Abstract
HpaG is a type III-secreted elicitor protein of Xanthomonas axonopodis pv. glycines. We have determined the critical amino acid residues important for hypersensitive response (HR) elicitation by random and site-directed mutagenesis of HpaG and its homolog XopA. A plasmid clone carrying hpaG was mutagenized by site-directed mutagenesis, hydroxylamine mutagenesis, and error-prone PCR. A total of 52 mutants were obtained, including 51 single missense mutants and 1 double missense mutant. The HR elicitation activity was abolished in the two missense mutants [HpaG(L50P) and HpaG(L43P/L50P)]. Seven single missense mutants showed reduced activity, and the HR elicitation activity of the rest of the mutants was similar to that of wild-type HpaG. Mutational and deletion analyses narrowed the region essential for elicitor activity to the 23-amino-acid peptide (H2N-NQGISEKQLDQLLTQLIMALLQQ-COOH). A synthetic peptide of this sequence possessed HR elicitor activity at the same concentration as the HpaG protein. This region has 78 and 74% homology with 23- and 27-amino-acid regions of the HrpW harpin domains, respectively, from Pseudomonas and Erwinia spp. The secondary structure of the peptide is predicted to be an α-helix, as is the HrpW region that is homologous to HpaG. The predicted α-helix of HpaG is probably critical for the elicitation of the HR in tobacco plants. In addition, mutagenesis of a xopA gene yielded two gain-of-function mutants: XopA(F48L) and XopA(F48L/M52L). These results indicate that the 12 amino acid residues between L39 and L50 of HpaG have critical roles in HR elicitation in tobacco plants.
In many interactions between gram-negative plant-pathogenic bacteria and plants, hrp (for hypersensitive reaction and pathogenicity) genes are required for pathogenicity in the host plant and induction of the hypersensitive response (HR) in nonhost plants (17). Regions that contain a cluster of hrp genes and other components required for pathogenicity are designated pathogenicity islands (PAIs) (3, 15). Most hrp genes that encode components of the type III protein secretion system mediate the translocation of effector proteins, such as Avr (avirulence) proteins, across the bacterial membrane and the walls and plasma membranes of plant cells (10).
HR is a highly localized plant cell death that occurs when nonhost plants or resistant cultivars of host plants are infiltrated with the plant pathogen or HR elicitor molecules, such as Avr proteins and harpins. HR is thought be a resistance reaction of plants to microbial pathogens (11). Harpins are a group of HR elicitors that are secreted by the type III secretion pathway and elicit HR when infiltrated into the apoplast of leaves of nonhost plants. Unlike Avr proteins, which must be delivered inside the cell to exert their functions, harpins can elicit HR when delivered to the intercellular space of plant cells (10). Since the first known harpin, HrpN, was identified from Erwinia amylovora, many harpins have been reported from Pseudomonas, Ralstonia, and Xanthomonas species (4, 8, 12, 14, 15, 27). Harpins are glycine-rich, heat stable, and lack cysteine, but the biochemical mechanisms of HR elicitation in nonhost plants are unclear. One reason for this is that the amino acid sequences of harpins do not share significant homology with other known proteins or among themselves.
The mode of action of harpins is still controversial. HrpZ of Pseudomonas syringae pv. syringae associates with the walls rather than the membranes of plant cells, and the protein elicits no response from protoplasts, which lack walls (13). However, HrpZ of P. syringae pv. phaseolicola binds to lipid bilayers and forms an ion-conducting pore (16). The N-terminal 109 amino acids and the C-terminal 216 amino acids of HrpZ are able to elicit HR to a level similar to full-length HrpZ (2). Kim et al. and Charkowski et al. showed that the HrpW harpins of E. amylovora and P. syringae pv. tomato are composed of two domains—the N-terminal harpin domain and C-terminal Pel (pectate lyase) domain—and proposed that HrpW acts in the cell wall (8, 14).
We previously published the first report of a harpin from Xanthomonas species, HpaG (15). At 13.4 kDa, HpaG is smaller than other known harpins (15). Four additional Xanthomonas HpaG homologs have been reported. HpaG shows a true harpin-like activity, and Hpa1 of X. oryzae pv. oryzae possesses HR elicitor activity at relatively high concentrations (i.e., >5 μM). However, XopA of X. campestris pv. vesicatoria does not induce HR (15). To understand the nature of the HR induction by HpaG homologs in nonhost plants, we performed detailed mutational analysis of hpaG, identifying 23 amino acid residues that are essential and sufficient for the elicitor activity of HpaG. Using site-directed mutagenesis, we determined the amino acid residues that have the most influence on the elicitor activity. Finally, we obtained a gain-of-function mutant of XopA.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in the present study are listed in Table 1. Escherichia coli cells were cultivated at 37°C in Luria broth (LB; USB) or on LB agar plates. X. campestris pv. vesicatoria strains were grown at 28°C in LB or on YDC (1% yeast extract, 2% calcium carbonate, 2% d-glucose) agar plates. Antibiotics were used in E. coli cultures at 100 μg/ml for ampicillin and 34 μg/ml for chloramphenicol.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| Escherichia coli | ||
| DH5α | F− φ80dlacZΔM15 Δ(lacZYA-argF)U169 endA1 deoR recA1 hsdR17(rK− mK+) phoA supE44 λ−thi-1 gyrA96 relA1 | Gibco-BRL |
| BL21(DE3) | F−ompT hsdSB(rB− mB−) gal dcm (DE3) | Novagen |
| Xanthomonas campestris pv. vesicatoria | ||
| 82-8 | Wild type, race 1 | R. E. Stall |
| E-3 | Wild type, race 2 | R. E. Stall |
| LS833 | Wild type, race 3 | S. Lee |
| Plasmids | ||
| pT7-7 | T7 promoter-based expression vector; Ampr | 24 |
| pET14b | T7 promoter-based expression vector; Ampr | Novagen |
| pLysS | Encoding T7 lysozyme gene; Cmr | Novagen |
| pBluescript II SK(+) | Phagemid, pUC derivative; Ampr | Stratagene |
| pBSN | pBluescript II SK(+) with NdeI site in β-galactosidase gene start codon | This study |
| pBHPAG | 0.4-kb NdeI-BamHI fragment, including entire hpaG gene, cloned into pBSN | This study |
| pHpaGMT | SmaI-BssHII fragment (T7 promoter region) of pBHPAG was deleted and religated; template for site-directed mutagenesis of hpaG | This study |
| pTJ1 | PCR-amplified hpaG in pT7-7; Ampr | This study |
| pJ14 | hpaG in pET14b; Ampr | This study |
| pHGM61 | hpaG(L4A) in pET14b; Ampr | This study |
| pHGM62 | hpaG(Q7A) in pET14b; Ampr | This study |
| pHGM63 | hpaG(G9A) in pET14b; Ampr | This study |
| pHGM64 | hpaG(F14D) in pET14b; Ampr | This study |
| pHGM65 | hpaG(Q16A) in pET14b; Ampr | This study |
| pHGM66 | hpaG(P19A) in pET14b; Ampr | This study |
| pHGM67 | hpaG(G20E) in pET14b; Ampr | This study |
| pHGM68 | hpaG(Q21A) in pET14b; Ampr | This study |
| pHGM69 | hpaG(Q24A) in pET14b; Ampr | This study |
| pHGM70 | hpaG(Q29A) in pET14b; Ampr | This study |
| pHGM71 | hpaG(G30E) in pET14b; Ampr | This study |
| pHGM72 | hpaG(Q32A) in pET14b; Ampr | This study |
| pHGM73 | hpaG(G33A) in pET14b; Ampr | This study |
| pHGM26 | hpaG(S35A) in pET14b; Ampr | This study |
| pHGM7 | hpaG(E36A) in pET14b; Ampr | This study |
| pHGM27 | hpaG(K37A) in pET14b; Ampr | This study |
| pHGM28 | hpaG(Q38A) in pET14b; Ampr | This study |
| pHGM46 | hpaG(L39A) in pET14b; Ampr | This study |
| pHGM74 | hpaG(L39P) in pET14b; Ampr | This study |
| pHGM6 | hpaG(D40A) in pET14b; Ampr | This study |
| pHGM29 | hpaG(Q41A) in pET14b; Ampr | This study |
| pHGM46 | hpaG(L42A) in pET14b; Ampr | This study |
| pHGM75 | hpaG(L42D) in pET14b; Ampr | This study |
| pHGM36 | hpaG(L43P) in pET14b; Ampr | This study |
| pHGM30 | hpaG(T44A) in pET14b; Ampr | This study |
| pHGM40 | hpaG(Q45A) in pET14b; Ampr | This study |
| pHGM32 | hpaG(L46A) in pET14b; Ampr | This study |
| pHGM41 | hpaG(I47A) in pET14b; Ampr | This study |
| pHGM11 | hpaG(L50A) in pET14b; Ampr | This study |
| pHGM76 | hpaG(L50P) in pET14b; Ampr | This study |
| pHGM12 | hpaG(L51A) in pET14b; Ampr | This study |
| pHGM13 | hpaG(Q52A) in pET14b; Ampr | This study |
| pHGM42 | hpaG(Q53A) in pET14b; Ampr | This study |
| pHGM15 | hpaG(S54A) in pET14b; Ampr | This study |
| pHGM16 | hpaG(N55A) in pET14b; Ampr | This study |
| pHGM18 | hpaG(E58A) in pET14b; Ampr | This study |
| pHGM8 | hpaG(Q59A) in pET14b; Ampr | This study |
| pHGM19 | hpaG(G60V) in pET14b; Ampr | This study |
| pHGM9 | hpaG(Q61A) in pET14b; Ampr | This study |
| pHGM20 | hpaG(G62V) in pET14b; Ampr | This study |
| pHGM21 | hpaG(Q63A) in pET14b; Ampr | This study |
| pHGM38 | hpaG(G64A) in pET14b; Ampr | This study |
| pHGM10 | hpaG(Q65A) in pET14b; Ampr | This study |
| pHGM22 | hpaG(G66A) in pET14b; Ampr | This study |
| pHGM23 | hpaG(G67V) in pET14b; Ampr | This study |
| pHGM5 | hpaG(D99A) in pET14b; Ampr | This study |
| pHGM17 | hpaG(I120T) in pET14b; Ampr | This study |
| pHGM37 | hpaG(L121P) in pET14b; Ampr | This study |
| pHGM2 | hpaG(A126T) in pET14b; Ampr | This study |
| pHGM34 | hpaG(A126V) in pET14b; Ampr | This study |
| pHGM3 | hpaG(S131L) in pET14b; Ampr | This study |
| pHGM77 | hpaG(L43P/L50P) in pET14b; Ampr | This study |
| pTHGC | hpaG(ΔN67) in pT7-7; Ampr | This study |
| pTHG67 | hpaG(ΔC66) in pET14b; Ampr | This study |
| pTHG58 | hpaG(ΔC75) in pET14b; Ampr | This study |
| pTHG56 | hpaG(ΔC77) in pET14b; Ampr | This study |
| pTHG54 | hpaG(ΔC79) in pET14b; Ampr | This study |
| pTHG52 | hpaG(ΔC81) in pET14b; Ampr | This study |
| pTHG50 | hpaG(ΔC83) in pET14b; Ampr | This study |
| pTXOPA | PCR product of xopA cloned into pET14b; Ampr | This study |
| pTXH2 | 120-bp NdeI-PvuII fragment of pTXOPA and 73-bp PvuII-BamHI fragment of pTHG67 coligated into pET14b; Ampr | This study |
| pTXM1 | xopA(F48L) in pET14b; Ampr | This study |
| pTXM2 | xopA(M52L) in pET14b; Ampr | This study |
| pTXM4 | xopA(F48L/M52L) in pET14b; Ampr | This study |
Ampr, ampicillin resistance; Cmr, chloramphenicol resistance.
DNA manipulations.
Standard methods were used for DNA cloning, restriction mapping, and gel electrophoresis (23). The vector DNA was treated with appropriate restriction enzymes, and extraction of DNA fragments from gels was carried out by using the QIAEX II gel extraction kit as described by the manufacturer (Qiagen, Valencia, Calif.). All other standard molecular biological methods were carried out as described by Sambrook et al. (23). The oligonucleotides used for the mutagenesis of hpaG and xopA are listed in Tables 2 and 3. All oligonucleotides were designed by using the PrimerSelect program (DNASTAR) to minimize secondary structure and dimer formation and were chemically synthesized by CoreBioSystem (Seoul, Korea).
TABLE 2.
Oligonucleotides used for deletion mutagenesis of hpaGa
| Oligonucleotide | Sequence; description |
|---|---|
| T7 promoter | 5′-TAATACGACTCACTATAGGG-3′ |
| hpaGΔC77 | 5′-GGATCCTAATTGTTGCTCTGCTGAAG-3′; mutagenic primer used to construct HpaGΔC77 |
| hpaGΔC79 | 5′-GGATCCTAGCTCTGCTGAAGCAGG-3′; mutagenic primer used to construct HpaGΔC79 |
| hpaGΔC81 | 5′-GGATCCTACTGAAGCAGGGCCATG-3′; mutagenic primer used to construct HpaGΔC81 |
| hpaGΔC83 | 5′-GGATCCTACAGTGCCATGATGAGCTGGGTC-3′; mutagenic primer used to construct HpaGΔC83 |
| hpaGΔN671 | 5′-CATATGGACTCTGGCGGTC-3′; mutagenic primer used to construct HpaGΔN67 |
| hpaGrev | 5′-GGATCCTTACTGCATCGATC-3′; mutagenic primer used to construct HpaGΔN67 |
Base differences from the wild-type sequence are shown in bold face italic. New BamHI and NdeI sites are indicated by underlined bases.
TABLE 3.
Oligonucleotides used for site-directed mutagenesisa
| Oligonucleotide | Sequence; description |
|---|---|
| M13-20 | 5′-GTAAAACGACGGCCAGTG-3′ |
| hpaGL4A | 5′-ATGAATTCTGCGAACACACAGCTCG-3′; mutagenic primer used to construct HpaG(L4A) |
| hpaGQ7A | 5′-CTTTGAACACAGCGCTCGGCGC-3′; mutagenic primer used to construct HpaG(Q7A) |
| hpaGG9A | 5′-CACAGCTCGCCGCCAACTCGTC-3′; mutagenic primer used to construct HpaG(G9A) |
| hpaGF14D | 5′-CCAACTCGTCCGACTTTCAGGTTGAC-3′; mutagenic primer used to construct HpaG(F14D) |
| hpaGQ16A | 5′-TCCTTCTTTGCGGTTGACCCCG-3′; mutagenic primer used to construct HpaG(Q16A) |
| hpaGP19A | 5′-CAGGTTGACGCCGGCCAGAAC-3′; mutagenic primer used to construct HpaG(P19A) |
| hpaGG20E | 5′-GTTGACCCCGAACAGAACACGC-3′; mutagenic primer used to construct HpaG(G20E) |
| hpaGQ21A | 5′-GACCCCGGCGCGAACACGC-3′; mutagenic primer used to construct HpaG(Q21A) |
| hpaGQ24A | 5′-CCAGAACACGGCATCTAGTCCGAAC-3′; mutagenic primer used to construct HpaG(Q24A) |
| hpaGQ29A | 5′-AGTCCGAACGCGGGCAACCAG-3′; mutagenic primer used to construct HpaG(Q29A) |
| hpaGG30E | 5′-CCGAACCAGGAAAACCAGGGCATC-3′; mutagenic primer used to construct HpaG(G30E) |
| hpaGQ32A | 5′-CAGGGCAACGCGGGCATCTC-3′; mutagenic primer used to construct HpaG(Q32A) |
| hpaGG33A | 5′-CAGGGCAACCAGGCCATCTCGG-3′; mutagenic primer used to construct HpaG(G33A) |
| hpaGL39P | 5′-TCGGAAAAGCAACCGGACCAGC-3′; mutagenic primer used to construct HpaG(L39P) |
| hpaGL42D | 5′-AACTGGACCAGGACCTGACCCAGCTCATC-3′; mutagenic primer used to construct HpaG(L42D) |
| hpaGL50P | 5′-CAGCTCATCATGGCACCGCTGCAGCAG-3′; mutagenic primer used to construct HpaG(L50P) |
| hpaGD99A | 5′-CGTCGGAGCCATTCTCCAG-3′; mutagenic primer used to construct HpaG(D99A) |
| xopAF48L | 5′-GACCCAGCTCATCTTTTCAATG-3′; mutagenic primer used to construct XopA(F48L) |
| xopAM52L | 5′-CTTTTCACTGCTTCTGCAGG-3′; mutagenic primer used to construct XopA(M52L) |
Base differences from the wild-type sequence are shown in boldface italic.
Random mutagenesis.
Random mutagenesis of hpaG was performed by using the error-prone PCR (25) and hydroxylamine mutagenesis (18) methods, with modifications. For the error-prone PCR, pTJ1 was used as a template, and the primers hpaGfrw (5′-GCGGCCATATGAATTCTTTGA-3′) and hpaGrev (5′-GGATCCTTACTGCATCGATC-3′) were used. The PCR products were cleaned, digested with NdeI and BamHI, separated by agarose-gel electrophoresis, purified from the gel, and fused between the NdeI and BamHI sites of the plasmid pET14b (Novagen, Madison, Wis.). For the hydroxylamine mutagenesis, 10 μg of pJ14 DNA were incubated in a reaction mixture containing 0.5 M hydroxylamine and 5 mM EDTA in 0.1 M potassium phosphate (pH 6.0) at 50°C for 4, 8, or 12 h or at 37°C for 12 or 24 h. After the treatment, the plasmids were diluted in TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA) and dialyzed overnight to remove the hydroxylamine. The dialyzed plasmid DNA was precipitated with ethanol and transformed into E. coli BL21(DE3).
Deletion mutagenesis.
Deletion derivatives of HpaG—HpaGΔN67, HpaGΔC66, HpaGΔC75, HpaGΔC77, HpaGΔC79, HpaGΔC81, and HpaGΔC83—were constructed by PCR with pTJ1 as a template. The upstream T7 promoter-specific primer was complementary to the template DNA upstream of the hpaG insert and included a unique NdeI site. The downstream primers were complementary to an internal region of hpaG and included a translational stop codon and a unique BamHI site. The downstream primers designed for each deletion mutant are listed in Table 2. Each 50-μl PCR contained 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 50 mM KCl, 20 ng of pTJ1 DNA, 200 μM concentrations of deoxynucleoside triphosphates, 2 μM concentrations of primers, and 1.2 U of Taq polymerase (TaKaRa Shuzo Co. Shiga, Japan). The reaction mixtures were heated for 2 min at 94°C and then amplified over 10 cycles of 1 min at 94°C, 1 min at 45°C, and 1 min at 72°C, followed by 20 cycles of 1 min at 94°C, 1 min at 50°C, and 1 min at 72°C. The PCR products were purified with phenol and chloroform extraction and the DNA was precipitated with ethanol. After digestion with NdeI and BamHI, the PCR products were fused between the corresponding sites in the pET14b vector.
Site-directed mutagenesis with megaprimer PCR.
Site-directed mutagenesis of hpaG was performed by using the PCR-mediated megaprimer method (5). In the first PCR amplification, the template DNA, pHpaGMT, was constructed as follows. The 0.4-kb NdeI-BamHI fragment from pTJ1 was fused between the corresponding sites in pBSN, generating pBHPAG. To delete the T7 promoter sequence region, pBHPAG was digested with SmaI and BssHII, blunt ended with the Klenow fragment (TaKaRa Shuzo Co.), and religated, generating pHpaGMT. In the first PCR, pHpaGMT was used as the template DNA, and the M13-20 primer and individual mutagenic primers were used (Table 3). The PCR conditions used were the same as in the deletion mutagenesis method described above. The PCR products were isolated from 1.2% agarose gels, purified by using the QIAEX II gel extraction kit, and resuspended in distilled water for use in subsequent PCRs as megaprimers. In the second PCR amplification, pTJ1 DNA was used as the template; the T7 promoter primer and the megaprimer of the gel elution product of first PCR were used as primers. The reaction conditions used were the same as in the first PCRs. The second PCR products were purified with phenol and chloroform extraction, followed by precipitation with ethanol. After digestion with NdeI and BamHI, the digested DNA was fused between the corresponding sites in pET14b. Site-directed mutagenesis of xopA was performed as described above. Mutants with single amino acid substitutions are denoted as the one-letter notation of the original amino acid and its position in the HpaG amino acid sequence, followed by the substituted amino acid.
DNA sequencing and data analysis.
Mutagenized hpaG and xopA DNA fragments in pET14b were sequenced to confirm the presence of the appropriate mutation. For DNA sequencing, plasmid DNAs containing the hpaG and xopA mutant clones were purified by using the QIAprep Spin Miniprep Kit (Qiagen). The T7 promoter primer and the T7 terminator primer (5′-CTAGTTATTGCTCAGCGGT-3′) were used in sequencing reactions. The reactions were carried out by using the ABI Prism BigDye terminator cycle sequencing kit (version 2.0; Perkin-Elmer Corp., Norwalk, Conn.) on an ABI 3700 DNA Analyzer (Applied Biosystems, Foster City, Calif.) at the National Instrumentation Center for Environmental Management, Seoul, Korea. DNA sequence data were analyzed by using the SEQMAN and MEGALIGN software (DNASTAR) and GENETYX-WIN software (Software Development, Tokyo, Japan).
Overexpression and purification of HpaG and HpaG mutant proteins.
The site-directed and deletion mutant clones were introduced into the E. coli strain BL21(DE3)(pLysS) for protein overexpression. Strains harboring each mutant clone were grown overnight in LB containing ampicillin and chloramphenicol, and the overnight cultures were diluted 100-fold in LB and grown at 37°C with agitation. At an optical density of 0.8 at 600 nm, IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 1 mM, the cultures were grown at 37°C for 2 h with agitation, and the cells were then harvested by centrifugation. The cells were concentrated 100-fold by resuspending the pellet in 20 mM Tris-HCl (pH 8.0), sonicated, and boiled for 10 min. After centrifugation, the protein in the partially purified samples was quantitated by using the Bradford method, with bovine serum albumin as the standard (7), and the protein samples were used for the primary analysis of HR elicitor activity on tobacco leaves. The proteins were also visualized by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis, followed by staining with Coomassie brilliant blue R.
N-terminal His-tagged proteins were used when more highly purified proteins were required. E. coli BL21(DE3)(pLysS) cells carrying the hpaG or xopA mutants fused into the pET14b vector were grown in LB broth, and the His-tagged proteins were expressed after the addition of IPTG. Cells were harvested and lysed by sonication in 0.5 ml of lysis buffer (10 mM imidazole, 20 mM Tris-HCl [pH 8.0]). After centrifugation at 10,000 × g for 20 min at room temperature to pellet the cellular debris, the supernatant was loaded onto a Ni-NTA spin column (Qiagen), binding His-tagged protein. The Ni-NTA matrix was centrifuged at 1,000 × g for 2 min at room temperature, and the matrix was then washed two times with washing buffer (20 mM imidazole, 20 mM Tris-HCl [pH 8.0]) to remove unbound protein. His-tagged protein was eluted by stepwise addition of 0.1 ml of elution buffer 1 (0.5 M imidazole, 20 mM Tris-HCl [pH 8.0]) and 0.1 ml of elution buffer 2 (1 M imidazole, 20 mM Tris-HCl [pH 8.0]). The eluted protein was dialyzed with 20 mM Tris-HCl (pH 8.0) to remove the imidazole, and the concentration of the purified protein was measured by the Bradford method with bovine serum albumin as the standard (7).
Plant assays.
For HR tests, tobacco (Nicotiana tabacum cv. Samsun NN) plants were inoculated with HpaG, HpaG derivatives, XopA, or XopA derivatives in 20 mM Tris-HCl (pH 8.0), and the responses of the plants were observed for 12 to 24 h after inoculation.
Immunodetection of HpaG mutants.
Purified HpaG mutant proteins were separated by SDS-polyacrylamide gel electrophoresis (on a 15% acrylamide gel) and then transferred to Hybond-P membrane (Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom) by electroblotting at 25 V for 60 min in transfer buffer (48 mM Tris, 39 mM glycine, 0.037% [wt/vol] SDS, 20% [vol/vol] methanol [pH 8.3]). For immunoblot detection, a rabbit polyclonal anti-HpaG antibody was used as the primary antibody and alkaline-phosphatase-conjugated goat anti-rabbit immunoglobulin G (Pierce Biotechnology, Rockford, Ill.) was used as the secondary antibody. Positive signals were detected by using One-Step NBT/BCIP solutions (Pierce).
Peptide synthesis.
The HpaG peptide (H2N-NQGISEKQLDQLLTQLIMALLQQ-COOH) and the HpaG(L50P) peptide (H2N-NQGISEKQLDQLLTQLIMAPLQQ-COOH) were synthesized by A&PEP (Chungnam, Korea).
Protein secondary structure prediction.
The secondary structures of HpaG, HpaG derivatives, XopA, and XopA derivatives were predicted by using the protein structure prediction server HNN Secondary Structure Prediction Method at Network Protein Sequence @nalysis (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_nn.html) (9).
RESULTS
Deletion analysis of hpaG.
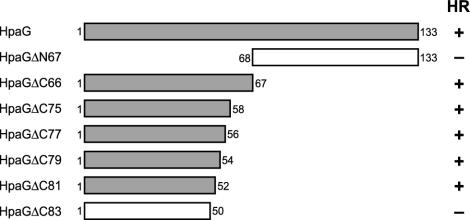
To determine the regions in HpaG that are critical for the induction of HR in nonhost plants, we constructed seven truncated HpaG derivatives and tested their ability to induce HR in tobacco plants. HpaGΔC66, HpaGΔC75, HpaGΔC77, HpaGΔC79, and HpaGΔC81 had elicitor activity equivalent to that of the wild-type HpaG, whereas HpaGΔN67 and HpaGΔC83 failed to induce HR even at concentrations greater than 10 μM (Fig. 1). These results indicate that the N-terminal 52 amino acids of HpaG are sufficient for elicitor activity and that the C-terminal 75 amino acid residues are not essential for elicitor activity.
FIG. 1.
Diagram of HpaG and truncated HpaG proteins used to test regions of the protein for elicitor activity. The C-terminal region of HpaG was not necessary to elicit HR on tobacco leaves. Open and closed bars represent fragments that did not elicit HR and fragments with full HR elicitation activity, respectively. The HR elicitor activities of each mutant in tobacco leaves are designated by “+” and “−” in the right-hand column. +, HR activity equivalent to that of wild-type HpaG; −, no HR observed.
Random and site-directed mutagenesis of the hpaG gene.
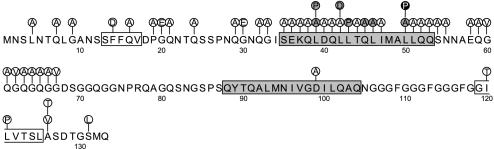
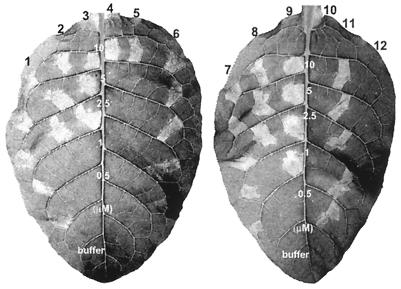
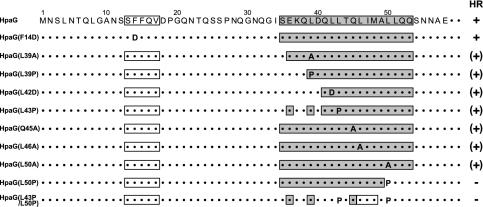
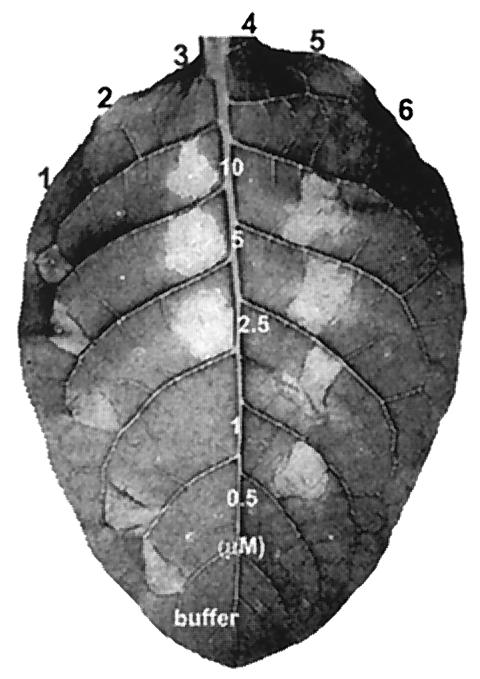
To determine the amino acid residues of HpaG that have critical roles in HR elicitor activity, we used site-directed mutagenesis to generate 46 HpaG derivatives with single amino acid substitutions and one mutant, HpaG(L43P/L50P), in which two amino acids were altered (Fig. 2). In addition, three mutants, HpaG(I120T), HpaG(L121P), and HpaG(A126V), were constructed by using error-prone PCR methods, and two mutants, HpaG(A126T) and HpaG(S131L), were generated by using hydroxylamine mutagenesis (Fig. 2). Among the 52 missense mutants, the HR elicitor activity of 43 mutants was the same as that of wild-type HpaG, but 7 mutants produced less HR activity than the wild-type (data not shown and Fig. 2). Two mutant proteins, HpaG(Q45A) and HpaG(L50A), elicited HR on tobacco leaves at 1 μM but failed to induce HR at 0.5 μM (Fig. 3). HpaG(L39A), HpaG(L39P), and HpaG(L46A) elicited HR on tobacco leaves at concentrations greater than 2.5 μM, and HpaG(L42D) and HpaG(L43P) elicited HR at concentrations greater than 5 μM (Fig. 3).
FIG. 2.
Amino acid substitutions in the HpaG amino acid sequence. Single amino acid substitutions in each mutant are indicated by closed circles, with the substituted amino acid residue shown in the circle. The HR elicitor activity of each mutant in tobacco leaves is represented by white circles for full HR activity, gray circles for reduced activity, and black circles for no HR activity. The predicted secondary structure of HpaG is indicated with open rectangles for predicted β-sheet regions and gray rectangles for predicted α-helical regions.
FIG. 3.
Comparison of the HR elicitor activity in tobacco leaves of HpaG mutant proteins, the HpaG peptide, and the HpaG(L50P) peptide. The proteins were infiltrated into tobacco leaves at concentrations of 10, 5, 2.5, 1, or 0.5 μM in 20 mM Tris-HCl (pH 8.0). Labeling: 1 and 7, HpaG; 2, HpaG(L39A); 3, HpaG(L39P); 4, HpaG(L42D); 5, HpaG(L43P); 6, HpaG(Q45A); 8, HpaG(L46A); 9, HpaG(L50A); 10, HpaG(L50P); 11, HpaG peptide; 12, HpaG(L50P) peptide; buffer, 20 mM Tris-HCl (pH 8.0). Tobacco (N. tabacum cv. Samsun NN) leaves were photographed 24 h after infiltration.
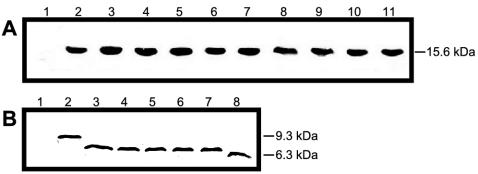
To confirm that the reduced elicitor activity of the mutant proteins was not due to protein instability, all of the expressed proteins were detected by using immunoblots. As shown in Fig. 4, each HpaG derivative was successfully expressed, exhibited wild-type heat stability, and cross-reacted with polyclonal anti-HpaG antibodies. This indicated that the loss of activity of the mutants was not due to instability of the proteins.
FIG. 4.
Immunodetection of HpaG mutant proteins. Production of the mutant proteins was induced with IPTG induction. After boiling and purification procedures, the proteins were analyzed by immunoblotting with a rabbit polyclonal anti-HpaG antibody. (A) Immunodetection of full-length HpaG mutants. Lanes: 1, pET14b vector control; 2, HpaG; 3, HpaG(L39A); 4, HpaG(L39P); 5, HpaG(L42D); 6, HpaG(L43P); 7, HpaG(Q45A); 8, HpaG(L46A); 9, HpaG(L50A); 10, HpaG(L50P); 11, HpaG(L43P/L50P). (B) Immunodetection of HpaG deletion mutants. Lanes: 1, pET14b vector control; 2, HpaGΔC66; 3, HpaGΔC75; 4, HpaGΔC77; 5, HpaGΔC79; 6, HpaGΔC81; 7, HpaGΔC83; 8, HpaGΔN67.
The predicted N-terminal α-helix is important for elicitor activity.
The amino acid substitutions that affected the HR elicitor activity of HpaG were clustered in the region from L39 to L50. We therefore investigated the relationship between elicitor activity and predicted secondary structures in HpaG. Computer-generated predictions of the HpaG secondary structure showed that the protein has two possible α-helices and two possible β-sheet regions (Fig. 2). The first predicted α-helix is formed by the 19 amino acid residues between S35 and Q53, and the second is formed by the 17 amino acid residues between Q88 and Q104 (Fig. 2). The first predicted β-sheet is in the five amino acid residues between S13 and V17, and the second is in the seven amino acid residues between G119 and L125 (Fig. 2). Since the terminal 81 amino acid residues of HpaG are not required for elicitor activity, the second predicted α-helix and β-sheet in the C terminus were not taken into consideration for the elicitor activity. The HpaG mutants with mutations in the first putative α-helix, including HpaG(L39A), HpaG(L39P), HpaG(L42D), and HpaG(L43P), were affected in the ability to induce HR, as shown in Fig. 5. However, no null mutant that lacked elicitor activity was obtained. The mutants described above are predicted to have altered secondary structures, going from the putative α-helix to the random coil form in this region. Therefore, changing this putative α-helix into the random coiled form might abolish the elicitor activity. The HNN secondary structure prediction method (available at http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_nn.html) was used to predict mutants in which the putative α-helix would be affected, revealing two potential mutants, HpaG(L50P) and HpaG(L43P/L50P). These two mutants were constructed by site-directed mutagenesis. HpaG(L50P) was predicted to have changes in the lower edge of the putative α-helix, and the normally most α-helical region in the double mutant HpaG(L43P/L50P) was predicted to assume a random coil form (Fig. 5). Infiltration of HpaG(L50P) and HpaG(L43P/L50P) into tobacco leaves at concentrations of 10 μM resulted in no elicitor activity (Fig. 3 and 5). HpaG(F14D), in which the region of the first predicted β-sheet was predicted to assume a random coil form, had HR elicitation activity equivalent to that of wild-type HpaG (Fig. 5). This result indicates that the 12 amino acid residues between L39 and L50 have important roles in HR elicitation in tobacco plants and that the leucine residue at position 50 is the most critical for the elicitor activity.
FIG. 5.
Predicted secondary structures in the N-terminal region of representative HpaG mutants. Predicted α-helices and β-sheets are indicated as gray and white rectangles, respectively. The HR elicitor activity of each mutant in tobacco leaves is indicated as follows: +, HR activity equivalent to that of wild-type HpaG; (+), reduced activity relative to wild-type HpaG; and −, no HR observed.
A synthetic peptide comprising 23 amino acid residues of HpaG is sufficient to elicit HR.
Based on the analysis of the HpaG mutants produced by deletion and site-directed mutagenesis, we proposed that the putative N-terminal α-helical region, composed of the 19 amino acid residues from S35 to Q53, has an important role in the HR elicitation by HpaG in tobacco plants. We therefore synthesized the HpaG peptide, composed of the 23 amino acid residues H2N-NQGISEKQLDQLLTQLIMALLQQ-COOH, and the HpaG(L50P) peptide, composed of 23 amino acid residues H2N-NQGISEKQLDQLLTQLIMAPLQQ-COOH, in which the L50 was changed to a proline residue. The peptides were dissolved in 20 mM Tris-HCl (pH 8.0), diluted to various concentrations, and then infiltrated into tobacco leaves. The HpaG peptide at 0.5 μM elicited HR on tobacco leaves, but the HpaG(L50P) peptide failed to elicit HR, even at 10 μM (Fig. 3). These results indicate that the HpaG peptide has elicitor activity equivalent to that of the wild-type HpaG and that L50 has a critical role in the elicitor activity of HpaG.
The putative N-terminal α-helical region is common in harpins.
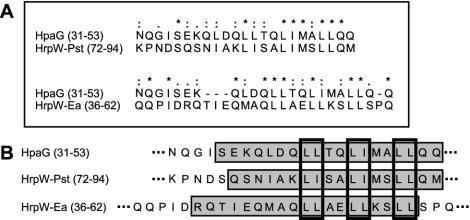
To determine whether the putative α-helical region in the N-terminal region of HpaG is present in other harpins, we compared the HpaG peptide sequence with the sequences of other harpins. The amino acid sequence of the HpaG peptide does not have homology with the sequences of HrpN, HrpZ, or PopA. However, the N-terminal harpin domain of the HrpW proteins from P. syringae pv. tomato and E. amylovora has some amino acid residues in common with the HpaG peptide region (Fig. 6A). However, the HpaG peptide shows no homology with the HrpW proteins of X. axonopodis pv. citri or X. campestris pv. campestris (data not shown). The HpaG peptide sequence has 78 and 74% homology with amino acid residues K72 to M94 of HrpW of P. syringae pv. tomato and residues Q36 to Q62 of HrpW of E. amylovora, respectively (Fig. 6A). Computer-based secondary structure analysis revealed that the two HrpW regions are predicted to have an α-helical region similar to the putative α-helix of the HpaG peptide (Fig. 6B). The leucine-rich motif found in the HpaG peptide (LLXXLIXXLL) was identified in the corresponding region of two HrpW proteins (Fig. 6B).
FIG. 6.
(A) Alignment of the HpaG peptide region (HpaG amino acids 31 to 53) with the corresponding region of the HrpW proteins from P. syringae pv. tomato (amino acids K72 to M94) and E. amylovora (amino acids Q36 to Q62). The alignment was produced by using the CLUSTAL X program. Asterisks (*), colons (:), and periods (.) indicate identical amino acid residues, conserved residues, and similar residues, respectively. (B) Computer-predicted secondary structures in the HpaG amino acid 31 to 53 region, amino acids 72 to 94 from HrpW of P. syringae pv. tomato, and amino acids 36 to 62 from HrpW of E. amylovora. Predicted α-helices are represented by gray rectangles. The Leu and Ile repeat regions are indicated by bold rectangles.
Gain-of-function mutants of XopA.
The harpin XopA does not elicit HR in tobacco plants. Since XopA lacks 16 amino acid residues that correspond to the region between positions 59 to 74 in HpaG, we engineered XopA as a chimeric protein to make the protein active. First, we constructed a chimeric protein, XopA-HpaG, by exchanging the C terminus of XopA with the C terminus of HpaG. This was performed by coligating the 120-bp NdeI-PvuII fragment of pTXOPA and the 73-bp PvuII-BamHI fragment of pTHG67 between the NdeI and BamHI sites of pET14b. The resulting plasmid, pTXH2, was sequenced to confirm the correct construction and then transformed into E. coli strain BL21(DE3). The XopA-HpaGΔC66 fusion protein contains the XopA domain from the start codon to the 41st amino acid residue and the HpaG domain from the 41st to the 67th amino acid residues. The resulting XopA-HpaG fusion protein does not contain the D40 residue of HpaG and did not exhibit elicitor activity (Fig. 7).
FIG. 7.
Comparison of the HR elicitor activity in tobacco leaves of the XopA mutant proteins and the XopA-HpaG swap protein. The proteins were injected into tobacco leaves at concentrations of 10, 5, 2.5, 1, or 0.5 μM in 20 mM Tris-HCl (pH 8.0). Labeling: 1, HpaG; 2, XopA; 3, XopA(F48L); 4, XopA(M52L); 5, XopA(F48L/M52L); 6, XopA-HpaG swap protein; buffer, 20 mM Tris-HCl (pH 8.0). Tobacco (N. tabacum cv. Samsun NN) leaves were photographed 24 h after injection.
Based on an alignment of the deduced amino acid sequences of XopA and HpaG, we mutagenized the xopA gene by site-directed mutagenesis. The F and M residues at positions 48 and 52 of XopA, respectively, differ from the leucine residues in these positions in HpaG. Therefore, the codons for residues F48 and M52 of XopA were mutated into leucine codons, both individually and in one clone containing both mutations (Fig. 8). XopA(M52L) was not able to elicit HR, but XopA(F48L) and XopA(F48L/M52L) elicited HR on tobacco leaves at 2.5 and 1 μM, respectively (Fig. 7).
FIG. 8.
Multiple alignment of the HpaG peptide region and the corresponding regions from XopA, the XopA derivatives, and Hpa1 from X. oryzae pv. oryzae. In the sequences of the HpaG homologs, the amino acid residues that differ from the HpaG peptide are indicated by shaded boxes. The Leu and Ile repeat motifs are indicated by bold rectangles. The relative HR activity of each protein and the minimum concentrations that elicit HR in tobacco leaves are shown in the column to the right.
To determine whether the inability of XopA to induce HR in tobacco plants is unique to X. campestris pv. vesicatoria strain 82-8 (race 1), we isolated and sequenced xopA genes from strains that represent races 2 and 3, X. campestris pv. vesicatoria strain E-3 (race 2) and LS833 (race 3). The DNA sequences of the isolated xopA genes were identical to that of X. campestris pv. vesicatoria strain 82-8 (race 1) (data not shown).
DISCUSSION
In this study, we investigated the critical amino acid residues that determine the HR elicitor activity of HpaG and XopA, by using random and site-directed mutagenesis. Initially, we found that XopA cannot elicit HR in tobacco leaves, even though the amino acid sequence of XopA is very similar to that of HpaG (15). The major difference between HpaG and XopA is that XopA lacks 16 amino acid residues that correspond to the region between positions 59 and 74 in HpaG (15). These sixteen amino acid residues, QGQGQGQGGDSGGQGG, are mainly glycine and glutamine residues. Of the 28 glycine residues in the HpaG protein, 9 are in this region. A prominent feature of most harpins is a high glycine content: 21% in HpaG, 22.6% in HrpN of E. amylovora, 13.2% in HrpZ of P. syringae pv. syringae, 13.9% in HrpW of E. amylovora, and 20.9% in PopA of R. solanacearum (4, 12, 14, 15, 27). However, XopA has a relatively low content of glycine residues (8% glycine), owing to the lack of the 16-amino-acid region. This observation suggested that the glycine-rich 16-amino-acid region of HpaG has an important role in the elicitor activity of this protein. However, HpaG deletion analysis showed that the 16 amino acid residues are dispensable for the elicitor activity and that the N-terminal 52 amino acids of HpaG are sufficient to maintain the elicitor activity and heat stability of the protein. This indicates that the inability of XopA to induce HR is not related to the lack of the 16 amino acid residues and that the residues that are important for the elicitor activity reside outside of this region. The fact that there are only four glycine residues in the N-terminal 52-amino-acid region suggests that the glycine richness of harpins is not important for the elicitor activity but that it may have other roles.
Site-directed mutagenesis analysis of the N-terminal 52 amino acid residues showed that amino acid substitutions that affect elicitor activity are located within the 19-amino-acid region SEKQLDQLLTQLIMALLQQ. These 19 amino acids clearly play important roles in inducing HR because the synthetic HpaG peptide, composed of 23 amino acid residues encompassing the 19 amino acid residues, possesses full elicitor activity. The amino acid composition of the peptide region contains unexpected features. In contrast to the other harpins, which are glycine-rich, the HpaG peptide is rich in glutamine and leucine. The HpaG peptide has only one glycine residue (4.3%), but six glutamine (26%) and six leucine (26%) residues. Since the five HpaG mutants with affected elicitor activity have substitutions at L39, L42, L43, L46, and L50, and one has a substitution at Q45, the leucine residues in the 23-amino-acid sequence probably have an important role in HR elicitation.
Interestingly, the 19 amino acid residues that affect elicitor activity are predicted to have an α-helical structure. There appears to be an important correlation between the α-helical feature of HpaG and its HR elicitor activity. Alfano et al. noted that HrpZ has nine probable α-helices; however, the relationship between the α-helices and the HR elicitor activity was not examined (2). The predicted α-helix that is conserved between HpaG and the HrpW harpins of Pseudomonas and Erwinia species suggests that the feature constitutes a key functional domain in harpin elicitor activity. However, there are no clues as to the specific mechanisms of the predicted α-helix in HR induction.
Comparing the two Xanthomonas HrpW proteins with the HrpW harpins of Pseudomonas and Erwinia species, the two Xanthomonas HrpW proteins were not reported to be HR elicitors. Therefore, we cannot consider the HrpW proteins of Xanthomonas species as harpins, based on the sequence homology. Comparison of the four HrpW proteins shows that the Xanthomonas HrpW proteins are considerably smaller than the Pseudomonas and Erwinia HrpW harpins. Moreover, the Xanthomonas HrpW proteins have high homology with the pectate lyase domains of the Pseudomonas and Erwinia HrpW harpins but not with the harpin domains of the HrpW harpins. The two Xanthomonas HrpW proteins also have a predicted α-helix in the N-terminal region, but we did not find any significant homology with the HpaG peptide sequence (data not shown).
The characteristics of other peptide elicitors were examined by way of comparison with those of HpaG. A 13-amino-acid oligopeptide, derived from the Phytophthora megasperma glycoprotein, was shown to be both necessary and sufficient for elicitor activity, as measured by phytoalexin accumulation in parsley (21); in addition, the AVR9 28-amino-acid oligopeptide of Cladosporium fulvum is able to induce a hypersensitive necrosis in tomato (26). However, there are no similarities in the amino acid sequences or predicted structures of these peptide elicitors with those of HpaG. This indicates that the importance of the predicted α-helix in harpin elicitor activity might be limited to bacterial pathogens.
Since the first report of a harpin, HrpN of E. amylovora, several HrpN homologs have been identified from E. carotovora, E. chrysanthemi, and Pantoea stewartii subsp. stewartii (1, 6, 19), and HrpZ homologs have been identified from P. syringae pv. syringae, P. syringae pv. glycinea, and P. syringae pv. tomato (12, 22). All of the HrpN and HrpZ homologs are true harpins because they can elicit HR. However, the HpaG homologs from Xanthomonas species differ in their abilities to elicit HR (15). Sequence differences in the critical 23 amino acid residues of the HpaG peptide and the corresponding regions of other HpaG homologs probably contribute to the differences in HR elicitation ability. Comparison of HpaG with Hpa1 from X. oryzae pv. oryzae reveals that T44 of HpaG is changed to a cysteine residue in Hpa1 and that M48 and Q53 of HpaG are both serine residues in the Hpa1 sequence. Likewise, in XopA, L46 and M48 of HpaG are changed to phenylalanine residues, and A49 and L50 of HpaG are changed to serine and methionine residues (Fig. 8). Therefore, it is reasonable to propose that changes in a few amino acid residues lead to different levels of elicitor activity. This idea is consistent with the results of the mutagenesis of XopA.
In spite of their different abilities to induce HR in nonhost plants, HpaG, Hpa1, and XopA are necessary for full virulence in their respective host plants (15, 20, 28), indicating that the HpaG homologs have common roles, which contribute to compatible interactions with host plants. The C-terminal region of HpaG is not essential for HR elicitation ability; however, this region in HpaG homologs may have an unknown role in disease progress within the host plant.
Acknowledgments
This study was supported by grant CG1412 from the Crop Functional Genomics Center of the 21st Century Frontier R&D Program of the Ministry of Science and Technology of the Republic of Korea. J.-G.K., E.J., and J.O. are recipients of graduate fellowships from the Ministry of Education as part of the Brain Korea 21 Project.
REFERENCES
- 1.Ahmad, M., D. R. Majerczak, S. Pike, M. E. Hoyos, A. Novacky, and D. L. Coplin. 2001. Biological activity of harpin produced by Pantoea stewartii subsp. stewartii. Mol. Plant-Microbe Interact. 14:1223-1234. [DOI] [PubMed] [Google Scholar]
- 2.Alfano, J. R., D. W. Bauer, T. M. Milos, and A. Collmer. 1996. Analysis of the role of the Pseudomonas syringae pv. syringae HrpZ harpin in elicitation of the hypersensitive response in tobacco using functionally non-polar hrpZ deletion mutations, truncated HrpZ fragments, and hrmA mutations. Mol. Microbiol. 19:715-728. [DOI] [PubMed] [Google Scholar]
- 3.Alfano, J. R., A. O. Charkowski, W. L. Deng, J. L. Badel, T. Petnicki-Ocwieja, K. van Dijk, and A. Collmer. 2000. The Pseudomonas syringae Hrp pathogenicity island has a tripartite mosaic structure composed of a cluster of type III secretion genes bounded by exchangeable effector and conserved effector loci that contribute to parasitic fitness and pathogenicity in plants. Proc. Natl. Acad. Sci. USA 97:4856-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arlat, M., F. Van Gijsegem, J. C. Huet, J. C. Pernollet, and C. A. Boucher. 1994. PopA1, a protein which induces a hypersensitivity-like response on specific Petunia genotypes, is secreted via the Hrp pathway of Pseudomonas solanacearum. EMBO J. 13:543-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barettino, D., M. Feigenbutz, R. Valcárcel, and H. G. Stunnenberg. 1994. Improved method for PCR-mediated site-directed mutagenesis. Nucleic Acids Res. 22:541-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bauer, D. W., Z.-M. Wei, S. V. Beer, and A. Collmer. 1995. Erwinia chrysanthemi HarpinEch: an elicitor of the hypersensitive response that contributes to soft-rot pathogenesis. Mol. Plant-Microbe Interact. 8:484-491. [DOI] [PubMed] [Google Scholar]
- 7.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 8.Charkowski, A. O., J. R. Alfano, G. Preston, J. Yuan, S. Y. He, and A. Collmer. 1998. The Pseudomonas syringae pv. tomato HrpW protein has domains similar to harpins and pectate lyases and can elicit the plant hypersensitive response and bind to pectate. J. Bacteriol. 180:5211-5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Combet, C., C. Blanchet, C. Geourjon, and G. Deleage. 2000. NPS@: network protein sequence analysis. Trends Biochem. Sci. 25:147-150. [DOI] [PubMed] [Google Scholar]
- 10.Galán, J. E., and A. Collmer. 1999. Type III secretion machines: bacterial devices for protein delivery into host cells. Science 284:1322-1328. [DOI] [PubMed] [Google Scholar]
- 11.Goodman, R. N., and A. J. Novacky. 1994. The hypersensitive reactions in plants to pathogens. APS Press, St. Paul, Minn.
- 12.He, S. Y., H.-C. Huang, and A. Collmer. 1993. Pseudomonas syringae pv. syringae HarpinPss: a protein that is secreted via the Hrp pathway and elicits the hypersensitive response in plants. Cell 73:1255-1266. [DOI] [PubMed] [Google Scholar]
- 13.Hoyos, M. E., C. M. Stanley, S. Y. He, S. Pike, X.-A. Pu, and A. Novacky. 1996. The interaction of HarpinPss, with plant cell walls. Mol. Plant-Microbe Interact. 9:608-616. [Google Scholar]
- 14.Kim, J. F., and S. V. Beer. 1998. HrpW of Erwinia amylovora, a new harpin that contains a domain homologous to pectate lyases of a distinct class. J. Bacteriol. 180:5203-5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim, J.-G., B. K. Park, C.-H. Yoo, E. Jeon, J. Oh, and I. Hwang. 2003. Characterization of the Xanthomonas axonopodis pv. glycines Hrp pathogenicity island. J. Bacteriol. 185:3155-3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee, J., B. Klüsener, G. Tsiamis, C. Stevens, C. Neyt, A. P. Tampakaki, N. J. Panopoulos, J. Nöller, E. W. Weiler, G. R. Cornelis, J. W. Mansfield, and T. Nürnberger. 2001. HrpZPsph from the plant pathogen Pseudomonas syringae pv. phaseolicola binds to lipid bilayers and forms an ion-conducting pore in vitro. Proc. Natl. Acad. Sci. USA 98:289-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindgren, P. B. 1997. The role of hrp genes during plant-bacterial interactions. Annu. Rev. Phytopathol. 35:129-152. [DOI] [PubMed] [Google Scholar]
- 18.Maloy, S. R. 1990. Hydroxylamine mutagenesis, p. 50-54. In S. R. Maloy (ed.), Experimental techniques in bacterial genetics. Jones & Bartlett, Boston, Mass.
- 19.Mukherjee, A., Y. Cui, Y. Liu, and A. K. Chatterjee. 1997. Molecular characterization and expression of the Erwinia carotovora hrpNEcc gene, which encodes an elicitor of the hypersensitive reaction. Mol. Plant-Microbe Interact. 10:462-471. [DOI] [PubMed] [Google Scholar]
- 20.Noël, L., F. Thieme, D. Nennstiel, and U. Bonas. 2002. Two novel type III-secreted proteins of Xanthomonas campestris pv. vesicatoria are encoded within the hrp pathogenicity island. J. Bacteriol. 184:1340-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nürnberger, T., D. Nennstiel, T. Jabs, W. R. Sacks, K. Hahlbrock, and D. Scheel. 1994. High affinity binding of a fungal oligopeptide elicitor to parsley plasma membranes triggers multiple defense responses. Cell 78:449-460. [DOI] [PubMed] [Google Scholar]
- 22.Preston, G., H.-C. Huang, S. Y. He, and A. Collmer. 1995. The HrpZ proteins of Pseudomonas syringae pvs. syringae, glycinea, and tomato are encoded by an operon containing Yersinia ysc homologs and elicit the hypersensitive response in tomato but not soybean. Mol. Plant-Microbe Interact. 8:717-732. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Tabor, S., and C. C. Richardson. 1985. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. USA 82:1074-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tarun, A. S., J. S. Lee, and A. Theologis. 1998. Random mutagenesis of 1-aminocyclopropane-1-carboxylate synthase: a key enzyme in ethylene biosynthesis. Proc. Natl. Acad. Sci. USA 95:9796-9801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Kan, J. A., G. F. van den Ackerveken, and P. J. de Wit. 1991. Cloning and characterization of cDNA of avirulence gene avr9 of the fungal pathogen Cladosporium fulvum, causal agent of tomato leaf mold. Mol. Plant-Microbe Interact. 4:52-59. [DOI] [PubMed] [Google Scholar]
- 27.Wei, Z.-M., R. J. Laby, C. H. Zumoff, D. W. Bauer, S. Y. He, A. Collmer, and S. V. Beer. 1992. Harpin, elicitor of the hypersensitive response produced by the plant pathogen Erwinia amylovora. Science 257:85-88. [DOI] [PubMed] [Google Scholar]
- 28.Zhu, W., M. M. Magbanua, and F. F. White. 2000. Identification of two novel hrp-associated genes in the hrp gene cluster of Xanthomonas oryzae pv. oryzae. J. Bacteriol. 182:1844-1853. [DOI] [PMC free article] [PubMed] [Google Scholar]