Abstract
In Escherichia coli, cell division is mediated by the concerted action of about 12 proteins that assemble at the division site to presumably form a complex called the divisome. Among these essential division proteins, the multimodular class B penicillin-binding protein 3 (PBP3), which is specifically involved in septal peptidoglycan synthesis, consists of a short intracellular M1-R23 peptide fused to a F24-L39 membrane anchor that is linked via a G40-S70 peptide to an R71-I236 noncatalytic module itself linked to a D237-V577 catalytic penicillin-binding module. On the basis of localization analyses of PBP3 mutants fused to green fluorescent protein by fluorescence microscopy, it appears that the first 56 amino acid residues of PBP3 containing the membrane anchor and the G40-E56 peptide contain the structural determinants required to target the protein to the cell division site and that none of the putative protein interaction sites present in the noncatalytic module are essential for the positioning of the protein to the division site. Based on the effects of increasing production of FtsQ or FtsW on the division of cells expressing PBP3 mutants, it is suggested that these proteins could interact. We postulate that FtsQ could play a role in regulating the assembly of these division proteins at the division site and the activity of the peptidoglycan assembly machineries within the divisome.
In Escherichia coli, cell division relies on the concerted action of at least 12 proteins, FtsZ, FtsA, ZipA, ZapA, FtsK, FtsQ, FtsL, FtsB, FtsW, penicillin-binding protein 3 (PBP3) (also called FtsI), FtsN, and AmiC. These proteins assemble at the division site in a specific order to presumably form a dynamic membrane-associated supramolecular complex called the divisome (1, 5, 7, 12, 15, 25). The earliest step is the polymerization of FtsZ, a tubulin-like protein, into an intracellular ring-shaped structure. This cytoskeletal ring recruits FtsA, an actin-like protein, ZapA, and ZipA (2, 17, 23). The other proteins, FtsK, FtsQ, FtsL with FtsB, FtsW, PBP3, FtsN, and AmiC, subsequently join the ring.
The functions of most of these proteins are not known. How the divisome works in term of protein-protein interaction is poorly understood. Until now, the only known direct interactions are those between FtsZ and FtsA or ZipA (17, 26, 29). The bitopic proteins FtsB and FtsL seem to interact in a coiled-coil structure through their periplasmic domain (6). Other protein-protein interactions should be involved in the functioning of the divisome.
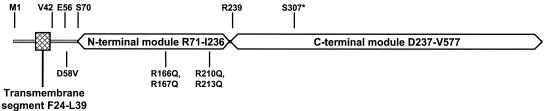
The multimodular class B PBP3 specifically catalyzes peptide cross-bridges of the septal cell wall peptidoglycan during cell division. It consists of a short intracellular M1-R23 peptide fused to an F24-L39 membrane anchor that is linked via a G40-S70 peptide to an R71-I236 noncatalytic module, itself linked to a D237-V577 catalytic penicillin-binding module. It has been proposed that the activity of the transpeptidase module of PBP3 is regulated by the interaction of its N-terminal noncatalytic module with other cell division proteins (11, 24). Previous experiments allowed identification of three peptide segments in the noncatalytic module of PBP3 that have protein-protein interaction potentials and specific functions. The G40-S70 sequence and the membrane anchor-containing module appear to play an important role in the proper insertion of the protein within the divisome at the division site (24, 30). It might interact with FtsW, which is essential for the recruitment of PBP3 at the cell septation site (25). The H160-G172 segment, located at the intermodule junction, seems to be involved in intramolecular interactions and plays an important role in the conformation of the protein. The E206-V217 segment, which is exposed at the surface of the noncatalytic module of PBP3, plays an important role in cell septation, presumably by interacting with other components of the divisome. FtsQ, FtsL, and FtsW are plausible partners (24).
In order to assess the role of the G40-S70 and E206-V217 segments and their interaction with other cell division proteins, the localization of PBP3 modified in these segments was analyzed. A search for suppressors of the dominant-negative effect due to mutations in the E206-V217 segment was carried out by analyzing the effect of overproduction of FtsQ, FtsL, or FtsW on E. coli cells producing a PBP3 mutant altered in this segment. The results reported hereafter show that the first 56 amino acid residues of PBP3 contain the structural determinants required for targeting of PBP3 at the cell division site and that none of the putative protein interaction sites in the noncatalytic module is essential for the positioning of the protein to the division site. FtsQ could play a role in regulating the assembly of the division proteins FtsL, FtsB, FtsW, and PBP3.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Strains were described in Table 1. The bacteria were grown in Luria-Bertani (LB) rich medium and minimal medium [6.33 g of K2HPO4 · 3H2O, 2.95 g of KH2PO4, 1.05 g of (NH4)2SO4, 0.10 g of MgSO4 · 7H2O, 0.28 mg of FeSO4 · 7H2O, 7.1 mg of Ca(NO3)2 · 4H2O, 4 mg of thiamine, 4 g of glucose, 50 μg of lysine per liter (pH 7.0)]. Ampicillin (100 μg ml−1), chloramphenicol (30 μg ml−1), and kanamycin (40 μg ml−1) were added when appropriate.
TABLE 1.
Strains, plasmids, and oligonucleotides
| Strain or DNA | Relevant genetic markers and featuresa | Source or reference |
|---|---|---|
| Strains | ||
| LMC500 | F−araD139 Δ(argF-lac)U169 deoC1 flbB5301 ptsF25 rbsR ΔrelA1 rpsL150 thi lysA1 | 28 |
| EC548 | F− ΔlacX74 galE galK thi rpsL ΔphoA(PvuII) ftsI::TnphoA173ΔIS50R(Kanr)/pDSW262 | 30 |
| TOP10 F′ | F′ {lacIq Tn10 (Tetr)} mcrA Δ(mrr-hsdRMS-mcrBC) φ80 lacZM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL(Strr) endA1 nupG | Invitrogen |
| Plasmids | ||
| pDSW234 | gfp-ftsI (WT) under control of a weakened trc promoter, Ampr, ftsI cloned in EcoRI-HindIII | 30 |
| pDSW262 | pBAD33-ftsI, arabinose regulation, Cmr | 30 |
| pHR275 | pINII-B2 carrying a truncated ftsI coding for M1-R239 peptide | 24 |
| pHR458 | pGB2 derivative coding D58V PBP3 mutant | 24 |
| pUCBM20/ftsI | Ampr, codes for wild-type or mutant PBP3 | 24 |
| pMCL210 | lac promoter, lacZα, Cmr, P15A replication origin | 27 |
| pDML995 | B. licheniformis 749 bla divergon on a pMK4 | 13 |
| pNB2 | ftsQ cloned in pBTacl vector, Ampr, Tetr | 4 |
| pET-28a | Six-His fusion proteins expression vector, T7 promoter, Kanr | Novagen |
| pBAD/His A | Six-His fusion proteins expression vector, PBAD promoter, Ampr | Invitrogen |
| pDML2480 | pDSW234 (D58V) | This study |
| pDML2481 | pDSW234 (R166Q, R167Q) | This study |
| pDML2482 | pDSW234 (R210Q, R213Q) | This study |
| pDML2483 | pDSW234 (D58V, R210Q, R213Q) | This study |
| pDML2484 | pDSW234 (K2-R239) | This study |
| pDML2485 | pDSW234 (K2-S70) | This study |
| pDML2486 | pDSW234 (K2-E56) | This study |
| pDML2487 | pDSW234 (K2-V42) | This study |
| pDML2488 | pDSW234 (K2-V42-BlaR) | This study |
| pDML2489 | pDSW234 (GFP) | This study |
| pDML2407 | pMCL210-ftsW | Pastoret et al.b |
| pDML2490 | pMCL210-ftsL | This study |
| pDML2491 | pMCL210-ftsQ | This study |
| pDML2494 | pET-28a/his-ftsI (WT) | This study |
| pDML2495 | pBAD/his-ftsI (WT) | This study |
| pDML2496 | pBAD/his-ftsI (R166Q, R167Q) | This study |
| pDML2497 | pBAD/his-ftsI (R210Q, R213Q) | This study |
| pDML2498 | pBAD/his-ftsI (D58V, R210Q, R213Q) | This study |
| Oligonucleotides | ||
| gfp-ftsI | 5′-CACGAATTCAACAACAACAAAGCAGCGGCGAAAACGC-3′ | |
| ftsI-NruI | 5′-GTTCAGTTCGCGATAAACC-3′ | |
| dupV42 | 5′-CGCGTATAAtaataaggatcca-3′ | |
| 3′-ATATTattattcctaggttcga-5′ | ||
| dupE56 | 5′-CGCGTAGCGTGGTTACAAGTTATCTCCCCGGATATGCTGGTGAAAGAGTAAta-3′ | |
| 3′-ATCGCACCAATGTTCAATAGAGGGGCCTATACGACCACTTTCTCATTattcga-5′ | ||
| MluI-blaR | 5′-CGAACGCGTATCTGTGTCTATATTGGCCATGCAAAAAG-3′ | |
| BlaR-HindIII | 5′-CGAAAGCTTATCGGGAAACGGAGGGATAAATCC-3′ | |
| BamHI-ftsL | 5′-CCGGATCCTTAGaggacgaatgcATGATCAGC-3′ | |
| ftsL-HindIII | 5′-cgaagcttgcgtcgcgtttatccTTATTTTTGC-3′ | |
| XbaI-ftsQ | 5′-GCTCTAGAATAAggcggactaatATGTCGCAGGC-3′ | |
| ftsQ-HindIII | 5′-cgaagcttgaTCATTGTTGTTCTGCCTGTGCC-3′ | |
| NheI-ftsIfwd | 5′-caaaaataaggataaacggctagcATGAAAGCAGCG-3′ | |
| NheI-ftsIrev | 5′-GCTGCTTTCATgctagccgtttatccttatttttgc-3′ | |
| ftsID58V | 5′-GGTGAAAGAGGGCGTCATGCGTTCTCTTCGCG-3′ | |
| ftsIcplD58V | 5′-CGCGAAGAGAACGCATGACGCCCTCTTTCACC-3′ |
For oligonucleotides, coding sequences are in capital letters, restriction sites are in italic, stop codons are bold, start codons are underlined, and mutations are bold and underlined.
S. Pastoret, C. Fraipont, T. den Blaauwen, B. Wolf, M. E. G. Aarsman, A. Piette, A. Thomas, R. Brasseur, and M. Nguyen-Distèche, submitted for publication.
E. coli LMC500 cells expressing green fluorescent protein (GFP) fusion proteins were grown in glucose minimal medium at 28°C without isopropyl-β-d-thiogalactopyranoside (IPTG) for more than 20 mass doublings before being harvested at an optical density at 450 nm (OD450) of 0.1.
E. coli EC548 cells expressing GFP fusion proteins were grown for two mass doublings in LB supplemented with 0.2% l-arabinose at 37°C from an overnight culture grown in the same conditions. They were then diluted 400-fold to an OD600 of 0.0025 in LB containing 0.2% glucose at 37°C and harvested after seven mass doubling periods (OD600 of ≈0.4) when long filaments were observed.
E. coli TOP10 F′ cells coexpressing PBP3 mutants and FtsL, FtsQ, or FtsW were grown at 37°C in LB medium inoculated with an overnight culture (inoculum 2%). When the OD600 reached a value of 0.3, 0.1 mM IPTG was added, and the culture was maintained for 30 min at 37°C before the addition of 0.005 or 0.01% l-arabinose (arabinose induction was omitted when GFP-PBP3 constructs controlled by a weakened trc promoter were used). Cells were harvested 3 h after induction.
Molecular biological procedures.
Plasmids and oligonucleotides are described in Table 1. Oligonucleotides were purchased from Amersham Biosciences and Eurogentec. All constructs were sequenced to verify their integrity, using an ALFexpress DNA sequencer (Amersham Biosciences).
Recombinant plasmids.
Construction of GFP fusions. Plasmid pDSW234 encoded the wild-type PBP3 fused to the C-terminal end of GFP. In this construct, the initiating methionine of PBP3 is absent from the fusion (30). The SacII-NruI fragment of pDSW234 was exchanged with the corresponding fragment of pUCBM20/ftsI(R166Q, R167Q), pUCBM20/ftsI(R210Q, R213Q), and pHR275 carrying the truncated ftsI gene encoding the M1-R239 PBP3 (24). The resulting plasmids, pDML2481, pDML2482, and pDML2484, encoded GFP-PBP3(R166Q, R167Q), GFP-PBP3(R210Q, R213Q), and GFP-PBP3(K2-R239), respectively, and the gene fusion was under the control of a weakened trc promoter (30).
The ftsI gene encoding the PBP3(D58V) mutant was amplified by PCR using pHR458 as a template, the gfp-ftsI primer bearing an EcoRI site at its 5′ end, and the ftsI-NruI primer. The PCR product was digested by EcoRI and NruI and inserted in the corresponding sites of pDSW234, digested by the same enzymes. The resulting plasmid, pDML2480, encoded GFP-PBP3(D58V). Plasmid pDML2483 was obtained by replacing the SacII-NruI fragment of pDML2480 with the corresponding fragment of pUCBM20/ftsI(R210Q, R213Q) and encoded the GFP-PBP3(D58V, R210Q, R213Q) triple mutant. To construct pDML2485, which encoded GFP-PBP3(K2-S70), the SacII-HindIII fragment of pDSW234 was excised. The plasmid was treated with T4 DNA polymerase to make blunt ends and then ligated on itself. Because of the construction, the encoded PBP3(K2-S70) had an additional heptapeptide (QLGCFGG) at the carboxy end. To generate GFP-PBP3(K2-E56) and GFP-PBP3(K2-V42), the MluI-HindIII fragment of pDSW234 encoding the R41-S588 sequence of PBP3 was replaced by double-stranded oligonucleotides dupE56 and dupV42 to give rise to pDML2486 and pDML2487, respectively.
To fuse GFP-PBP3(K2-V42) to the penicillin-binding domain of BlaR, MluI and HindIII restriction sites were added, respectively, upstream and downstream of the sequence encoding the carboxy-terminal domain of the Bacillus licheniformis BlaR protein by PCR, using pDML995 as the template and the oligonucleotides MluI-blaR and blaR-HindIII as primers. The resulting PCR product was then digested by MluI and HindIII and cloned in the corresponding sites of pDSW234 to give rise to pDML2488. pDSW234 was digested by EcoRI and HindIII and treated with the mung bean nuclease before being circularized to give pDML2489, which encoded GFP under the control of a weakened trc promoter.
Construction of plasmids coding for PBP3, FtsL, and FtsQ.
An NheI site was first inserted upstream of the ftsI gene by site-directed mutagenesis with the QuikChange kit (Stratagene) using pUCBM20/ftsI as template and the primers NheI-ftsIfwd and NheI-ftsIrev. The PCR product was then restricted with NheI and EcoRI and cloned in the NheI-EcoRI sites of pET-28a to give pDML2494, which encoded the His-tagged PBP3. In order to place this fused gene under the control of the arabinose promoter, the NcoI-EcoRI fragment was excised from pDML2494 and cloned into the NcoI-EcoRI sites of pBAD/His A to create pDML2495. The SacII-PstI fragments containing the double mutations (R166Q R167Q or R210Q R213Q) were excised from pUCBM20/ftsI recombinants and exchanged with the unmodified SacII-PstI fragment of pDML2495 to give rise to pDML2496 and pDML2497, respectively. The PBP3(D58V, R210Q, R213Q) triple mutant was obtained by site-directed mutagenesis (QuikChange kit from Stratagene), using pDML2497 as a template and the primers ftsID58V and ftsIcplD58V. The PCR product was digested with SacII and PstI and exchanged with the unmodified fragment of pDML2495 to create pDML2498.
The ftsL and ftsQ genes were amplified by PCR, using as templates plasmids pUCBM20/ftsI and pNB2, respectively, and the primers BamHI-ftsL and ftsL-HindIII (for ftsL) and XbaI-ftsQ and ftsQ-HindIII (for ftsQ). The PCR products were digested with BamHI and HindIII (for ftsL) and XbaI and HindIII (for ftsQ) and then ligated into the corresponding sites of pMCL210 to give pDML2490 and pDML2491, respectively. In these constructs the ftsL and ftsQ genes were under the control of the lac promoter.
Microscopy and image analysis.
Cells were fixed in culture medium with 2.8% formaldehyde and 0.04% glutaraldehyde for 15 min at room temperature, collected by centrifugation at 4,500 × g for 5 min, washed twice in phosphate-buffered saline (140 mM NaCl, 27 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4 [pH 7.2]), suspended in water, and immobilized on agarose slides as described by Koppelman et al. (20). They were photographed with a Photometrics CoolSNAPfx CCD camera (Roper Scientific) mounted on an Olympus BX60 microscope through a UPlanFI ×100/1.3 oil-immersion objective, both in phase contrast and fluorescence (with U-MNB filter cube, 470- to 490-nm band-pass excitation filter, >515-nm long-pass emission filter). Images were analyzed by using the public-domain software Object-Image 2.08 by N. Vischer (University of Amsterdam http://simon.bio.uva.nl/object-image.html), based on NIHImage by W. Rasband.
Membrane preparation and fluorescent labeling of penicillin-binding protein.
Cells were harvested by centrifugation and suspended in a solution containing 20 mM Tris-HCl (pH 8), 5 mM EDTA, and 100 μg of lysozyme ml−1 on ice for 30 min. MgCl2 (final concentration, 15 mM), Benzonase (4 U ml−1; Merck), and 10−4 M phenylmethylsulfonyl fluoride were then added, and the cell suspension was frozen and thawed three times. Membranes were spun down at 30,000 × g for 30 min and resuspended in a solution containing 10 mM Tris-HCl (pH 8), 500 mM NaCl, 10% glycerol, and 10% ethylene glycol.
Membranes were diluted in 20 mM phosphate (pH 7.6)-150 mM NaCl and incubated for 10 min at 30°C with 5 × 10−5 M β-iodopenicillanic acid to inactivate any residual β-lactamase. 5′-Fluorescein-ampicillin was then added to a final concentration of 5 × 10−5 M, and membranes were further incubated for 25 min at 30°C (22). Proteins were then boiled for 5 min in sodium dodecyl sulfate (SDS) sample buffer to denature GFP.
Separation and detection of proteins.
Proteins were solubilized and separated by standard SDS-polyacrylamide gel electrophoresis (PAGE) methods (21). For immunological detection of GFP, proteins were electroblotted to a polyvinylidene difluoride (PVDF) membrane and revealed with mouse anti-GFP monoclonal antibodies and horseradish peroxidase-coupled goat anti-mouse immunoglobulin G antibodies. Detection was performed using the ECL chemiluminescence reagent (Amersham Biosciences).
Fluorescence detection after SDS-PAGE was done with a Molecular Imager FX (Bio-Rad Laboratories) instrument using the parameter set designed for fluorescein isothiocyanate detection (excitation by 488-nm Ar-ion laser, 515- to 545-nm band-pass emission filter). For GFP fluorescence, samples were not boiled but solubilized for 30 min at 37°C in SDS sample buffer. Quantification was performed with the Quantity One 4.1 software (Bio-Rad Laboratories).
RESULTS
Localization of PBP3 altered in putative interaction sites.
The characterization of PBP3 mutants in the noncatalytic module of the protein (Table 2; Fig. 1) allowed identification of amphiphilic peptide segments that appeared to be involved in protein-protein interaction and to perform specific functions (24). The simultaneous exchange of Arg 210 and Arg 213 with Gln in the E206-V217 segment leads to a PBP3 that still binds penicillin and that has a dominant-negative phenotype [formation of filaments in the ftsI(Ts) strain or lengthening of ftsI+ cells] similar to that of the active-site serine S307C/A mutant, indicating that PBP3 mutants compete with the resident PBP3, preventing it from being functional (3, 24). This dominant-negative effect is reversed by replacement of the Asp58 with Val in the G57-Q66 segment of the PBP3(R210Q, R213Q) mutant and PBP3(S307C/A) mutant. A similar effect was noted by substituting the uncleavable lipoprotein signal peptide for the M1-L39 membrane anchor of the PBP3(S307C) mutant (19). Alteration of the membrane anchor or G57-Q66 segment would thus prevent these PBP3 mutants from competing with the wild-type protein. The modification of Arg 166 and Arg 167 into Gln in the H160-G172 segment at the intermodule junction gave rise to a protein that lacked cell division activity, lacked penicillin binding activity, and showed no dominant-negative effect (24). It was suggested that the PBP3(R166Q, R167Q) mutant did not compete with wild-type PBP3, probably because the G57-Q66 segment of this PBP3 mutant is no longer capable of targeting the protein to the division site.
TABLE 2.
Properties of E. coli PBP3 variantsa
| Description of GFP-PBP3 construct | Cell septation activity | Penicillin binding | Dominant-negative effect |
ftsI+ strain LMC500
|
PBP3-depleted strain EC548
|
||
|---|---|---|---|---|---|---|---|
| Length (μm) (no. of measured cells) | % of cells with fluorescent ring | Length (μm) (no. of measured cells) | % of cells with fluorescent ring | ||||
| No GFP fusion | 2.7 ± 0.6 (467) | ||||||
| WTb | +c | +c | −c | 3.6 ± 1.0 (583) | 64 | 4.6 ± 1.1 (125) | 77 |
| R210Q, R213Q | −c | +c | +c | 5.2 ± 2.4 (353) | 80 | 41.1 ± 20.3 (46) | 70 |
| D58V, R210Q, R213Q | −c | NDd | −c | 4.8 ± 1.8 (532) | 75 | 53.7 ± 14.9 (20) | 75 |
| D58V | −c | +c | −c | 4.5 ± 1.2 (528) | 60 | 38.7 ± 19.1 (54) | 94 |
| R166Q, R167Q | −c | −c | −c | 3.2 ± 0.7 (293) | 49 | 18.4 ± 12.0 (231) | 65 |
| K2-R239 | −c | −c | +c | 2.9 ± 0.7 (315) | 4 | 25.4 ± 12.4 (82) | 88 |
| K2-S70 | − | − | ND | 2.6 ± 0.5 (266) | 6 | 21.9 ± 11.6 (73) | 80 |
| K2-E56 | − | − | ND | 2.4 ± 0.6 (351) | 0 | 24.1 ± 13.6 (152) | 76 |
| K2-V42 | − | − | ND | 2.3 ± 0.5 (202) | 0 | 35.4 ± 13.1 (121) | 24 |
| K2-V42-BlaR | − | +e | ND | 2.3 ± 0.5 (335) | 0 | 21.5 ± 13.0 (107) | 20 |
| GFP alone | − | − | − | 2.7 ± 0.6 (271) | 0 | 47.5 ± 18.0 (20) | 0 |
E. coli strain ftsI+ LMC500 was grown in glucose minimal medium at 28°C and strain ftsI-null EC548 depleted of endogenous PBP3 was grown in LB medium at 37°C.
Wild type.
See reference 24.
ND, not determined.
Due to the activity of BlaR.
FIG. 1.
Schematic view of PBP3 showing the positions of the relevant amino acid residues.
In order to determine whether mutations in segments that have protein-protein interaction potentials affect PBP3 localization, PBP3 mutants were fused to the C terminus of GFP. The protein fusions were placed under the control of a weakened trc promoter (30). Plasmids pDML2480, pDML2481, pDML2482, and pDML2483 (Table 1) encoding the GFP-PBP3(D58V), GFP-PBP3(R166Q, R167Q), GFP-PBP3(R210Q, R213Q), and GFP-PBP3(D58V, R210Q, R213Q) mutants, respectively, were used to transform the wild-type ftsI+ strain LMC500 and the conditional ftsI-null strain EC548. The ftsI+ transformants were grown under conditions close to steady state in glucose minimal medium at 28°C, whereas the ftsI-null transformants were grown at 37°C in LB medium in the presence of glucose to repress the expression of native PBP3, which is under the control of the PBAD promoter (see Materials and Methods).
As derived from Western blotting of the plasma membrane isolated from LMC500 transformants, the amount of the GFP-PBP3 variants was estimated at about four times that of the natural chromosomally encoded PBP3 (results not shown).
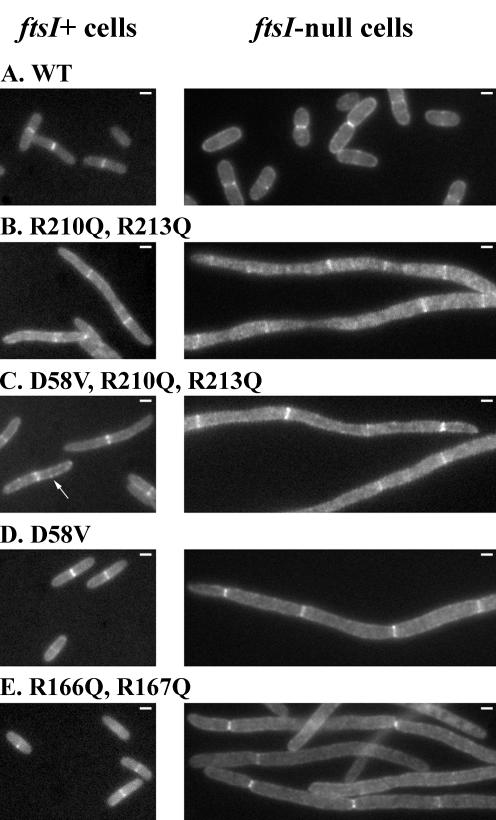
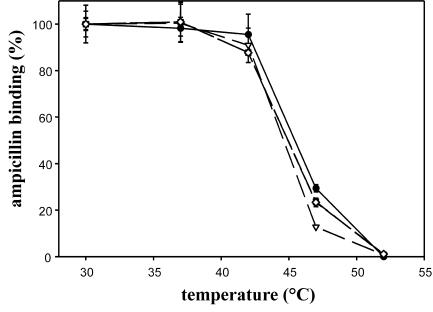
As shown in Fig. 2, each of the PBP3 mutants localized at the division site of both strains. In LMC500 transformants, PBP3 fluorescent rings were observed in 49 to 80% of the cells, and the average cell length ranged from 3.2 to 5.2 μm, depending on the mutant (Table 2; Fig. 1). The increase of the cell length reflects a delay in the division process. This was also observed with transformants producing the native GFP-PBP3 fusion. A number of cells expressing GFP-PBP3(D58V) showed some fluorescent aggregates in their membranes (Fig. 2C). To assess if this could reflect an instability of the protein, its thermostability was measured by incubating the membranes isolated from the transformants for 10 min at different temperatures and then by measuring the amount of PBP3 left in an active form by the subsequent binding with fluorescent ampicillin for 25 min at 30°C. GFP-PBP3(D58V) showed the same thermostability as wild-type PBP3, PBP3(R210Q, R213Q), and PBP3(D58, R210Q, R213Q) under the conditions used (Fig. 3). Interestingly, the PBP3(R166Q, R167Q) mutant, which does not bind penicillin and does not show a dominant-negative effect, was targeted to the septal site (Fig. 2E). These results indicate that the segments containing D58 and R210 and R213 residues that have protein-protein interaction potentials are not involved in septal targeting of PBP3 and that this targeting does not rely on the proper folding of the catalytic module.
FIG. 2.
Localization of GFP-PBP3 mutants in ftsI+ LMC500 transformants grown in glucose minimal medium at 28°C (left) and in ftsI-null EC548 transformants depleted of wild-type PBP3 grown in rich medium at 37°C (right). The white bars in the upper right corners of the pictures represent 1 μm.
FIG. 3.
Thermostability of GFP-PBP3 mutants. The amounts of PBP retained in an active form after 10 min of incubation at various temperatures were estimated by fluorescent ampicillin labeling at 30°C, as measured by SDS-PAGE and fluorescence quantification. The results are expressed as percentages of labeling obtained after the 10-min preincubation of the samples at 30°C. •, GFP-PBP3; ▪, GFP-PBP3(D58V); ▿, GFP-PBP3(R210Q, R213Q); ⋄, GFP-PBP3(D58V, R210Q, R213Q).
Minimal structural determinant required for septal recruitment of PBP3.
In order to define the minimum structural determinant required for septal recruitment of PBP3, the size of the protein was reduced. GFP was fused to the truncated PBP3(K2-R239) lacking the C-terminal penicillin-binding module. PBP3(M1-R239) has been previously reported to exhibit a dominant-negative phenotype when overexpressed (Table 2; Fig. 1) (24). We also made GFP fusions with PBP3(K2-S70) bearing the G57-Q66 segment, the PBP3(K2-E56) lacking this segment, and the PBP3(K2-V42) comprising only the cytoplasmic domain, the membrane-spanning segment, and the first three periplasmic amino acid residues. To facilitate the insertion of PBP3(K2-V42) in the plasma membrane, the truncated protein was also fused to the N terminus of the penicillin-binding domain of the BlaR protein from B. licheniformis, which is involved in β-lactamase induction (18) and is assumed to be neutral in regard to E. coli cell division. GFP-PBP3(K2-R239), GFP-PBP3(K2-S70), GFP-PBP3(K2-E56), GFP-PBP3(K2-V42), and GFP-PBP3(K2-V42)-BlaR were encoded by plasmids pDML2484, pDML2485, pDML2486, pDML2487, and pDML2488, respectively (see Materials and Methods and Table 1). The ftsI+ strain LMC500 and ftsI-null strain EC548 were transformed with these plasmids and grown as described above.
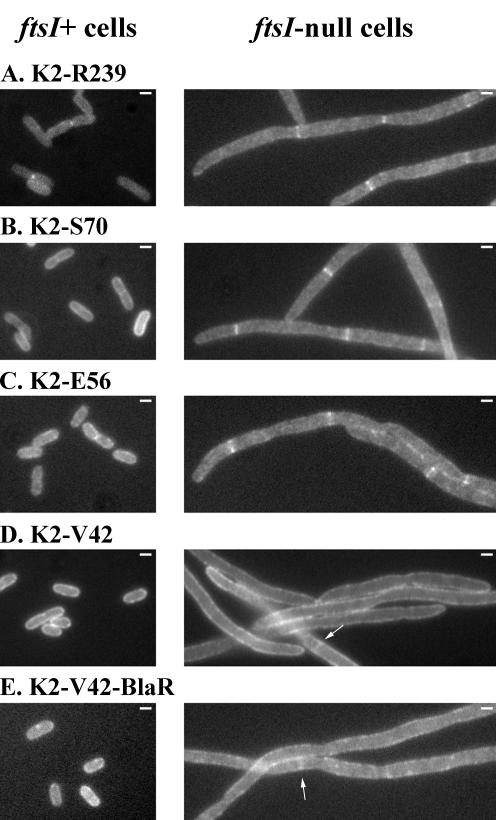
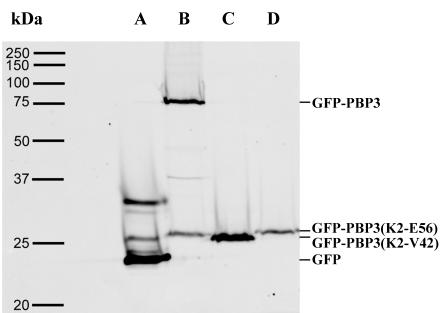
As shown in Fig. 4 and Table 2, GFP-PBP3(K2-R239), GFP-PBP3(K2-S70), and GFP-PBP3(K2-E56) localized in the septal region of 75 to 87% of PBP3-depleted cells, whereas the shortest form, GFP-PBP3(K2-V42), whether fused to BlaR or not, localized in only 20 to 25% of the cells, with a weak fluorescence signal compared to other truncated forms. The size of EC548 expressing the nonfunctional GFP-PBP3 derivatives varied from 18 to 53 μm, illustrating the depletion of functional PBP3. The variability in length could reflect differences in the growth rate as well as in the dominant-negative effect of each GFP-PBP3 derivative. For ftsI+ LMC500, a faint fluorescence signal was observed at the division site of a low percentage of cells expressing GFP-PBP3(K2-R239) or GFP-PBP3(K2-S70) (Fig. 4 and Table 2). The other shorter forms did not localize in the wild-type strain. As shown by SDS-PAGE and fluorescence measurements of membrane proteins prepared from E. coli LMC500 expressing these derivatives, production of GFP-PBP3(K2-E56) is four times less than that of wild-type GFP-PBP3, whereas GFP-PBP3(K2-V42) is produced in larger amounts than the wild-type fusion (Fig. 5). These results suggest that the truncated forms of PBP3 did not compete or did not compete well with the native protein for the recruiting factor but did keep an affinity for the division site. The affinity of the shortest form, comprising the membrane spanning-segment and the first three periplasmic amino acid residues, was particularly low. These results indicate that the structural determinants required to target PBP3 at the septation site not only lie in the transmembrane segment but extend a little further within the first 17 periplasmic amino acid residues. They also indicate that the other domains of the protein are not essential in this process.
FIG. 4.
Localization of truncated forms of PBP3 fused to the C terminus of GFP in ftsI+ LMC500 transformants grown in glucose minimal medium at 28°C (left) and in ftsI-null EC548 transformants depleted of wild-type PBP3 grown in rich medium at 37°C (right). The white bars in the upper right corners of the pictures represent 1 μm. Arrows point to weakly fluorescent rings.
FIG. 5.
Production of GFP (A), GFP-PBP3 (B), GFP-PBP3(K2-V42) (C), and GFP-PBP3(K2-E56) (D) in ftsI+ LMC500 cells grown in glucose minimal medium at 28°C. Fluorescence was detected after SDS-PAGE of nonboiled samples.
Effect of the overproduction of FtsQ, FtsL, or FtsW on the morphological phenotype of E. coli producing PBP3 variants.
PBP3 is a late recruit to the cell division site. In order to investigate the interaction of PBP3 with the other cell division proteins, the effects of FtsQ, FtsL, and FtsW on the phenotype of E. coli expressing PBP3 mutants were analyzed.
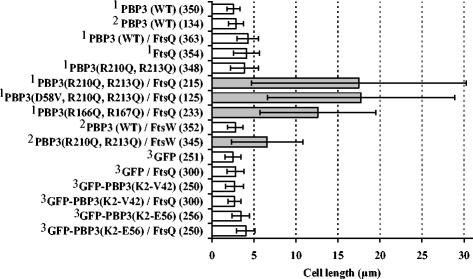
E. coli TOP10 F′ cells were cotransformed with plasmids pDML2497, bearing the modified ftsI gene encoding PBP3(R210Q, R213Q), and pDML2407, pDML2490, or pDML2491, encoding FtsW, FtsL, or FtsQ, respectively. The PBP3 double mutant was under the control of the PBAD promoter, whereas the other cell division proteins were under the control of the lac promoter. Transformants were grown at 37°C in LB medium containing 100 μM IPTG and 0.005 or 0.01% arabinose as described in experimental procedures. Figure 6 shows the results. The cells producing the PBP3 double mutant were slightly longer than those producing wild-type PBP3, indicating a weak dominant-negative effect under the conditions used. The coexpression of native PBP3 and FtsW had no effect on the size of the cells, whereas that of the native PBP3 and FtsQ increased the length of the cells 1.7-fold. It was observed previously that overexpression of FtsQ led to the formation of filaments (10). When the cells expressed the PBP3 double mutant and FtsQ, they were 4 to 4.5 times longer than those expressing both FtsQ and the native PBP3 or the PBP3 mutant alone. The cells coproducing FtsW and the PBP3 double mutant were 1.5 times longer than the control cells. FtsL had no effect (data not shown). Thus, FtsQ and FtsW to a lesser extent exacerbated the dominant-negative effect of the PBP3 double mutant.
FIG. 6.
Effect of the overproduction of FtsQ or FtsW on the morphological phenotype of E. coli Top10 F′ cells producing PBP3 variants. Transformants were grown at 37°C in LB medium containing 0.005% arabinose (1) or 0.01% arabinose (2) to induce expression of PBP3 variants and 0.1 mM IPTG to induce expression of FtsW or FtsQ. (3) GFP-, FtsQ-, and GFP-PBP3-encoding genes were under the control of the lac promoter and were induced with 0.1 mM IPTG. Numbers of measured cells are in parentheses.
The effect of FtsQ overexpression was then analyzed with E. coli TOP10 F′, producing PBP3(D58V, R210Q, R213Q) (from pDML2498), which did not show a dominant-negative effect, and PBP3(R166Q, R167Q) (from pDML2496), which, in addition, did not bind penicillin. Surprisingly, both transformants were three- to fourfold longer than those producing FtsQ alone, a result similar to that obtained with the PBP3(R210Q, R213Q) mutant (Fig. 6).
Finally, the effect of FtsQ was examined with cells expressing truncated PBP3(K2-E56) or PBP3(K2-V42), both fused to GFP and under the control of the trc promoter (from plasmids pDML2486 and pDML2487, respectively). Transformants were grown at 37°C in LB medium in the presence of 100 μM IPTG. Under these conditions, FtsQ had no appreciable effect on the size of the cells expressing these constructs.
DISCUSSION
The membrane anchor-containing peptide M1-L39 is required for the functioning of E. coli PBP3, and its alteration prevents the protein from reaching the midcell region (14, 16, 30). Our results show that this region is necessary but not sufficient to target the protein to the division site, since GFP-PBP3(K2-V42) localizes poorly to the division site, even when it is slightly overexpressed, in cells depleted of wild-type PBP3. By contrast, PBP3(K2-E56), containing the membrane anchor and the G40-E56 peptide, is targeted just like the wild-type protein to the division site of PBP3-depleted cells and thus bears the key structural determinants required for this function.
None of the previously predicted interaction sites that are present in the noncatalytic module of PBP3 appear to be essential for the insertion of the protein at midcell, because all PBP3 mutants including the inactive PBP3(R166Q, R167Q) mutant localized to the septum. These sites may be involved in interactions with the other cell division proteins and the peptidoglycan-metabolizing enzymes and thus stabilize the PBP3 once it has reached its proper location and take part in its function.
Recently, Wissel and Weiss published results converging with ours. They showed by random mutagenesis the importance of residues R23, L39, and Q46 for the localization of PBP3 and of residues G57, S61, L62, and R210, each located in our previously predicted interaction sites, for the recruitment of FtsN (31).
The mechanism by which the K2-E56 peptide directs the protein to the septum is not known. FtsW is required for the recruitment of PBP3 at the division site (25). Overexpression of FtsW enhanced slightly the dominant-negative effect of the PBP3(R210Q, R213Q) mutant, suggesting an interaction between FtsW and PBP3.
FtsW depends on FtsL and FtsB for its localization to the septum. Overexpression of FtsL had no effect on E. coli cells producing the PBP3(R210Q, R213Q) mutant, probably because FtsB, which seems to interact with FtsL and stabilize it (6), is not present in sufficient amounts. By contrast, overexpression of FtsQ exacerbated the dominant-negative effect of the PBP3(R210Q, R213Q) double mutant. In addition, overexpression of FtsQ blocks division of cells producing the PBP3(R166Q, R167Q) mutant, which is completely inactive and probably misfolded. A similar effect had already been observed by Dai and Lutkenhaus when FtsQ is produced in the thermosensitive ftsI23 strain at the permissive temperature (8). These results suggest a functional interaction between these cell division proteins that might be direct or indirect.
These suggestions were in agreement with results obtained by using an E. coli two-hybrid system that showed interactions between FtsW and PBP3 and between FtsQ, FtsW, and PBP3 (9).
Mutations in the noncatalytic module of E. coli PBP3 result in perturbation of cell growth and cell division (24). Overexpression of FtsQ inhibits division of ftsI+ cells, producing these PBP3 mutants. FtsQ might promote the interaction of the inactive protein with the division machinery at the expense of wild-type PBP3. These data show the important role of the noncatalytic module of PBP3 in peptidoglycan assembly at the septum through protein-protein interaction. They also point out the importance of the ratio between components of the divisome. The divisome is also sensitive to overproduction of FtsQ when it contains thermosensitive FtsZ, FtsA, or PBP3 mutants (8) at the permissive temperature. Thus, FtsQ could play a role in regulating the assembly of the division proteins and in the control of the peptidoglycan assembly machineries within the divisome.
One could ask why the inactive and probably misfolded PBP3(R166Q, R167Q) mutant does not disturb cell division when it localizes as native PBP3 in the presence of endogenous PBP3, while the PBP3(R210Q, R213Q) mutant and PBP3(S307A) do. One explanation is that the divisome is a dynamic structure. Exchanges between the inactive PBP3 mutant and the wild-type protein could occur once PBP3 is located at the division site due to its first 56 amino acid residues. Depending on the affinity of PBP3 mutants for the other cell division proteins, PBP1b, lipid II, and/or peptidoglycan, the dominant-negative effect is more or less strong. Indeed, the modified PBP3s, which showed a severe dominant-negative effect in our previous report when they were expressed in large amounts or in ftsI(Ts) cells, caused only a reduced lengthening when slightly expressed in a wild-type background. This was also noticed by Wissel and Weiss (31). This work supports the hypothesis of the existence of multiple interactions between PBP3 and other division proteins that are involved in distinct functions.
Acknowledgments
This work was supported in part by the Belgian program on Interuniversity Poles of Attraction initiated by the Belgian State, Prime Minister's Office, Services Fédéraux des Affaires Scientifiques, Techniques et Culturelles (PAI no. P5/33), the Fonds de la Recherche Fondamentale Collective (contract no. 2.4521.01), the Actions de Recherche Concertées 03/08-297, and a Vernieuwingsimpuls grant (T.D.B., 016.001.024) from The Netherlands Organization for Scientific Research (NWO).
A.P. is a Fellow of the Fonds pour la Formation à la Recherche dans l'Industrie et dans l'Agriculture, Brussels.
We thank D. S. Weiss (University of Iowa) for the gift of plasmid pDSW234, S. Blacher (University of Liège) for technical assistance in phase-contrast microscopy, and J. Coyette (University of Liège) for fruitful discussions.
REFERENCES
- 1.Bernhardt, T. G., and P. A. de Boer. 2003. The Escherichia coli amidase AmiC is a periplasmic septal ring component exported via the twin-arginine transport pathway. Mol. Microbiol. 48:1171-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bramhill, D., and C. M. Thompson. 1994. GTP-dependent polymerization of Escherichia coli FtsZ protein to form tubules. Proc. Natl. Acad. Sci. USA 91:5813-5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Broome-Smith, J. K., P. J. Hedge, and B. G. Spratt. 1985. Production of thiol-penicillin-binding protein 3 of Escherichia coli using a two primer method of site-directed mutagenesis. EMBO J. 4:231-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buddelmeijer, N., M. E. Aarsman, A. H. Kolk, M. Vicente, and N. Nanninga. 1998. Localization of cell division protein FtsQ by immunofluorescence microscopy in dividing and nondividing cells of Escherichia coli. J. Bacteriol. 180:6107-6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buddelmeijer, N., and J. Beckwith. 2002. Assembly of cell division proteins at the E. coli cell center. Curr. Opin. Microbiol. 5:553-557. [DOI] [PubMed] [Google Scholar]
- 6.Buddelmeijer, N., N. Judson, D. Boyd, J. J. Mekalanos, and J. Beckwith. 2002. YgbQ, a cell division protein in Escherichia coli and Vibrio cholerae, localizes in codependent fashion with FtsL to the division site. Proc. Natl. Acad. Sci. USA 99:6316-6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, J. C., M. Minev, and J. Beckwith. 2002. Analysis of ftsQ mutant alleles in Escherichia coli: complementation, septal localization, and recruitment of downstream cell division proteins. J. Bacteriol. 184:695-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dai, K., and J. Lutkenhaus. 1992. The proper ratio of FtsZ to FtsA is required for cell division to occur in Escherichia coli. J. Bacteriol. 174:6145-6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DiLallo, G., M. Fagioli, D. Barionovi, P. Ghelardini, and L. Paolozzi. 2003. Use of a two-hybrid assay to study the assembly of a complex multicomponent protein machinery: bacterial septosome differentiation. Microbiology 149:3353-3359. [DOI] [PubMed] [Google Scholar]
- 10.Dopazo, A., P. Palacios, M. Sanchez, J. Pla, and M. Vicente. 1992. An amino-proximal domain required for the localization of FtsQ in the cytoplasmic membrane, and for its biological function in Escherichia coli. Mol. Microbiol. 6:715-722. [DOI] [PubMed] [Google Scholar]
- 11.Eberhardt, C., L. Kuerschner, and D. S. Weiss. 2003. Probing the catalytic activity of a cell division-specific transpeptidase in vivo with beta-lactams. J. Bacteriol. 185:3726-3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Errington, J., R. A. Daniel, and D. J. Scheffers. 2003. Cytokinesis in bacteria. Microbiol. Mol. Biol. Rev. 67:52-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Filée, P., K. Benlafya, M. Delmarcelle, G. Moutzourelis, J. M. Frère, A. Brans, and B. Joris. 2002. The fate of the BlaI repressor during the induction of the Bacillus licheniformis BlaP beta-lactamase. Mol. Microbiol. 44:685-694. [DOI] [PubMed] [Google Scholar]
- 14.Fraipont, C., M. Adam, M. Nguyen-Distèche, W. Keck, J. Van Beeumen, J. A. Ayala, B. Granier, H. Hara, and J. M. Ghuysen. 1994. Engineering and overexpression of periplasmic forms of the penicillin-binding protein 3 of Escherichia coli. Biochem. J. 298:189-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gueiros-Filho, F. J., and R. Losick. 2002. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev. 16:2544-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guzman, L. M., D. S. Weiss, and J. Beckwith. 1997. Domain-swapping analysis of FtsI, FtsL, and FtsQ, bitopic membrane proteins essential for cell division in Escherichia coli. J. Bacteriol. 179:5094-5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hale, C. A., and P. A. de Boer. 1997. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell 88:175-185. [DOI] [PubMed] [Google Scholar]
- 18.Hardt, K., B. Joris, S. Lepage, R. Brasseur, J. O. Lampen, J. M. Frère, A. L. Fink, and J. M. Ghuysen. 1997. The penicillin sensory transducer, BlaR, involved in the inducibility of beta-lactamase synthesis in Bacillus licheniformis is embedded in the plasma membrane via a four-alpha-helix bundle. Mol. Microbiol. 23:935-944. [DOI] [PubMed] [Google Scholar]
- 19.Houba-Herin, N., H. Hara, M. Inouye, and Y. Hirota. 1985. Binding of penicillin to thiol-penicillin-binding protein 3 of Escherichia coli: identification of its active site. Mol. Gen. Genet. 201:499-504. [DOI] [PubMed] [Google Scholar]
- 20.Koppelman, C. M., M. E. G. Aarsman, J. Postmus, E. Pas, A. O. Muijsers, D. J. Scheffers, N. Nanninga, and T. den Blaauwen. 2004. R174 of Escherichia coli FtsZ is involved in membrane interaction and protofilament bundling, and is essential for cell division. Mol. Microbiol. 51:645-657. [DOI] [PubMed] [Google Scholar]
- 21.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 22.Lakaye, B., C. Damblon, M. Jamin, M. Galleni, S. Lepage, B. Joris, J. Marchand-Brynaert, C. Frydrych, and J. M. Frère. 1994. Synthesis, purification and kinetic properties of fluorescein-labelled penicillins. Biochem. J. 300:141-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma, X., and W. Margolin. 1999. Genetic and functional analyses of the conserved C-terminal core domain of Escherichia coli FtsZ. J. Bacteriol. 181:7531-7544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marrec-Fairley, M., A. Piette, X. Gallet, R. Brasseur, H. Hara, C. Fraipont, J. M. Ghuysen, and M. Nguyen-Distèche. 2000. Differential functionalities of amphiphilic peptide segments of the cell-septation penicillin-binding protein 3 of Escherichia coli. Mol. Microbiol. 37:1019-1031. [DOI] [PubMed] [Google Scholar]
- 25.Mercer, K. L., and D. S. Weiss. 2002. The Escherichia coli cell division protein FtsW is required to recruit its cognate transpeptidase, FtsI (PBP3), to the division site. J. Bacteriol. 184:904-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mosyak, L., Y. Zhang, E. Glasfeld, S. Haney, M. Stahl, J. Seehra, and W. S. Somers. 2000. The bacterial cell-division protein ZipA and its interaction with an FtsZ fragment revealed by X-ray crystallography. EMBO J. 19:3179-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakano, Y., Y. Yoshida, Y. Yamashita, and T. Koga. 1995. Construction of a series of pACYC-derived plasmid vectors. Gene 162:157-158. [DOI] [PubMed] [Google Scholar]
- 28.Taschner, P. E., N. Ypenburg, B. G. Spratt, and C. L. Woldringh. 1988. An amino acid substitution in penicillin-binding protein 3 creates pointed polar caps in Escherichia coli. J. Bacteriol. 170:4828-4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang, X., J. Huang, A. Mukherjee, C. Cao, and J. Lutkenhaus. 1997. Analysis of the interaction of FtsZ with itself, GTP, and FtsA. J. Bacteriol. 179:5551-5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weiss, D. S., J. C. Chen, J. M. Ghigo, D. Boyd, and J. Beckwith. 1999. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J. Bacteriol. 181:508-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wissel, M. C., and D. S. Weiss. 2004. Genetic analysis of the cell division protein FtsI (PBP3): amino acid substitutions that impair septal localization of FtsI and recruitment of FtsN. J. Bacteriol. 186:490-502. [DOI] [PMC free article] [PubMed] [Google Scholar]