Abstract
In Sinorhizobium meliloti, choline is the direct precursor of phosphatidylcholine, a major lipid membrane component in the Rhizobiaceae family, and glycine betaine, an important osmoprotectant. Moreover, choline is an efficient energy source which supports growth. Using a PCR strategy, we identified three chromosomal genes (choXWV) which encode components of an ABC transporter: ChoX (binding protein), ChoW (permease), and ChoV (ATPase). Whereas the best homology scores were obtained with components of betaine ProU-like systems, Cho is not involved in betaine transport. Site-directed mutagenesis of choX strongly reduced (60 to 75%) the choline uptake activity, and purification of ChoX, together with analysis of the ligand-binding specificity, showed that ChoX binds choline with a high affinity (KD, 2.7 μM) and acetylcholine with a low affinity (KD, 145 μM) but binds none of the betaines. Uptake competition experiments also revealed that ectoine, various betaines, and choline derivatives were not effective competitors for Cho-mediated choline transport. Thus, Cho is a highly specific high-affinity choline transporter. Choline transport activity and ChoX expression were induced by choline but not by salt stress. Western blotting experiments with antibodies raised against ChoX demonstrated the presence of ChoX in bacteroids isolated from nitrogen-fixing nodules obtained from Medicago sativa roots. The choX mutation did not have an effect on growth under standard conditions, and neither Nod nor Fix phenotypes were impaired in the mutant, suggesting that the remaining choline uptake system(s) still present in the mutant strain can compensate for the lack of Cho transporter.
Choline is a common constituent of eukaryotic membranes in the form of phosphatidylcholine (PC) and therefore should be widespread in different environments, including the soil and the rhizosphere. Indeed, significant amounts of choline are readily liberated into the environment from plant and animal residues (15). Sinorhizobium meliloti, a plant root-associated bacterium, possesses distinct transport activities for choline uptake (27) and has the ability to oxidize choline to glycine betaine via the bet operon (34, 24). In contrast to Escherichia coli and Bacillus subtilis (25, 2), S. meliloti can use choline for growth. This depends on a functional bet locus (34, 24) associated with catabolism of glycine betaine which is absent in E. coli and B. subtilis. This catabolism is reduced under hyperosmotic conditions, and under these conditions glycine betaine accumulation is favored (34). Moreover, due to the presence of a PC synthase in S. meliloti, which directly condenses choline to CDP-diacylglyceride, choline is a direct precursor of PC, as recently demonstrated for other bacteria, including Agrobacterium, Brucella, and Pseudomonas (6, 19). In addition to this PC synthase pathway, S. meliloti possesses a methylation pathway for PC biosynthesis which functions by threefold methylation of phosphatidylethanolamine with S-adenosylmethionine as a methyl donor (5). Whereas the presence of PC is rather unusual in bacterial membranes, PC is a major lipid membrane component in the Rhizobiaceae family, and PC biosynthesis is required for normal growth of S. meliloti (5). To fulfill the requirement for choline, S. meliloti requires effective transport systems to take up this trimethylammonium compound from exogenous sources.
In E. coli, import of choline is mediated at a low concentration by the high-affinity BetT transporter and at a high substrate concentration by the low-affinity multicomponent ABC uptake system ProU (36, 16). In B. subtilis, choline uptake occurs by two evolutionarily highly conserved ABC transporters, OpuB and OpuC, that probably evolved through the duplication of a primordial gene cluster. Despite the close sequence relatedness of the two systems, these high-affinity transporters exhibit very different substrate specificities (13). In S. meliloti, three kinetically distinct transport activities for choline uptake have been identified; one constitutive activity has low affinity, and two activities have high affinity, and they are either inducible by choline or constitutively expressed (27). While choline has multiple functions in this symbiotic bacterium, nothing is currently known at the molecular level about the route of choline transport. The present study was initiated to gain some understanding of the mechanisms of choline uptake in S. meliloti, and our results provide the first identification and detailed analysis of a high-affinity choline-binding protein-dependent transport system (Cho) in this species. We also demonstrated the high level of specificity of the binding protein and its expression in bacteroids from nodules of Medicago sativa, the host plant of S. meliloti.
MATERIALS AND METHODS
Chemicals.
Choline and glycine betaine were purchased from Sigma-Aldrich (Saint Quentin Fallavier, France), and proline betaine was purchased from Extrasynthèse (Genay, France). Radioactive [methyl-14C]choline (2.04 GBq/mmol) and [methyl-14C]acetylcholine (0.68 GBq/mmol) were purchased from Amersham Corp. (Little Chalfont, United Kingdom) and Sigma-Aldrich, respectively. [methyl-14C]glycine betaine was prepared from [methyl-14C]choline as previously described (24), and [U-14C]proline betaine (4.6 GBq/mmol) was obtained from the Commissariat à l'Energie Atomique (Gif-sur-Yvette, France). Ni2+-nitrolotriacetic acid resin was obtained from QIAGEN (Courtaboeuf, France). Rabbit anti-ChoX antibody was prepared by Eurogentec (Angers, France), and rabbit anti-immunoglobulin G alkaline phosphatase conjugate was obtained from Sigma-Aldrich.
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. The E. coli DH5α and MT616 strains were used for subcloning of the pF1 insert and as a helper strain for triparental mating, respectively. The E. coli BL21(DE3)(pLysS) strain was used for overexpression of the choX gene from the T7 promoter in plasmids pETNE and pETNX. S. meliloti strains were routinely grown at 30°C in Luria-Bertani (LB) medium containing 5 g of NaCl per liter, 2.5 mM MgSO4,and 2.5 mM CaCl2. For uptake experiments and periplasmic protein extraction, cells were grown in MCAA medium containing 0.1% sodium malate, 0.1% Casamino Acids (technical), and minerals as described previously (34). For physiological analysis of the role of Cho and for the choX expression study (Western blotting), cells were grown in M9 minimal medium (20) supplemented with 0.2% mannitol or 0.2% choline as a carbon source. The osmolarities of the various media were increased by addition of 0.3 M NaCl. When necessary, glycine betaine was added at a concentration of 1 mM, and choline was used at a concentration of 7 mM, which allowed maximal stimulation of choline oxidase (34). The antibiotics ampicillin, tetracycline, chloramphenicol, and spectinomycin were used in E. coli cultures at final concentrations of 100, 20, 20, and 100 μg/ml, respectively. Rifampin and spectinomycin were used in S. meliloti cultures at final concentrations of 20 and 100 μg/ml, respectively.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Escherichia coli strains | ||
| DH5α | FsupE44 ΔlacU169(φ80dlacZΔM15) hsdR17(rK− mK+) recA1 endA1 gyrA96 thi-I relA1 | Bethesda Research Laboratories |
| MT616 | MT607(pRK600) | 8 |
| BL21(DE3)(pLysS) | F ompT hasdSB (rB− mB−) gal dcm (DE3) pLysS (Cmr) | Novagen |
| Sinorhizobium meliloti strains | ||
| RCR2011 | SU47, wild type | 30 |
| 1021 | Derivative of RCR2011, Strr | 20 |
| GM1211 | Derivative of RCR2011, Strr Lac | 23 |
| 5000 | Derivative or RCR2011, Rifr | 8 |
| M1A | Rm5000 choX::Ω, Rifr Spr | This study |
| Plasmids | ||
| PLAFR3 | IncP cosmid cloning vector, Tcr | 10 |
| pRK600 | ColE1 replicon with RK2 transfer region, Cmr | 8 |
| pBluescriptSK(−) | Derivative of pUC19 with f1(−)oriR, Ampr | Stratagene |
| pGEM T-3Zf(+) | Cloning vector | Promega |
| pET20-b(+) | Derivative of pBR322, T7 promoter, 3′ His codons, Ampr | Novagen |
| pSUP202 | ColE1, Mob+ Tcr Ampr Cmr | 33 |
| pHP45-Ω | Apr, pBR322 derivative with interposon Ω Smr/Spr | 28 |
| pGQ5 | pGEMT vector with bup1-pro4-amplified fragment | This study |
| pF1 | pLAFR3 with an S. meliloti 16-kb insert containing cho locus | This study |
| p1.2E | 1.2-kb EcoRI fragment from pF1 cloned into pBSSK− vector | This study |
| p1.5E | 1.5-kb EcoRI fragment from pF1 cloned into pBSSK− vector | This study |
| p3.4HE | 3.4-kb HindIII-EcoRI fragment from pF1 cloned into pBSSK vector | This study |
| p6.5H | 6.5-kb HindIII fragment from pF1 cloned into pBSSK vector | This study |
| pSUP6.5H | 6.5-kb HindIII fragment from pF1 cloned into pSUP202 vector | This study |
| pXΩ | pSUP6.5H, choX::Ω (BglII insertion) | This study |
| pETNE | pET20b, Nde1-EcoRI fragment (choX without His codon fusion) | This study |
| pETNX | pET20b, Nde1-Xho1 fragment (choX with 3′ His codon fusion) | This study |
DNA manipulation and cloning of the cho locus.
Restriction analysis, ligation, transformation, plasmid DNA extraction, and Southern hybridization were carried out by standard techniques (21, 31). DNA probes were labeled by using the Prime-a-gene random priming system (Promega, Charbonnières, France) and [α-32P]dCTP (Amersham Corp.). A PCR strategy was used to amplify an internal fragment of the choV gene. The PCR mixtures containing each degenerate primer and Rm5000 genomic DNA were cycled automatically by using a Biometra thermocycler (T gradient model; Biometra GmbH, Göttingen, Germany) through temperature and time cycles as follows: denaturation at 95°C for 1 min, annealing at 40°C for 1 min, and extension at 70°C for 1 min. The sequences of the two degenerate primers used were 5′-GAR ATI TTY GTI ATI ATG GG-3′ (bup1) and 5′-CAT DAT IGC DAT ICK RTC ICC-3′ (pro4). The resulting fragment, which was the expected size (564 bp), was cloned into pGEMT to obtain pGQ5 and was sequenced. The latter plasmid was used as a probe to screen, by colony hybridization, a genomic DNA library of S. meliloti obtained by partial Sau3A digestion of S. meliloti 2011 DNA cloned into pLAFR3 and kindly provided by D. Kahn (Laboratoire de Biologie moléculaire des Relations Plantes-Microorganismes, CNRS-INRA, Castanet-Tolosan, France). One clone containing a recombinant cosmid, designated pF1, strongly hybridized with the probe. The 16-kb insert of pF1 was subcloned, and the region of interest was sequenced by MWG Biotech (Ebersberg, Germany) by using the fluorescent ABI dye-labeled deoxy terminator method. DNA and protein sequences were analyzed by using BLAST protocols (1).
Mutagenesis of S. meliloti.
The choX gene was mutated by insertion of a BamHI-digested Ω interposon (Spr/Smr) into the BglII restriction site of pSUP6.5H, which corresponded to a 6.5-kb HindIII fragment from pF1 cloned at the HindIII site of the suicide vector pSUP202 (Table 1). Triparental spot mating was used to introduce recombinant plasmids from E. coli into S. meliloti as previously described (7, 8). The Ω insertion was finally recombined into the S. meliloti Rm5000 genome, and correct recombination of the interposon in the genomic choX gene was verified by Southern hybridization.
Overproduction and purification of ChoX.
The choX gene under the control of the T7 promoter from pET20-b(+) was overexpressed in E. coli BL21(DE3)(pLysS) (Table 1). Two constructs were made. The first construct (pETNE), which allowed overproduction of a native ChoX protein without any extra amino acid residues, resulted from a PCR fragment digested by NdeI and EcoRI and cloned into the pET20b vector restricted with the same two restriction enzymes. The PCR fragment was obtained by using S. meliloti RCR2011 DNA as the template, Pfu polymerase, and primers PxNde (5′-AGG GGA ACG ACG CAT ATG ATA AGG A-3′; yielding an NdeI site) and PxEco (5′-AGT CAG GAA TTC CAC GAA ACA GGG T-3′; overlapping an EcoRI site). The second construct (pETNX), which allowed overproduction of a ChoX protein with a C-terminal His6 tag, resulted from a PCR fragment obtained by amplification of RCR2011 DNA with primers PxNde and PxXho (5′-TGC CGC CGA CTC GAG GCC GAG G-3′), which created a XhoI site. This PCR fragment, digested with NdeI and XhoI, was cloned into pET20b. The purification steps used for ChoX overexpressed from pETNX in the E. coli BL21(DE3)(pLysS) strain were those described by Novagen (Merck KGaA, Darmstadt, Germany). Briefly, E. coli recombinant cells were grown at 37°C in LB medium (200 ml) with ampicillin (50 μg/ml) and chloramphenicol (30 μg/ml) until the A600 was 0.8, and this was followed by a 2-h expression period initiated by addition of 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG). The cells were centrifuged, resuspended in 4 ml of lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole [pH 8.0], 1 mg of lysozyme per ml), and incubated at 4°C for 30 min before sonication on ice (eight times for 30 s each time). After centrifugation, the protein content of the soluble fraction (supernatant) was determined by the Bradford method (3). Purification by Ni2+-nitrilotriacetic acid affinity chromatography was performed as described by the supplier (QIAGEN). ChoX was eluted from the column with 200 mM imidazole in buffer A (50 mM NaH2PO4, 300 mM NaCl; pH 8.0).
Transport assays.
Cells were harvested at an A600 of 1.5, washed twice in the fresh medium used for the culture, and diluted to obtain a final A600 of 0.5. All assays were carried out at 30°C with 1 ml of cell suspension for 1 min, and radioactive substrates (100,000 dpm) were used at the following concentrations: 10 μM for choline, 1 and 40 μM for glycine betaine, and 10 μM for proline betaine and proline. Uptake was determined by rapid filtration through GF/F glass microfiber filters (Whatman, Maidstone, United Kingdom), which were rinsed with 3 ml of the corresponding medium. The radioactivity remaining on the filters was determined with a liquid scintillation spectrometer (model LS6000SC; Beckman Instruments, Villepinte, France). The transport rate was linear during the 1-min assay, and there was no inhibition by the intracellular choline accumulated by the cells, in agreement with previous results (27). For competition experiments, cold competitors were added at a final concentration of 100 μM or 1 mM to a 10 μM [14C]choline solution (100,000 dpm). Competition uptake experiments were performed with a 1-min incubation period before filtration.
Periplasmic protein extraction and binding assays.
E. coli BL21(DE3)(pLysS) was grown to an A600 of 0.6 in LB medium containing 0.4 mM IPTG, and S. meliloti Rm5000 and M1A were grown to an A600 of 1.5 in MCAA medium supplemented with 7 mM choline. Cells were collected by centrifugation (10,000 × g, 10 min, 20°C) and resuspended in 10 mM Tris-HCl (pH 7.5). Periplasmic proteins were released by cold osmotic shock as described by Neu and Heppel (22) and were concentrated by ultrafiltration by using the standard procedure (17). To determine binding activities, 100 μg of periplasmic proteins was incubated overnight with 5 nmol of [14C]choline (500,000 dpm) in 10 mM Tris-HCl buffer (pH 7.5) at 4°C, separated by nondenaturing polyacrylamide gel electrophoresis (PAGE), and autoradiographed, as described previously (17). For determination of the substrate-binding affinities for choline and acetylcholine, binding assays with the purified ChoX were performed by using ammonium sulfate precipitation (29). Samples containing 8 μg of protein (final concentration, 5 μM) were incubated at 25°C for 15 min with various concentrations of 14C-labeled substrates (1 to 40 μM choline and 1 to 120 μM acetylcholine) in 50-μl reaction mixtures containing 10 mM Tris-HCl buffer (pH 7.4). The proteins were precipitated by adding 950 μl of an ice-cold saturated ammonium sulfate solution, and after incubation for 15 min on ice, the precipitated ChoX protein was collected by rapid filtration onto GF/F glass microfiber filters (Whatman). Each filter was then washed twice with 3 ml of an ice-cold ammonium sulfate solution, and the radioactivity retained by ChoX was determined by scintillation counting. For each substrate concentration, measurements were obtained in triplicate in order to determine the binding constant. Alternatively, an analysis of the specificity of the binding activity of ChoX was performed by gel filtration. A 100-fold excess of unlabeled competitors was added into the binding assay mixture, and the radioactivity retained by ChoX was separated from the unbound [14C]choline on a gel filtration column (Sephadex G-25; Amersham Biosciences Europe GmbH, Orsay, France) that was eluted with 100 mM Tris-HCl buffer (pH 7.5).
Immunological analysis.
Total cell proteins, periplasmic proteins, and purified ChoX protein were separated by sodium dodecyl sulfate (SDS)-PAGE and transferred to nitrocellulose membranes (Hybond protein; pore size, 0.2 μm) by electroblotting. Immunoblotting was performed by using a 1/20,000 dilution of a polyclonal serum raised against the purified ChoX protein of S. meliloti. The immunoblots were developed with rabbit anti-immunoglobulin G alkaline phosphatase conjugate (Sigma-Aldrich), as instructed by the manufacturer.
Nodulation, nitrogen fixation assays, and bacteroid preparation.
The symbiotic proficiency of S. meliloti strains was assayed by using alfalfa (M. sativa L. cv. Europe) seedlings grown in a sterilized mixture of vermiculite and sand and inoculated with the appropriate strains 1 week after sowing, as described previously (18). The number and the mass of nodules were determined 3, 5, 6, and 7 weeks after inoculation. Nitrogen fixation capacity was determined by C2H2 reduction by using a gas chromatograph (38). Freshly harvested nodules (4 weeks old) were used to isolate bacteroids as described by Trinchant et al. (38), and proteins were extracted for SDS-PAGE analysis.
Nucleotide sequence accession number.
The nucleotide sequence of the cho locus has been deposited in the GenBank database under accession number AF360731.
RESULTS
Cloning strategy and genetic organization of the cho locus.
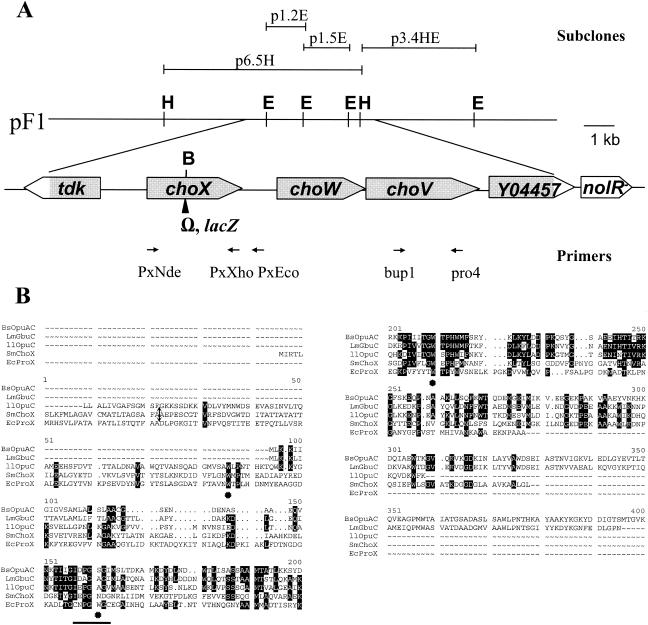
The presence of periplasmic glycine betaine and choline-binding proteins in S. meliloti (4, 17) suggests that ABC-type transporters might contribute to trimethylammonium uptake in this species. In order to isolate such transporters, a PCR strategy based on domains specifically conserved in ATPases of well-characterized bacterial ABC ProU-like transporters and ProV-like orthologues present in databases was used. Two stretches of amino acids, EIFVIMG (positions 65 to 71) located upstream of the Walker A motif, and GDRIVLM (positions 243 to 249) located downstream of the Walker B motif in the OpuAA system from B. subtilis, were selected to design degenerate primers, which were designated bup1 and pro4 (see Materials and Methods). A PCR fragment of the expected size was obtained by using Rm5000 genomic DNA as the template and was cloned into the pGEMT vector before a genomic DNA library of S. meliloti 2011 was screened. A recombinant cosmid (pF1) bearing a 16-kb EcoRI insert was isolated by colony hybridization. By using restriction analysis and Southern hybridization, the region homologous to the PCR-amplified fragment was subcloned in pBS-SK (Fig. 1A) and sequenced. Analysis of the sequenced DNA (4,311 bp) revealed the presence of five open reading frames (accession number AF360731). The genetic organization of this region (Fig. 1A) indicated that there were two divergently transcribed sets of genes, with the tdk gene encoding a putative thymidine kinase located 288 bp from the choXWV genes and open reading frame Y04457 having an unknown function. Analysis of the S. meliloti genome showed that these genes are present on the chromosome (SMc02737 to SMc02739) and are located upstream of the nolR gene (http://sequence.toulouse.inra.fr/meliloti.html). Within the cho locus, the first gene, choX, is separated from choW by a large 160-bp noncoding region, while choW and choV overlap by 4 bp in the ATGA sequence, suggesting that there is translational coupling between these two genes. The choXWV genes had the characteristics of genes encoding an ABC transporter, and the genetic organization of the cho locus followed the binding protein first rule, which is mainly encountered in a binding protein transport operon. The choX gene encodes a hydrophilic protein containing 309 amino acid residues (Mr, 32,904) which exhibited the characteristic signature of a signal peptide for secretion into the periplasm (26). ChoX has a positively charged N-terminal sequence (MIR+TLSLK+), followed by a hydrophobic stretch of amino acids (MLAGAVCMATLTA) and the motif GSAFA, in agreement with the consensus GXAXA sequence of a signal peptidase I site (Fig. 1B). Comparison of the ChoX protein sequence with protein sequences in the data libraries revealed that the highest amino acid identity (28%) was obtained with the glycine betaine-binding proteins BusAB, GbuC, and OpuAC from Lactococcus lactis, Listeria monocytogenes, and B. subtilis, respectively (Fig. 1B). As usually observed for the ligand-binding proteins involved in ABC transporters (37), ChoX is the less conserved protein encoded by the cho locus. ChoW (Mr, 30,323; 281 amino acids) is highly hydrophobic, with six putative transmembrane domains, and corresponds to the integral inner membrane permease component of the transporter. ChoW is homologous to the glycine betaine permease BusAB of L. lactis, OpuAB of B. subtilis, and ProW of E. coli (45 to 50% amino acid identity). However, ChoW is considerably smaller than ProW (354 amino acids), mostly as the consequence of the lack of the N-terminal region of ProW thought to be exposed in the periplasmic space (39). It should also be mentioned that the choW start codon codes for a valine. The last gene, choV, encodes a 349-amino-acid hydrophilic protein with a predicted Mr of 37,615. The ChoV protein is likely the ATPase of the transporter and has motifs characteristic of such proteins (i.e., Walker A and B boxes, a linker peptide, and a switch motif). The highest levels of homology (about 50% amino acid identity) of ChoV with proteins in the databases were the levels of homology with ATPases of glycine betaine ProU-type systems, including ProV of E. coli, BusAA of L. lactis, OpuAA of B. subtilis, and GbuA of L. monocytogenes.
FIG. 1.
Organization of the cho locus and ChoX protein homology. (A) Genetic and physical maps of the cho locus. A partial restriction map of the 16-kb pF1 insert with derived subclones is shown. The positions of the bup1, pro4, PxNde, PxXho, and PxEco primers are indicated below the map. The genes deduced from the nucleotide sequence analysis are represented by large arrows, and the limits of the sequenced region shown in the genetic map are indicated by shading. The open parts of the arrows correspond to the adjacent sequences available in the S. meliloti database (http://sequence.toulouse.inra.fr/meliloti.html). The position of the Ω insertion or the lacZ fusion is indicated by an arrowhead. Abbreviations for restriction enzymes: B, BglII; E, EcoRI; H, HindIII. Open reading frames: tdk, thymidine kinase gene; YO4457, unknown function; nolR−, negative regulator of nod gene defective in the S. meliloti 1021 strain. (B) Comparison of the amino acid sequences of the S. meliloti ChoX protein (SmChoX) and the glycine betaine-binding proteins OpuAC of B. subtilis (BsOpuAC) (accession number U17292), GbuC of L. monocytogenes (LmGbuC) (accession number AF039835), BusC of L. lactis (LlBusC) (accession number AF139575), and ProX of E. coli (EcProX) (accession number M24856). The position of the signal peptidase I site is indicated by an arrow. The tryptophan residues which are key determinants of high-affinity binding of glycine betaine by E. coli ProX are indicated by asterisks under the sequence, and the motif conserved in ProX homologues is underlined. The numbers above the alignment indicate the amino acid residues in ProX.
cho encodes a choline transporter.
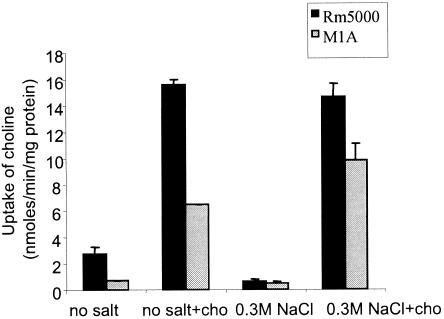
The highest levels of homology for cho-encoded proteins were obtained with the components of glycine betaine-proline betaine ProU-like systems, which suggested that the S. meliloti transporter is involved in trimethylammonium uptake. In order to precisely define the substrate of Cho, transport activities for glycine betaine, proline betaine, and choline, the major quaternary ammonium compounds, were measured in wild-type strain Rm5000 and mutant strain M1A obtained by reciprocal recombination with a choX::Ω construction (Fig. 1A). Surprisingly, there were no significant differences in betaine uptake activities between the two strains (data not shown), whatever the growth medium composition (with or without added 0.3 M NaCl, in the presence or the absence of 1 mM glycine betaine, or a combination of the two conditions), the growth phase of the culture (exponential or stationary phase), and the substrate concentration used for uptake measurement (1 or 40 μM). In contrast, when 10 μM [methyl-14C]choline was used as a substrate in transport experiments, clear differences in uptake activities were observed between strains Rm5000 and M1A grown until the stationary phase (Fig. 2). The mutation in choX reduced by 75 and 60% the choline uptake activity in cells grown in control MCAA medium and in MCAA medium containing 7 mM choline, respectively. It is also interesting that in both strains, choline uptake was induced about sixfold when choline was added to the growth medium, whereas addition of 0.3 M NaCl alone resulted in a very low level of choline accumulation. In choline-induced cells, addition of salt did not result in uptake inhibition, and a mutation in ChoX reduced by 35% choline uptake activity at high osmolarity (0.3 M NaCl). These results suggested that Cho is a high-affinity choline ABC transporter, which has an overall activity that is stimulated by choline but not by salt stress. Interestingly, the presence of choline in the growth medium alleviated the inhibition observed in salt-stressed cells. In addition, Cho activity might be growth phase dependent since no difference in uptake activity was detected between the wild-type and mutant strains when exponentially grown cells were used (data not shown). The remaining choline transport activity in mutant strain M1A indicated that there is another system or other systems for choline uptake in S. meliloti.
FIG. 2.
Choline uptake activity in S. meliloti wild-type strain Rm5000 and the choX M1A mutant. Uptake of [methyl-14C]choline (10 μM) was assayed in cells grown until the stationary phase in MCAA medium supplemented or not supplemented with 7 mM choline (cho) and maintained at low osmolarity (no salt) or high osmolarity (0.3 M NaCl). The values are means for triplicate experiments from four independent cultures, and the error bars indicate standard deviations.
The specificity of choline transport in the wild-type strain was assessed in competition experiments performed in the presence of 10- and 100-fold molar excesses of potential competitors. Different compounds known to be compatible solutes in soil bacteria (ectoine, glycine betaine, proline betaine, γ-butyrobetaine, and carnitine) were tested together with choline derivatives (acetylcholine, phosphorylcholine, and choline-O-sulfate). Dimethylthetine, an S-methyl homologue of glycine betaine, trigonelline, and spermidine were also tested. The polyamine was chosen since it is transported by E. coli via the PotA system, whose ATPase (14) shows significant homology with ChoV (43% identical amino acid residues). At a competitor/substrate ratio of 10/1, acetylcholine was the only competitor of choline uptake; 53% inhibition and 32% inhibition were observed in cells grown in the presence of choline and maintained at low and high osmolarities, respectively. Increasing the competitor/substrate ratio to 100/1 showed that acetylcholine was the only substrate that competed with choline uptake in the wild-type strain. In order to evaluate the role of Cho in acetylcholine uptake, [14C]acetylcholine transport activity was measured in the wild-type and M1A mutant strains grown at low osmolarity in the presence of choline and collected at the stationary phase. At substrate concentrations of 10 and 100 μM, no significant difference was observed (data not shown), indicating that under the growth conditions tested, Cho might not be involved in acetylcholine transport. Thus, the Cho transporter seems highly specific for choline and is not involved in glycine betaine or proline betaine uptake, despite its high levels of homology with betaine transporters.
ChoX purification and binding activity.
The results presented above clearly showed that S. meliloti Cho behaved like a choline transporter. To subsequently investigate the role of the periplasmic binding protein, ChoX was overproduced and purified. The choX gene was overexpressed in E. coli BL21(DE3)(pLysS) under the T7 promoter of pET20-b(+) in a fusion with a His tag-specifying sequence. The recombinant plasmid pETNX (Table 1) allowed overproduction of ChoX, which was purified by Ni2+ chelate affinity chromatography as described in Materials and Methods. SDS-PAGE analysis of the purified fraction revealed a single band corresponding to a protein with an apparent molecular mass of 33 kDa, in agreement with the expected size of the His6-tagged ChoX without its peptide signal (Mr, 32,188). Overall, when we started with an extract containing 63 mg of soluble proteins, 1.3 mg of the highly purified periplasmic form of ChoX was obtained.
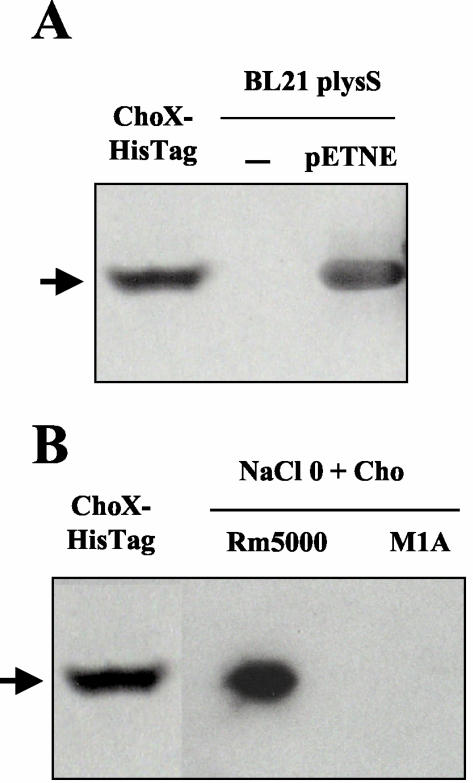
This purified protein was used for binding assays performed with 14C-labeled substrates, and the complex formed between ChoX and the substrate was separated from the unbound substrate by gel filtration. Of the four compounds tested (choline, acetylcholine, glycine betaine, and proline betaine), only choline and acetylcholine were bound to ChoX. In the presence of [methyl-14C]choline as the substrate, addition of a 100-fold excess of unlabeled choline was followed by total disappearance of the label associated with ChoX, demonstrating the specificity of the binding phenomenon (data not shown). Addition of a 100-fold excess of unlabeled acetylcholine significantly decreased the intensity of the labeling by about 70%, whereas, as expected, addition of unlabeled glycine betaine and proline betaine had no effect, confirming that the specificity of the binding phenomenon is very narrow. The maximal binding capacity for choline, as determined by ammonium sulfate precipitation, was 4.8 nmol/mg of protein with a free ligand concentration of 40 μM. The calculated KD for choline was 2.7 μM, whereas it was much higher (145 μM) for acetylcholine. The binding activity was also detected by direct PAGE of the 14C-labeled ligand-protein binding complex in nondenaturing conditions as described previously (17). Briefly, the purified ChoX protein and periplasmic fractions from the E. coli BL21 and BL21(pETNE) strains were incubated with [methyl-14C]choline and subjected to PAGE, followed by autoradiography (Fig. 3A). Since high-affinity choline uptake in E. coli depends only on the betaine choline carnitine transporter BetT (16), no choline-binding activity was detected in periplasmic proteins from E. coli strain BL21. However, induction of choX gene expression in the E. coli BL21 strain carrying the choX gene on the recombinant plasmid pETNE (Table 1) directed the synthesis of a [14C]choline-binding protein. Thus, ChoX was expressed and translocated to the periplasm, meaning that its signal peptide was successfully recognized by the secretion machinery of E. coli. In addition, it is interesting that the presence of a C-terminal His tag on ChoX had no effect on the choline-binding activity since the 14C-labeled purified protein showed the same electrophoretic mobility as the 14C-labeled untagged native overproduced ChoX protein produced in E. coli complemented with the recombinant plasmid pETNE. These binding assays indicated that the narrow range of substrates transported by Cho seems to be linked to the high binding specificity of ChoX. While ChoX can bind [14C]acetylcholine in vitro, results presented above suggested that acetylcholine was not transported by Cho. This discrepancy might be explained by the very low affinity of ChoX for acetylcholine (KD, 145 μM), whereas uptake experiments were performed with a substrate concentration of 100 μM.
FIG. 3.
Choline-binding activity of ChoX and periplasmic proteins as determined by nondenaturing PAGE. (A) Autoradiography of E. coli proteins from recombinant strain BL21(DE3)(pLysS) expressing ChoX from S. meliloti. Purified His-tagged ChoX (40 μg of protein) and periplasmic proteins (100 μg of protein) from E. coli BL21(DE3)(pLysS) carrying or not carrying the recombinant plasmid pETNE were incubated overnight with [methyl-14C]choline (5.5 kBq; 3.25 nmol), subjected to gel electrophoresis (10% polyacrylamide), and then autoradiographed. E. coli cells were grown in LB medium containing 0.4 mM IPTG and were collected at an optical density at 600 nm of 0.6. (B) Autoradiography of periplasmic proteins from S. meliloti. Cells from wild-type strain Rm5000 and mutant M1A were grown in MCAA medium containing 7 mM choline (Cho) until the stationary phase. Purified ChoX (40 μg) and periplasmic proteins (100 μg) were incubated overnight with [methyl-14C]choline (5.5 kBq; 3.25 nmol) and analyzed as described above. The arrow indicates the position of the ChoX-[14C]choline complex.
ChoX is a periplasmic choline-binding protein present in free-living and symbiotic forms of S. meliloti.
The results presented above clearly show that heterologous expression of the choX gene from S. meliloti results in a choline-binding protein. Using nondenaturing gel electrophoresis followed by autoradiography, we subsequently investigated the presence of such a protein in periplasmic extracts of S. meliloti wild-type strain Rm5000 grown at low osmolarity in the presence of choline. A strong radioactive band corresponding to the [14C]choline-choline-binding protein complex and having the same electrophoretic mobility as the [14C]choline-purified ChoX protein complex was detected in periplasmic extracts from the Rm5000 strain. In contrast, analysis of crude periplasmic extracts from the M1A mutant strain grown in the same conditions did not show that any label was bound (Fig. 3B). Thus, in the growth conditions used here, ChoX was the only choline-binding protein present in the periplasm of S. meliloti.
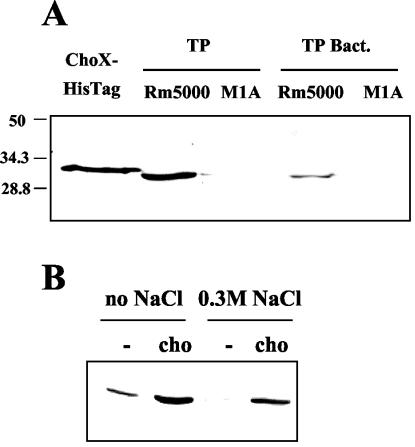
To analyze the effects of growth conditions on the presence of ChoX in S. meliloti and to identify ChoX in bacteroids, the differentiated symbiotic form of the bacterium present in alfalfa nitrogen-fixing nodules, a polyclonal antibody specifically raised against purified ChoX was produced. By immunoblotting, this anti-ChoX antibody was able to detect a 32-kDa protein in total extracts from wild-type strain Rm5000 grown in MCAA medium supplemented with choline (Fig. 4A). As expected, this protein was slightly smaller than the purified recombinant His6-tagged ChoX. No signal was detected with extracts from the choX::Ω M1A mutant strain. While we could not totally eliminate the presence of a ChoX homologue in S. meliloti Rm5000 which could not react with the ChoX antibody, the data are in full agreement with the results presented above and obtained after radiography of the nondenaturing gel (Fig. 3B). Since choline is a ubiquitous molecule in plants and has been identified in alfalfa nodules (18), we wanted to determine whether this transporter was present in bacteroids. Therefore, bacteroids from nodules produced on M. sativa roots by strains Rm5000 and M1A were purified, and the ChoX protein level was estimated by immunoblotting. As shown in Fig. 4A, a protein of the expected size, which corresponded to ChoX, was present in wild-type bacteroids, whereas no signal was detected in the mutant choX bacteroids. This result demonstrated that the ChoX protein is synthesized by the symbiotic form of S. meliloti and suggested that the whole Cho system is probably functional in bacteroids.
FIG. 4.
Immunodetection of the ChoX protein in S. meliloti. (A) Purified recombinant His6-tagged ChoX, total proteins (TP) from wild-type Rm5000 and mutant M1A cells grown in MCAA medium supplemented with 7 mM choline, and total proteins from bacteroids (TP Bact.) isolated from 4-week-old nodules produced on M. sativa roots by strains Rm5000 and M1A were detected by Western blot analysis with anti-ChoX antibody from S. meliloti. The sizes (in kilodaltons) of individual prestained marker proteins (Bio-Rad) are indicated on the left. (B) Immunodetection of ChoX in total protein extracts from GMI211 cells grown in M9 minimal medium containing 0.2% mannitol and maintained at low osmolarity (no salt) or high osmolarity (0.3 M NaCl) in the presence (cho) or absence (−) of choline (7 mM).
Uptake experiments have indicated that the rather low constitutive Cho activity was significantly induced by the presence of the substrate in the growth medium, independent of the osmolarity (Fig. 2). To get a better understanding of the regulation of this system, immunodetection of ChoX in total protein extracts of S. meliloti grown in M9 medium after addition of choline (7 mM) or salt (0.3 M NaCl) was performed (Fig. 4B). In the absence of choline, a very low level of ChoX was observed, particularly in cells grown in the presence of 0.3 M NaCl, in which the choline-binding protein could not be detected. Addition of choline, independent of the presence of 0.3 M NaCl, led to an increase in the ChoX level. Thus, the availability of choline highly induced ChoX expression, whereas salt stress alone had a negative effect on ChoX synthesis. The level of ChoX detected in this experiment is in good agreement with the choline uptake activity of Cho (Fig. 2). Additional experiments showed that a choline concentration of 1 mM was sufficient to induce ChoX biosynthesis (data not shown).
Phenotypes of S. meliloti choX mutant.
In order to precisely define the phenotype of a choX-deficient S. meliloti mutant, the growth properties of the free-living heterotrophic bacterium and the efficiency of the endosymbiotic form were evaluated. Since S. meliloti can use choline as an osmoprotectant and as an energy source after it is converted to glycine betaine (34), the growth parameters were studied in high-osmolarity medium (0.3 to 0.7 M NaCl) supplemented or not supplemented with choline (10 μM, 100 μM, and 7 mM) and in minimal medium containing choline (14 mM) as the only carbon and/or nitrogen source. Significant differences in growth rates and final cell yields between wild-type strain Rm5000 and mutant strain M1A were never observed (data not shown). Obviously, other choline uptake system(s) can compensate for the lack of the Cho transporter. Indeed, in the presence of choline in the growth medium, 65 and 40% of the choline uptake activity of the wild-type strain still remained in the mutant strain grown at high and low osmolarities, respectively (Fig. 2). Thus, it is not very surprising that the choX mutation did not have an effect under standard laboratory growth conditions.
The effects of the choX mutation on nodulation of the alfalfa host plant and on nitrogen fixation activity were also tested. Seedlings were inoculated with the wild-type and mutant strains, and the nodulation efficiency was monitored for 7.5 weeks. No difference was observed in the kinetics of nodulation between the Rm5000 and M1A strains, and the weights of 7.5-week-old nodules obtained with the two strains were comparable (data not shown). In addition, the acetylene reduction activities measured at various times after bacterial infection (4, 6, and 7.5 weeks) for M1A-nodulated plants and Rm5000-nodulated plants indicated that the nitrogen fixation activity was not altered (data not shown). Thus, the nodulation and nitrogen-fixing phenotypes of the M1A mutant strain were Nod+ and Fix+, and the maximum acetylene reduction activity was 25 nmol of ethylene per h per mg (fresh weight) of nodules. As observed for the free-living bacterium, the absence of a functional Cho system is not crucial for the endosymbiotic form of S. meliloti.
DISCUSSION
Because of the competition among soil bacteria for carbon and nitrogen sources, the symbiotic bacterium S. meliloti is subjected to major nutrition challenges. Consequently, this organism is equipped with a very large number of transport systems, which are probably efficiently regulated. As shown by the entire annotated sequence of S. meliloti (11), genes encoding transport systems constitute the largest class of genes (12%), and most of these genes are ABC transporters, which are still uncharacterized. The data presented here describe the first isolation and characterization of an S. meliloti locus which encodes a trimethylammonium, high-affinity, multicomponent, binding protein-dependent transport system, Cho. While Cho is closely related to glycine betaine ProU-like transport systems, its substrate is not betaine but rather choline. It has been shown previously that choline is an efficient carbon and nitrogen source for this bacterium, after it is converted into glycine betaine, which is further catabolized (34). However, the mechanisms of choline transport are not known, and no genes have been assigned to these functions yet. In this study, we isolated and sequenced an S. meliloti DNA fragment composed of three genes, choXWV, which encode a typical ABC transporter with a soluble, ligand-binding, periplasmic protein (ChoX), an integral inner membrane component (ChoW), and an ATPase linked to the membrane (ChoV). Inactivation of the choX gene had no effect on high-affinity glycine betaine or proline betaine uptake activity, whereas it caused a strong reduction in high-affinity choline transport. Since choW- and choV-deficient mutants were not constructed, the corresponding genes were designated based on in silico data and their physiological proximity to choX. The remaining choline uptake activity in the choX-deficient mutant indicated that there is at least one other transporter for this quaternary ammonium compound, in agreement with previous results showing distinct transport activities for choline uptake in S. meliloti (27). Indeed, on the basis of the annotated sequence and considering the homology with previously characterized choline transporters (BetT from E. coli and OpuB from B. subtilis), analysis of the entire genomic sequence revealed the presence of other potential choline transporters, either an ABC system or a symporter. Purified ChoX binds selectively and with high affinity (KD, 2.7 μM) to choline, but none of the betaines appear to be recognized. Only acetylcholine can also be bound by ChoX, but it is bound with a very low affinity (KD, 145 μM). Since the periplasmic protein is thought to ensure the substrate specificity and directionality of the overall transport reaction, Cho can be defined as a strict high-affinity choline transporter. Surprisingly, while ChoX binds choline, no significant homology was found with either OpuBC or OpuCC, the choline-binding proteins of B. subtilis. In contrast, the highest levels of homology for ChoX were found with glycine betaine-binding proteins of the L. lactis BusA, L. monocytogenes Gbu, and B. subtilis OpuA transporters and to a lesser extent with ProX, the periplasmic protein of the E. coli glycine betaine ProU transporter (Fig. 1B). The high-resolution crystal structure of the ProX protein, in complex with each of its ligands (i.e., ProX∼glycine betaine and ProX∼proline betaine), was just determined recently (32). The binding pocket is formed by the indole groups of three tryptophan residues, Trp65, Trp140, and Trp188. This crystallographic study revealed that cation-π interactions between the positive charges of the quaternary amines of the ligands and the indole groups of the three tryptophan residues are the key determinants of the high-affinity binding of betaines by ProX. In addition, the entire motif C136XPGWGC142 is strictly conserved among several close homologues of ProX from various bacteria. While the overall structure of the S. meliloti choline-binding protein is still unknown, it is more likely that choline interacts with ChoX by using the positive charge of the quaternary amine group. However, if two tryptophan residues from ProX, at positions 65 and 188, are well conserved in ChoX, the Trp residue at position 140 is replaced by an Asn residue, and the two ProX cysteine residues, Cys136 and Cys142, are absent in ChoX (Fig. 1B). Thus, the arrangement of the binding site for choline, which possesses a hydroxylic group, is obviously different from the arrangement of the binding site for glycine betaine, which has a carboxylic group. A crystallographic study of ChoX would be very informative and should allow us to precisely define the structure of the choline-binding site.
Characterized ABC choline transporters are rather scarce. To our knowledge, the only choline ABC transporters in bacteria that have been fully characterized are the OpuB and the OpuC systems from B. subtilis (13, 12). These two systems are closely related and evolved from a primordial gene cluster duplication. Regardless of the identity, but considering the functionality, Cho is physiologically more similar to OpuB, which is highly specific for choline, than to OpuC, which is involved in the entry of a large variety of compounds, including choline, choline-O-sulfate, glycine betaine, γ-butyrobetaine, crotonobetaine, ectoine, carnitine, and probably some other substrates. Western blotting experiments with a polyclonal antiserum cross-reacting with the presumed substrate-binding proteins from both the OpuB and OpuC transporters have suggested that expression of the opuB and opuC operons is regulated in response to increasing osmolarity of the growth medium (13). Our studies show that Cho activity is strongly stimulated by the presence of choline in the growth medium, whereas elevated osmolarity has no effect (Fig. 2). Such results are in full agreement with immunodetection of ChoX, which indicated that there was clear induction in choline-grown cells and a very low level in NaCl-grown cells (Fig. 4B). In contrast to B. subtilis, S. meliloti uses choline as a carbon and nitrogen source, and expression of the cho and opuB genes is obviously regulated differently. Whereas in the gram-positive bacterium OpuB contributes to osmotic adjustment (13), it is more likely that choline taken up by S. meliloti via Cho is catabolized after subsequent conversion into glycine betaine and/or is used as a direct precursor of PC. This phospholipid is crucial for S. meliloti since it is required for normal growth (5) and also for a successful interaction with the host plant, alfalfa (35). In this context, it is interesting to highlight the presence of ChoX in differentiated bacteroids (Fig. 4A). Choline is indeed available in alfalfa, and significant amounts of choline have recently been found in the cytosol of nodule cells, in the peribacteroid space of the symbiosome, and also in bacteroids (18). The choline concentration in bacteroids was estimated to be approximately 1 mM, a concentration sufficient to induce ChoX synthesis. In addition, these results indicate that choline provided by the host plant is transported into symbiosomes and bacteroids through the peribacteroid membrane. In fact, preliminary experiments with purified symbiosomes confirmed that there is choline transport through this membrane (data not shown), and previous data have indicated that there is choline transport activity in isolated bacteroids (9).
At present, the physiological effect of the choX mutation on the phenotype of S. meliloti, either as a free-living cell or as an endosymbiotic form, is not clear. Significant residual levels of choline transport activity in the mutant suggest that there must be an alternative route(s) for choline uptake. In B. subtilis, for example, double mutations in the opuB and opuC loci are required to abolish osmoprotection by choline, since each of the ABC transporters, OpuB and OpuC, is able to provide the cell with enough choline to sustain growth under unfavorable circumstances (13). Thus, identification of the other choline transporter system(s) should help workers evaluate the importance of choline for S. meliloti, both as a heterotrophic soil bacterium and during the establishment and maintenance of symbiosis.
Acknowledgments
Financial support for this study was provided by the Centre National de la Recherche Scientifique.
We are grateful to the colleagues who generously provided strains and the S. meliloti genomic bank used in this study. We thank R. Krämer for the gift of cold ectoine.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Boch, J., B. Kempf, and E. Bremer. 1994. Osmoregulation in Bacillus subtilis: synthesis of the osmoprotectant glycine betaine from exogenously provided choline. J. Bacteriol. 176:5364-5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Brhada, F., M. C. Poggi, and D. Le Rudulier. 1997. Choline and glycine betaine uptake in various strains of rhizobia isolated from nodules of Vicia faba var. major and Cicer arietinum L.: modulation by salt, choline, and glycine betaine. Curr. Microbiol. 34:167-172. [DOI] [PubMed] [Google Scholar]
- 5.de Rudder, K. E., I. M. Lopez-Lara, and O. Geiger. 2000. Inactivation of the gene for phospholipid N-methyltransferase in Sinorhizobium meliloti: phosphatidylcholine is required for normal growth. Mol. Microbiol. 37:763-772. [DOI] [PubMed] [Google Scholar]
- 6.de Rudder, K. E., C. Sohlenkamp, and O. Geiger. 1999. Plant-exuded choline is used for rhizobial membrane lipid biosynthesis by phosphatidylcholine synthase. J. Biol. Chem. 274:20011-20016. [DOI] [PubMed] [Google Scholar]
- 7.Ditta, G., S. Stanfield, D. Corbin, and D. R. Helinski. 1980. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 77:7347-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finan, T. M., E. Hartweig, K. LeMieux, K. Bergman, G. C. Walker, and E. R. Signer. 1984. General transduction in Rhizobium meliloti. J. Bacteriol. 159:120-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fougère, F., and D. Le Rudulier. 1990. Uptake of glycine betaine and its analogues by bacteroids of Rhizobium meliloti. J. Gen. Microbiol. 136:157-163. [DOI] [PubMed] [Google Scholar]
- 10.Friedman, A. M., S. R. Long, S. E. Brown, W. J. Buikema, and F. M. Ausubel. 1982. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene 18:289-296. [DOI] [PubMed] [Google Scholar]
- 11.Galibert, F., T. M. Finan, S. R. Long, A. Puhler, P. Abola, F. Ampe, F. Barloy-Hubler, M. J. Barnett, A. Becker, P. Boistard, G. Bothe, M. Boutry, L. Bowser, J. Buhrmester, E. Cadieu, D. Capela, P. Chain, A. Cowie, R. W. Davis, S. Dreano, N. A. Federspiel, R. F. Fisher, S. Gloux, T. Godrie, A. Goffeau, B. Golding, J. Gouzy, M. Gurjal, I. Hernandez-Lucas, A. Hong, L. Huizar, R. W. Hyman, T. Jones, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, E. Kiss, C. Komp, V. Lelaure, D. Masuy, C. Palm, M. C. Peck, T. M. Pohl, D. Portetelle, B. Purnelle, U. Ramsperger, R. Surzycki, P. Thebault, M. Vandenbol, F. J. Vorholter, S. Weidner, D. H. Wells, K. Wong, K. C. Yeh, and J. Batut. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293:668-672. [DOI] [PubMed] [Google Scholar]
- 12.Kappes, R. M., B. Kempf, and E. Bremer. 1996. Three transport systems for the osmoprotectant glycine betaine operate in Bacillus subtilis: characterization of OpuD. J. Bacteriol. 178:5071-5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kappes, R. M., B. Kempf, S. Kneip, J. Boch, J. Gade, J. Meier-Wagner, and E. Bremer. 1999. Two evolutionarily closely related ABC transporters mediate the uptake of choline for synthesis of the osmoprotectant glycine betaine in Bacillus subtilis. Mol. Microbiol. 32:203-216. [DOI] [PubMed] [Google Scholar]
- 14.Kashiwagi, K., S. Miyamoto, E. Nukui, H. Kobayashi, and K. Igarashi. 1993. Functions of PotA and PotD proteins in spermidine-preferential uptake system in Escherichia coli. J. Biol. Chem. 268:19358-19363. [PubMed] [Google Scholar]
- 15.Kortstee, G. J. 1970. The aerobic decomposition of choline by microorganisms. I. The ability of aerobic organisms, particularly coryneform bacteria, to utilize choline as the sole carbon and nitrogen source. Arch. Mikrobiol. 71:235-244. [PubMed] [Google Scholar]
- 16.Lamark, T., I. Kaasen, M. W. Eshoo, P. Falkenberg, J. McDougall, and A. R. Strøm. 1991. DNA sequence and analysis of the bet genes encoding the osmoregulatory choline-glycine betaine pathway of Escherichia coli. Mol. Microbiol. 5:1049-1064. [DOI] [PubMed] [Google Scholar]
- 17.Le Rudulier, D., K. Gloux, and N. Riou. 1991. Identification of an osmotically induced periplasmic glycine betaine-binding protein from Rhizobium meliloti. Biochim. Biophys. Acta 1061:197-205. [DOI] [PubMed] [Google Scholar]
- 18.Mandon, K., M. Østerås, E. Boncompagni, J. C. Trinchant, G. Spennato, M. C. Poggi, and D. Le Rudulier. 2003. The Sinorhizobium meliloti glycine betaine biosynthetic genes (betCBA) are induced by choline and highly expressed in bacteroids. Mol. Plant-Microbe Interact. 16:709-719. [DOI] [PubMed] [Google Scholar]
- 19.Martinez-Morales, F., M. Schobert, I. M. Lopez-Lara, and O. Geiger. 2003. Pathways for phosphatidylcholine biosynthesis in bacteria. Microbiology 149:3461-3471. [DOI] [PubMed] [Google Scholar]
- 20.Meade, H. M., S. R. Long, G. B. Ruvkun, S. E. Brown, and F. M. Ausubel. 1982. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J. Bacteriol. 149:114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 22.Neu, H. C., and L. A. Heppel. 1965. The release of enzymes from Escherichia coli by osmotic shock and during the formation of spheroplasts. J. Biol. Chem. 240:3685-3692. [PubMed] [Google Scholar]
- 23.Niel, C., J. B. Guillaume, and M. Bechet. 1977. Demonstration of two enzymes with beta-galactosidase activity in Rhizobium meliloti. Can. J. Microbiol. 23:1178-1181. [PubMed] [Google Scholar]
- 24.Østerås, M., E. Boncompagni, N. Vincent, M. C. Poggi, and D. Le Rudulier. 1998. Presence of a gene encoding choline sulfatase in Sinorhizobium meliloti bet operon: choline-O-sulfate is metabolized into glycine betaine. Proc. Natl. Acad. Sci. USA 95:11394-11399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perroud, B., and D. Le Rudulier. 1985. Glycine betaine transport in Escherichia coli: osmotic modulation. J. Bacteriol. 161:393-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Platzer, J., W. Sterr, M. Hausmann, and R. Schmitt. 1997. Three genes of a motility operon and their role in flagellar rotary speed variation in Rhizobium meliloti. J. Bacteriol. 179:6391-6399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pocard, J. A., T. Bernard, L. T. Smith, and D. Le Rudulier. 1989. Characterization of three choline transport activities in Rhizobium meliloti: modulation by choline and osmotic stress. J. Bacteriol. 171:531-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 29.Richarme, G., and A. Kepes. 1983. Study of binding protein-ligand interaction by ammonium sulfate-assisted adsorption on cellulose esters filters. Biochim. Biophys. Acta 742:16-24. [DOI] [PubMed] [Google Scholar]
- 30.Rosenberg, C., P. Boistard, J. Dénarié, and F. Casse-Delbart. 1981. Genes controlling early and late functions in symbiosis are located on a megaplasmid in Rhizobium meliloti. Mol. Gen. Genet. 184:326-333. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 32.Schiefner, A., J. Breed, L. Bosser, S. Kneip, J. Gade, G. Holtmann, K. Diederichs, W. Welte, and E. Bremer. 2004. Cation- interactions as determinants for binding of the compatible solutes glycine betaine and proline betaine by the periplasmic ligand-binding protein ProX from Escherichia coli. J. Biol. Chem. 279:5588-5596. [DOI] [PubMed] [Google Scholar]
- 33.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 34.Smith, L. T., J. A. Pocard, T. Bernard, and D. Le Rudulier. 1988. Osmotic control of glycine betaine biosynthesis and degradation in Rhizobium meliloti. J. Bacteriol. 170:3142-3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sohlenkamp, C., I. M. López-Lara, and O. Geiger. 2003. Biosynthesis of phosphatidylcholine in bacteria. Prog. Lipid Res. 42:115-162. [DOI] [PubMed] [Google Scholar]
- 36.Styrvold, O. B., P. Falkenberg, B. Landfald, M. W. Eshoo, T. Bjornsen, and A. R. Strøm. 1986. Selection, mapping, and characterization of osmoregulatory mutants of Escherichia coli blocked in the choline-glycine betaine pathway. J. Bacteriol. 165:856-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tam, R., and M. H. Saier, Jr. 1993. Structural, functional, and evolutionary relationships among extracellular solute-binding receptors of bacteria. Microbiol. Rev. 57:320-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trinchant, J. C., V. Guérin, and J. Rigaud. 1994. Acetylene reduction by symbiosomes and free bacteroids from broad bean (Vicia faba L.) nodules (role of oxalate). Plant Physiol. 105:555-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whitley, P., T. Zander, M. Ehrmann, M. Haardt, E. Bremer, and G. von Heijne. 1994. Sec-independent translocation of a 100-residue periplasmic N-terminal tail in the E. coli inner membrane protein ProW. EMBO J. 13:4653-4661. [DOI] [PMC free article] [PubMed] [Google Scholar]