Abstract
Mycothiol (MSH), a functional analogue of glutathione (GSH) that is found exclusively in actinomycetes, reacts with electrophiles and toxins to form MSH-toxin conjugates. Mycothiol S-conjugate amidase (Mca) then catalyzes the hydrolysis of an amide bond in the S conjugates, producing a mercapturic acid of the toxin, which is excreted from the bacterium, and glucosaminyl inositol, which is recycled back to MSH. In this study, we have generated and characterized an allelic exchange mutant of the mca gene of Mycobacterium smegmatis. The mca mutant accumulates the S conjugates of the thiol-specific alkylating agent monobromobimane and the antibiotic rifamycin S. Introduction of M. tuberculosis mca epichromosomally or introduction of M. smegmatis mca integratively resulted in complementation of Mca activity and reduced levels of S conjugates. The mutation in mca renders the mutant strain more susceptible to electrophilic toxins, such as N-ethylmalemide, iodoacetamide, and chlorodinitrobenzene, and to several oxidants, such as menadione and plumbagin. Additionally we have shown that the mca mutant is also more susceptible to the antituberculous antibiotic streptomycin. Mutants disrupted in genes belonging to MSH biosynthesis are also more susceptible to streptomycin, providing further evidence that Mca detoxifies streptomycin in the mycobacterial cell in an MSH-dependent manner.
After AIDS, tuberculosis (TB), caused by Mycobacterium tuberculosis, is the second leading cause of death from infectious agents worldwide (26). The resurgence of TB as a potential publichealth threat due to its synergy with human immunodeficiency virus and the emergence of multidrug-resistant TB strains has resulted in renewed interest in this gram-positive actinomycete. M. tuberculosis is a facultative intracellular pathogen residing in macrophages and in the granuloma, where it is faced with constant assault from toxic agents. Survival in such hostile environments is dependent on detoxification mechanisms that allow M. tuberculosis to persist in the host despite the host immune response.
Detoxification processes rely on either phase I or phase II detoxification reactions, or both. In phase I reactions, cytochrome P450, a mixed-function oxidase, catalyzes the incorporation of an oxygen atom from O2 into a xenobiotic substrate, making the toxin more hydrophilic. If the product of the cytochrome P450-mediated reaction is not sufficiently hydrophilic but is thiol reactive, a subsequent reaction where the toxin is conjugated to glutathione (GSH) occurs, an important process in the mammalian liver. In mammals, GS toxin conjugates are acted upon by a γ-glutamyl transpeptidase that cleaves the glutamic acid group from the GSH molecule followed by hydrolysis of the glycine moiety by a peptidase resulting in a cysteine toxin conjugate in a multiorgan process. This conjugate is acetylated in the kidney by an acetyl-coenzyme A-dependent N-acetyltransferase to form a mercapturic acid that is excreted (8).
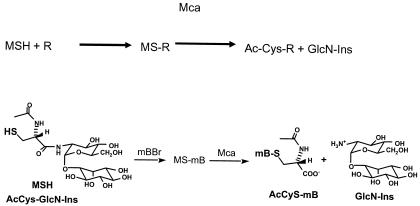
Actinomycetes like M. tuberculosis do not produce GSH but instead synthesize a novel low-molecular-weight thiol, mycothiol (MSH) (13), a conjugate of N-acetyl-l-cysteine amide linked to the psuedodisaccharide comprised of d-glucosamine α (1-1) linked to myo-inositol. Mycobacterium smegmatis mutants lacking MSH are more sensitive to oxidizing agents, electrophiles, and several antibiotics (18, 21, 22), indicating that MSH-dependent detoxification mechanisms exist in the mycobacterial cell. Indeed, a novel MSH-dependent detoxification mechanism where MSH reacts with a toxin, such as the fluorescent alkylating agent monobromobimane (mBBr), to form an MS toxin conjugate (designated MS-mB) which is acted upon by a mycothiol S-conjugate amidase (Mca) to yield a mercapturic acid (AcCyS-mB) and 1d-myo-inosityl 2-amino-2-deoxy-glucopyranoside (GlcN-Ins) (Fig. 1) was described by Newton et al. (14). The substrates for this enzyme include the S conjugates of mBBr, iodoacetamide, and N-ethylmalemide (NEM). Interestingly, one of the substrates for this enzyme is the S conjugate of cerulenin, an antibiotic from the actinomycete Cephalosporin ceruleans. Recently, the mycothiol S conjugate of rifamycin S, from which the semisynthetic antituberculous drug rifampin is derived, was demonstrated to be a substrate for Mca (25). Maynes et al. (11) recently published the crystal structure of MshB, a homolog of Mca that catalyzes the second step of MSH biosynthesis, cleavage of the amide bond in GlcNAc-Ins. They modeled the catalytic domain of Mca from this crystal structure and found that amino acid residues involved in binding the active-site zinc and in catalysis aligned perfectly, with the exception of Lys19 in Mca in place of Ser20 in MshB.
FIG. 1.
Schematic representation of the mycothiol-dependent amidase-catalyzed detoxification reaction.
A search of sequence databases revealed that homologs of Mca are present in all other mycobacteria. Homologs of this gene have also been reported in several antibiotic biosynthetic operons, including the erythromycin biosynthetic operon. Furthermore, mercapturic acids of antibiotics have been found in the broth of some antibiotic-producing species (16). For these reasons, we have postulated that this enzyme, in conjunction with mycothiol, may play a major role in detoxification of antibiotics. To better understand the role of Mca and MSH in xenobiotic detoxification, a targeted mutant of the mca gene in the saprophytic mycobacteria M. smegmatis was constructed. In this study we describe the generation and characterization of this M. smegmatis mca mutant.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Escherichia coli strain DH5α was used as the host strain for cloning experiments. E. coli was grown in Luria-Bertani broth and on Luria-Bertani solid media. M. smegmatis mc2155 was the parent wild-type strain used for construction of knockout mutants. M. smegmatis was grown in Middlebrook 7H9 broth (Difco) with 0.05% Tween and was supplemented with either Middlebrook oleic acid-albumin-dextrose-catalase (OADC) supplement or 1% glucose. M. smegmatis was also grown on Middlebrook 7H10 solid medium (Difco) supplemented with OADC or 1% glucose. Ampicillin (100 μg ml−1 for E. coli), gentamicin (15 μg ml−1 for E. coli and 10 μg ml−1 for M. smegmatis), kanamycin (100 μg ml−1 for E. coli and 25 μg ml−1 for M. smegmatis), and hygromycin (100 μg ml−1 for E. coli and 50 μg ml−1 for M. smegmatis) were added as needed. Complements of the mutant harboring the recombinant pALACE vector were grown on Middlebrook 7H10 solid medium supplemented with OADC and 1% acetamide for induction of the cloned gene.
Molecular biology techniques.
Genomic DNA was isolated from M. smegmatis cultures as described by Hatfull and Jacobs (7). M. smegmatis transformations were carried out using a Bio-Rad Gene Pulser with mycobacterial cells prepared as described by Snapper et al. (24). Standard recombinant DNA techniques, such as restriction digestion, ligation, and transformation, were carried out as described by Sambrook et al. (23). Probes for Southern blotting were labeled with digoxigenin (DIG)-labeled deoxynucleoside triphosphates by using a Roche DIG labeling kit, and membranes were developed according to the manufacturer's instructions (Roche Diagnostics). The list of strains, plasmids, and oligonucleotides used in this study are described in Table 1.
TABLE 1.
Strains, plasmids, and oligonucleotides used in this study
| Strain, plasmid, or oligonucleotide | Characteristic(s)a | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | F recA1 hsdR17 thi-1 gyrA96 supE44 endA1 relA1 recA1 deoR Δ(lacZYA-argF)U169 (φ80 lacZΔM15) | |
| M. smegmatis | ||
| mc2155 | Parental strain | 24 |
| Ami37 | Mutant in mca | This study |
| Ami40 | Mutant in mca | This study |
| ami37phint | Ami37 complemented with M. smegmatis mca | This study |
| ami37palace | Ami37 complemented with M. tuberculosis mca | This study |
| Plasmids | ||
| pMR1082K | Knockout construct with M. smegmatis mca disrupted with gentamicin resistance cassette | This study |
| pALACE | Hygr | 17 |
| pHINT | Hygr | K. Downing |
| pGINT | Genr | K. Downing |
| pCR2.1 | TA cloning vector, Ampr Kanr | Invitrogen |
| pMK1082 | PCR-amplified M. smegmatis mca cloned into pCR 2.1 | This study |
| pHINTmk1082 | M. smegmatis mca cloned into pHINT | This study |
| pMR4 | M. tuberculosis mca cloned into pALACE vector | 22 |
| Oligonucleotides | ||
| Pgint1082i-5′ | TTAAGCTTGCCGAAGCGCTGATACG | This study |
| Pgint1082i-3′ | TTGATATCGGCTCGTCGGCCAGAAG | This study |
Ampr, ampicillin resistance; Kanr, kanamycin resistance; Genr, gentamicin resistance; Hgr, hygromycin resistance.
Bioinformatic analyses.
The genomic sequence and the predicted genes were obtained for M. tuberculosis H37Rv (4) from the Tuberculist website (http://www.genolist.pasteur.fr/Tuberculist). Sequence data for M. smegmatis were obtained from the Institute for Genomic Research (http://www.tigr.org). BLAST searches were performed according to the methods of Altschul et al. (1). PFAM database information was obtained from http://www.sanger.ac.uk/Software/Pfam/, and PROSITE database information was obtained at http://www.expasy.ch/prosite.
Targeted mutagenesis of mca.
A recombination cassette was constructed in order to disrupt mca on the M. smegmatis chromosome. The M. smegmatis mca gene fragment, spanning from 300 bp upstream to 40 bp downstream, was PCR amplified using primers Pgint1082I-5′ and Pgint1082I-3′, listed in Table 1. The amplified gene was cloned into PCR cloning vector pCR 2.1 to yield pMK1082. pMK1082 was digested with PmaCI, because there is a unique restriction site for this enzyme in the middle of the M. smegmatis mca gene. A gentamicin resistance cassette was prepared by restriction digestion of pGINT vector with SacI. The appropriate SacI fragment containing the gentamicin resistance gene was agarose gel purified and treated with Klenow fragment of DNA polymerase to yield blunt ends. This fragment was ligated with pMK1082 restriction digested with PmaCI such that the resulting plasmid, pMR1082K, contained the gentamicin resistance gene disrupting the mca open reading frame. pMR1082K also has a kanamycin resistance marker in the pCR2.1 vector backbone that provides for negative selection. M. smegmatis mc2155 was transformed with pMR1082K, and gentamicin-resistant, kanamycin-sensitive transformants were chosen for further analysis. In addition, M. smegmatis mc2155 was transformed with empty vector pGINT to serve as a control for further characterization studies.
Sensitivity assays for antibiotics, alkylating agents, and oxidative agents.
Three different methods were used for sensitivity assays. E-test strips (AB Biodisk) were used to determine the MICs of isoniazid, rifampin, and vancomycin for the different strains (21). Disk diffusion assays were performed for vancomycin, cerulenin, lincomycin, erythromycin, rifampin, and streptomycin according to Rawat et al. (22). In addition, the following alkylating agents and oxidants were tested: NEM, iodoacetamide, chlorodinitrobenzene (CDNB), mBBr, menadione, plumbagin, nitrofurantoin, cumene hydroperoxide, and hydrogen peroxide. For the preceding two methods, 7H10 solid media supplemented with 1% glucose were used, and these plates were incubated for 48 to 72 h. The third method was the minimal broth dilution assay, where the toxin or drug was serially diluted in 1 ml of 7H9 medium supplemented with 1% glucose and cells were added to each tube in the serial dilution, with an optical density at 600 nm (OD600) of 0.05. After 2 to 3 days of incubation at 37°C the tubes were checked visually for growth. All assays were performed in triplicate at least three times.
Determination of mycothiol levels and mycothiol amidase activity.
Derivatization of cell extracts with mBBr and high-performance liquid chromatography (HPLC) analysis of the derivatized samples to determine the thiol content were performed essentially as described earlier (2). Control samples treated with NEM and then with mBBr were also analyzed. Results are reported as micromoles per gram of residual dry weight measured on the pellet obtained from the 50% acetonitrile-water extraction.
Mca activity was tested essentially as described by Newton et al. (14). Briefly, samples to be tested were pelleted and brought up in 200 μl of 25 mM HEPES, pH 7.5. The cells were sonicated and centrifuged for 3 min at 14,000 × g. The cell extract protein concentration was measured using the Bio-Rad protein assay. A reaction volume of 50 μl consisted of 100 μg of protein sample in 25 mM HEPES (pH 7.5), 30 μM mycothiol-mBBr adduct (MS-mB), and 3 mM 2-mercaptoethanol. The samples were incubated for 30 min at 30°C, and then the reaction was stopped by adding 50 μl of 40 mM methanesulfonic acid. The samples were subjected to HPLC analysis as described earlier (14).
Analysis of M. smegmatis mc2155 and Ami37 (Δmca::gentr) treated with mBBr and rifamycin S in culture.
Duplicate samples of 10 ml of late-log-phase M. smegmatis mc2155 cells and Ami37, the mca knockout mutant, were incubated on ice for 20 min. To the iced cells, 180 mM mBBr in acetonitrile was added to a final concentration of 0.5 mM mBBr, and the cells were incubated on ice for an additional 30 min. Excess 2-mercaptoethanol (1.0 mM) was added to scavenge unreacted mBBr and was allowed to react with the cells for 10 min on ice. The cells were then harvested by centrifugation at 4°C, and the supernatant was retained for analysis of thiol-mB derivatives in the medium. The cells were extracted with 50% acetonitrile in water. After incubation at 60°C for 10 min, the cells were acidified and centrifuged to remove cell debris. The supernatant was analyzed after dilution with 10 mM methanesulfonic acid using the same conditions described above for the MSH assay.
For rifamycin S treatment, four liters of M. smegmatis mc2155 and Ami37 was cultured to an OD600 of 1. The cells were then harvested and placed in 200 ml of ice-cold 7H9 Middlebrook medium containing 1% glucose. The cells were incubated with 1 mg of rifamycin S/ml for 1 h on ice and then were washed two times with ice-cold medium and resuspended in 20 ml of ice-cold medium. The cells were then diluted into 180 ml of 37°C medium and sampled at 1 min, 10 min, 60 min, 2 h, and 16 h. As described by Steffek et al. (25), the cells were extracted and analyzed for rifamycin S conjugates. Cellular rifamycin S conjugates were quantified by HPLC at 315 nm, an isosbestic wavelength for the rifamycin S and rifamycin SV (RifSV) forms, using an approximate extinction coefficient of 24,000 M−1 cm−1 and MS-RifSV standards.
Complementation of Ami37 with M. tuberculosis mca and M. smegmatis mca homolog.
The M. tuberculosis mca gene was amplified from M. tuberculosis genomic DNA and cloned into a replicative vector, pALACE, as described earlier (22) to yield pM4. The mca mutant was transformed with pM4, and transformants were selected on 7H10 agar plates supplemented with OADC, 50 μg of hygromycin ml−1, and 10 μg of gentamicin ml−1. To ascertain that the transformants contained the plasmid, the plasmids were rescued. Protein expression was checked on sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels after induction of protein expression by growth in 1% acetamide. One transformant, ami37palace, was selected for further detailed study.
For complementation with the M. smegmatis mca gene, the pMK1082 plasmid described earlier and the pHINT vector were restriction digested with HindIII. The pHINT fragment was dephosphorylated and then ligated to the pMK1082 fragment to create pHINTMK1082. Ami37 was transformed with pHINTMK1082, and the cells were selected on 7H10 agar plates supplemented with OADC, 50 μg of hygromycin ml−1, and 10 μg of gentamicin ml−1. One transformant, ami37phint, was selected for further detailed study.
RESULTS
Bioinformatic analyses.
M. tuberculosis mca is an 864-bp gene that codes for a protein of 288 amino acids that has a molecular mass of 32.7 kDa and a pI of 5.1. M. smegmatis mca is also an 864-bp gene that codes for a 288-amino-acid protein. Fasta analysis of the two proteins revealed 78% similarity between 227 out of 288 of the amino acids. Both genes have a PF02585 PFAM signature, which has been described as a lmbE-like protein signature. The lmbE gene from Streptomyces lincolnensis is a gene of unknown function present in the lincomycin biosynthesis cluster (19).
Homologs of mca are present in all mycobacterial species sequenced so far, and homologs are also found in other antibiotic biosynthesis clusters, such as leinamycin biosynthesis genes of Streptomyces atroolivaceus, rifamycin biosynthesis genes of Amycolatopsis mediterranei, erythromycin biosynthesis genes of Streptomyces erythrae, and streptothricin biosynthesis genes of Streptomyces rochei. Three homologs are found in Streptomyces coelicolor, and two homologs are found in Streptomyces avermitilis. The M. tuberculosis genome also has two paralogs, Rv1170, which codes for MshB (3, 11, 15, 22), and Rv0323c, a gene that is present only in pathogenic mycobacteria and thus is absent in nonpathogenic M. smegmatis.
The mca gene is the first gene in an operon of three genes in M. tuberculosis. The second gene in the operon, Rv1083, 88 bp in length, encodes a 9.2-kDa protein of unknown function. Rv1083 has homology to the gene ML2390 (57% similarity in 59 out of 101 of the amino acids) from M. leprae. ML2390 has an N-terminal signal sequence and is thus possibly secreted. The last gene in the operon, Rv1084, 673 bp in length, encodes a 71-kDa protein of unknown function. Interestingly, the Rv1084 gene contains a carboxypeptidase zinc binding domain (Prosite entry PS00133) and a thioredoxin conserved domain, COG1331. The organization of the operon in M. smegmatis is the same as that of M. tuberculosis.
Ami37 is a knockout mutant of mca.
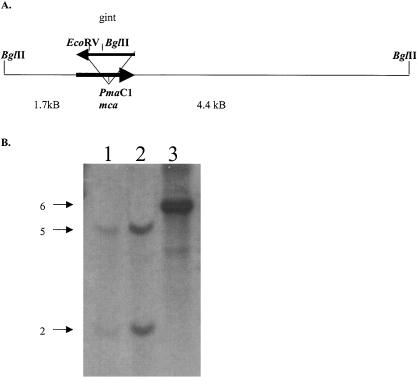
Genomic DNA, extracted from both the parental strain and putative mutants, was digested with BglII and was subjected to Southern hybridization using a DIG-labeled probe (Fig. 2B). The probe hybridized to a BglII fragment of approximately 6 kb in the parent strain (Fig. 2B, lane 3); however, disruption of mca by the gentamicin cassette introduced a second BglII site in the mutant DNA (Fig. 2A). Restriction digestion with BglII should result in two bands, one 2 kb in size and another approximately 5 kb in size. Two of the putative mutants, Ami37 (Fig. 2B, lane 2) and Ami40 (Fig. 2B, lane 1), showed two bands of 2 and 5 kb, indicating that the mca gene has been disrupted by the gentamicin resistance gene in these two clones. Mutant Ami37 was shown to have a MSH content equivalent to the parental strain (10 ± 2 μmol/g) and was chosen for further analysis.
FIG. 2.
Schematic representation of the mca inactivation in M. smegmatis and Southern analysis of mutants versus the parent strain, mc2155. (A) Site of disruption of the Ami37 mca mutant. (B) Southern analysis of BglII-digested genomic DNA from mc2155 (lane 3) and the mutants Ami37 (lane 2) and Ami40 (lane 1). Ten micrograms of DNA was loaded on the gel. The gel was hybridized with a probe for M. smegmatis mca that was PCR DIG-labeled using primers Pgint1082I-5′ and Pgint1082I-3′. The probe hybridized to a 6-kb BglII fragment in the parent strain. The disruption of mca in the mutants introduced a BglII site into the fragment which resulted in the probe hybridizing to two bands of 2 and 5 kb.
Mutant Ami37 does not possess Mca activity.
The mutant Ami37 was compared to the wild-type parental strain in terms of its ability to detoxify mBBr and to secrete mercapturic acids essentially as described previously (14). Extracts of the mutant lack amidase activity with MS-mB (<0.0015 nmol/min/mg) and therefore do not produce the mercapturic acid product AcCys-mB. Under the same conditions, extracts of the parental strain exhibit high activity with MS-mB (25 nmol/min/mg). Thus, amidase activity toward MS-mB is effectively eliminated in Ami37.
A close homolog of mca is mshB, which codes for MshB deacetylase, one of the enzymes involved in the biosynthesis of mycothiol (15). This enzyme is known to possess Mca activity as well as GlcNAc-Ins deacetylase activity, albeit at a much lower level than that of Mca. To examine whether MshB activity is upregulated in the mutant to compensate for loss of Mca, we assayed the parent and mutant strains for MshB amidase activity. The mBBr adduct of Cys-GlcN-Ins (CySmB-GlcN-Ins) is known to be a very poor substrate for Mca (25) but is still the best known amidase substrate for MshB (G. Newton and R. C. Fahey, unpublished data). Assay of cellular extracts with CySmB-GlcN-Ins (0.1 mM) gave specific activities of 0.12 and 0.16 nmol min−1 mg−l for the parent and mutant strains, respectively. This indicates that only a small increase in MshB activity is observed in the mca mutant and cannot significantly compensate for the loss of Mca activity.
Analysis of M. smegmatis mc2155 and Ami37 treated with mBBr in culture.
Following an experiment which examined the fate of mycothiol and mBBr in M. smegmatis cells treated with mBBr (14), we sought to determine whether the mutant exhibited a phenotype in culture different from that of the parent wild-type strain. When the parent strain mc2155 was treated with 0.5 mM mBBr in vivo for 30 min on ice, both MS-mB and the product of the Mca reaction (AcCyS-mB) were found within the cells at a ratio of approximately 1:3. AcCyS-mB was present in the media at low levels (∼5% of the total intracellular content) (Table 2). In contrast, in Ami37 only the conjugate MS-mB was detected intracellularly. There was also no trace of AcCys-SmB in media for the mutant, confirming that the mutant is unable to cleave the MS-mB conjugate (Table 2).
TABLE 2.
Amount of substrate MS-mB and product AcCyS-mB of Mca present intracellularly and extracellularly in the parent strain and the mca mutant Ami37 following reaction with mBBr
| Substrate | Amt of substrate for strain:
|
|
|---|---|---|
| mc2155 | Ami37 | |
| Intracellular | ||
| MS-mB (μmol/g) | 2.8 ± 0.02 | 16.6 ± 0.5 |
| AcCyS-mB (μmol/g) | 9.9 ± 0.2 | <0.02 |
| Total | 12.7 | 16.6 ± 0.5 |
| Extracellular | ||
| AcCyS-mB (μM) | 0.75 ± 0.04 | <0.1 |
Analysis of M. smegmatis mc2155 and Ami37 treated with rifamycin S in culture.
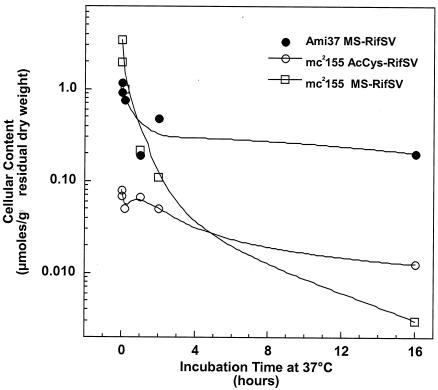
Steffek et al. (25) demonstrated that MSH could form a conjugate with rifamycin S that served as a substrate for Mca. When M. smegmatis mc2155 was treated with rifamycin S on ice for 1 h, the cells washed with cold medium, and a cold suspension of the cells diluted into warm medium, initial analysis showed that approximately 30% of the normal mycothiol content was present as the MS-RifSV conjugate (Fig. 3). MS-RifSV declined three orders of magnitude to the limit of detection at 16 h (Fig. 3). The Ami37 mutant had a lesser amount of MS-RifSV initially, and it declined only threefold over 16 h. In the parent strain, the S conjugate of N-acetylcysteine with rifamycin S (AcCyS-RifSV), a mercapturic acid, was evident in the cells, demonstrating the activity of the endogenous amidase on the MS-RifSV conjugate. No AcCyS-RifSV was found above the limit of detection (<0.02 μmol/g) in Ami37, consistent with the disruption of the mca gene. The loss of MS-RifSV from mc2155 could not be accounted for in terms of products excreted into the medium, as analysis of the medium revealed maximal levels of AcCyS-RifSV and MS-RifSV equivalent to only 4 and 30%, respectively, of the MS-RifSV lost from the cells. Other components with similar UV absorption were noted in the HPLC analysis of both cells and medium and may represent alternative metabolites. Ribosylation at the C-23 hydroxyl has been identified as a mechanism for resistance to rifampin in M. smegmatis (5, 20) and might also occur with rifamycin S, MS-RifSV, or AcCyS-RifSV.
FIG. 3.
Analysis of rifamycin S conjugates in mca mutant. Four liters of M. smegmatis mc2155 and Ami37 at ODs of 1 were harvested, transferred to 200 ml of ice-cold medium with 1 mg of rifamycin S/ml, incubated on ice for 1 h, washed two times with ice-cold medium, and resuspended in 20 ml of ice-cold medium. The cells were then diluted into 180 ml of 37°C medium and were sampled at 1 min, 10 min, 60 min, 2 h, and 16 h. For each sample the cells were extracted and analyzed for rifamycin S conjugates. Cellular rifamycin S conjugates were quantified by HPLC at 315 nm using an approximate extinction coefficient of 24,000 M−1 cm−1.
Complementation of Ami37 with M. tuberculosis and M. smegmatis mca restores Mca activity.
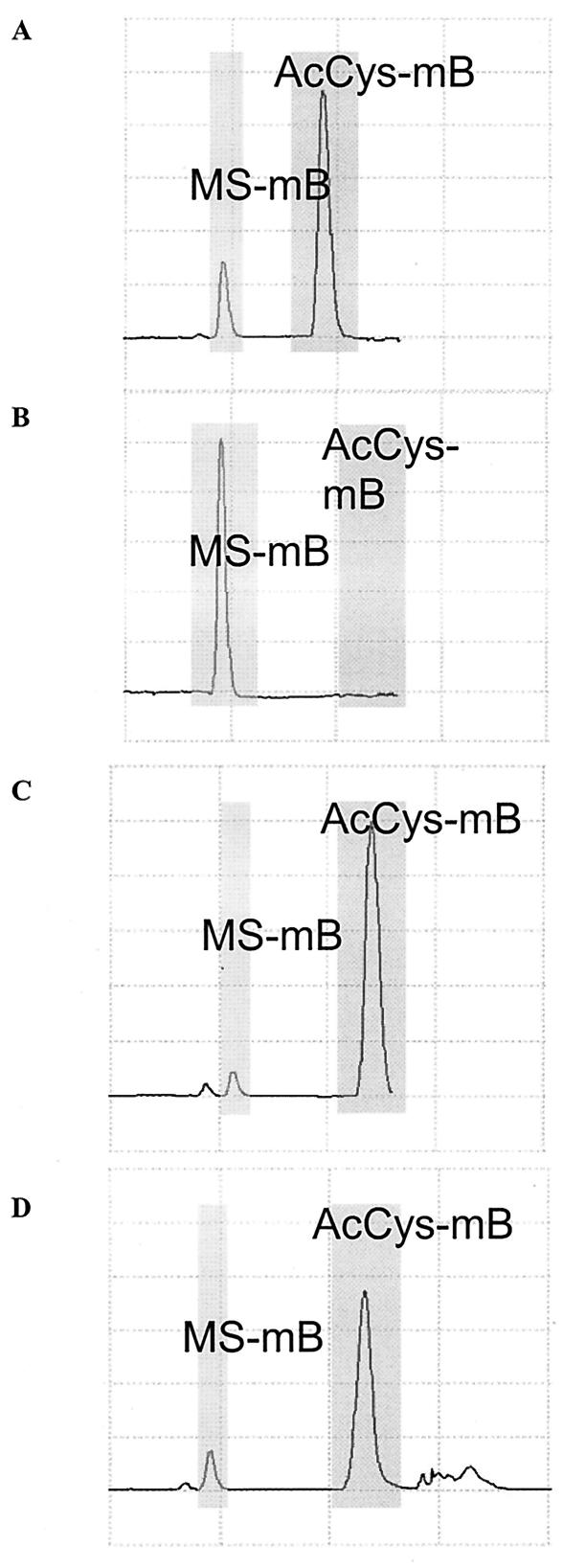
To confirm that the decrease in mycothiol was caused by the disruption in mca, mca was reintroduced into Ami37. The amidase activity for 100 μg of cell extracts was determined using MS-mB as a substrate. In Fig. 4A, most of the MS-mB has been converted to the mercapturic acid AcCyS-mB in the parent strain, while in Ami37 cell extracts none of the substrate has been converted to mercapturic acid AcCyS-mB (Fig. 4B). Introduction of M. tuberculosis mca epichromosomally or introduction of M. smegmatis mca integratively resulted in the complementation of the activity as both proteins are able to catalyze the conversion of MS-mB to AcCyS-mB (Fig. 4C and D, respectively), confirming that the M. tuberculosis and M. smegmatis proteins are functionally the same.
FIG. 4.
Amidase assay of M. smegmatis mca mutant and complement. The cells were pelleted and sonicated in 25 mM HEPES, pH 7.5. Protein analysis was performed, and 100 μg of protein sample from each strain was reacted with 30 μM MS-mB and 3 mM β-mercaptoethanol in a volume of 50 μl for 30 min. The reaction was stopped and thiols were analyzed. (A) mc2155; (B) Ami37; (C) ami37palace; (D) ami37phint.
Antibiotic sensitivity of Ami37.
It has been demonstrated that Mca can react with a broad range of mycothiol S conjugates, including two different classes of antibiotics, exemplified by cerulenin (14) and rifamycin S (25). Both adducts are cleaved to produce a mercapturic acid and GlcN-Ins. Consequently, Ami37 was tested for sensitivity to cerulenin and rifampin, a derivative of rifamycin S that is used as an antituberculous drug. In the cerulenin and rifampin disk diffusion assay there was no difference in clearing between Ami37 and mc2155gint (data not shown). For cerulenin, this result was confirmed with the minimal broth dilution assay where there was no difference in the MIC between the two strains (Table 3), indicating that the Mca-dependent detoxification pathway does not play a major role in the detoxification of cerulenin, although cerulenin can form an S conjugate with mycothiol. The MIC of rifampin for mc2155gint was fourfold higher than that for mc2155, as determined by both the minimal broth dilution assay and E-test (Table 3).
TABLE 3.
MICs for mc2155gint and Ami37
| Antibiotic | MIC (μg/ml) of antibiotic for strain:
|
|
|---|---|---|
| mc2155gint | Ami37 | |
| Rifampin | 25-50 | 6.3-12.5 |
| Rifampina | 32 | 8 |
| Erythromycin | 50 | 25-50 |
| Erythromycina | 12 | 2 |
| Lincomycin | >90 | >90 |
| Cerulenin | 0.63 | 0.63 |
| Streptomycin | 1.25-2.5 | 0.13-0.25 |
| Isoniazida | 1-4 | 1 |
| Vancomycin | 50-100 | 25-50 |
| Vancomycina | 6 | 3 |
E-test.
Because Mca homologs have been identified in the antibiotic biosynthesis operons of S. erythrae (erythromycin biosynthesis) and S. lincolnensis (lincomycin biosynthesis) (16), the sensitivities of Ami37 and the parent strain to erythromycin and lincomycin were also checked. In disk diffusion assays no significant difference was found in the zone of clearing for erythromycin and lincomycin between Ami37 and mc2155gint (data not shown). When minimal broth dilution assay was performed, there was still no difference in the MIC of lincomycin for Ami37 and mc2155gint. In fact, both Ami37 and mc2155gint were able to grow in 90 μg of lincomycin/ml (Table 3). The MIC of erythromycin was perhaps twofold higher for the parent strain as determined by the minimal broth dilution assay and was sixfold higher as determined by the E-test.
Recent studies have reported that mutants deficient in mycothiol are more sensitive to vancomycin and are more resistant to isoniazid (3, 10, 17, 18, 21). To determine whether this sensitivity is also dependent on Mca, the sensitivity of Ami37 to these two antibiotics was checked by disk diffusion assay and E-test, followed by minimal broth dilution assay for vancomycin only. No difference in sensitivity was detected with the disk diffusion assay for vancomycin (data not shown), although a twofold difference in vancomycin MIC values between Ami37 and mc2155gint was detected in the E-test and minimal broth dilution assays (Table 3). As with erythromycin, the MICs determined by minimal broth dilution assay were considerably higher than those of the E-test. For isoniazid there was no difference in zone of inhibition (data not shown) or the MIC, indicating that Mca plays no role in the resistance of mutants lacking mycothiol to isoniazid (Table 3).
Sensitivity of Ami37 and mc2155gint for a whole range of other antibiotics was checked by disk diffusion assays (data not shown). Sensitivity to streptomycin, a second-line drug for M. tuberculosis treatment, was shown to increase in Ami37. Complementation of the mutant with M. smegmatis mca and M. tuberculosis mca (Table 4) restored the parental strain sensitivity to streptomycin. The observed MIC of streptomycin was 10-fold higher for mc2155gint than for Ami37 (Table 3). As sensitivity to streptomycin for mutants lacking MSH has not been previously reported and as MSH is a crucial part of the amidase-mediated detoxification reaction, mutants in all steps of the mycothiol biosynthetic pathway were tested for sensitivity to streptomycin. As seen in Table 5, mutants disrupted in mycothiol biosynthetic genes with the exception of the mshB mutant are also more sensitive to streptomycin. The lack of difference in sensitivity between the mshB mutant and the parent strain is not unexpected, because we have previously reported that the M. smegmatis mshB mutant has 10% of the mycothiol content of the parent strain and is not as sensitive as the other mutants to oxidants, toxins, and antibiotics (22).
TABLE 4.
Sensitivity of mca mutants and their complements to streptomycin and iodoacetamide
| Amt of drug (μg) | Zone of clearing (diameter in mm) of strain:
|
|||
|---|---|---|---|---|
| mc2155gint | Ami37 | ami37phint | ami37palace | |
| Streptomycin | ||||
| 5 | 26 ± 1 | 32 ± 1 | 24 ± 1 | 23 ± 3 |
| Iodoacetamide | ||||
| 0.01 | 33 ± 1 | 53 ± 3 | 40 ± 0 | 31 ± 3.0 |
| 0.1 | 62 ± 2 | 84 ± 2 | 78 ± 0 | 61 ± 9.0 |
TABLE 5.
Sensitivity to streptomycin of mutants disrupted in genes involved in mycothiol biosynthesis
| Amt (μg) | Zone of clearing (diam in mm) for strain:
|
||||
|---|---|---|---|---|---|
| mc2155 | A1 (mshA mutant) | myco504 (mshB mutant) | I64 (mshC mutant) | mshD (mshD mutant) | |
| 1 | 15 ± 0 | 21 ± 1 | 18 ± 0 | 18 ± 0 | 25 ± 1 |
| 5 | 27 ± 0 | 35 ± 1 | 26 ± 0 | 32 ± 0 | 37 ± 0 |
| 50 | 41 ± 0 | 49 ± 0 | 38 ± 0 | 50 ± 0 | 50 ± 0 |
Sensitivity of mca mutant and complement to toxins and oxidants.
It was shown previously that sensitivity to iodoacetamide is MSH dependent, as all mutants in MSH biosynthesis are more sensitive than the parent strain to iodoacetamide (21). It has also been previously shown that mycothiol reacts with iodoacetamide to form a conjugate that is a substrate for Mca (14). The relative activity of Mca for MS-acetamide compared to that of the MS-mB adduct is only 0.5%. Nevertheless, Ami37 is more sensitive than mc2155gint to iodoacetamide as determined by the disk inhibition assay. Furthermore, this sensitivity is reversible upon complementation (Table 4). Similarly, Ami37 is more sensitive to NEM (Table 6), the MSH-NEM adduct being another substrate of Mca. The relative activity for MS-NEM is 2.1% compared to that of MS-mB, which is greater than that of iodoacetamide but still substantially less than that reported for cerulenin. Ami37 is also more sensitive to CDNB, a common substrate for GSH transferases, than mc2155gint (Table 6). In contrast, there is no significant difference in sensitivity to mBBr, the substrate for which the highest amidase activity has been reported, as determined by the disk inhibition assay. Even in the minimal broth dilution assay, the mBBr MIC for Ami37 is 0.0063 μmol/ml, twofold less than 0.0125 μmol/ml, the mBBr MIC for mc2155gint.
TABLE 6.
Sensitivity to toxins and oxidants
| Reagent | Amt (μmol) | Zone of clearing (diam in mm) for strain:
|
|
|---|---|---|---|
| mc2155gint | Ami37 | ||
| NEM | 0.01 | 0 | 14 ± 1 |
| mBBr | 0.1 | 14 ± 1 | 14 ± 2 |
| 0.5 | 25 ± 0 | 26 ± 1 | |
| CDNB | 0.05 | 12 ± 0 | 19 ± 0 |
| Plumbagin | 0.002 | 23 ± 4 | Clear |
| Menadione | 0.05 | 11 ± 1 | 21 ± 3 |
| 0.1 | 44 ± 1 | Clear | |
| Nitrofurantoin | 0.5 | 10 ± 0 | 9 ± 0 |
| Hydrogen peroxide | 1.0 | 15 ± 0 | 16 ± 1 |
| 10.0 | 40 ± 1 | 39 ± 0 | |
| Cumene hydroperoxide | 0.5 | 13 ± 1 | 16 ± 1 |
It was previously shown that mutants lacking mycothiol are more sensitive to redox cycling agents, menadione, plumbagin, and nitrofurantoin (21). To investigate the role of Mca in the detoxification of oxidants, sensitivity to oxidants was determined by the disk inhibition assay. In Table 5 it can be seen that Ami37 is more sensitive than mc2155gint to plumbagin and menadione. Such compounds are known to form conjugates with thiols (6). In contrast, there is no difference in sensitivity to other oxidative stress inducers, such as hydrogen peroxide, cumene hydrogen peroxide, and nitrofurantoin (Table 6), that do not form conjugates with mycothiol.
DISCUSSION
The mca gene codes for an Mca that plays a key role in detoxification of electrophiles. Mca is a zinc metalloenzyme (25) and has been shown to react with the mycothiol S conjugates of alkylating agents and the antibiotics cerulenin and rifamycin S (14, 25). In this reaction, mycothiol reacts with toxins to form conjugates similar to glutathione toxin conjugates and the amidase cleaves an amide bond in the mycothiol moiety of the conjugate to release a mercapturic acid and GlcN-Ins. In this report, we describe the generation and characterization of an mca-specific mutant, Ami37, disrupted in the mca gene of M. smegmatis. We show that the mca gene, either from M. smegmatis or M. tuberculosis, is able to restore the wild-type phenotype associated with the mutation in M. smegmatis.
Mercapturic acids, products of the Mca-catalyzed reaction, have been found in fermentation broths of actinomycetes, such as granaticin A metabolite, Ws009A, seongomycin, cysfluoretin, and phenoxazinone metabolite (16). It was also previously demonstrated that mutants in the mycothiol biosynthetic pathway are more sensitive to antibiotics, toxins, and oxidants. This sensitivity of mutants lacking mycothiol may depend in part on the mycothiol-dependent amidase-catalyzed detoxification reaction (Fig. 1). We considered first the role of Mca in detoxification of antibiotics. If the amidase plays a role in the detoxification of an antibiotic, then the mca mutant should be more sensitive to this antibiotic.
We demonstrated that the mca mutant has enhanced sensitivity to streptomycin, a second-line drug employed in treatment for M. tuberculosis, as demonstrated by the increase in the zone of inhibition for Ami37 (Table 4) and the 10-fold decrease in the MIC for Ami37 compared to that for the control strain (Table 3). Moreover, the complements ami37palace, where the M. tuberculosis mca is overexpressed, and ami37phint, with the M. smegmatis mca, exhibited reversion to wild-type levels of sensitivity to streptomycin. Furthermore, the mutants disrupted in the mycothiol biosynthetic pathway, with the exception of the mshB mutant, are also more susceptible to streptomycin, as would be expected if the detoxification was dependent on Mca activity (Table 5). Examination of the structure of streptomycin reveals the presence of a thiol-reactive aldehyde that likely reacts with MSH to form a thiohemiacetal adduct. This reaction is reversible, and how it could be involved in detoxification is not immediately obvious. Other metabolic modifications of the streptomycin moiety may be involved.
The sensitivity to several antibiotics in addition to streptomycin appears to depend upon Mca. It was previously reported that mutants lacking mycothiol are sensitive to vancomycin, erythromycin, and rifampin (21). Additionally, in the case of rifampin, the S conjugate of rifamycin, the parent antibiotic from which rifampin is derived, has been demonstrated to be a substrate for Mca (25). No differences were seen in the zone of inhibition in the disk diffusion assay for any of these antibiotics; thus, the MICs were checked by E-strips and by minimal broth dilution assay for all these antibiotics. The MICs of rifampin, erythromycin, and vancomycin were fourfold higher, less than two fold higher, and twofold higher, respectively, for mc2155 than for Ami37 (Table 3). Thus, Mca cleavage of a MSH conjugate of these antibiotics may be important in the overall detoxification, but elucidation of the detailed chemistry involved will require further studies.
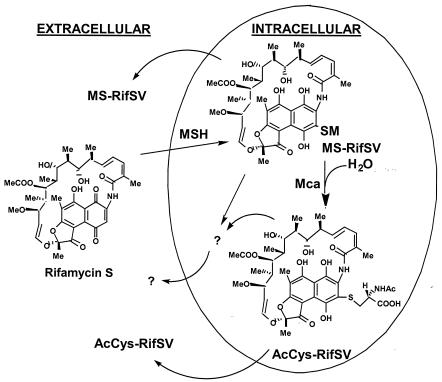
Examination of the M. smegmatis parent strain and mutant Ami37 treated with rifamycin S showed that a substantial quantity of the cellular MSH is converted to the MS-RifSV conjugate. In the parent strain, some of this is released to the medium and some is cleaved by Mca to produce AcCyS-RifSV, which is also found in the medium (Fig. 3 and 5). However, a significant fraction of the cellular MS-RifSV conjugate produced in the cell subsequently disappeared and could not be accounted for in terms of cellular AcCyS-RifSV or identified secreted forms. HPLC analysis indicated that significant levels of additional rifamycin derivatives are produced in the cells and are released into the medium (Fig. 5). Prime candidates would be ribosylated forms of rifamycin S or its MSH-derived metabolites, because ribosylation is an established pathway for inactivation of the related antibiotic rifampin (5, 20). Further studies are needed to elaborate the pathways involved.
FIG. 5.
Scheme showing the structures of rifamycin S and its MSH-derived metabolites and summarizing the role of mycothiol in detoxifying rifamycin S in M. smegmatis.
Other antibiotics are not dependent upon Mca. Lincomycin also has a gene homologous to mca in its antibiotic biosynthesis cluster; however, Ami37 and mc2155gint were able to survive and grow in 90 μg of lincomycin/ml (Table 3). In addition, cerulenin, another antibiotic which forms an S conjugate with MSH that can serve as a substrate for Mca (14), produced no difference in sensitivity as measured by the disk diffusion assay or the MIC as measured by the minimal broth dilution assay between Ami37 and mc2155gint (Table 3). The lack of sensitivity of Ami37 to cerulenin and lincomycin suggests that formation of the mycothiol S conjugate suffices to detoxify this compound and that accumulation of the S conjugate within the cell had little or no adverse consequence.
Electrophiles severely damage biological molecules, such as DNA bases and protein sulfhydryl groups. The mutant disrupted in mca is more sensitive to NEM, iodoacetamide, and CDNB than the control strain (Table 5). Because a number of natural products with structures related to the maleimide ring (e.g., maleimycin, showdomycin, and pencolide) are produced by bacteria and fungi, the MSH-dependent Mca detoxification system may protect the cell against electrophilic assault from such toxins. Moreover, in E. coli a GSH-dependent detoxification pathway has been described (12) where the GSH-NEM adduct produced in the cells serves as a substrate for an imidase which catalyzes the hydrolytic cleavage of the imide bond and converts the NEM to maleamic acid that is secreted into the medium while the GSH is recycled. In contrast, in E. coli the GSH-CDNB adduct is excreted and the GSH fails to be recovered (9).
The results with the alkylating agent mBBr were unexpected. No difference in sensitivity was measured by disk diffusion assay (Table 6), and only a twofold decrease in the MIC of mBBr for Ami37 was determined (data not shown). The mBBr adduct, MS-mB, is the best substrate known for the Mca enzyme. Indeed, even when M. smegmatis was incubated on ice with mBBr, the conjugate MS-mB formed spontaneously and the amide bond in this conjugate was easily cleaved to yield AcCyS-mB, such that AcCyS-mB represented 78% of the total of MS-mB and AcCyS-mB within the cell (Table 2). Moreover, the cells had already started to excrete AcCyS-mB into the medium (Table 2). Thus, while the MSH-Mca system is clearly involved in eliminating bimane from the cell, mycothiol itself and not Mca is the essential moiety in detoxifying mBBr.
Oxidants, such as plumbagin, a napthoquinone, and menadione, 2-methyl-1,4-naphthoquinone, can rapidly form conjugates with thiols, such as mycothiol. Indeed, Zadzinski et al. (27) demonstrated that incubation with 0.5 mM menadione results in a decrease in GSH concentration in yeast cells, as GS-menadione conjugates are formed and exported. In a similar manner, menadione and plumbagin may form MS-menadione and MS-plumbagin conjugates in mycobacteria that are cleaved by Mca to form mercapturic acids that are exported. Indeed, the disruption of mca results in an increase in sensitivity to these oxidants, as does the lack of mycothiol in mutants disrupted in the mycothiol biosynthetic pathway (21). In contrast to plumbagin and menadione, Ami37 is not sensitive to other oxidants, such as hydrogen peroxide, cumene hydrogen peroxide, and nitrofurantoin (Table 6), although sensitivity is exhibited by mutants lacking mycothiol (21). Thus, oxidants that can form stable S conjugates, such as plumbagin and menadione, may require Mca for detoxification, while the superoxide that they generate as redox cycling agents may be detoxified by other mycothiol-dependent or antioxidant mechanisms.
In conclusion, we have shown that an M. smegmatis Mca mutant is susceptible to several antibiotics, streptomycin in particular. The mutant is also more susceptible to electrophiles and oxidants that react with mycothiol directly. Thus, these findings support a major role for mycothiol and Mca in detoxification. The isolation of mercapturic acids and mycothiol adducts of these antibiotics and toxins will solidify the significance of this detoxification mechanism in mycobacteria.
Acknowledgments
This work was supported by grants to Y.A. from the TB Veterans Association and the Canadian Institute of Health Research, Institute of Infection and Immunity, MOP 64950, and grant MCB-0235705 to R.C.F. from the National Science Foundation. Y.A. is a Canadian Institute of Health Research and British Columbia Lung Association Scholar.
We thank Mary Ko, Rayken Chow, and Teresa Koledin for technical assistance and the Institute for Genomic Research for providing access to M. smegmatis sequence data.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderberg, S. J., G. L. Newton, and R. C. Fahey. 1998. Mycothiol biosynthesis and metabolism. Cellular levels of potential intermediates in the biosynthesis and degradation of mycothiol in mycobacterium smegmatis. J. Biol. Chem. 273:30391-30397. [DOI] [PubMed] [Google Scholar]
- 3.Buchmeier, N. A., G. L. Newton, T. Koledin, and R. C. Fahey. 2003. Association of mycothiol with protection of Mycobacterium tuberculosis from toxic oxidants and antibiotics. Mol. Microbiol. 47:1723-1732. [DOI] [PubMed] [Google Scholar]
- 4.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, B. G. Barrell, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 5.Dabbs, E. R., K. Yazawa, Y. Mikami, M. Miyaji, N. Morisaki, S. Iwasaki, and K. Furihata. 1995. Ribosylation by mycobacterial strains as a new mechanism of rifampin inactivation. Antimicrob. Agents Chemother. 39:1007-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedman, M. 1973. The chemistry and biochemistry of the sulfhydryl group in amino acids, peptides and proteins. Pergamon Press, New York, N.Y.
- 7.Hatfull, G. F., and W. R. Jacobs, Jr. 2000. Molecular genetics of mycobacteria. American Society for Microbiology, Washington, D.C.
- 8.Hinchman, C. A., and N. Ballatori. 1994. Glutathione conjugation and conversion to mercapturic acids can occur as an intrahepatic process. J. Toxicol. Environ. Health. 41:387-409. [DOI] [PubMed] [Google Scholar]
- 9.Kaluzna, A., and G. Bartosz. 1997. Transport of glutathione S-conjugates in Escherichia coli. Biochem. Mol. Biol. Int. 43:161-171. [DOI] [PubMed] [Google Scholar]
- 10.Koledin, T., G. L. Newton, and R. C. Fahey. 2002. Identification of the mycothiol synthase gene (mshD) encoding the acetyltransferase producing mycothiol in actinomycetes. Arch. Microbiol. 178:331-337. [DOI] [PubMed] [Google Scholar]
- 11.Maynes, J. T., C. Garen, M. M. Cherney, G. Newton, D. Arad, Y. Av-Gay, R. C. Fahey, and M. N. James. 2003. The crystal structure of 1-D-myo-inosityl 2-acetamido-2-deoxy-alpha-D-glucopyranoside deacetylase (MshB) from Mycobacterium tuberculosis reveals a zinc hydrolase with a lactate dehydrogenase fold. J. Biol. Chem. 278:47166-47170. [DOI] [PubMed] [Google Scholar]
- 12.McLaggan, D., H. Rufino, M. Jaspars, and I. R. Booth. 2000. Glutathione-dependent conversion of N-ethylmaleimide to the maleamic acid by Escherichia coli: an intracellular detoxification process. Appl. Environ. Microbiol. 66:1393-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newton, G. L., K. Arnold, M. S. Price, C. Sherrill, S. B. Delcardayre, Y. Aharonowitz, G. Cohen, J. Davies, R. C. Fahey, and C. Davis. 1996. Distribution of thiols in microorganisms: mycothiol is a major thiol in most actinomycetes. J. Bacteriol. 178:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newton, G. L., Y. Av-Gay, and R. C. Fahey. 2000. A novel mycothiol-dependent detoxification pathway in mycobacteria involving mycothiol S-conjugate amidase. Biochemistry 39:10739-10746. [DOI] [PubMed] [Google Scholar]
- 15.Newton, G. L., Y. Av-Gay, and R. C. Fahey. 2000. N-acetyl-1-d-myo-inosityl-2-amino-2-deoxy-alpha-d-glucopyranoside deacetylase (MshB) is a key enzyme in mycothiol biosynthesis. J. Bacteriol. 182:6958-6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newton, G. L., and R. C. Fahey. 2002. Mycothiol biochemistry. Arch. Microbiol. 178:388-394. [DOI] [PubMed] [Google Scholar]
- 17.Newton, G. L., T. Koledin, B. Gorovitz, M. Rawat, R. C. Fahey, and Y. Av-Gay. 2003. The glycosyltransferase gene encoding the enzyme catalyzing the first step of mycothiol biosynthesis (mshA). J. Bacteriol. 185:3476-3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newton, G. L., M. D. Unson, S. J. Anderberg, J. A. Aguilera, N. N. Oh, S. B. delCardayre, Y. Av-Gay, and R. C. Fahey. 1999. Characterization of Mycobacterium smegmatis mutants defective in 1-d-myo-inosityl-2-amino-2-deoxy-alpha-d-glucopyranoside and mycothiol biosynthesis. Biochem. Biophys. Res. Commun. 255:239-244. [DOI] [PubMed] [Google Scholar]
- 19.Peschke, U., H. Schmidt, H. Z. Zhang, and W. Piepersberg. 1995. Molecular characterization of the lincomycin-production gene cluster of Streptomyces lincolnensis 78-11. Mol. Microbiol. 16:1137-1156. [DOI] [PubMed] [Google Scholar]
- 20.Quan, S., H. Venter, and E. R. Dabbs. 1997. Ribosylative inactivation of rifampin by Mycobacterium smegmatis is a principal contributor to its low susceptibility to this antibiotic. Antimicrob. Agents Chemother. 41:2456-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rawat, M., G. L. Newton, M. Ko, G. J. Martinez, R. C. Fahey, and Y. Av-Gay. 2002. Mycothiol-deficient Mycobacterium smegmatis mutants are hypersensitive to alkylating agents, free radicals, and antibiotics. Antimicrob. Agents Chemother. 46:3348-3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rawat, M., S. Kovacevic, H. Billman-Jacobe, and Y. Av-Gay. 2003. Inactivation of mshB, a key gene in the mycothiol biosynthesis pathway in Mycobacterium smegmatis. Microbiology 149:1341-1349. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed.. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 24.Snapper, S. B., L. Lugosi, A. Jekkel, R. E. Melton, T. Kieser, B. R. Bloom, and W. R. Jacobs Jr. 1988. Lysogeny and transformation in mycobacteria: stable expression of foreign genes. Proc. Natl. Acad. Sci. USA 85:6987-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steffek, M., G. L. Newton, Y. Av-Gay, and R. C. Fahey. 2003. Characterization of Mycobacterium tuberculosis mycothiol S-conjugate amidase. Biochemistry 42:12067-12076. [DOI] [PubMed] [Google Scholar]
- 26.World Health Organization. 2002. W.H.O. fact sheet no. 104.
- 27.Zadzinski, R., A. Fortuniak, T. Bilinski, M. Grey, and G. Bartosz. 1998. Menadione toxicity in Saccharomyces cerevisiae cells: activation by conjugation with glutathione. Biochem. Mol. Biol. Int. 44:747-759. [DOI] [PubMed] [Google Scholar]