Abstract
The role of the putative P450 monooxygenase OxyD and the chlorination time point in the biosynthesis of the glycopeptide antibiotic balhimycin produced by Amycolatopsis balhimycina were analyzed. The oxyD gene is located directly downstream of the bhp (perhydrolase) and bpsD (nonribosomal peptide synthetase D) genes, which are involved in the synthesis of the balhimycin building block β-hydroxytyrosine (β-HT). Reverse transcriptase experiments revealed that bhp, bpsD, and oxyD form an operon. oxyD was inactivated by an in-frame deletion, and the resulting mutant was unable to produce an active compound. Balhimycin production could be restored (i) by complementation with an oxyD gene, (ii) in cross-feeding studies using A. balhimycina JR1 (a null mutant with a block in the biosynthesis pathway of the building blocks hydroxy- and dihydroxyphenylglycine) as an excretor of the missing precursor, and (iii) by supplementation of β-HT in the growth medium. These data demonstrated an essential role of OxyD in the formation pathway of this amino acid. Liquid chromatography-electrospray ionization-mass spectrometry analysis indicated the biosynthesis of completely chlorinated balhimycin by the oxyD mutant when culture filtrates were supplemented with nonchlorinated β-HT. In contrast, supplementation with 3-chloro-β-HT did not restore balhimycin production. These results indicated that the chlorination time point was later than the stage of free β-HT, most likely during heptapeptide synthesis.
In the past several decades, the glycopeptide antibiotic vancomycin became the antibiotic of last resort for the treatment of infections caused by multiresistant gram-positive bacteria such as methicillin-resistant Staphylococcus aureus strains (37). However, the occurrence of vancomycin-resistant bacteria (3) and the expected increase in resistance may limit the medical use of even vancomycin in the near future. The search for new glycopeptide antibiotics is essential to overcome this problem. One strategy for obtaining new glycopeptides is to genetically manipulate the producer strains. This approach requires a molecular understanding of glycopeptide biosynthesis.
In order to study the biosynthesis of glycopeptide antibiotics and the functions of the relevant genes (6, 22, 31), we chose the balhimycin producer strain Amycolatopsis balhimycina DSM5908 (36) as a model system. A. balhimycina belongs to the order of Actinomycetales and was formerly described as Amycolatopsis mediterranei (8, 18). It was isolated from an Indian soil sample originating from the Himalayas (18). A. balhimycina shows the typical growth characteristics of actinomycetes and forms an orange substrate mycelium but no spores when cultivated on solid media (20). The in vitro and in vivo activities of balhimycin are comparable to those of vancomycin (34), but balhimycin shows a slight increase in antibiotic activity toward anaerobic bacteria (for example, clostridia) (8).
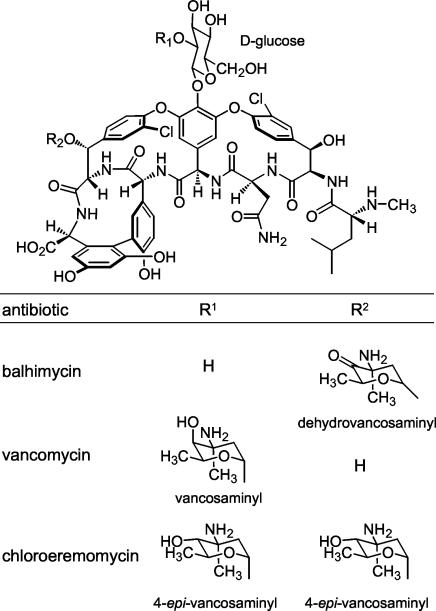
The chemical structure of vancomycin-type antibiotics (Fig. 1) such as balhimycin is based on a central heptapeptide core. This peptide core contains five aromatic amino acids. In the case of vancomycin and balhimycin, the nonproteinogenic amino acids 4-hydroxyphenylglycine (HPG; positions four and five), 3,5-dihydroxyphenylglycine (DPG; position seven), and β-hydroxytyrosine (β-HT; positions two and six) are incorporated. These aromatic acid side chains are linked to each other to form two diaryl ether rings and one biaryl ring, and the aglycone thus formed is modified by sugar substituents. The formation pathways of DPG and HPG have been studied in detail (11, 15, 23). Furthermore, investigations of the synthesis of β-HT revealed the participation of the perhydrolase Bhp (25) and the nonribosomal peptide synthetase module BpsD (27). As an additional modification of the peptide core, both β-HT residues of balhimycin are chlorinated (Fig. 1). In general, chlorine atoms as well as glycosyl groups have a strong influence on the antibiotic activities of glycopeptides (12, 13, 19), most likely by stabilizing the dimerization of these compounds (4, 12, 16).
FIG. 1.
Structure of glycopeptide antibiotics balhimycin (A. balhimycina DSM5908), vancomycin (Amycolatopsis orientalis C329.4), and chloroeremomycin (A. orientalis A82846).
Recently, the NADH/FAD-dependent halogenase BhaA was identified as essential for the chlorination reaction of both β-HT residues, at positions 2 and 6 of the glycopeptide aglycone (25). However, the substrate of BhaA and therefore the chlorination time point during balhimycin biosynthesis remained unclear.
Here we report that the putative P450 monooxygenase OxyD, together with perhydrolase Bhp and the nonribosomal peptide synthetase BpsD, is required for the formation of the nonproteinogenic amino acid β-HT. In addition, we present evidence that the chlorination of the β-HT residues does not take place either during precursor synthesis or at the aglycone.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The strains and plasmids used for this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used for this study
| Strain or plasmid | Relevant featuresa | Source or reference |
|---|---|---|
| Strains | ||
| E. coli XL1-Blue | General cloning host | 7 |
| A. balhimycina DSM5908 | Balhimycin-producing wild type | 18 |
| A. balhimycina OP696 | Nonproducing mutant, in frame deletion in the bhp gene | 25 |
| A. balhimycina BpsD-cat | Nonproducing mutant, replacement of a part of the bpsD gene with a cat cassette | 26 |
| A. balhimycina JR1 | Nonproducing mutant, in-frame deletion in the pgat gene | 23 |
| A. balhimycina OP090 | Nonproducing mutant, in-frame deletion in the oxyD gene | This study |
| A. balhimycina OP090v | A. balhimycina with pOP2 integrated into the chromosome | This study |
| A. balhimycina OP090k | A. balhimycina OP090 complemented with an additional oxyD gene | This study |
| Plasmids | ||
| pSP1 | Gene disruption vector; Eryr | 20 |
| pSET152 | Amr; integration system of the phage ΦC31 | 5 |
| pJOE890 | Apr | 1 |
| pUC18ermEp1 | Apr | 25 |
| Cosmid 16.1 | Contains part of the balhimycin biosynthetic gene cluster | 26 |
| pOP2 | pSP1 derivative containing a part of the balhimycin biosynthetic gene cluster; including the oxyD gene with a 390-bp in-frame deletion | This study |
| pJOEOP3 | pJOE890 derivative containing the fragment frOP3 (see Materials and Methods) | This study |
| pJOEOP4 | pJOE890 derivative containing the fragment frOP4 (see Materials and Methods) | This study |
| pSPOPb | pSP1 derivative containing the fragment frOP3 (see Materials and Methods) | This study |
| pUC-oxyD | pUC18ermEp1 derivative containing the oxyD gene and its ribosomal binding site | This study |
| pSET-oxyD | pSET152 derivative, contains oxyD upstream of ermE*p | This study |
Amr, ampicillin resistant; Apr, apramycin resistant; Eryr, erythromycin resistant.
Media and culture conditions.
Escherichia coli strains were grown in Luria broth (28) supplemented with 150 μg of ampicillin ml−1 or 100 μg of apramycin ml−1 when necessary to maintain plasmids. A. balhimycina strains were grown in R5 medium (14) at 30°C. Liquid and solid media were supplemented with 50 μg of erythromycin ml−1 or 50 μg of apramycin ml−1 to select for strains carrying integrated antibiotic resistance genes.
Cultivation of OP090 in the presence of β-HT derivatives.
OP090 (Table 1) was incubated under standard conditions in 20 ml of R5 medium (14). After 24 h of growth, β-HT or chloro-β-HT (CHT) was added (1 mg/ml). The supernatants were harvested at different time points, and 20 μl of each was used to determine the production of balhimycin in a bioassay with Bacillus subtilis.
Preparation of A. balhimycina RNA.
A. balhimycina was cultivated in 100 ml of R5 medium for 3 days. The cells were then harvested and shock frozen at −70°C. An aliquot was resuspended in 100 μl of P buffer (32) containing 10 mg of lysozyme and then incubated for 7 min at 37°C. The RNA was extracted by use of an RNeasy mini kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions.
RT-PCR analysis.
RNA prepared from A. balhimycina was treated with 3 U of RNase-free DNase I (Promega, Madison, Wis.) and precipitated according to standard protocols (28). The RNA concentration was photometrically determined with a Genequant fixed-wavelength photometer (Pharmacia, Freiburg, Germany). Reverse transcription (RT) reactions were performed by use of an Omniscript RT kit (Qiagen) according to the manufacturer's instructions. The primers used for RT of the bhp-bpsD and bpsD-oxyD overlapping mRNA sequences were cotraorfYTGA (5′-TCAGCGTGGTGGTCCCCATC-3′) and cotraoxyDTGA (5′-CCAGAAGCCGGAGGGGGAAC-3′), respectively. PCRs were carried out in a programmable thermal controller (MJ Research, Inc., La Jolla, Calif.) under the following conditions: initial denaturation (95°C for 2 min); 25 cycles of denaturation (95°C for 20 s), annealing (60°C for 30 s), and polymerization (72°C for 40 s); and finally, an additional polymerization step (72°C for 7 min). Each PCR mixture (25 μl) contained a 1-μl aliquot of RT reaction product, 100 pmol of each primer, deoxyribonucleoside 5′-triphosphates at a final concentration (each) of 20 μM (DNA polymerization mix; Pharmacia), 10× reaction buffer (Qiagen), 5× Q solution (Qiagen), and 3.5 U of Taq DNA polymerase (Qiagen). The following oligonucleotide primer pairs were used: cotraorfYATG (5′-AGGAGCTGGCCGCCGTGATC-3′) and cotraorfYTGA (5′-TCAGCGTGGTGGTCCCCATC-3′), for amplification of the bhp-bpsD overlapping fragment, and cotraoxyDATG (5′-CGGAAGTGCTCGGTGTCAGC-3′) and cotraoxyDTGA (5′-CCAGAAGCCGGAGGGGGAAC-3′), for amplification of the bpsD-oxyD overlapping fragment. The PCR products were analyzed by agarose gel electrophoresis (1.0%).
Preparation and manipulation of DNA.
The methods used for the isolation and manipulation of DNA were described by Sambrook et al. (28) and Hopwood et al. (14). PCR fragments were isolated from agarose gels with a Qiaquick kit (Qiagen). Restriction endonucleases were obtained from various suppliers and were used according to their specifications.
PCR protocols for amplification of fragments frOP3, frOP4, and the oxyD gene and for characterization of OP090k.
PCRs were performed with a programmable thermal controller (MJ Research, Inc.). Each PCR mixture (100 μl) contained 100 pmol of each primer, 1.0 μg of template DNA (cosmid 16.1), deoxyribonucleoside 5′-triphosphates at a final concentration (each) of 20 μM (DNA polymerization mix; Pharmacia), 10× reaction buffer (Qiagen), 5× Q solution (Qiagen), and 3.5 U of Taq DNA polymerase. Dimethyl sulfoxide (Stratagene) was added to the reaction mixture at a final concentration of 3% to enhance the specificity of hybridization. For amplification of the fragments frOP3 and frOP4, which are part of the deletion plasmid pOP2 (see below), the following PCR conditions were used: initial denaturation (95°C for 2 min) before the addition of the polymerase; 30 cycles of denaturation (95°C for 20 s), annealing (68°C for 30 s), and polymerization (72°C for 1 min 45 s); and an additional polymerization step (72°C for 7 min) at the end. The primers used were as follows: for amplification of the fragment frOP3, ΔXI (5′-GGTCTGATCGCCCGCGGTTACCTGCACCGGCCG-3′) and ΔXII (5′-GGTCTAGAGATATCGGTGTGCGCCTGCCGCGGGGTCATCC-3′); and for amplification of the fragment frOP4, ΔXIII (5′-GGGATATCGACGACCCGGACACCTTCCTGCCCGG-3′) and ΔXVI (5′-GGGATATCGCACGTTCGTCGACCGCAGGTCGTCC-3′). For amplification of the oxyD gene, the annealing step was done at 60°C for 30 s and the polymerization step was done at 72°C for 1 min 30 s. The sequences of the primers were as follows: oxyDlow, 5′-GAGATCTTGGAGACCCTGATGCAGACG-3′; and oxyDup, 5′-GAGATCTGGTCAGCGCCCGGTGAACC-3′. For confirmation of the integration of pSET-oxyD (see below) into the genome of OP090k outside of the oxyD locus, an annealing temperature of 55°C (30 s) was used. The polymerization step was done at 72°C for 30 s. The sequences of the primers, which amplified a 1,078-bp fragment of the balhimycin gene cluster containing the entire ΔoxyD gene construct, were as follows: oxyDex1, 5′-GAGGACAGCTTCTTCGAGGTCG-3′; and oxyDex2, 5′-CGCATCAACGGTGTCAGCTT-3′.
Construction of plasmids pOP2 and pSET-oxyD.
Plasmids were constructed for an internal deletion of the P450 monooxygenase gene oxyD (pOP2) as well as for complementation of the oxyD deletion mutant strain A. balhimycina OP090 (pSET-oxyD).
(i) pOP2.
The 1,300-bp fragment frOP3, including a sequence encoding 188 amino acids of the N terminus of the P450 monooxygenase gene oxyD (1,191 bp), and the 1,287-bp fragment frOP4, including a sequence encoding 76 amino acids of the C terminus of oxyD, were ligated into the EcoRV site of the vector pJOE890, resulting in the plasmids pJOEOP3 and pJOEOP4, respectively. frOP3 was then ligated as an XbaI fragment into the single XbaI site of the vector pSP1, resulting in the plasmid pSPOPb. To obtain the plasmid pOP2, containing a partly deleted oxyD gene, we ligated frOP4 as an EcoRV fragment into the EcoRV site of pSPOPb.
(ii) pSET-oxyD.
A 1,217-bp fragment consisting of oxyD and its ribosomal binding site was ligated as a blunt-ended fragment into the vector pJOE890 and then integrated into the BamHI cleavage site of the vector pUC18ermEp1 as a BglII fragment, resulting in the plasmid pUC-oxyD. The ermE*p-oxyD expression construct was then ligated as an EcoRI-XbaI fragment into the single EcoRI-XbaI site of the vector pSET152, resulting in the complementation plasmid pSET-oxyD.
Cross-feeding studies with different A. balhimycina strains.
To investigate whether different null mutant strains were blocked in the same biosynthetic pathway, we performed cross-feeding experiments. For these studies, the strains of interest were plated on an R5 agar plate, with a small cell-free region (approximately 0.5 cm) left between them. After 5 days of incubation (30°C), an agar strip containing both mutants was cut out and analyzed in a bioassay with B. subtilis to determine whether the diffusional exchange of accumulated intermediates restored the balhimycin production ability of the tested null mutants.
Determination of β-HT and CHT uptake by A. balhimycina.
To determine the uptake of β-HT and CHT by A. balhimycina, we measured the concentrations of the amino acids in the medium at different time points by reversed-phase high-performance liquid chromatography (HPLC). The cell-free supernatant (sample injection volume, 20 μl) was separated at a flow rate of 2 ml/min on a Nucleosil C18 column (12.5 cm by 0.4 cm by 5 μm) via a gradient elution using the ThermoSeparation spectrum system (pump, model P200; automatic probe injector, model AS3000; UV detector, model UV3000HR; Thermo Request Systems, Egelsbach, Germany). The following gradient was used: at t = 0 min, 100% A; at t = 10 min, 80% A and 20% B; at t = 13 min, 100% B; at t = 16 min, 100% A (solvent A, 0.1% phosphoric acid; solvent B, acetonitrile).
Determination of balhimycin biosynthesis by HPLC-ESI-MS.
Balhimycin production was determined with bioassays using cell-free supernatants of Amycolatopsis strains grown on R5 medium, with B. subtilis ATCC 6633 as a test organism (14). Investigations of the balhimycin variants in culture broth were performed by HPLC-electrospray ionization-mass spectrometry (HPLC-ESI-MS). Culture broth was prepared by centrifugation and filtration to obtain particle-free samples. LC-ESI-MS experiments were performed on a Bruker Esquire 3000+ instrument coupled to an Agilent 1100 HPLC system (Bruker-Franzen, Bremen, Germany). Separations were performed on a Nucleosil C18 column (2 mm by 100 mm by 5 μm) (Grom, Herrenberg, Germany) at a flow rate of 200 μl min−1. The following gradient was used: at t = 0 min, 95% A and 5% B; at t = 1 min, 83% A and 17% B; at t = 15 min, 80% A and 20% B; at t = 17 min, 100% B (solvent A, 0.1% trifluoroacetic acid in water; solvent B, 0.1% trifluoroacetic acid in acetonitrile).
Nucleotide sequence accession number.
The nucleotide sequences of the balhimycin biosynthetic genes reported in this paper are available from the EMBL data library under accession number Y16952.
RESULTS
The P450 monooxygenase gene oxyD is part of an operon including the genes bhp and bpsD.
In the balhimycin biosynthetic gene cluster, altogether four genes (oxyA to -D) have been identified whose gene products show significant similarities to P450 monooxygenases. OxyA-, -B, and -C show higher sequence homologies to each other (41 to 46% similarity; 55 to 64% identity) than to OxyD (26 to 31% similarity; 38 to 48% identity). For the vancomycin producer, it has been shown that the homologous oxygenases are P450 monooxygenases (38). OxyA, -B, and -C in A. balhimycina catalyze the cross-linking steps between the aromatic rings within the balhimycin peptide backbone in a defined order (6). In contrast, the function of OxyD remained unclear. The genes oxyA to -C are clustered in a region approximately 12.7 kb upstream of oxyD, which lies directly downstream of the perhydrolase gene bhp and the gene bpsD, which codes for a nonribosomal peptide synthetase (NRPS). Since Bhp and BpsD are involved in β-HT formation (25, 27), it was assumed that OxyD also participates in this pathway. No termination signals were detectable in the intergenic DNA sequences of bhp, bpsD, and oxyD, indicating cotranscription of the three genes. In order to prove the operon structure of the bhp-bpsD-oxyD region, we performed an RT-PCR analysis. Using primer pairs corresponding to (i) the 3′ region of bhp and the 5′ region of bpsD and (ii) the 3′ region of bpsD and the 5′ region of oxyD, we amplified (i) a bhp-bpsD overlapping fragment (346 bp; contains 155 bp of the bhp end region and 105 bp of the bpsD start region) and (ii) a bpsD-oxyD overlapping fragment (341 bp; contains 185 bp of the bpsD end region and 137 bp of the oxyD start region) (Fig. 2). Thus, the existence of transcription termination sites between bhp and bpsD as well as between bpsD and oxyD could be excluded, and therefore the three genes oxyD, bhp, and bpsD are part of one operon.
FIG. 2.
RT-PCR investigations of oxyD cotranscription with the genes bhp and bpsD. The amplified regions are indicated with black bars. DNA, positive control with total DNA used as the PCR template; RNA, negative control with total RNA used as the PCR template; M, marker (100-bp ladder).
Inactivation of P450 monooxygenase gene oxyD.
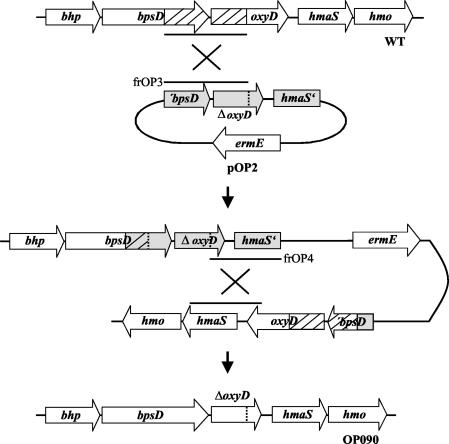
The fact that oxyD is cotranscribed with bhp and bpsD underscored the possibility of a coordinated function of these genes. To prove the participation of OxyD in β-HT synthesis, we constructed a null mutant of A. balhimycina with an in-frame deletion within oxyD (OP090). The gene replacement plasmid pOP2 (for construction details, see Materials and Methods), containing the oxyD gene with a 390-bp in-frame deletion, was used to transform the A. balhimycina wild-type strain by means of a modified direct transformation method (21). About 150 erythromycin-resistant transformants were obtained, indicating an integration of pOP2 via a first homologous recombination process. Nine randomly selected resistant colonies were tested for the ability to produce balhimycin in a bioassay. One of these colonies (OP090v) lacked production of an active compound. In this case, the homologous fragment frOP3 most likely was used for the integration, resulting in a bhp-bpsD-oxyD operon with an oxyD gene with an in-frame deletion (Fig. 3). Obviously, the intact oxyD gene copy downstream was inactive, most likely because of the missing natural promoter.
FIG. 3.
Construction of oxyD in-frame deletion strain OP090 by using plasmid pOP2 via homologous recombination. WT, A. balhimycina wild type; ermE, erythromycin resistance gene; frOP3 and frOP4, see Materials and Methods.
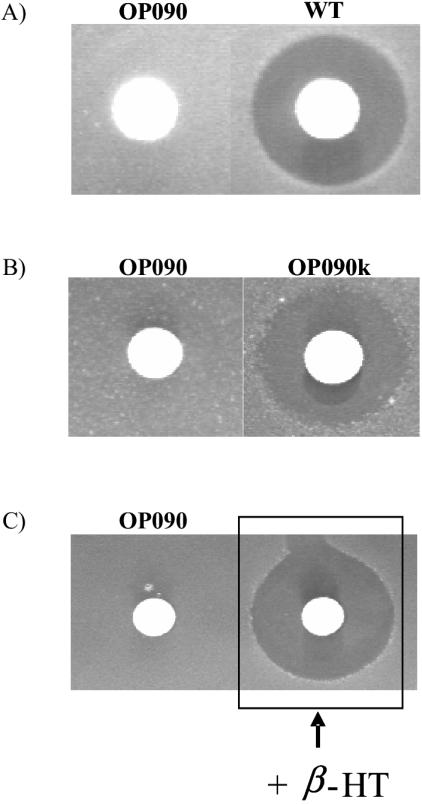
To obtain a deletion mutant, a second homologous recombination process was essential (Fig. 3). To provoke a second recombination, we placed strain OP090v under stress conditions as described previously (25), using temperature shifts and ultrasound treatment. After the application of the stress protocol, 500 colonies were examined on R5 plates with and without erythromycin. Four of the tested colonies lacked erythromycin resistance, indicating the loss of pOP2. The balhimycin production abilities of these colonies were tested in a bioassay. One colony (OP266) was able to produce balhimycin again, indicating a crossover event in the same homologous region of pOP2 as that used for the integration. In contrast, the other three colonies (OP090, OP163, and OP364) were unable to produce an active compound, indicating an exchange of the wild-type allele with the deleted oxyD gene as a result of the second recombination event (Fig. 4A).
FIG. 4.
(A) Bioassay with culture filtrates of oxyD mutant strain OP090 and A. balhimycina wild type (WT). (B) Bioassay with culture filtrates of OP090k and OP090. (C) Bioassay of feeding experiment with OP090 and β-HT. In each assay, 20 μl of culture filtrate was analyzed. The inhibition zones indicate growth inhibition of the B. subtilis test organism due to balhimycin production.
In the case of the mutant strain OP090, the in-frame deletion of oxyD was verified by PCR analysis, with total DNA used as a template (data not shown).
OP090 can be complemented by an additional oxyD gene copy.
To demonstrate that the loss of balhimycin production in OP090 was the result of only the deletion of oxyD and not of any additional mutational event, we introduced a complete copy of oxyD into the genome of OP090 by using the integrative vector pSET152. The integration of the complementation plasmid pSET-oxyD (for construction details, see Materials and Methods) into the OP090 chromosome resulted in the complemented mutant OP090k. PCR experiments with the total DNA of OP090k revealed that the integration of pSET-oxyD occurred at a neutral position in the genome, most likely at a ΦC31 attachment site, and not via homologous recombination into the chromosomal oxyD locus (data not shown). A bioassay with the supernatant of OP090k demonstrated the restoration of balhimycin production (Fig. 4B).
This result confirmed that the failure of OP090 to produce balhimycin was a result of only the deletion of oxyD. Therefore, OxyD plays an essential role in the balhimycin biosynthesis process. In further LC-ESI-MS investigations, no intermediates or variants of a higher molecular mass than 200 Da were detected in the culture filtrate of OP090 (data not shown), indicating the participation of OxyD in an early biosynthesis step.
OxyD participates in the β-HT formation pathway.
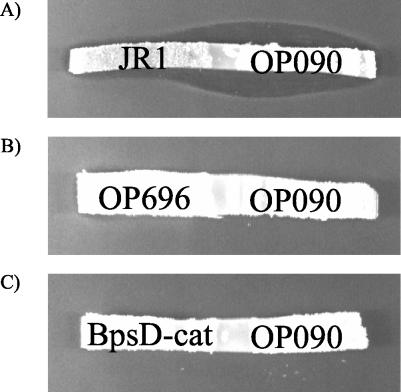
The previous data showed an essential function of OxyD in an early reaction of balhimycin biosynthesis, most likely within the β-HT formation pathway. To examine the involvement of OxyD in the synthesis of β-HT, we performed cross-feeding experiments (see Materials and Methods) with OP090, the bhp deletion mutant OP696 (25), and the bpsD disruption mutant BpsD-cat (27). In a first control experiment, the cross-feeding property between OP090 and the null mutant JR1 (blocked in HPG and DPG synthesis [23]) was investigated. Successful cross feeding was demonstrated by the appearance of an inhibition zone in both cases (Fig. 5A). The ability of OP090 to produce balhimycin in the neighborhood of JR1 demonstrated that OP090 had taken up an intermediate that was excreted by JR1 and converted it to balhimycin. This intermediate was likely a low-molecular-weight compound, since A. balhimycina is not able to take up intermediates with high molecular weights, such as, for example, the linear heptapeptide or the aglycone.
FIG. 5.
Cross-feeding experiments with strains OP090 and JR1 (A), OP090 and OP696 (B), and OP090 and BpsD-cat (C). In these bioassays, agar strips from R5 agar plates containing the mutants grown for 5 days at 30°C were used. The inhibition zones in panel A indicate the restoration of balhimycin biosynthesis by JR1 and OP090 due to the diffusional exchange of accumulated intermediates. No cross-feeding was detectable in panels B and C.
In further studies, the combinations OP090-OP696 and OP090-BpsD-cat were tested. No inhibition zones were detectable in the bioassays (Fig. 5B and C). The lack of cross feeding in these cases demonstrated that OP090 is blocked in the same pathway as OP696 and BpsD-cat, namely, β-HT synthesis. To further prove the inhibition within the β-HT formation pathway, we incubated OP090 in liquid medium containing β-HT dissolved at a concentration of 1 mg/ml. The harvested supernatant was then tested in a bioassay. An inhibition zone indicated the production of an active compound by OP090 in the presence of β-HT (Fig. 4C).
LC-ESI-MS studies confirmed that this active compound in the supernatant was balhimycin (Fig. 6). The data from cross-feeding experiments and β-HT feeding experiments clearly demonstrate that OP090 is blocked in β-HT synthesis.
FIG. 6.
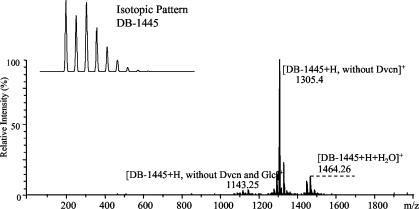
LC-ESI-MS analysis of OP090 complementation by feeding with nonchlorinated β-HT. A mass spectrum indicating production of the main metabolite balhimycin (DB-1445) and its variants and the isotopic pattern of DB-1445 typical for complete chlorination is shown. Glc, glucose; Dvcn, dehydrovancosamin.
Determination of chlorination time point.
A previous analysis of bhaA deletion mutants demonstrated the chlorination activity of BhaA in balhimycin biosynthesis (25). Attempts to establish an in vitro assay for the halogenating enzyme BhaA were not successful, probably because the natural substrate of BhaA is not available (K. H. van Pée, personal communication). We therefore intended to define the time point of chlorination (and thereby the substrate of BhaA) by using different mutants that were affected in balhimycin production.
Chlorination of balhimycin does not occur before or during formation of β-HT.
To exclude possible chlorination before or during β-HT formation, we used the oxyD null mutant strain OP090 (see above), which is blocked in β-HT synthesis, in feeding studies with nonchlorinated β-HT. After 5 days of growth, the supernatant was harvested and investigated by LC-ESI-MS analysis. The resulting mass spectrum unambiguously showed the production of balhimycin (molecular mass, 1,445 Da) (Fig. 6) and of variants that were also found in the culture filtrate of the wild-type strain (data not shown). To confirm that chlorinated balhimycin was synthesized, we measured the isotopic patterns (Fig. 6). They were identical to the theoretically calculated pattern of twofold chlorinated balhimycins.
These data clearly demonstrate that the halogenase BhaA is able to chlorinate either β-HT or an intermediate derived from it. In the case of a tyrosine precursor as a natural substrate of BhaA, a conversion of β-HT to chlorinated balhimycin should not have taken place. Therefore, a chlorination reaction at a biosynthetic stage earlier than free β-HT can be excluded.
Chlorination of balhimycin does not occur with free β-HT as a substrate of the halogenase.
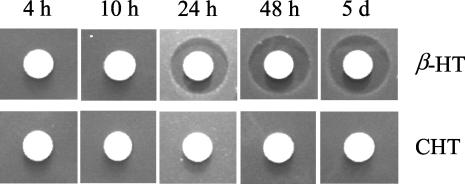
The data obtained from feeding studies with nonchlorinated β-HT revealed that the earliest time point of chlorination is the release of β-HT from BpsD. Therefore, we investigated whether the halogenase BhaA can use free β-HT as a substrate. In this case, CHT should represent a natural building block for heptapeptide backbone synthesis by the NRPS modules. To investigate whether CHT can be activated and introduced into the peptide core by peptide synthetase modules two and six, we fed CHT to strain OP090. Subsequently, we used a bioassay to analyze the biological activity of the supernatant after 4, 10, 24, and 48 h and after 5 days of incubation. No activity was detectable at any of the tested time points (Fig. 7). Whereas the supplementation of nonchlorinated β-HT in a control experiment led to the biosynthesis of active balhimycin (Fig. 7) after 24 h, the bioassay data revealed that the block of β-HT synthesis in OP090 cannot be complemented by CHT. These data were independently confirmed by LC-ESI-MS studies of culture filtrates.
FIG. 7.
Bioassay of CHT feeding studies with OP090. (Top) Control experiment with β-HT supplemented in the medium. The inhibition zones in the controls at 24 h, 48 h, and 5 days indicate the availability of the supplemented β-HT for balhimycin biosynthesis. (Bottom) Supplemented CHT does not restore the synthesis of balhimycin.
One reason for this could have been the general inability of A. balhimycina cells to take up the CHT dissolved in the medium. To exclude this possibility, we investigated the CHT concentration of the isolated supernatant probes by HPLC. Time-dependent monitoring of culture filtrates with HPLC revealed a distinct decrease in the CHT concentration during the incubation time, comparable to the decrease in the β-HT concentration in the control experiment (Fig. 8). In contrast, the concentrations of both amino acids were stable in cell-free medium, excluding spontaneous degradation as the reason for the reduction of dissolved CHT and β-HT.
FIG. 8.
Uptake of CHT and β-HT by A. balhimycina. Growth medium containing dissolved CHT (A) or β-HT (B) was incubated for 5 days in the absence (▪) or presence (▴) of growing A. balhimycina cells. The concentrations of CHT and β-HT were measured by HPLC.
Thus, like β-HT, CHT is certainly taken up, but it cannot be used as a substrate for peptide synthesis. Therefore, we can exclude the possibility that free chlorinated β-HT is a naturally occurring precursor. This fact clearly points to a chlorination time point later than the stage of free β-HT, most likely during the nonribosomal synthesis of the balhimycin heptapeptide core.
DISCUSSION
For our studies on the role of the putative P450 monooxygenase OxyD, we constructed the oxyD in-frame deletion mutant A. balhimycina OP090. Bioassays showed that OP090 lacked the ability to produce active balhimycin. The observed defect was restored by the integration of an intact oxyD gene. Additional mutations or polar effects on the genes downstream of oxyD could therefore be excluded as a putative cause of the OP090 phenotype. Thus, the production of active balhimycin by OP090 in the presence of β-HT clearly identified OxyD as an enzyme that is, like Bhp and BpsD, essential for β-HT synthesis. The functional cooperation of these genes is also reflected on the DNA level: the three genes are part of one common operon, which guarantees coordinated expression.
The cotranscription of genes whose enzyme products form a functional unit can also be found in other biosynthetic gene clusters. cloQ and cloR in the clorobiocin cluster of Streptomyces roseochromogenes DS 12.976 as well as novQ and novR in the novobiocin cluster of Streptomyces spheroides are likely to form an operon. These enzymes are involved in the biosynthesis of the prenylated 4-hydroxybenzoate moiety (RingA) derived from tyrosine (24). Furthermore, polyene antibiotic gene clusters contain very large genes encoding polyketide synthases, some of which seem to be cotranscribed. Such transcripts could be extremely long, e.g., encompassing >47 kb synthesized from the nysA (encodes the ketosynthase-acyl transferase-dehydratase-acyl carrier protein), nysB (encodes a bimodular protein which catalyzes the first two cycles of chain extension), and nysC (encodes extension modules 3 to 8, organized into hexamodular proteins) genes in Streptomyces noursei or from the corresponding amph genes in Streptomyces nodosus, the producers of nystatin and amphotericin, respectively (2). However, the coexpression of genes forming a functional unit is not a general observation. For example, expression studies of biosynthetic genes of the macrolide antibiotic tylosin revealed no coexpression and even no coregulation of the five genes coding for the polyketide synthases (30).
OxyD resembles OxyA, OxyB, and OxyC, which participate in linkage of the aromatic residues (6), but it has more significant similarity to the P450 monooxygenases NovI (56% similarity; 42% identity) and NikQ (52% similarity; 34% identity). NovI is responsible for the β-hydroxylation of a tyrosine intermediate covalently bound to a NRPS protein in the biosynthesis of the aminocoumarin antibiotic novobiocin (9). NikQ is a hydroxylating enzyme in the synthesis of β-hydroxyhistidine as a precursor of nikkomycin antibiotics (10).
In accordance with these reactions, we propose a hydroxylating function for OxyD, with a tyrosine bound to the NRPS BpsD as a substrate. This speculation is confirmed by the presence of the tyrosine-specific adenylation domain in BpsD (9). The inactivation of OxyD in A. balhimycina OP090 prevented the formation of β-HT, which is an important building block of the balhimycin heptapeptide backbone. The oxyD mutant, like mutants defective in bpsD and bhp, is an in-frame null mutant and does not produce balhimycin precursors since no active compound was detectable in the bioassay and no higher-molecular-weight balhimycin variant could be identified by the LC-ESI-MS analysis of the OP090 supernatant. Obviously, no naturally occurring alternative amino acid of β-HT, for example, tyrosine, can be incorporated at positions two and six of the heptapeptide backbone. This is in accordance with the results of tyrosine feeding studies with the bhp-deficient strain A. balhimycina OP696 (35).
In further studies, we used the oxyD mutant strain OP090 as a suitable tool for analyzing the chlorination time point in balhimycin biosynthesis. Even though BhaA, which belongs to the group of NADH/FAD-dependent halogenases, was identified as the enzyme catalyzing the chlorination of balhimycin at both positions (25), the substrate of this reaction has not been identified yet. The first hints that chlorination is not a tailoring reaction at a very late stage of glycopeptide biosynthesis were obtained with the mutant A. balhimycina SP1-1, which accumulated fully chlorinated linear heptapeptides as natural intermediates (22, 31). This means that the chlorination reaction occurs at a time point prior to oxidative cyclization through oxygenases OxyA/B and -C and subsequent glycosylation-methylation reactions. Since many identified NADH/FAD-dependent halogenases probably use substrates of a low molecular weight, for example, phenols and pyrrols (33), one might have speculated that chlorination in balhimycin biosynthesis occurs with tyrosine or β-HT as the natural substrate of BhaA. However, the incorporation and subsequent conversion of fed β-HT as well as the failure of the NRPSs BpsA and BpsB to use CHT as a building block for heptapeptide synthesis clearly exclude this possibility. Heptapeptide synthesis itself and further modification reactions such as epimerization are catalyzed by the NRPSs BpsA, BpsB, and BpsC (26) according to well-known rules (for reviews, see references 17 and 29).
The data obtained in this study are the first evidence that the chlorination process must be a reaction during heptapeptide synthesis, similar to those normally catalyzed by domains incorporated in the NRPS. Therefore, a close association of the halogenase with the NRPS machinery must be postulated.
Acknowledgments
This work was supported by grants from the European Union (MEGATOP, QLK3-1999-00650; and COMBIG-TOP, LSHG-CT-2003-503491) and the Deutsche Forschungsgemeinschaft (DFG) (Wo485/3-3 and SU 239/3-3). The work of R. D. Süssmuth was supported by an Emmy-Noether-Fellowship for young investigators of the DFG (SU 239/2-1).
We thank E. Takano for a critical reading of the manuscript.
REFERENCES
- 1.Altenbuchner, J., P. Viell, and I. Pelletier. 1992. Positive selection vectors based on palindromic DNA sequences. Methods Enzymol. 216:457-466. [DOI] [PubMed] [Google Scholar]
- 2.Aparicio, J. F., P. Caffrey, J. A. Gil, and S. B. Zotchev. 2002. Polyene antibiotic biosynthesis gene clusters. Appl. Microbiol. Biotechnol. 61:179-188. [DOI] [PubMed] [Google Scholar]
- 3.Arthur, M., F. Depardieu, P. Reynolds, and P. Courvalin. 1996. Quantitative analysis of the metabolism of soluble cytoplasmic peptidoglycan precursors of glycopeptide-resistant enterococci. Mol. Microbiol. 21:33-44. [DOI] [PubMed] [Google Scholar]
- 4.Beauregard, D. A., D. H. Williams, M. N. Gwynn, and D. J. C. Knowles. 1995. Dimerization and membrane anchors in extracellular targeting of vancomycin group antibiotics. Antimicrob. Agents Chemother. 39:781-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 116:43-49. [DOI] [PubMed] [Google Scholar]
- 6.Bischoff, D., S. Pelzer, B. Bister, G. J. Nicholson, S. Stockert, M. Schirle, W. Wohlleben, G. Jung, and R. D. Süβmuth. 2001. The biosynthesis of vancomycin-type glycopeptide antibiotics—the order of the cyclization steps. Angew. Chem. 40:4688-4691. [DOI] [PubMed] [Google Scholar]
- 7.Bullock, W. O., J. M. Fernandez, and J. M. Short. 1987. XL1-Blue, a high efficiency plasmid transforming recA Escherichia coli strain with beta galactosidase selection. Focus 5:376-378. [Google Scholar]
- 8.Chatterjee, S., E. K. S. Vijayakumar, S. R. Nadkarni, M. V. Patel, J. Blumbach, and B. N. Ganguli. 1994. Balhimycin, a new glycopeptide antibiotic with an unusual hydrated 3-amino-4-oxoaldopyranose sugar moiety. J. Org. Chem. 59:3480-3484. [Google Scholar]
- 9.Chen, H., and C. T. Walsh. 2001. Coumarin formation in novobiocin biosynthesis: β-hydroxylation of the aminoacyl enzyme tyrosyl-S-NovH by a cytochrome P450 NovI. Chem. Biol. 8:301-312. [DOI] [PubMed] [Google Scholar]
- 10.Chen, H., B. K. Hubbard, S. E. O'Connor, and C. T. Walsh. 2002. Formation of β-hydroxy histidine in the biosynthesis of nikkomycin antibiotics. Chem. Biol. 9:103-112. [DOI] [PubMed] [Google Scholar]
- 11.Choroba, O. W., D. H. Williams, and J. B. Spencer. 2000. Biosynthesis of the vancomycin group of antibiotics: involvement of an unusual dioxygenase in the pathway to (S)-4-hydroxyphenylglycine. J. Am. Chem. Soc. 122:5389-5390. [Google Scholar]
- 12.Gerhard, U., J. P. Mackay, R. A. Maplestone, and D. H. Williams. 1993. The role of the sugar and chlorine substituents in the dimerization of vancomycin antibiotics. J. Am. Chem. Soc. 115:232-237. [Google Scholar]
- 13.Harris, C. M., R. Kannan, H. Kopecka, and T. M. Harris. 1985. The role of the chlorine substituents in the antibiotic vancomycin: preparation and characterization of mono- and didechloro-vancomycin. J. Am. Chem. Soc. 107:6652-6658. [Google Scholar]
- 14.Hopwood, D. A., J. M. Bibb, K. F. Chater, T. Kieser, C. J. Bruton, H. M. Kieser, D. J. Lydiate, C. P. Smith, J. M. Ward, and H. Schrempf. 1985. Genetic manipulation of Streptomyces: a laboratory manual. The John Innes Foundation, Norwich, United Kingdom.
- 15.Hubbard, B. K., M. G. Thomas, and C. T. Walsh. 2000. Biosynthesis of l-p-hydroxy-phenylglycine, a non-proteinogenic amino acid constituent of peptide antibiotics. Chem. Biol. 7:931-942. [DOI] [PubMed] [Google Scholar]
- 16.Mackay, J. P., U. Gerhard, D. A. Beauregard, M. S. Westwell, M. S. Searle, and D. H. Williams. 1994. Glycopeptide antibiotic activity and the possible role of dimerization: a model for biological signaling. J. Am. Chem. Soc. 116:4581-4590. [Google Scholar]
- 17.Marahiel, M. A., T. Stachelhaus, and H. D. Mootz. 1997. Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem. Rev. 97:2651-2673. [DOI] [PubMed] [Google Scholar]
- 18.Nadkarni, S. R., M. V. Patel, S. Chatterjee, E. K. Vijayakumar, K. R. Desikan, J. Blumbach, and B. N. Ganguli. 1994. Balhimycin, a new glycopeptide antibiotic produced by Amycolatopsis sp. Y-86,21022. J. Antibiot. 47:334-341. [DOI] [PubMed] [Google Scholar]
- 19.Nagarajan, S. R. 1993. Structure-activity relationships of vancomycin-type glycopeptide antibiotics. J. Antibiot. 46:1181-1195. [DOI] [PubMed] [Google Scholar]
- 20.Pelzer, S. 1997. Etablierung eines wirts-vektor-systems und genetische Analyse des Balhimycin-Produzenten Amycolatopsis mediterranei DSM5908. Ph.D. thesis. University of Tübingen, Tübingen, Germany.
- 21.Pelzer, S., W. Reichert, M. Huppert, D. Heckmann, and W. Wohlleben. 1997. Cloning and analysis of a peptide synthetase gene of the balhimycin producer Amycolatopsis mediterranei DSM5908 and development of a gene disruption/replacement system. J. Biotechnol. 56:115-128. [DOI] [PubMed] [Google Scholar]
- 22.Pelzer, S., R. Süβmuth, D. Heckmann, J. Recktenwald, P. Huber, G. Jung, and W. Wohlleben. 1999. Identification and analysis of the balhimycin biosynthetic gene cluster and its use for manipulating glycopeptide biosynthesis in the producing organism Amycolatopsis mediterranei DSM5908. Antimicrob. Agents Chemother. 41:1565-1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pfeifer, V., G. J. Nicholson, J. Ries, J. Recktenwald, A. B. Schefer, R. M. Shawky, J. Schröder, W. Wohlleben, and S. Pelzer. 2001. A polyketide synthase in glycopeptide biosynthesis: the biosynthesis of the non-proteinogenic amino acid (S)-3,5-dihydroxyphenylglycine. J. Biol. Chem. 276:38370-38377. [DOI] [PubMed] [Google Scholar]
- 24.Pojer, F., S. M. Li, and L. Heide. 2002. Molecular cloning and sequence analysis of the clorobiocin biosynthetic gene cluster: new insights into the biosynthesis of aminocoumarin antibiotics. Microbiology 148:3901-3911. [DOI] [PubMed] [Google Scholar]
- 25.Puk, O., P. Huber, D. Bischoff, R. Recktenwald, G. Jung, R. D. Süβmuth, K.-H. van Pée, W. Wohlleben, and S. Pelzer. 2002. Glycopeptide biosynthesis in Amycolatopsis mediterranei DSM5908: function of a halogenase and a haloperoxidase/perhydrolase. Chem. Biol. 9:225-235. [DOI] [PubMed] [Google Scholar]
- 26.Recktenwald, J. 1999. Molekulare Untersuchungen zur Synthese des Peptid-rückgrats von Balhimycin. Ph.D. thesis. University of Tübingen, Tübingen, Germany.
- 27.Recktenwald, J., R. M. Shawky, O. Puk, F. Pfennig, U. Keller, W. Wohlleben, and S. Pelzer. 2002. Nonribosomal biosynthesis of vancomycin-type antibiotics: a heptapeptide backbone and eight peptide synthetase modules. Microbiology 148:1105-1118. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Sieber, S. A., and M. A. Marahiel. 2003. Learning from nature's drug factories: nonribosomal synthesis of macrocyclic peptides. J. Bacteriol. 185:7036-7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stratigopoulos, G., and E. Cundliffe. 2002. Expression analysis of the tylosin-biosynthetic gene cluster: pivotal regulatory role of the tylQ product. Chem. Biol. 9:71-78. [DOI] [PubMed] [Google Scholar]
- 31.Süßmuth, R., S. Pelzer, G. Nicholson, T. Walk, W. Wohlleben, and G. Jung. 1999. New advances in the biosynthesis of glycopeptide antibiotics of the vancomycin type from Amycolatopsis mediterranei. Angew. Chem. 38:1976-1979. [DOI] [PubMed] [Google Scholar]
- 32.Thompson, C. J., T. Kieser, J. M. Ward, and D. A. Hopwood. 1982. Physical analysis of antibiotic-resistance genes from Streptomyces and their use in vector construction. Gene 20:51-62. [DOI] [PubMed] [Google Scholar]
- 33.van Pée, K. H. 2001. Microbial biosynthesis of halometabolites. Arch. Microbiol. 175:250-258. [DOI] [PubMed] [Google Scholar]
- 34.Vértesy, L., H. W. Fehlhaber, H. Kogler, and M. Limbert. 1996. New 4-oxovancosamine-containing glycopeptide antibiotics from Amycolatopsis sp. Y-86,20122. J. Antibiot. 49:115-118. [DOI] [PubMed] [Google Scholar]
- 35.Weist, S., B. Bister, O. Puk, D. Bischoff, G. Nicholson, S. Stockert, W. Wohlleben, G. Jung, and R. D. Süβmuth. 2002. Fluorobalhimycin—a new chapter in glycopeptide antibiotic research. Angew. Chem. 41:3383-3385. [DOI] [PubMed] [Google Scholar]
- 36.Wink, J., R. M. Kroppenstedt, B. N. Ganguli, S. R. Nadkarni, P. Schumann, G. Seibert, and E. Stackebrandt. 2003. Three new antibiotic producing species of the genus Amycolatopsis, Amycolatopsis balhimycina sp. nov., A. tolypomycina sp. nov., A. vancoresmycina sp. nov., and description of Amycolatopsis keratiniphila subsp. keratiniphila subsp. nov. and A. keratiniphila subsp. nogabecina subsp. nov. Syst. Appl. Microbiol. 26:38-46. [DOI] [PubMed] [Google Scholar]
- 37.Yao, R. C., and L. W. Crandal. 1994. Glycopeptides: classification, occurrence, and discovery, p. 1-28. In R. Nagarajan (ed.), Glycopeptide antibiotics. Marcel Dekker Inc., New York, N.Y.
- 38.Zerbe, K., O. Pylypenko, F. Vitali, W. Zhang, S. Rouset, M. Heck, J. W. Vrijbloed, D. Bischoff, B. Bister, R. D. Süβmuth, S. Pelzer, W. Wohlleben, J. A. Robinson, and I. Schlichting. 2002. Crystal structure of OxyB, a cytochrome P450 implicated in an oxidative phenol coupling reaction during vancomycin biosynthesis. J. Biol. Chem. 277:47476-47485. [DOI] [PubMed] [Google Scholar]