Abstract
Four orthologous genes (TK1108, TK1404, TK1777, and TK2185) that can be annotated as phosphomannomutase (PMM) genes (COG1109) have been identified in the genome of the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. We previously found that TK1777 actually encodes a phosphopentomutase. In order to determine which of the remaining three orthologues encodes a phosphoglucomutase (PGM), we examined the PGM activity in T. kodakaraensis cells and identified the gene responsible for this activity. Heterologous gene expression and purification and characterization of the recombinant protein indicated that TK1108 encoded a protein with high levels of PGM activity (690 U mg−1), along with high levels of PMM activity (401 U mg−1). Similar analyses of the remaining two orthologues revealed that their protein products exhibited neither PGM nor PMM activity. PGM activity and transcription of TK1108 in T. kodakaraensis were found to be higher in cells grown on starch than in cells grown on pyruvate. Our results clearly indicate that, among the four PMM gene orthologues in T. kodakaraensis, only one gene, TK1108, actually encodes a protein with PGM and PMM activities.
Genome sequencing has provided an enormous amount of information on the primary structure and number of genes in a particular organism (5, 11). Based on the assumption that genes with high levels of similarity encode proteins that have the same function, the presence or absence of various orthologues is often used in estimating whether a specific metabolic pathway is present. This approach, however, has its limitations. When an orthologue of an expected enzyme is not found, the gene must be identified through classical biochemical and cloning methods (13, 14). On the other hand, when multiple orthologues are present on the genome, each gene product must be carefully examined in order to distinguish the enzymatic activities or functions in the cell. Analyses of the expression levels of the genes also contribute to elucidating their functions (17).
We recently determined the entire genome sequence of a hyperthermophilic archaeon, Thermococcus kodakaraensis KOD1 (unpublished data). This strain was originally isolated from a solfatara on Kodakara Island, Kagoshima, Japan (2, 9). It displays heterotrophic growth on a variety of organic substrates, such as amino acids, pyruvate, and starch, and we have taken an interest in the metabolic pathways involved in the assimilation of these carbon compounds. Through genome annotation, it has been possible to identify orthologues for a majority of the enzymes involved in glycolysis and gluconeogenesis, including the archaeal ADP-dependent glucose kinase, the ADP-dependent phosphofructokinase, and the archaeal class IA type fructose 1,6-bisphosphate aldolase (7). An orthologue of the key enzyme of gluconeogenesis, fructose 1,6-bisphosphatase, was not present, and a novel, archaeal-type fructose 1,6-bisphosphatase from T. kodakaraensis was subsequently identified and characterized (14).
Although phosphoglucomutase (PGM) (EC 5.4.2.2) is not a member of the glycolytic enzyme group, we also noticed that the gene encoding this enzyme has not been identified in Archaea. PGM catalyzes the interconversion of glucose 6-phosphate and glucose 1-phosphate, and it plays a vital role in carbohydrate metabolism in a wide range of microorganisms, as well as in plant and animal cells (6, 8, 10, 15). From a catabolic point of view, PGM provides the glycolytic intermediate glucose 6-phosphate from glucose 1-phosphate, which is often the product of intracellular polysaccharide degradation by various glycan phosphorylases (20). A well-known example is the glycolytic reentry of glucose that has been stored as energy in the form of glycogen or trehalose (15). On the other hand, glucose 1-phosphate is the precursor of sugar nucleotides that are necessary in the synthesis of various glucose-containing polysaccharides. Therefore, PGM also has an important biosynthetic role, supplying the glucose 1-phosphate from glucose 6-phosphate that is produced through glycolysis or gluconeogenesis (15).
PGM activity has been detected in crude extracts of several archaeal strains (21, 22). In the genome databases, many open reading frames have been annotated as genes encoding putative phosphomannomutases (PMMs) and can be considered likely candidates to encode archaeal PGMs. However, most genomes harbor more than one open reading frame designated a PMM gene. For example, Pyrococcus furiosus, Pyrococcus abyssi, and Pyrococcus horikoshii have three orthologues, while two orthologues have been found in the Methanococcus jannaschii and Aeropyrum pernix genomes. Like the genomes of other hyperthermophilic archaea, the T. kodakaraensis genome harbors more than one open reading frame that is annotated as a PMM gene; actually, it harbors four such open reading frames (TK1108, TK1404, TK1777, and TK2185). While all four of these open reading frames are members of cluster 1109 of orthologous genes (COG1109), it has been found previously that TK1777 actually encodes a phosphopentomutase (13). This was unexpected, as all previously identified genes encoding phosphopentomutases were members of COG1015 and had a distinct primary structure. Therefore, as mentioned above, biochemical analyses of the remaining three orthologues is necessary to accurately determine their individual functions. In this study, we obtained biochemical evidence that allowed us to identify the true archaeal PGM gene in T. kodakaraensis.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
T. kodakaraensis KOD1 was isolated from a solfataric hot spring at a wharf on Kodakara Island, Kagoshima, Japan (2, 9). T. kodakaraensis cells were grown in either a nutrient-rich MA-YT medium (13) or minimal ASW-AA medium (16). Escherichia coli strain DH5α was used for subcloning of the gene fragments and DNA manipulation. E. coli strain BL21(DE3) (Novagen, Madison, Wis.) was used as a host, and pET-21a (Novagen) was used as a vector for gene expression.
Chemicals and enzymes.
Glucose 1-phosphate, glucose 6-phosphate, glucose 1,6-bisphosphate, mannose 1-phosphate, mannose 6-phosphate, fructose 1-phosphate, fructose 6-phosphate, glucosamine 1-phosphate, glucosamine 6-phosphate, N-acetylglucosamine 1-phosphate, N-acetylglucosamine 6-phosphate, 2-deoxyribose 1-phosphate, 2-deoxyribose 5-phosphate, 3-phosphoglyceric acid, 2-phosphoglyceric acid, 2,3-diphosphoglyceric acid, phosphoglucoisomerase, phosphomannoisomerase, glucose 6-phosphate dehydrogenase, enolase, pyruvate kinase, and lactate dehydrogenase were purchased from Sigma (St. Louis, Mo.). Alcohol dehydrogenase, NADP, and NADH were purchased from Oriental Yeast (Tokyo, Japan). Deoxyribose 5-phosphate aldolase (13) and glucosamine 6-phosphate deaminase (unpublished data) were purified from T. kodakaraensis. Other chemicals and components of various media were purchased from Wako Pure Chemicals (Osaka, Japan) or Nacalai Tesque (Kyoto, Japan).
DNA manipulation and sequencing.
Restriction enzymes and DNA polymerase were purchased from Toyobo (Osaka, Japan) and Takara Shuzo (Kyoto, Japan). Genomic and plasmid DNAs were isolated by using QIAGEN genomic and plasmid DNA isolation kits, respectively (QIAGEN, Hilden, Germany). DNA ligation was performed by using a DNA ligation kit (Toyobo). A QIAEX gel extraction kit (QIAGEN) was used to recover DNA fragments from agarose gels. DNA sequencing was performed with an ABI PRISM BigDye terminator cycle sequencing Ready Reaction kit (Applied Biosystems, Foster City, Calif.). Database homology searches were performed by using the Basic Local Alignment Search Tool (BLAST) program (1). Sequence analyses were performed by using DNASIS software (Hitachi Software, Yokohama, Japan). Multiple-alignment and phylogenetic analyses were performed by using the ClustalW program (18) provided by the DNA Data Bank of Japan (DDBJ). The phylogenetic tree was constructed by the neighbor-joining method after alignment.
Partial purification of PGM activity from strain KOD1.
T. kodakaraensis cells were cultivated at 85°C for 16 h in MA-YT medium (13) containing 0.5% soluble starch (Nacalai Tesque). The cells were harvested and disrupted by sonication in ice water. All purification steps were performed at room temperature unless indicated otherwise. The membrane and cytosolic fractions from the cell lysate were separated by ultracentrifugation at 110,000 × g for 70 min at 4°C. The cytosolic fraction, containing the PGM activity, was loaded onto Resource Q (Amersham Biosciences, Little Chalfont, Buckinghamshire, United Kingdom) which had been equilibrated with buffer A (50 mM sodium phosphate buffer, pH 7.0). The proteins were eluted with a linear gradient of 0 to 1 M sodium chloride in buffer A. Then the fractions with PGM activity were dialyzed against buffer A and loaded onto Mono Q HR 5/5 (Amersham Biosciences) which had been equilibrated with buffer A. The proteins were eluted with a linear gradient of 0 to 1 M sodium chloride in buffer A, and the fractions carrying PGM activity were dialyzed against 2 M ammonium sulfate and loaded onto Resource ISO (Amersham Biosciences) which had been equilibrated with 2 M ammonium sulfate (pH 7.0). The proteins were eluted with a linear gradient of 2 to 0 M ammonium sulfate (pH 7.0). Then the fractions carrying PGM activity were dialyzed against buffer A and loaded onto a hydroxyapatite column (Bio-Scale CHT-I; Bio-Rad, Hercules, Calif.). Fractions exhibiting PGM activity were concentrated by using Centricon YM-30 (Millipore Corporation, Bedford, Mass.) and were further purified on a gel filtration column (Superdex 200 HR 10/30; Amersham Biosciences) which had been equilibrated with buffer A containing 150 mM sodium chloride. The protein concentration was determined with a bicinchoninic acid protein assay kit (Pierce, Rockford, Ill.) used according to the manufacturer's instructions; bovine serum albumin was used as the standard. N-terminal amino acid residues were determined with a protein sequencer (model 491 cLC; Applied Biosystems). The partially purified protein was also analyzed by matrix-assisted laser desorption ionization—time of flight mass spectrometry at Hitachi Science Systems (Hitachinaka, Japan).
Expression of the TK1108 gene and purification of the recombinant protein.
NdeI and BamHI sites were introduced into the N and C termini of the TK1108 gene, respectively, by PCR. The DNA fragment was inserted into pET-21a at the NdeI and BamH I sites, resulting in pET-1108. E. coli strain BL21(DE3) carrying pET-1108 was grown at 37°C in Luria-Bertani medium containing ampicillin (50 μg/ml) until an optical density at 660 nm of 0.5 was reached. Gene expression was induced by addition of 0.2 mM (final concentration) isopropyl-β-d-thiogalactopyranoside (IPTG), and the preparation was incubated for another 4 h at 37°C. The cells were harvested by centrifugation at 6,000 × g for 10 min at 4°C and washed with 50 mM potassium phosphate buffer (pH 7.0). Then the cells were resuspended in the same buffer and disrupted by sonication on ice. The supernatant after centrifugation (15,000 × g for 30 min at 4°C), containing the recombinant TK1108, was incubated at 85°C for 20 min. Heat-labile proteins were removed by centrifugation (15,000 × g for 30 min at 4°C). Recombinant TK1108 was purified to homogeneity with Resource Q, Resource ISO, and Superdex 200HR10/30 columns by using the methods as described above for partial purification of PGM activity from T. kodakaraensis. Recombinant TK1404 and TK2185 were also expressed and purified by using the same method. The apparent homogeneity of the proteins was examined by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE). Molecular mass estimates were obtained by gel filtration (Superdex 200 HR 10/30) by using HMW and LMW gel filtration calibration kits (Amersham Biosciences).
Enzyme activity assay.
PGM activity was measured by a discontinuous assay coupled with glucose 6-phosphate dehydrogenase. Formation of glucose 6-phosphate from glucose 1-phosphate was measured by monitoring NADPH formation with glucose 6-phosphate dehydrogenase. The initial reaction mixture (200 μl) consisted of 100 mM Tris-HCl (pH 7.0), 10 mM MgCl2, 50 μM glucose 1,6-bisphosphate, 5 mM glucose 1-phosphate, and enzyme solution. After incubation at the desired temperature for 1 min (for activity measurements, 60 to 95°C) or 5 min (30 to 50°C), the reaction mixture was cooled on ice for 5 min, and the amount of glucose 6-phosphate produced was measured by addition of 800 μl of water containing 0.5 mM NADP and 2 U of glucose 6-phosphate dehydrogenase. After incubation at 25°C for 3 min, the amount of NADPH was measured at 340 nm. The amount of glucose 6-phosphate produced by the enzyme at 25°C during a 3-min period was subtracted. The product formation was proportional to the incubation time under these conditions. One unit was defined as the amount of activity that produced 1 μmol of glucose 6-phosphate from glucose 1-phosphate per min. All other enzyme activities were measured as described previously (13).
When the effect of pH on the enzyme activity was examined, the reaction was carried out by using 100-μl reaction mixtures containing the following buffers at a concentration of 20 mM: MES (morpholineethanesulfonic acid) buffer (pH 4.5 to 6.5), HEPES buffer (pH 6.5 to 7.5), and bicine buffer (pH 7.5 to 8.5). All of the buffers were prepared so that the pH reflected accurate values at 90°C. After the first reaction, 100 μl of 1 M Tris-HCl (pH 8.0) was added to the reaction mixture to bring the pH of the mixture to 8.0 (the optimal pH for the coupling enzyme). To examine the effects of the various metal ions on enzyme activity, the first reaction mixture was incubated with the metal cations. After incubation the mixture was cooled in ice water, and as glucose 6-phosphate dehydrogenase is an Mg2+-dependent enzyme, the coupling reaction was initiated by adding 10 mM Mg2+, NADP+, and the exogenous enzymes.
RNA isolation and dot blot analysis.
RNA was isolated from cells grown in MA-YT medium supplemented with either starch (0.5%) or sodium pyruvate (0.5%). Cells were harvested in the early log phase (optical density at 660 nm, 0.1). RNA was isolated with an RNeasy Midi kit (QIAGEN). For dot blot analysis, 1 μg of total RNA was denatured by heat treatment at 65°C for 15 min and spotted onto a nylon membrane (Hybond-N+; Amersham Biosciences). Digoxigenin labeling of DNA fragments, hybridization, and washing of the membranes were performed according to the instructions of the manufacturer (Roche Diagnostics, Basel, Switzerland). A DNA fragment corresponding to the entire TK1108 coding region was used as a probe. A control experiment was performed by using the DNA ligase gene from strain KOD1 as a probe.
Nucleotide sequence accession numbers.
The nucleotide sequences of the TK1777, TK2185, TK1108, and TK1404 genes determined in this study have been deposited in the DDBJ, EMBL, and GenBank DNA databases under accession numbers AB126239, AB126240, AB126241, and AB126242, respectively.
RESULTS
Presence of four putative PMM genes in the genome of T. kodakaraensis.
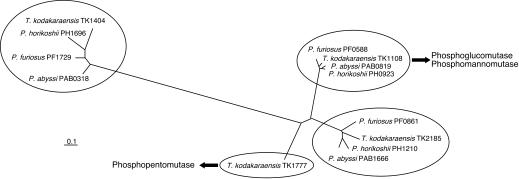
Three of the four orthologues, TK1108 (encoding 456 amino acid residues with a molecular weight of 49,850), TK1777 (encoding 450 amino acid residues with a molecular weight of 49,529), and TK2185 (encoding 449 amino acid residues with a molecular weight of 48,659), encoded proteins having similar molecular weights, while TK1404 encoded a smaller protein (236 amino acids with a molecular weight of 26,728). The three larger proteins displayed high levels of similarity with one another (TK2185 and TK1108, 44% identical; TK1777 and TK2185, 43% identical; TK1108 and TK1777, 38% identical) and also displayed equivalent levels of similarity with the biochemically characterized PGM from E. coli (TK1777, 24%; TK2185, 24%; TK1108, 25%). Two motifs conserved in various phosphosugar mutases (3, 19), TXSHNP containing the active site serine residue and DXDXDR involved in metal binding, were also conserved among these three proteins (Fig. 1). Therefore, it is quite difficult, if not impossible, to distinguish the functions of these enzymes from the sequence data. We also noticed that highly related orthologues of TK2185, TK1108, and TK1404 are present in the genomes of P. furiosus, P. horikoshii, and P. abyssi. The high levels of similarity of the corresponding genes in the Thermococcales strains allowed us to classify these genes on the basis of their primary structure. A phylogenetic tree of the orthologues is shown in Fig. 2. TK1777, the orthologue unique to T. kodakaraensis, actually encoded a phosphopentomutase and not a phosphohexomutase (13).
FIG. 1.
Primary structure comparison of the protein products of the four PMM orthologues of T. kodakaraensis. The asterisks beneath the sequences indicate the identical amino acid residues in the four proteins. The gene numbers are indicated on the left, whereas the residue numbers are indicated on the right. The bars above the alignment indicate the four conserved motifs identified in various phosphosugar mutases. The multiple-sequence alignment analysis was performed by using the ClustalW program provided by DDBJ.
FIG. 2.
Unrooted phylogenetic tree of PMM orthologues from different species of the Thermococcales. An optimal amino acid alignment was created with the program ClustalW provided by DDBJ, and a phylogenetic tree was constructed. The tree was displayed with the TreeView program. The DDBJ/EMBL/GenBank protein accession numbers for the sequences are as follows: P. abyssi PAB0318, CAB49399; P. abyssi PAB0819, CAB50141; P. abyssi PAB1666, CAB49971; P. furiosus PF0588, AAL80712; P. furiosus PF0861, AAL80985; P. furiosus PF1729, AAL81853; P. horikoshii PH0923, BAA30019; P. horikoshii PH1210, BAA30310; P. horikoshii PH1696, BAA30809; T. kodakaraensis TK1108, AB126241; T. kodakaraensis TK1404, AB126242; T. kodakaraensis TK1777, AB126239; and T. kodakaraensis TK2185, AB126240.
PGM activity in T. kodakaraensis and partial purification of PGM.
In order to identify the gene(s) that actually encodes a PGM, the enzyme activity in extracts of T. kodakaraensis KOD1 cells grown in the presence of 0.5% yeast extract and 0.5% tryptone along with 0.5% starch was examined. PGM activity was detected in the cell extracts, and the specific activity was 0.8 U mg−1. After the enzyme activity was determined, we partially purified the PGM from the cell extract. PGM was purified 14-fold by anion-exchange, hydrophobic, hydroxyapatite, and gel filtration column chromatography. A 50-kDa protein was found to correspond well with the results of activity measurements through each purification step. During this process, we did not observe PGM activity in fractions other than those used in the purification procedure. We analyzed the N-terminal amino acid sequence of the protein and found that it corresponded to the sequence encoded by TK1777. This was unexpected, as this protein is a phosphopentomutase with only trace levels of PGM activity (13). We then subjected the band to matrix-assisted laser desorption ionization—time of flight mass spectroscopy (12) and found that it was a mixture of two proteins, the proteins encoded by TK1777 and TK1108. Therefore, we analyzed TK1108.
Heterologous expression of the TK1108 gene and purification of the recombinant protein.
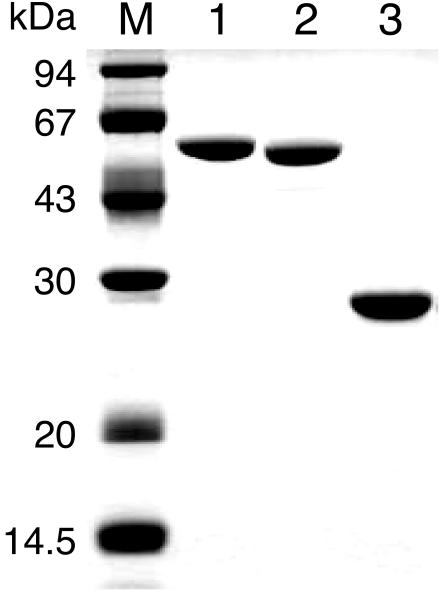
We expressed the TK1108 gene in E. coli and obtained the recombinant protein in a soluble form. The recombinant protein was purified to apparent homogeneity by heat treatment at 85°C for 20 min, followed by anion-exchange, hydrophobic, and gel filtration column chromatography (Fig. 3). The molecular mass of the protein estimated by SDS-PAGE agreed with the value calculated from the deduced amino acid sequence. Furthermore, the six N-terminal amino acid residues of the purified recombinant protein were identical to the deduced amino acid sequence encoded by the gene. The purified protein exhibited high levels of PGM activity (420 U mg−1) in the presence of 10 mM MgCl2 and 5 mM glucose 1-phosphate at 90°C and was designated PGMTk. Gel filtration chromatography indicated that the molecular mass of PGMTk was approximately 210 kDa. Taking into account the molecular mass of the subunit (49.8 kDa), this result indicates that PGMTk exists in a tetrameric form.
FIG. 3.
SDS-PAGE analysis of the purified protein products of TK1108, TK2185, and TK1404. Lane M, molecular mass markers; lane 1, purified recombinant TK1108; lane 2, purified recombinant TK2185; lane 3, purified recombinant TK1404.
Effects of metal cations, pH, and temperature on PGM activity.
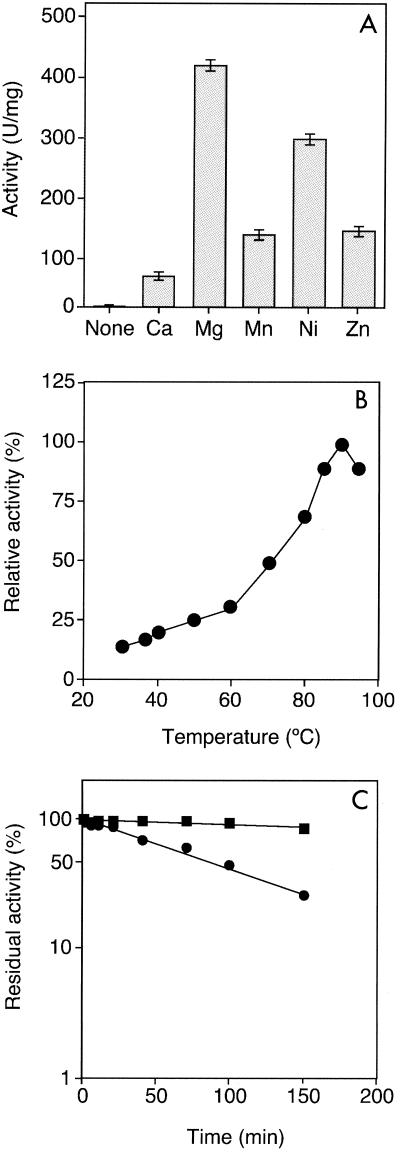
Purified PGMTk was dialyzed against 25 mM Tris-HCl buffer (pH 8.0) containing 5 mM EDTA and used for further analysis. Activity measurements were performed by using a linked assay coupled with glucose 6-phosphate dehydrogenase. PGMTk did not display detectable PGM activity in the absence of metal ions, indicating that the activity was metal ion dependent. Addition of MgCl2 (0.5 to 10 mM) led to significant levels of PGM activity, which was saturated at 1 mM (data not shown). Besides Mg2+, we also found that Ni2+, Mn2+ and Zn2+ at a concentration of 1 mM could also support PGM activity, although to a lesser extent (Fig. 4A).
FIG. 4.
(A) Effect of metal cations on PGMTk enzyme activity. A chloride salt of each metal cation was added at a final concentration of 1 mM, and PGM activity was examined at 90°C. (B) Temperature profile of PGMTk activity. PGM activity was examined at pH 7.0 at various temperatures. (C) Thermostability of PGMTk. PGMTk was heated in Tris-HCl buffer (pH 7.0) at 90°C (▪) and at 100°C (•) for various times, and residual activity was examined at 90°C. An activity of 100% corresponded to 420 U/mg.
We examined the effects of pH and temperature on the PGMTk activity in the presence of 10 mM Mg2+. At a fixed temperature of 90°C, PGMTk displayed maximal activity at pH 7. The temperature profile of the enzyme indicated that the optimal temperature was 90°C under our assay conditions (Fig. 4B). The thermostability of the recombinant protein was monitored in the presence of 10 mM Mg2+, and the protein was found to be highly stable even at 100°C. The enzyme displayed a half-life of ∼85 min in boiling water (Fig. 4C). A kinetic analysis was also carried out, and PGMTk catalyzed the reaction with Michaelis-Menten kinetics; the Km with glucose 1-phosphate was 3.0 mM, and the kcat was 575 s−1 subunit−1 at 90°C (Table 1).
TABLE 1.
Kinetic parameters of PGMTk with various substratesa
| Substrate | Vmax (μmol min−1 mg−1) | Km (mM) | kcat (s−1 subunit−1) | kcat/Km (s−1 subunit−1 mM−1) |
|---|---|---|---|---|
| Glucose 1-phosphate | 690 ± 32 | 3.0 ± 0.5 | 575 ± 25 | 192 |
| Mannose 1-phosphate | 401 ± 19 | 3.2 ± 0.3 | 330 ± 16 | 103 |
| 2-Deoxyribose 1-phosphate | 230 ± 26 | 3.5 ± 0.8 | 190 ± 20 | 54 |
Activity was measured at 90°C.
Activity with various substrates.
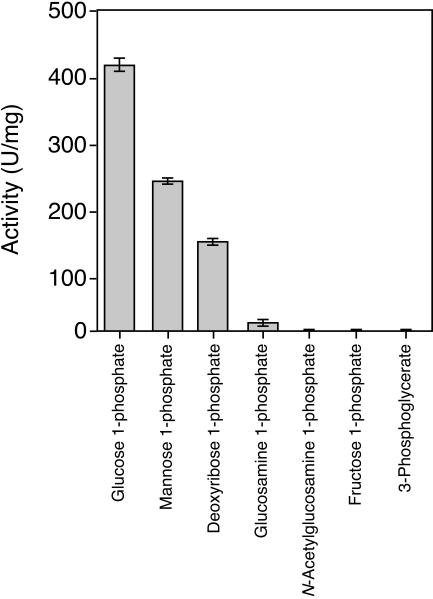
The mutase activities of PGMTk with various phosphorylated compounds (5 mM) were examined (Fig. 5). Among the substrates, PGMTk exhibited high levels of mutase activity with glucose 1-phosphate and mannose 1-phosphate. The enzyme also displayed relatively low activity with deoxyribose 1-phosphate and glucosamine 1-phosphate (Fig. 5). We carried out a kinetic analysis of the PMM activity of PGMTk and found that the reaction followed Michaelis-Menten kinetics with a Km of 3.2 mM and a kcat of 330 s−1 subunit−1 at 90°C (Table 1). We also examined the activity with 2-deoxyribose 1-phosphate and obtained a Km of 3.5 mM and a kcat of 190 s−1 subunit−1 (Table 1). No mutase activity was detected with fructose 1-phosphate, N-acetylglucosamine 1-phosphate, and 3-phosphoglycerate.
FIG. 5.
Mutase activity of PGMTk with various phosphorylated compounds. 2-Deoxyribose 1-phosphate, α-d-glucose 1-phosphate, α-d-mannose 1-phosphate, d-fructose 1-phosphate, d-glucosamine 1-phosphate, N-acetyl-d-glucosamine 1-phosphate, and 3-phospho-d-glyceric acid were independently used as substrates at a final concentration of 5 mM. Activity was measured as described elsewhere (13).
PGM activity in T. kodakaraensis KOD1.
We examined the PGM activity in T. kodakaraensis KOD1 cells grown in a synthetic medium based on amino acids (ASW-AA medium) (16). This medium meets the minimal requirements for growth of strain KOD1. To the ASW-AA medium, we also added either 0.5% starch (a glycolytic substrate) or 0.5% sodium pyruvate (a gluconeogenic substrate). PGM activity was detected in all cell extracts, and the levels of activity were 0.16 ± 0.01 U mg−1 in cells grown on ASW-AA medium, 0.42 ± 0.05 U mg−1 in cells grown with pyruvate, and 1.34 ± 0.04 U mg−1 in cells grown with starch. PGM activity seemed to be induced in the presence of abundant levels of glucose (starch). This tendency was also confirmed in nutrient-rich MA-YT medium; we found that there was a 2.7-fold increase in PGM activity in cells grown on starch (0.5%) compared to cells grown on sodium pyruvate (0.5%). Using RNA isolated from cells grown in these media, we also performed dot blot experiments and found that transcription levels were higher in starch-grown cells (data not shown), which is consistent with the induction of PGM activity observed in cells grown on starch.
Absence of PGM and PMM activities in the protein products encoded by TK2185 and TK1404.
Our biochemical analysis showed that the protein encoded by TK1108 (PGMTk) undoubtedly exhibits PGM and PMM activities, which provided strong evidence that the protein represents the true PGM/PMM in T. kodakaraensis. It has been shown previously that TK1777 encodes a phosphopentomutase with only trace levels of PGM and PMM activity (13). In order to determine whether the remaining two genes, TK2185 and TK1404, encoded a PGM and/or PMM, we expressed the genes in E. coli and purified the recombinant proteins (Fig. 3). We found that neither protein exhibited PGM or PMM activity, even when high substrate concentrations (30 mM) were used. Altogether, our results revealed that among the four PMM orthologues in T. kodakaraensis, only one gene actually encodes a highly active PGM/PMM.
DISCUSSION
Here, using biochemical analysis, we showed that only one (TK1108) of four putative PMM genes present in the genome of T. kodakaraensis actually encodes a protein with significant PGM activity. This is the first report in which an archaeal PGM gene has been experimentally identified. The protein product also exhibited comparable levels of PMM activity, an activity that also could not be detected in the other orthologue proteins. During purification from T. kodakaraensis cells, we could not detect any PGM activities other than that derived from the TK1108 protein. Although we cannot rule out the possibility that there are other PGMs and/or PMMs that were not expressed under the conditions examined, it is most likely that TK1108 is responsible for both the PGM and PMM activities in T. kodakaraensis.
Another observation that supports our conclusions is that the levels of PGM activity found in T. kodakaraensis cells agreed well with the transcription levels of the PGMTk gene. Both levels were higher in cells grown on starch than in pyruvate-grown cells. This may reflect a role of the enzyme in starch degradation, in which glucose 1-phosphate is produced by the function of starch phosphorylases. Another possibility is that the enzyme is involved in intracellular glycogen synthesis. We also found putative ADP-glucose synthase (ADP-glucose pyrophosphorylase) genes and a glycogen synthase gene in the T. kodakaraensis genome. When abundant, sugars may be stored in the cells in the form of glycogen.
The phylogenetic tree of PMM genes from Thermococcales (Fig. 2) allowed us to identify the corresponding genes in three Pyrococcus genomes. TK1108 was closely related to PF0588, PH0923, and PAB0819, suggesting that the latter three genes may encode the PGM/PMMs in their organisms. Interestingly, all three Pyrococcus genes formed gene clusters with the genes encoding a putative mannose-1-phosphate guanylyl transferase, a putative mannosyl 3-phosphoglycerate phosphatase, and mannosyl 3-phosphoglycerate synthase. In particular, the protein products of the latter two genes from P. horikoshii have been biochemically characterized and have been clearly shown to exhibit the expected enzyme activities in a biosynthetic pathway for mannoglycerate, a compatible solute (4). The results obtained in this study, namely, the significant PMM activity of PGMTk, agree well with the proposal that genes clustered in the immediate vicinity of the mannosyl 3-phosphoglycerate phosphatase and mannosyl 3-phosphoglycerate synthase genes are involved in mannoglycerate biosynthesis.
Although PGMTk displays phosphopentomutase activity (Table 1), it seems unlikely that PGMTk is the major phosphopentomutase in T. kodakaraensis. The Km of the TK1777 product with deoxyribose 1-phosphate is slightly lower than that of PGMTk, and the kcat/Km value is also higher. It has been reported previously that phosphopentomutase activity is nearly equivalent in T. kodakaraensis cells grown on pyruvate or starch (13), and this was consistent with the transcription levels of TK1777 (data not shown). In contrast, we found here that PGM activity and transcription of TK1108 are both upregulated in the presence of starch. With these results, it is reasonable to conclude that TK1108 is the PGM/PMM gene and that TK1777 is the phosphopentomutase gene of T. kodakaraensis.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Atomi, H., T. Fukui, T. Kanai, M. Morikawa, and T. Imanaka. Description of Thermococcus kodakaraensis sp. nov., a well studied hyperthermophilic archaeon previously reported as Pyrococcus sp. KOD1. Archaea, in press. [DOI] [PMC free article] [PubMed]
- 3.Dai, J. B., Y. Liu, W. J. Ray, Jr., and M. Konno. 1992. The crystal structure of muscle phosphoglucomutase refined at 2.7-angstrom resolution. J. Biol. Chem. 267:6322-6337. [PubMed] [Google Scholar]
- 4.Empadinhas, N., J. D. Marugg, N. Borges, H. Santos, and M. S. da Costa. 2001. Pathway for the synthesis of mannosylglycerate in the hyperthermophilic archaeon Pyrococcus horikoshii. Biochemical and genetic characterization of key enzymes. J. Biol. Chem. 276:43580-43588. [DOI] [PubMed] [Google Scholar]
- 5.Fraser, C. M., J. A. Eisen, K. E. Nelson, I. T. Paulsen, and S. L. Salzberg. 2002. The value of complete microbial genome sequencing (you get what you pay for). J. Bacteriol. 184:6403-6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hammond, K. D., and D. Balinsky. 1978. Isozyme studies of several enzymes of carbohydrate metabolism in human adult and fetal tissues, tumor tissues, and cell cultures. Cancer Res. 38:1323-1328. [PubMed] [Google Scholar]
- 7.Imanaka, H., T. Fukui, H. Atomi, and T. Imanaka. 2002. Gene cloning and characterization of fructose-1,6-bisphosphate aldolase from the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Biosci. Bioeng. 94:237-243. [DOI] [PubMed] [Google Scholar]
- 8.Lytovchenko, A., L. Sweetlove, M. Pauly, and A. R. Fernie. 2002. The influence of cytosolic phosphoglucomutase on photosynthetic carbohydrate metabolism. Planta 215:1013-1021. [DOI] [PubMed] [Google Scholar]
- 9.Morikawa, M., Y. Izawa, N. Rashid, T. Hoaki, and T. Imanaka. 1994. Purification and characterization of a thermostable thiol protease from a newly isolated hyperthermophilic Pyrococcus sp. Appl. Environ. Microbiol. 60:4559-4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naught, L. E., and P. A. Tipton. 2001. Kinetic mechanism and pH dependence of the kinetic parameters of Pseudomonas aeruginosa phosphomannomutase/phosphoglucomutase. Arch. Biochem. Biophys. 396:111-118. [DOI] [PubMed] [Google Scholar]
- 11.Nelson, K. E., I. T. Paulsen, J. F. Heidelberg, and C. M. Fraser. 2000. Status of genome projects for nonpathogenic bacteria and archaea. Nat. Biotechnol. 18:1049-1054. [DOI] [PubMed] [Google Scholar]
- 12.Pappin, D. J. 1997. Peptide mass fingerprinting using MALDI-TOF mass spectrometry. Methods Mol. Biol. 64:165-173. [DOI] [PubMed] [Google Scholar]
- 13.Rashid, N., H. Imanaka, T. Fukui, H. Atomi, and T. Imanaka. 2004. Presence of a novel phosphopentomutase and a 2-deoxyribose 5-phosphate aldolase reveals a metabolic link between pentoses and central carbon metabolism in the hyperthermophilic archaeon Thermococcus kodakaraensis. J. Bacteriol. 186:4185-4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rashid, N., H. Imanaka, T. Kanai, T. Fukui, H. Atomi, and T. Imanaka. 2002. A novel candidate for the true fructose-1,6-bisphosphatase in archaea. J. Biol. Chem. 277:30649-30655. [DOI] [PubMed] [Google Scholar]
- 15.Ray, W. J. J., and E. J. J. Peck. 1972. Phosphomutases, p. 407-458. In P. D. Boyer (ed.), The enzymes, 3rd ed., vol. 6. Academic Press, New York, N.Y. [Google Scholar]
- 16.Sato, T., T. Fukui, H. Atomi, and T. Imanaka. 2003. Targeted gene disruption by homologous recombination in the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Bacteriol. 185:210-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schut, G. J., S. D. Brehm, S. Datta, and M. W. Adams. 2003. Whole-genome DNA microarray analysis of a hyperthermophile and an archaeon: Pyrococcus furiosus grown on carbohydrates or peptides. J. Bacteriol. 185:3935-3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Videira, P. A., L. L. Cortes, A. M. Fialho, and I. Sá-Correia. 2000. Identification of the pgmG gene, encoding a bifunctional protein with phosphoglucomutase and phosphomannomutase activities, in the gellan gum-producing strain Sphingomonas paucimobilis ATCC 31461. Appl. Environ. Microbiol. 66:2252-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weinhäusel, A., R. Griessler, A. Krebs, P. Zipper, D. Haltrich, K. D. Kulbe, and B. Nidetzky. 1997. α-1,4-d-Glucan phosphorylase of gram-positive Corynebacterium callunae: isolation, biochemical properties and molecular shape of the enzyme from solution X-ray scattering. Biochem. J. 326:773-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xavier, K. B., R. Peist, M. Kossmann, W. Boos, and H. Santos. 1999. Maltose metabolism in the hyperthermophilic archaeon Thermococcus litoralis: purification and characterization of key enzymes. J. Bacteriol. 181:3358-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu, J. -P., J. Ladapo, and W. B. Whitman. 1994. Pathway of glycogen metabolism in Methanococcus maripaludis. J. Bacteriol. 176:325-332. [DOI] [PMC free article] [PubMed] [Google Scholar]