBnorCZ | Derivative of pRK415 with B. neotomae norC-lacZ transcriptional fusion (Tcr Kmr) | This study |
| pBnarKZ | Derivative of pRK415 with B. neotomae narK-lacZ transcriptional fusion (Tcr Kmr) | This study |
| pBnnrA | Derivative of pBBR1MCS-5 with open reading frame of B. melitensis nnrA and its promoter region (Gmr) | This study |
| pBnnrB | Derivative of pBBR1MCS-5 with open reading frame of B. neotomae nnrB and its promoter region (Gmr) | This study |
| pBnirKV | Derivative of pBBR1MCS-5 with open reading frames of B. melitensis nirK, nirV, and their promoter regions (Gmr) | This study |
| pBgap | Derivative of pBBR1MCS-5 with open reading frames of B. melitensis nirK, nirV, nnrR, and their promoter regions (Gmr) | This study |
B. neotomae was typically grown on brucella broth at 30°C. Microaerobic growth was achieved by growing cells in 15-ml serum vials filled with 10 ml of medium. After cells were added, the vials were sealed to prevent oxygen exchange, ensuring that oxygen levels decreased during growth. When necessary, kanamycin or gentamicin was added to B. neotomae cultures at 25 μg ml−1. Nitrate was added to a final concentration of 11 mM. Agrobacterium tumefaciens C58 (Table 1) was grown on Sistrom's medium, and microaerobic conditions were achieved as previously described (21). Kanamycin and tetracycline were added to A. tumefaciens cultures at 100 and 3 μg ml−1, respectively. An A. tumefaciens strain (A011) containing an inactivated nnrR was constructed by insertion of a streptomycin-spectinomycin resistance cassette into nnrR (S.-H. Baek and J. P. Shapleigh, unpublished data). Escherichia coli strain DH5α was used as a maintenance strain for plasmids. E. coli S17-1 (29) was used as a donor for conjugative transfer of plasmids. E. coli strains were grown on Luria-Bertani medium.
Plasmids were mobilized into all strains by conjugation using standard protocols (31). Brucella recipients were plated onto medium containing brucella selective supplement (Sigma-Aldrich, Inc.) and any antibiotics required to select for plasmids. Exconjugants were selected after 5 days of incubation at 30°C. Wild-type B. neotomae was incapable of growth on this medium. Exconjugants were maintained on medium containing selective supplement through several rounds of isolation and then grown in liquid medium containing selective supplement, and an aliquot of this was saved for future use. All subsequent growth was done without selective supplement in the medium.
Construction of plasmids for sequencing, complementation, and fusions.
All plasmids used in this study are shown in Table 1. The oligonucleotides used to amplify fragments in this study are shown in Table 2. Oligonucleotides were designed using the available Brucella genomic sequences. All amplified fragments from B. neotomae were cloned into pUC19 (36) for sequencing and to facilitate further cloning. For construction of the lacZ fusions, the fragments were moved into the broad-host-range vector pRK415 (12) followed by introduction of the lacZ-Kanr cassette from pKOK6 (17). After proper orientation of the lacZ cassette was confirmed, the plasmids were transformed into S17 and then conjugated into Brucella or other strains. For complementation analyses, fragments were cloned into pRK415 or pBBR1MCS-5 (18) and then transformed into S17.
TABLE 2.
Oligonucleotides used in this study
| Genotype or description | Primera |
|---|---|
| nirK from B. neotomae | 5′-GCAGAATTCACAACATCGATTTCCATG-3′ (F) |
| 5′-CGCGGATCCTCGATCAGGTTATGA TTG-3′ (R) | |
| norB from B. neotomae | 5′-CGCGGTACCGAGTTCCTTGAACAG CCC-3′ (F) |
| 5′-CGCGAATTCCCGTGGGTGTAGT AATTG-3′ (R) | |
| Region downstream of nnrA from B. neotomae | 5′-CGCGAATTCCGACGATCTATGATCTTTC-3′ (F) |
| 5′-GCGCTGCAGATAAGCTTCGGCATGATG-3′ (R) | |
| Fragment containing the nirK and nnrA deletions from B. neotomae | 5′-CGGGAATTCGCCTCTATCGTTATGTGC-3′ (F) |
| 5′-CCGGGTACCTAGCCTGACTGATACGC-3′ (R) | |
| nnrB from B. neotomae | 5′-CGGCTGCAGGCGGGTTTCCAGTTTGAG-3′ (F) |
| 5′-CGCGGATCCCCTTTGTTGAGC ATATGG-3′ (R) | |
| norC-lacZ transcriptional fusion | 5′-GCGGAATTCTATGCCAAGGCCTGC TTC-3′ (F) |
| 5′-GCCGGATCCGCTGTTCAAGGAACTCGC-3′ (R) | |
| narK-lacZ transcriptional fusion | 5′-GCCGAATTCTCAGTTCC TGACAAGACC-3′ (F) |
| 5′-GGCGGATCCGATGGGCGTGCTAACAAG-3′ (R) | |
| nnrA from B. melitensis | 5′-GCGGAATTCGTTCTGATGCGGTTGAAG-3′ (F) |
| 5′-CGCGGATCCGATGTGCATACCCTCTCC-3′ (R) | |
| nirK-nnrA region from B. melitensis | 5′-CGCGGATCCGGTGGTGAATTTCTATTTC-3′ (F) |
| 5′-CGCGGTACCCACAAATAAACAGCTCGTC-3′ (R) |
Underlined regions in the sequences indicate a restriction site introduced to facilitate cloning. F, forward; R, reverse.
Biochemical assays.
Nitrite was measured using a standard colorimetric protocol (30). β-Galactosidase assays were carried out as described previously (19). For all β-galactosidase assays, cells were grown under microaerobic conditions. Results are reported from cells in late log phase, since this is when denitrifier promoter fusions have the highest activity (2). For experiments with sodium nitroprusside (SNP), a 200 mM stock of SNP was prepared in Sistrom's medium. Using a syringe, SNP was added to a final concentration of 2 mM, cultures were incubated an additional 4 h, and then cells were removed for β-galactosidase analysis as previously described (19). Nir activity assays were performed as described previously (30).
Mouse infection.
IRF-1−/− mice, originally produced by Matsuyama et al. from C57BL/6 (H-2b) mice, were kindly donated by Tak W. Mak, Amgen Institute, Ontario Cancer Institute, University of Toronto, Toronto, Ontario, Canada (22). These mouse strains were heterozygously bred in the Department of Animal Health and Biomedical Sciences animal care facilities, University of Wisconsin, and 6- to 9-week-old mice were used for experimental infection. Prior to infection, IRF-1−/− mice were genotyped by PCR (28). IRF-1−/− mice (n = 10/group) were injected intraperitoneally with 107 B. neotomae or B. neotomae/pBgap. To determine CFU in livers and spleens, two mice were killed from each group at different time points and samples were homogenized in phosphate-buffered saline and plated on brucella agar. Brucella colonies were counted after 3 days of incubation at 37°C with 5% CO2.
RESULTS
Absence of nirK in B. neotomae.
B. neotomae was found unable to grow under anoxic conditions on solid medium supplemented with nitrate. A similar result has been observed with R. sphaeroides 2.4.3, which is a complete denitrifier, and so the inability of B. neotomae to grow under these conditions might not reflect an inability to reduce nitrogen oxides (21). Since genes for denitrification can be induced in cells grown under oxygen-limited conditions, cells were cultured in nitrate-containing medium in sealed vials. The cells grew to an optical density at 600 nm of about 0.5 under these conditions. Analysis of the culture medium after oxygen-limited growth indicated that there was 332 ± 27 μg of nitrite/ml in the medium. This nitrite did not disappear even after the cells were incubated for 5 to 7 days. Moreover, cells grown under these conditions did not have any detectable Nir activity (Table 3). These results suggest that B. neotomae either lacks the gene coding for Nir or it is present but not expressed under the culture conditions used in these experiments.
TABLE 3.
Nitrite reductase activity of B. neotomae wild type or B. neotomae carrying plasmids containing nirK and nirV (pBnirKV) or nirK, nirV, and nnrA (pBgap)
| Strain | Nitrite reductase activity (units) |
|---|---|
| B. neotomae (nitrate)a | 0 |
| B. neotomae + pBnirKV (nitrate) | 0.4 ± 0.1 |
| B. neotomae + pBgap (nitrate) | 6.7 ± 3.8 |
| B. neotomae + pBgap (no nitrate)b | 5.1 ± 2.6 |
Medium was supplemented with 11 mM nitrate.
Medium was not supplemented with nitrate.
To determine if the B. neotomae genome contains nirK, the Nir structural gene, an attempt was made to amplify a fragment of the gene from B. neotomae genomic DNA using oligonucleotides that are targeted to regions encoding conserved areas of the protein (Table 2). No product of the correct size was amplified from B. neotomae genomic DNA. However, these oligonucleotides could be used to amplify a product of the correct size from R. sphaeroides 2.4.3 genomic DNA despite there being two mismatches in each oligonucleotide (data not shown).
The accumulation of nitrite during microaerobic growth in nitrate-supplemented medium and the inability to amplify nirK suggest the B. neotomae genome contains the genes for nitrate reductase (Nar) but not Nir. A partial loss of denitrification genes has also been observed in other α-proteobacteria, such as R. sphaeroides 2.4.1. In strain 2.4.1, nirK and nirV have been deleted but the deletion is limited to these genes (20). Assuming a similar type of deletion has occurred in B. neotomae, an attempt was made to amplify the closest gene downstream of nirK whose product is not involved in denitrification (Fig. 1). Oligonucleotides targeting this gene amplified a DNA fragment of the predicted size (data not shown). Sequencing of this fragment revealed >99% identity to the same region of DNA from B. suis and Brucella melitensis (data not shown). This suggests any deletion in this region is confined to genes involved in denitrification.
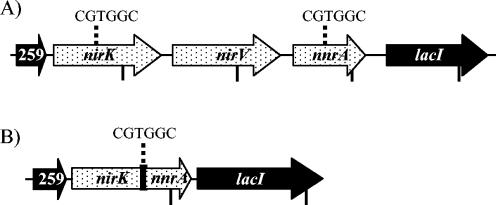
FIG. 1.
Comparison of the nirK-encoding regions of the B. suis and B. neotomae genomes. Arrows indicate locations of genes and their direction of transcription. Stippled arrows represent genes whose products are involved in denitrification. Vertical ticks mark distances of 1,000 bp. BRA0259 encodes a hypothetical protein, and its designation was assigned during genomic analysis of B. suis (24). (A) B. suis. The locations of the CGTGGC repeats are indicated. (B) B. neotomae. The region between the two CGTGGC sequences in panel A is not present, and the lone CGTGGC sequence in this region is indicated. nirK and nnrA are both partially deleted. The deletion removed the 3′ region of nirK, and so it is terminated by a line instead of an arrow. nnrA lacks its transcription start, and so it begins with a vertical mark.
To determine the extent of DNA loss in this region, oligonucleotides were designed to amplify a region starting about 1.3 kb upstream of nirK and ending within the previously sequenced gene immediately downstream of nirK. Amplification gave only a single band of about 2.2 kb (data not shown). In other Brucella species, amplification with these oligonucleotides resulted in a 4.4-kb fragment in length that included nirK, nirV, and a gene encoding an Fnr/CRP-type transcriptional regulator (data not shown) (Fig. 1). Since there are two genes in the Brucella denitrification gene cluster that encode Fnr/CRP-type regulatory proteins, the one near nirK has been designated nnrA and the one adjacent to the nitrous oxide synthase (nos) gene cluster has been designated nnrB. The nnr designation was chosen since a similar terminology has been used in related bacteria, such as P. denitrificans and R. sphaeroides, to identify genes whose products regulate nitrite and nitric oxide reductases (31, 32).
Sequencing of the 2.2-kb fragment amplified from B. neotomae revealed a deletion that affects nirK, nirV, and nnrA. Using the B. suis genome as a reference, the genes designated BRA0258 and BRA0259, which are the two genes immediately upstream of nirK, were present in their entirety in B. neotomae (Fig. 1). Downstream of the BRA0259 gene in B. neotomae is a fragment of nirK, comprising 541 bases from the 5′ region of the open reading frame (ORF) with the remaining 587 bases of nirK missing. The region between the 3′ end of BRA0259 and the end of the remaining nirK fragment has >99% identity with other Brucella genomes. The nirV ORF is completely absent in B. neotomae. The 5′ end of nnrA is also absent, but 229 bases of the 3′ region of nnrA are intact (Fig. 1). As expected, the gene encoding a LacI-type regulator followed nnrA, and the sequence was >99% identical to that of other Brucella genomes.
Characterization and expression of norCB in B. neotomae.
In B. suis and B. melitensis, the genes encoding all four nitrogen oxide reductases and required assembly factors are located within an ∼60-kb region of the small chromosome (6, 24). The presence of nitrate reductase and fragments of nirK and nnrA indicates there has been no large-scale deletion involving the entire denitrification cluster in B. neotomae. However, a possible consequence of the loss of nirK is that Nor is no longer essential. Oligonucleotides targeting sequences that encode conserved regions of NorB (Table 2) amplified a single fragment with >99% identity to the norB gene from other Brucella species (data not shown). Further sequencing of the nor region of the chromosome including the nor promoter region showed >99% identity to the same region from other sequenced Brucella species.
The organization of the nor genes in Brucella is similar to that of other denitrifiers, suggesting the genes norC through norD form an operon (26). The region upstream of norC would therefore contain all the regulatory motifs required for expression of the operon. To determine if the nor region was expressed, a norC promoter fusion was constructed with lacZ as the reporter. There was no detectable β-galactosidase activity in B. neotomae cells containing the fusion when cells were grown in nitrate-supplemented medium under microaerobic conditions (Table 4). An increase in incubation temperature to 37 or 42°C or changes in oxygen levels did not result in detectable β-galactosidase activity. However, the B. neotomae norC-lacZ fusion was expressed in A. tumefaciens, indicating that the lack of expression was not due to the vector itself (Table 4).
TABLE 4.
β-Galactosidase activity of B. neotomae narK-lacZ and B. neotomae norC-lacZ in A. tumefaciens and B. neotomae strains under various conditions
| Strain | Fusion | β-Galactosidase activity (Miller units)
|
||
|---|---|---|---|---|
| Aerobic | Microaerobic | Microaerobic + NO3− | ||
| B. neotomae | B. neotomae norC-lacZ | 0 | 0 | 0 |
| B. neotomae | B. neotomae narK-lacZ | NDa | 463 ± 93 | 889 ± 33 |
| A. tumefaciens | B. neotomae norC-lacZ | 4 ± 2 | 46 ± 6 | 204 ± 13 |
| A011b + pBnnrA | B. neotomae norC-lacZ | 7 ± 3 | 80 ± 9 | 159 ± 13 |
| A011 + pBnnrB | B. neotomae norC-lacZ | 4 ± 2 | 29 ± 5 | 38 ± 8 |
No measurement taken under this condition.
NnrR-deficient strain of A. tumefaciens.
In phenotypic tests, the only nitrogen oxide reductase shown to be expressed was nitrate reductase. Available genome sequence from other Brucella species suggests narK, which encodes a nitrite extrusion protein, is the first gene in an operon with other nar genes (6, 24). Therefore, the promoter region of narK from B. neotomae was fused to lacZ to serve as a positive control for promoter fusion experiments (Table 1). As expected, β-galactosidase activity was detected in B. neotomae containing the narK-lacZ fusion, and its activity showed a nitrate-dependent effect (Table 4).
Characterization of nnrB.
One possible explanation for the absence of norC-lacZ expression in B. neotomae is that the lack of Nir activity prevents endogenous NO production, which has been shown to be required for expression of both nirK and nor in other denitrifiers (19, 32). Since norC-lacZ expression in B. neotomae was not detected even in the presence of SNP, an NO generator, the presence of exogenous NO is not sufficient to restore norC-lacZ expression (data not shown). Moreover, in other denitrifiers lacking Nir activity the expression of nirK and nor does increase when the cells are grown with nitrate, due to the chemical production of NO from the high levels of nitrite that accumulate in the medium (19). The presence of nitrite in the medium did not lead to any detectable expression of norC-lacZ in B. neotomae, further indicating that NO was not limiting expression.
Inactivation of nnrA may account for the lack of expression of the norC-lacZ fusion in B. neotomae. However, there is an nnrA paralog, nnrB, in other Brucella species. The nnrB gene is immediately upstream of a gene encoding pseudoazurin (paz), which is involved in electron transfer to nitrogen oxide reductases in other denitrifiers, and a gene designated nnrS, which has been shown to be under NnrR control in R. sphaeroides 2.4.3 (3, 25). Amplification using oligonucleotides that targeted B. neotomae nnrB (Table 2) resulted in a product of the expected size which showed >99% sequence identity to the B. suis and B. melitensis sequences (data not shown). The deduced NnrB sequence of B. neotomae was 56 and 37% identical to NnrRs from A. tumefaciens and R. sphaeroides, respectively.
To test whether NnrB could activate norC expression, pBnnrB, which contains nnrB from B. neotomae and pBnorCZ, which carries the norC-lacZ promoter fusion, were mobilized into an NnrR-deficient strain of A. tumefaciens, A011 (Table 1). A011 carrying pBnnrB exhibited an increase in β-galactosidase activity when oxygen was restricted, but there was no further increase when nitrate was added to the medium (Table 4). The weak expression of the norC-lacZ fusion could be due to NnrB not being expressed. To test if NnrB was expressed, its ability to complement an NnrR-deficient phenotype in A. tumefaciens was studied. Complementation was assessed by monitoring nitrite accumulation and Nir activity of the nnrB-containing strain grown under limiting oxygen conditions in nitrate-supplemented medium. Under this condition the presence of nnrB did prevent nitrite accumulation (data not shown). However, it was observed that this strain grew slower than the wild-type strain upon reaching an optical density at 600 nm of about 0.50, which is the point in the growth curve where the denitrification genes are initially expressed (data not shown). Cells with NnrB eventually reached a density similar to that of the wild type.
Complementation of NnrR deficiency by nnrA.
Since NnrB only showed a limited ability to activate norC expression in the heterologous host A. tumefaciens, NnrA was tested under the same conditions to see if it was better able to activate expression. The nnrA from B. melitensis was amplified and cloned into a broad-host-range vector to make pBnnrA (Table 1). As with nnrB-containing strains, NnrR-deficient strains of A. tumefaciens with pBnnrA in trans did not accumulate nitrite when grown microaerobically in nitrate-supplemented medium. However, unlike with nnrB, A011 cells containing pBnnrA grew at near-wild-type rates during all phases of growth (data not shown). Expression of the B. neotomae norC promoter fusion in the presence of nnrA showed a nitrate-dependent increase and was three- to fivefold higher than was measured when nnrB was present (Table 4). The level of β-galactosidase activity in the A011 cells containing nnrA was similar to that measured in wild-type A. tumefaciens cells containing only the B. neotomae norC-lacZ promoter fusion and grown under identical conditions (Table 4).
Restoration of denitrification in B. neotomae.
Data from the complementation studies with nnrA and nnrB suggest that nnrA may be required for expression of some of the denitrification genes in B. neotomae. To test this, the entire nirK-nnrA region from B. melitensis was cloned into a broad-host-range vector to make pBgap, which was conjugated into B. neotomae (Table 1). When B. neotomae cells carrying pBgap were grown under oxygen-limiting conditions in nitrate-amended medium, significantly less nitrite accumulated in the medium compared to wild-type cells grown under identical conditions (34 ± 19 versus 332 ± 27 μg/ml, respectively). Nir assays revealed the B. neotomae cells containing pBgap had detectable Nir activity, consistent with the decrease in nitrite accumulation in the medium (Table 3). Cells grown in medium that had not been supplemented with nitrate also had significant levels of Nir activity (Table 3). This result has been observed in other denitrifiers (19) and is likely due to residual nitrate in the unsupplemented medium.
To test whether nnrA was required for expression of nirK, pBnirKV, which contains only nirK and nirV from B. melitensis, was mobilized into B. neotomae. When B. neotomae with pBnirKV was grown under limiting oxygen in medium containing nitrate, nitrite accumulation in the medium was slightly lower than with wild type but significantly higher than in medium from cells with pBgap (256 ± 31 μg/ml, versus 332 ± 27 μg/ml with the wild type). Consistent with this observation, B. neotomae with pBnirKV grown in medium with added nitrate had much lower levels of Nir activity than B. neotomae containing pBgap (Table 3), indicating that NnrB is a weak activator of nirK and, by extension, nor expression.
Loss of nirK and nnrA impacts in vivo growth.
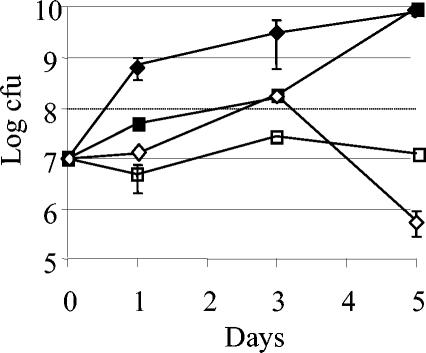
Given the importance of NO as part of the infection response, it is of interest to test whether the loss of the nirK-nnrA region has had an impact on the pathogenicity of B. neotomae. To test whether the deletion affects virulence, B. neotomae wild-type and pBgap-complemented strains were injected into IRF-1−/− mice. IRF-1−/− mice, unlike wild-type mice, are highly susceptible to Brucella infection. Brucella infection in IRF-1−/− mice is lethal, and the mortality is dependent on the virulence of the Brucella organisms (13, 14). Therefore, IRF-1−/− mice serve as a rapid indicator system to assess virulence of Brucella strains. IRF-1−/− mice were injected with either wild-type B. neotomae or B. neotomae/pBgap. At day 1 and every other day thereafter, two mice from each group were killed, and homogenized material from livers and spleens was cultured for CFU. At day 5 the mice injected with the wild-type strain began to die, and no more mice were killed in either group. By day 10 all the mice remaining in the wild-type group had died, while three of the four mice injected with the B. neotomae/pBgap survived for greater than 2 months. There was a general trend of higher CFU in both the liver and spleen in the wild-type strain than in the strain containing pBgap. The difference in CFU was greatest on day 5, with the numbers of bacteria in the liver and spleen being at least 2 logs higher with the wild-type strain than with B. neotomae/pBgap (Fig. 2). The differences in CFU were consistent with the eventual outcome of the experiment, since the mice injected with wild-type B. neotomae died soon after day 5. Similar levels of CFU were obtained when material from the liver or spleen was plated on medium containing gentamicin to select for cells containing pBgap (data not shown). This indicates that the presence of pBgap was not so detrimental to the survival of B. neotomae as to result in its loss during infection.
FIG. 2.
CFU counts from livers and spleens of IRF-1−/− mice infected with either B. neotomae or B. neotomae/pBgap. CFU counts were log transformed, and the data are an average from two mice at each time point. Material was plated on medium without any additional antibiotics. Diamonds, average liver CFU; squares, average CFU from spleens. Filled symbols are CFU from mice injected with wild-type B. neotomae; open symbols are CFU from mice injected with B. neotomae/pBgap. Error bars represent the range of CFU of the samples from each time point.
DISCUSSION
A number of α-proteobacteria form intimate associations with hosts, either as symbionts or as pathogens. The role of denitrification in these associations has been poorly studied. This is the first report describing the role of denitrification in an α-proteobacterial animal pathogen. In contrast to B. suis and B. melitensis, B. neotomae is a partial denitrifier, capable of nitrate reduction but incapable of reducing nitrite. Nitrite cannot be reduced since the structural gene for Nir has been partially deleted. The truncated Nir cannot form an active protein, since the remaining fragment is missing ligands for type 2 copper (9). The deletion of nirK in B. neotomae is accompanied by the loss of nirV, a gene of unknown function but likely having some role in Nir assembly or activity (10). The genes required for Nor production and assembly are present in B. neotomae.
Previously, the loss of portions of the denitrification pathway has been reported in some but not all strains of R. sphaeroides (20). Analysis of genomes of denitrifying and nondenitrifying strains of R. sphaeroides suggests that 2.4.1 and 2.4.3 had a common ancestor that was a denitrifier but that 2.4.1, and most other R. sphaeroides strains, lost the ability to reduce nitrite (19). However, strains did not lose the ability to reduce NO, since the nor operon and nnrR, which encodes a transcriptional regulator required for nirK and nor expression, are present and have been shown to be functional (20). In contrast, B. neotomae is unlikely to reduce NO, since a gene encoding an NnrR ortholog, nnrA, has been partially deleted along with nirK and nirV. B. neotomae cells lacking nnrA but containing nirK and nirV from B. melitensis exhibited only weak Nir activity (Table 3). If nnrA was present along with nirK and nirV, Nir activity increased significantly, demonstrating the importance of nnrA (Table 3).
In all brucellae characterized to date, there are two genes encoding NnrR-related proteins in the denitrification gene cluster. The occurrence of NnrR paralogs has not been previously described in other denitrifiers. Sequence comparisons have demonstrated that Brucella NnrA is more similar to NnrRs from other denitrifiers than is NnrB. NnrA is also a better activator of expression of B. neotomae norC and nor genes from A. tumefaciens than is NnrB (Table 4 and data not shown). The physiological function of NnrB is not clear; however, NnrB could be used to activate a subset of denitrification genes since it clusters with two genes, nnrS and paz, indirectly involved in nitrogen oxide reduction.
Examination of the B. suis and B. melitensis genomes in the region where the deletion occurred in the B. neotomae genome revealed a 6-base repeat at the start and end of the region that was deleted (Fig. 1). In pairwise comparisons of B. neotomae with B. suis and B. melitensis genomes, the nirK ORF is conserved until the sequence CGTGGC, bases 536 to 541 of nirK. The remaining fragment of nnrA in B. neotomae starts at base 465, which is preceded by the 6-bp sequence CGTGGC. The CGTGGC sequence does not occur between nirK and nnrA in other Brucella species, suggesting that a recombination event across these repeated sequences may have caused the deletion (4).
Since other species of Brucella have no similar deletions in their denitrification gene clusters, the deletion in B. neotomae occurred after the strain diverged and adapted to its current lifestyle. While the role of denitrification in Brucella is unclear, it has been hypothesized that the ability to reduce NO would provide an effective means of dealing with the NO produced in response to infection (34). Previous work has shown that NO production can increase long-term survival of Brucella abortus, a complete denitrifier, in macrophages (34). However, the survival of denitrification-compromised strains was not tested. In this study, it was shown that there was a significant difference in the lethality of B. neotomae strains with and without the genes encoding Nir and NnrA. Infection with 107 wild-type B. neotomae organisms proved lethal to all mice within 10 days. The majority of the mice (three out of four) survived when injected with B. neotomae carrying the nirK-nnrA region in trans. Infection persisted in mice injected with the nirK-nnrA strain, as indicated by the numbers of CFU found in the spleen (Fig. 2). The number of hepatic CFU decreased at day 5, indicating that the mice were able to control hepatic B. neotomae. These results demonstrate that denitrification, particularly the capacity to reduce nitrite and NO, is important in modulating the interaction of Brucella with its host. It is likely that the capacity to denitrify is physiologically significant, because it allows Brucella to use nitrate, nitrite, and NO as terminal oxidants in the oxygen-poor environment inside the macrophage (27). NO has also been reported to inhibit oxygen respiration in intracellular pathogens, and so the capacity to reduce NO may also have the paradoxical effect of allowing cells to use both nitrogen oxides and oxygen as terminal oxidants (33). Irrespective of the exact mechanism, utilization of nitrogen oxides by Brucella may enhance its long-term survival in the macrophage. In wild-type B. neotomae, the stress induced by limiting levels of terminal oxidants in the mouse macrophage may induce a stress response that causes the cells to have a more deleterious effect on their host. If this were the case, then it is not clear why nirK and nnrA has been lost from the B. neotomae genome, since the resulting phenotypic change would seem to be disadvantageous. One possible explanation is that wood rat macrophages may produce minimal NO. This would mitigate the physiological requirement for Nor, and so the phenotypic changes arising as a result of nirK, nirV, and nnrA inactivation would not be as deleterious. The deletion of these denitrifying genes might, however, have resulted in a limitation in the host range of B. neotomae. Future studies are required to determine whether the Brucella denitrification genes contribute to host range.
Acknowledgments
We thank Sigrid Holmgren for initiating this collaboration and David Glover for helping with infection of mice.
This work was supported by National Institutes of Health grant R01AI048490 and BARD-US 2968-98C (to G.S.) and the Department of Energy grant 95ER20206 (to J.P.S.).
REFERENCES
- 1.Anjum, M. F., T. M. Stevanin, R. C. Read, and J. W. Moir. 2002. Nitric oxide metabolism in Neisseria meningitidis. J. Bacteriol. 184:2987-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartnikas, T. B., I. E. Tosques, W. P. Laratta, J. Shi, and J. P. Shapleigh. 1997. Characterization of the region encoding the nitric oxide reductase of Rhodobacter sphaeroides 2.4.3. J. Bacteriol. 179:3534-3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartnikas, T. B., Y. Wang, T. Bobo, A. Veselov, C. P. Scholes, and J. P. Shapleigh. 2002. Characterization of a member of the NnrR regulon in Rhodobacter sphaeroides 2.4.3 encoding a heme-copper protein. Microbiology 148:825-833. [DOI] [PubMed] [Google Scholar]
- 4.Bzymek, M., and S. T. Lovett. 2001. Instability of repetitive DNA sequences: the role of replication in multiple mechanisms. Proc. Natl. Acad. Sci. USA 98:8319-8325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corbel, M. J. 1977. Isolation and partial characterization of a phage receptor from “Brucella neotomae” 5K33. Ann. Sclavo. 19:131-142. [PubMed] [Google Scholar]
- 6.DelVecchio, V. G., V. Kapatral, R. J. Redkar, G. Patra, C. Mujer, T. Los, N. Ivanova, I. Anderson, A. Bhattacharyya, A. Lykidis, G. Reznik, L. Jablonski, N. Larsen, M. D'Souza, A. Bernal, M. Mazur, E. Goltsman, E. Selkov, P. H. Elzer, S. Hagius, D. O'Callaghan, J. J. Letesson, R. Haselkorn, N. Kyrpides, and R. Overbeek. 2002. The genome sequence of the facultative intracellular pathogen Brucella melitensis. Proc. Natl. Acad. Sci. USA 99:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fennell, D. E., A. B. Carroll, Gossett, J. M., and S. H. Zinder. 2001. Assessment of indigenous reductive dechlorinating potential at a TCE-contaminated site using microcosms, polymerase chain reaction analysis, and site data. Environ. Sci. Technol. 35:1830-1839. [DOI] [PubMed] [Google Scholar]
- 8.Gandara, B., A. L. Merino, M. A. Rogel, and E. Martinez-Romero. 2001. Limited genetic diversity of Brucella spp. J. Clin. Microbiol. 39:235-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Godden, J. W., S. Turley, D. C. Teller, E. T. Adman, M. Y. Liu, W. J. Payne, and J. Legall. 1991. The 2.3 angstrom X-ray structure of nitrite reductase from Achromobacter cycloclastes. Science 253:438-442. [DOI] [PubMed] [Google Scholar]
- 10.Jain, R., and J. P. Shapleigh. 2001. Characterization of nirV and a gene encoding a novel pseudoazurin in Rhodobacter sphaeroides 2.4.3. Microbiology 147:2505-2515. [DOI] [PubMed] [Google Scholar]
- 11.Johnson, J. L. 1994. Similarity analysis of rRNAs, p. 683-700. In P. Gerhardt, R. G. E. Murray, W. A. Wood, and N. R. Krieg (ed.), Methods for general and molecular microbiology. American Society for Microbiology, Washington, D.C.
- 12.Keen, N. T., S. Tamaki, D. Kobayashi, and D. Trollinger. 1988. Improved broad-host-range plasmids for DNA cloning in gram-negative bacteria. Gene 70:191-197. [DOI] [PubMed] [Google Scholar]
- 13.Ko, J., A. Gendron-Fitzpatrick, T. A. Ficht, and G. A. Splitter. 2002. Virulence criteria for Brucella abortus strains as determined by interferon regulatory factor 1-deficient mice. Infect. Immun. 70:7004-7012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ko, J., A. Gendron-Fitzpatrick, and G. A. Splitter. 2002. Susceptibility of IFN regulatory factor-1 and IFN consensus sequence binding protein-deficient mice to brucellosis. J. Immunol. 168:2433-2440. [DOI] [PubMed] [Google Scholar]
- 15.Ko, J., and G. A. Splitter. 2003. Molecular host-pathogen interaction in brucellosis: current understanding and future approaches to vaccine development for mice and humans. Clin. Microbiol. Rev. 16:65-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kohler, S., V. Foulongne, S. Ouahrani-Bettache, G. Bourg, J. Teyssier, M. Ramuz, and J. P. Liautard. 2002. The analysis of the intramacrophagic virulome of Brucella suis deciphers the environment encountered by the pathogen inside the macrophage host cell. Proc. Natl. Acad. Sci. USA 99:15711-15716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kokotek, W., and W. Lotz. 1989. Construction of a lacZ-kanamycin-resistance cassette, useful for site-directed mutagenesis and as a promoter probe. Gene 84:467-471. [DOI] [PubMed] [Google Scholar]
- 18.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 19.Kwiatkowski, A., and J. P. Shapleigh. 1996. Requirement of nitric oxide for induction of genes whose products are involved in nitric oxide metabolism in Rhodobacter sphaeroides 2.4.3. J. Biol. Chem. 271:24382-24388. [DOI] [PubMed] [Google Scholar]
- 20.Kwiatkowski, A. V., W. P. Laratta, A. Toffanin, and J. P. Shapleigh. 1997. Analysis of the role of the nnrR gene product in the response of Rhodobacter sphaeroides 2.4.1 to exogenous nitric oxide. J. Bacteriol. 179:5618-5620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laratta, W. P., P. S. Choi, I. E. Tosques, and J. P. Shapleigh. 2002. Involvement of the PrrB/PrrA two-component system in nitrite respiration in Rhodobacter sphaeroides 2.4.3: evidence for transcriptional regulation. J. Bacteriol. 184:3521-3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsuyama, T., T. Kimura, M. Kitagawa, K. Pfeffer, T. Kawakami, N. Watanabe, T. M. Kundig, R. Amakawa, K. Kishihara, A. Wakeham, J. Potter, C. L. Furlonger, A. Narendran, H. Suzuki, P. S. Ohashi, C. J. Paige, T. Taniguchi, and T. W. Mak. 1993. Targeted disruption of IRF-1 or IRF-2 results in abnormal type I IFN gene induction and aberrant lymphocyte development. Cell 75:83-97. [PubMed] [Google Scholar]
- 23.Moreno, E., A. Cloeckaert, and I. Moriyon. 2002. Brucella evolution and taxonomy. Vet. Microbiol. 90:209-227. [DOI] [PubMed] [Google Scholar]
- 24.Paulsen, I. T., R. Seshadri, K. E. Nelson, J. A. Eisen, J. F. Heidelberg, T. D. Read, R. J. Dodson, L. Umayam, L. M. Brinkac, M. J. Beanan, S. C. Daugherty, R. T. Deboy, A. S. Durkin, J. F. Kolonay, R. Madupu, W. C. Nelson, B. Ayodeji, M. Kraul, J. Shetty, J. Malek, S. E. Van Aken, S. Riedmuller, H. Tettelin, S. R. Gill, O. White, S. L. Salzberg, D. L. Hoover, L. E. Lindler, S. M. Halling, S. M. Boyle, and C. M. Fraser. 2002. The Brucella suis genome reveals fundamental similarities between animal and plant pathogens and symbionts. Proc. Natl. Acad. Sci. USA 99:13148-13153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pearson, I. V., M. D. Page, R. J. van Spanning, and S. J. Ferguson. 2003. A mutant of Paracoccus denitrificans with disrupted genes coding for cytochrome c550 and pseudoazurin establishes these two proteins as the in vivo electron donors to cytochrome cd1 nitrite reductase. J. Bacteriol. 185:6308-6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Philippot, L. 2002. Denitrifying genes in bacterial and archaeal genomes. Biochim. Biophys. Acta 1577:355-376. [DOI] [PubMed] [Google Scholar]
- 27.Schnappinger, D., S. Ehrt, M. I. Voskuil, Y. Liu, J. A. Mangan, I. M. Monahan, G. Dolganov, B. Efron, P. D. Butcher, C. Nathan, and G. K. Schoolnik. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198:693-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Senaldi, G., C. L. Shaklee, J. Guo, L. Martin, T. Boone, T. W. Mak, and T. R. Ulich. 1999. Protection against the mortality associated with disease models mediated by TNF and IFN-gamma in mice lacking IFN regulatory factor-1. J. Immunol. 163:6820-6826. [PubMed] [Google Scholar]
- 29.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 30.Stewart, V., and J. Parales, Jr. 1988. Identification and expression of genes narL and narX of the nar (nitrate reductase) locus in Escherichia coli K-12. J. Bacteriol. 170:1589-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tosques, I. E., J. Shi, and J. P. Shapleigh. 1996. Cloning and characterization of nnrR, whose product is required for the expression of proteins involved in nitric oxide metabolism in Rhodobacter sphaeroides 2.4.3. J. Bacteriol. 178:4958-4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Spanning, R. J., E. Houben, W. N. Reijnders, S. Spiro, H. V. Westerhoff, and N. Saunders. 1999. Nitric oxide is a signal for NNR-mediated transcription activation in Paracoccus denitrificans. J. Bacteriol. 181:4129-4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voskuil, M. I., D. Schnappinger, K. C. Visconti, M. I. Harrell, G. M. Dolganov, D. R. Sherman, and G. K. Schoolnik. 2003. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J. Exp. Med. 198:705-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang, M., N. Qureshi, N. Soeurt, and G. Splitter. 2001. High levels of nitric oxide production decrease early but increase late survival of Brucella abortus in macrophages. Microb. Pathog. 31:221-230. [DOI] [PubMed] [Google Scholar]
- 35.Woodcock, D. M., P. J. Crowther, J. Doherty, S. Jefferson, E. DeCruz, M. Noyer-Weidner, S. S. Smith, M. Z. Michael, and M. W. Graham. 1989. Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 17:3469-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequence for the M13 mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]
- 37.Zumft, W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533-616. [DOI] [PMC free article] [PubMed] [Google Scholar]