Abstract
DsrA RNA is a small (87-nucleotide) regulatory RNA of Escherichia coli that acts by RNA-RNA interactions to control translation and turnover of specific mRNAs. Two targets of DsrA regulation are RpoS, the stationary-phase and stress response sigma factor (σs), and H-NS, a histone-like nucleoid protein and global transcription repressor. Genes regulated globally by RpoS and H-NS include stress response proteins and virulence factors for pathogenic E. coli. Here, by using transcription profiling via DNA arrays, we have identified genes induced by DsrA. Steady-state levels of mRNAs from many genes increased with DsrA overproduction, including multiple acid resistance genes of E. coli. Quantitative primer extension analysis verified the induction of individual acid resistance genes in the hdeAB, gadAX, and gadBC operons. E. coli K-12 strains, as well as pathogenic E. coli O157:H7, exhibited compromised acid resistance in dsrA mutants. Conversely, overproduction of DsrA from a plasmid rendered the acid-sensitive dsrA mutant extremely acid resistant. Thus, DsrA RNA plays a regulatory role in acid resistance. Whether DsrA targets acid resistance genes directly by base pairing or indirectly via perturbation of RpoS and/or H-NS is not known, but in either event, our results suggest that DsrA RNA may enhance the virulence of pathogenic E. coli.
Regulation by RNA, termed riboregulation, plays a substantial role in modulating gene expression in bacteria (reviewed in references 14, 17, 26, 33, 42, 43, and 44). In addition to their classically studied role in plasmid maintenance (reviewed in reference 42), Escherichia coli small RNAs act to change the conformation of target mRNAs (DsrA and RprA), block mRNA translation by occlusion of Shine-Dalgarno sequences (MicF, OxyS, Spf, and RyhB), degrade target mRNAs (DsrA, Oop, and RyhB), and titrate specific protein factors (OxyS and CsrB RNAs, 6S RNA), sometimes in combination.
Several E. coli small RNAs coordinate stress responses or virulence factors (reviewed in reference 14). A principal advantage of small RNAs as regulators is that they are not translated and therefore cost less energy to produce than do proteins. Also, many bacterial small RNAs are relatively stable and can persist to target transcripts with high specificity by antisense interactions (reviewed in references 17 and 43). Some small RNAs are degraded together with their target mRNAs (22).
One such RNA, DsrA RNA, is a small (87 nucleotides), multifunctional genetic regulator of E. coli. DsrA RNA modulates the levels of two global transcription regulators, RpoS (σs, the product of the rpoS gene) and H-NS (a nucleoid protein and transcription silencer in bacteria, produced from the hns gene). DsrA acts by sequence-specific RNA-RNA interactions to enhance translation of rpoS RNA and to stabilize rpoS message (18-20, 35). In addition to its role at rpoS, DsrA also binds hns mRNA by specific base-pairing interactions and blocks H-NS translation as it sharply increases hns mRNA turnover (18). The first stem-loop region of DsrA melts out to contact rpoS mRNA, whereas the second stem-loop region of DsrA base pairs with hns mRNA. This conformational change within DsrA acts to switch the translation state of two different mRNA targets (reviewed in references 1 and 17). Like that of many small, noncoding regulatory RNAs, direct DsrA activity on mRNAs requires Hfq, an Sm domain RNA-binding protein and putative RNA chaperone (reviewed in reference 43).
DsrA perturbation of H-NS and RpoS results in increased transcription of downstream genes repressed by H-NS or activated by RpoS (19, 35). H-NS and RpoS also act in concert to permit the transcription of a number of stress response and virulence factor proteins (reviewed in references 3 and 15). Many genes require additional regulatory proteins, in conjunction with H-NS and RpoS, to tailor specific responses to particular environmental stresses (reviewed in reference 15). The downstream effect of DsrA is therefore predicted to be the induction of a pleiotropic stress response.
Despite the study of key components of the DsrA regulatory network, the phenotype of DsrA activity in the cell has remained elusive. Here we used a genomics approach to define the downstream effects of DsrA in E. coli. DNA array-based transcriptome analysis suggests that DsrA stimulates acid resistance, which is known to enhance the virulence of pathogenic E. coli strains (9 and references therein). Both the hdeAB and glutamate-dependent (gad) acid resistance systems were induced, although the arginine-dependent (adi) genes were apparently not induced by DsrA. Furthermore, both nonvirulent (K-12) and pathogenic (O157:H7) strains of E. coli had compromised acid resistance in dsrA-null mutants. DsrA therefore plays a role in cellular acid resistance, an important feature for the survival of enteric bacteria in low-pH environments and for the virulence of pathogenic E. coli.
MATERIALS AND METHODS
Media, strains, and plasmids.
Cells were grown in Luria-Bertani (LB) medium (28). Where appropriate, antibiotics were used at the following concentrations: ampicillin, 100 μg/ml; chloramphenicol, 25 μg/ml; tetracycline, 10 μg/ml; kanamycin, 50 μg/ml; streptomycin, 50 μg/ml. E. coli strain M182 is described elsewhere (8). E. coli O157:H7 strain 1957 is a clinical isolate from stool or fecal material from 2001, provided by the Wadsworth Center Bacteriology Laboratory. Strain M182 dsrA::cat was made by P1 transduction of the cat gene from C600 dsrA::cat, which was provided by Susan Gottesman (National Institutes of Health, Bethesda, Md.). E. coli strain SM10 tra+ Kanr (λpir) (32) was provided by Kelynne Reed (Austin College, Sherman, Tex.).
Strain O157 dsrA::cat was constructed as follows. The dsrA::cat allele, along with ca. 900 bp of the flanking chromosomal DNA sequence, was amplified by PCR with primers W1748 (GCT CTA GAA AGA GAC AAC GAT AAC CTC G) and W1749 (GCT CTA GAG CGT AAT CCA TTA CCT CCA G) and was cloned by blunt-end ligation into λpir-dependent gene replacement vector pCVD442 (ori-R6K mob+ Ampr sacB) (11) at a filled-in XbaI restriction site. The recombinant plasmid DNA was used to transform DH5α (λpir) and plated on LB medium containing chloramphenicol and ampicillin. The resulting plasmid, pCVD442 dsrA::cat, was screened by restriction analysis and then used to transform E. coli SM10. Separately, a streptomycin-resistant variant of O157:H7 was selected by plating on LB medium containing streptomycin. This O157 (Strr) strain was mated with SM10 Kanr/pCVD442dsrA::cat in liquid culture for 2 h at 37°C. E. coli O157 Strr/pCVD-dsrA::cat was selected on plates (LB medium containing streptomycin and chloramphenicol). Selected clones were checked for kanamycin sensitivity and ampicillin resistance by patching of colonies onto plates. To prepare dsrA::cat chromosomal integrants, gene replacement was performed by growing cultures of O157 Strr/pCVDdsrA::cat and plating on LB medium containing 5% sucrose and chloramphenicol. Most sucrose-resistant colonies were Amps, indicating loss of the pir-dependent gene replacement vector. The exchange of dsrA for the dsrA::cat chromosomal allele was verified by chloramphenicol resistance and by PCR with primers W1748 and W1749.
Plasmids pBRdsrA and pBRdsrA*H were constructed by cloning the BamHI fragment of pDDS164 (34) or pDsrA*H (19) into the BamHI site within the Tetr gene of pBR322. Potential clones were selected for ampicillin resistance and then screened for insertional inactivation of the Tetr gene (Tets) on LB medium containing tetracycline. Clones were confirmed by restriction analysis and DNA sequencing.
Transcription profiling.
Freshly streaked cells of strain M182 containing either pBR322 or pBRdsrA were grown overnight in LB broth plus ampicillin at 37°C. Cells were then diluted 1:100 into 30 ml of LB broth plus ampicillin for growth at 30°C to induce DsrA (35). Cells were grown with vigorous shaking to an optical density at 600 nm of 0.3 to 0.4. Total cellular RNA was prepared and used to make labeled cDNA by reverse transcription in the presence of [α-33P]dATP (NEN). An oligonucleotide mixture complementary to every E. coli mRNA 3′ end (Sigma-Genosys) was used to prime cDNA synthesis. The 33P-labeled total cellular cDNA was used to probe filter-based DNA arrays (Sigma-Genosys) as previously described (39). Filters were then exposed to phosphorimager screens (Molecular Dynamics, Inc.) and scanned. Images were quantified with Arrayvision software (Imaging Research, St. Catharines, Ontario, Canada). The experiment was performed twice. Hybridization signals were normalized to total genomic DNA standards present on each filter. Differential expression for each set of experiments was determined for individual genes by dividing the signal from the DsrA-overproducing strain by that of the plasmid control. For images of arrays and spot data files, see the supplemental material.
Primer extension analysis.
RNA was extracted and primer extension was performed as previously described (5, 18). Primer sequences are as follows: adiA + 74, CTT GAT GGA GAA ACT CGC TTT CAA C; adiY + 83, AGT TCT CGC TAA AGC AAA GCG ATA C; gadA + 48, CGT TAA CAG CTT CTG GTC CAT TTC G; gadB + 64, GTT CCG ACC TTA AAT CCG TTA CTT G; gadX + 89, GGT GAG AAT ATA TTT ATG TCT TGC; nfnB + 37, TAT CCA TAA AGA CTC CAT GTG AAA G; W538dsrA, GAA ACT TGC TTA AGC AAG AAG C. The hns- and stpA-specific primers (48) and the hdeA-specific primer (2) are as previously described. All DNA oligonucleotides were purified via elution from polyacrylamide gels. The DNA oligonucleotides were end labeled with [γ-32P]ATP (Perkin-Elmer/NEN) and T4 polynucleotide kinase (New England Biolabs) as previously described (28), extracted with 1 volume each of phenol and then with chloroform-isoamyl alcohol (24:1), and purified by TE-10 spin column chromatography (Clontech).
Acid resistance assays.
Cultures of strains M182 and O157 and their respective dsrA mutants and merodiploid strains were grown overnight at 37°C and tested by dilution into LB medium at pH 2.0 as previously described (13), except that samples were taken each hour for plating for up to 6 h of acid treatment at 37°C. Cells were diluted in 10 mM Tris-HCl (pH 7.5)-1 mM magnesium chloride prior to plating. Percent survival is given as the titer of the CFU of acid-tested cells compared to that of a zero-time, untreated control sample.
RESULTS
Global mRNA analysis.
To determine the downstream effects of DsrA, we induced DsrA expression from a plasmid. Changes in the expression of total cellular RNA from cultures with and without DsrA overproduction at 30°C were compared by using genomic DNA filter arrays. Whereas transcript levels from many genes increased or decreased severalfold (Table 1), a number of genes related to acid resistance were strongly induced by DsrA overproduction (Table 1, top). The hdeAB operon, which encodes the H-NS-repressed acid resistance proteins HdeA and HdeB (12, 46, 47), produced 10- to 26-fold higher hdeA transcript levels and 12- to 43-fold higher hdeB transcript levels when DsrA was overproduced (Table 1, lines 1 and 2). Other H-NS- and RpoS-regulated acid resistance operon elements were also induced. For example, the gadAX operon, which encodes glutamate decarboxylase, GadA, and its positive regulator, GadX (40), was induced as much as threefold (Table 1, lines 4 and 5). Similarly, a gene that encodes a positive regulator of arginine-dependent acid resistance, adiY, was induced ca. fivefold (Table 1, line 7) (38).
TABLE 1.
DsrA functional genomics in E. coli K-12
| Category and genes | Location (min)a | Function | Fold change
|
|
|---|---|---|---|---|
| Filter arrayb | Primer extensionc | |||
| Acid resistance | ||||
| 1 hdeA | 78.76 | Acid resistance | 10-26 | 26 |
| 2 hdeB | 78.75 | Acid resistance | 12-43 | — |
| 3 hdeD | 78.78 | Acid resistance | 2.2-3.4 | — |
| 4 gadA | 78.98 | Acid resistance | 2.3-3.0 | 3.5 |
| 5 gadX (alias yhiX) | 78.95 | Acid resistance-virulence regulator | 2.3-3.2 | 2.6 |
| 6 gadB | 33.81 | Acid resistance | 1 | 3.5 |
| 7 adiY | 93.44 | Acid resistance regulator | 3.7-5.7 | NS |
| 8 adiA | 93.46 | Acid resistance | 1 | NS |
| Membrane or transport | ||||
| 9 proX | 60.47 | Osmotic shock | 3.3-3.9 | — |
| 10 ompX | 18.32 | Outer membrane protein | 2.3-4.5 | — |
| 11 ompA | 21.95 | Outer membrane protein | 2-2.6 | — |
| 12 ompF | 21.23 | Outer membrane protein | 2-2.5 | — |
| 13 ompT | 12.59 | Outer membrane protein | 1.9-3.1 | — |
| 14 rbsB | 84.80 | Periplasmic ribose-binding proteind | −2.6-3.4 | — |
| Regulatory | ||||
| 15 malT | 76.54 | Activator-maltose regulon | −2.3-2.5 | — |
| 16 rpoE | 58.36 | Activator-sigma factor | −2-2.5 | — |
| 17 hns | 27.84 | Repressor-nucleoid | 1 | 1.3e |
| 18 stpA | 60.27 | Repressor-nucleoid-RNA chaperone | 1.2-2.3 | 3.1 |
| 19 rcsA | 43.58 | Activator-capsule synthesis | 1.3-1.4 | ∼10f |
| 20 dps | 18.27 | DNA protection | 1.9-3.4 | — |
| Motility | ||||
| 21 fimA | 97.88 | Fimbriae | 2.2-3.5 | — |
| 22 fliA | 43.09 | Flagella | −2.1 | — |
| 23 fliC | 43.11 | Flagella | −2.6-7.3 | — |
| Other | ||||
| 24 ftn | 42.83 | Ferritin (iron storage) | 2.8-3.6 | — |
| 25 xylA | 80.34 | d-Xylose isomerase | 2.5-2.9 | — |
| 26 sucA | 16.34 | 2-Ketoglutarate dehydrogenase | −2-2.3 | — |
| 27 groS (alias mopB) | 94.16 | Chaperonin | −2.6-2.9 | — |
| 28 garR (alias yhaE) | 70.48 | Tartronate semialdehyde reductase | −2.3-3.2 | — |
| 29 sgbU (alias yiaR) | 80.77 | Hexulose 6-phosphate isomerase (putative) | −2.4-3.4 | — |
| 30 hchA (alias yedU) | 43.84 | Heat-inducible chaperone | 4.7-5.9 | — |
| 31 rihC (alias yaaF) | 0.59 | Ribonucleoside hydrolase | −2-2.2 | — |
| 32 nfnBg | 13.02 | Nitroreductase (control)g | 1 | 1.2g |
| Hypothetical protein | ||||
| 33 yagU | 6.51 | Alias b0287 | 2.6-2.8 | — |
| 34 yncC | 32.73 | Alias b1450 | 2.1-4.4 | — |
| 35 yeaK | 40.34 | Alias b1787 | 2-2.9 | — |
| 36 yeaP | 40.43 | Alias b1794 | 2-2.2 | — |
| 37 yeaQ | 40.46 | Alias b1795 | 2.9-3.3 | — |
| 38 yedW | 43.89 | Alias b1969 | 2.3-4 | — |
| 39 ybfE | 15.32 | Alias b2253 | 2.1-3.2 | — |
Sigma-Genosys nylon membrane gene array. Fold changes for each experiment (n = 2) are given as ranges. All values that decrease are so indicated by minus signs.
Average fold effect (n = 3 or 4). NS, no extension product seen. —, primer extension not performed.
DsrA also contains an antisense sequence complementary to rbsD in the rbsDACBK operon (19).
The hns mRNA levels do not change, because of H-NS autoregulation, while H-NS protein levels decrease. Values shown here are comparable to those seen previously for primer extension quantitation (19). RpoS mRNA is stabilized, and levels slightly increased (18).
The rcsA transcript levels were determined previously (34). Visual estimate of primer extension was ∼10-fold. Activity of a reporter gene fusion was elevated 13-fold.
The nfnB gene was unaffected by DsrA overproduction and was used as a control.
Primer extension analysis.
To verify and quantitate induction of the specific genes mentioned above, as well as to check for the overexpression of several genes expected to be induced that were not seen by array analysis, we measured the levels of key transcripts by gene-specific primer extension analysis. Cells containing plasmid vector were compared to cells overproducing either DsrA or a mutant DsrA variant from a plasmid. Identical growth conditions were used for the array and primer extension analyses. The mRNA levels of the acid resistance genes hdeA, gadA, gadX, gadB, adiY, and adiA were analyzed (Fig. 1; quantitation in Table 1, column five; data for adiY and adiA not shown). As a positive control, we determined the transcript levels of stpA. StpA is an H-NS paralog that is induced in hns mutants (48) and that registered a 1.2- to 2.3-fold increase in the array analysis (Table 1, line 18). As a negative control, levels of the unrelated nfnB nitroreductase mRNA were also checked (4).
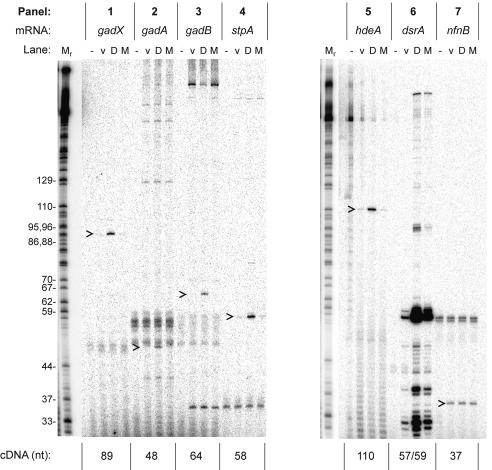
FIG. 1.
Quantitative primer extension analysis of acid resistance genes. RNA was extracted from cells with a vector plasmid (lanes v), a DsrA-overproducing plasmid (lanes D), or a plasmid overproducing inactive mutant DsrA (lanes M). A minus sign indicates a no-RNA control lane. The labeled cDNA products of primer extension were analyzed by polyacrylamide gel electrophoresis; representative gel data are shown. The size of the major cDNA product is given in nucleotides below the panel. The name of each mRNA (top) corresponds to transcripts originating from the major promoter of each gene. The nfnB gene was tested as an unregulated control. Carets indicate relevant primer extension products. A dideoxy-GTP sequencing ladder of an unrelated RNA (Tetrahymena thermophila L-21 IVS) was used as a size marker. The values on the left are sizes in nucleotides. nt, nucleotides.
The data confirm the induction of several types of acid resistance genes by DsrA, and the lengths of all cDNAs are consistent with transcription starting at the major promoter of each gene. The levels of hdeA transcript were increased 26-fold, corresponding exactly to those determined by the array analysis (Fig. 1, panel 5, and Table 1, line 1). The levels of gadA mRNA were also increased, by a factor of 3.5, again providing correspondence between the two methods (Fig. 1, panel 2, and Table 1, line 4), and stpA mRNA was increased 3.1-fold (Fig. 1, panel 4). A modest 2.6-fold increase observed for gadX mRNA was again corroborative (Fig. 1, panel 1, and Table 1, line 5). Interestingly, we also found a 3.5-fold increase in gadB mRNA, whereas such induction had been undetectable by array analysis (Fig. 1, panel 3, and Table 1, line 6). We attribute the differences between the two methods to the fact that oligonucleotide cDNA primers for the array analysis were optimized for cloning of the PCR products displayed on the arrays (25) and not necessarily for annealing to the specific mRNAs. Conversely, although the adiY transcript was 3.7- to 5.7-fold elevated in the array analysis (Table 1, line 7), we were unable to detect either adiY or adiA transcripts by primer extension analysis (data not shown). Regardless, since gadB and gadC form an operon and since gadC is required for glutamate decarboxylase-based acid resistance (9), we can conclude that both the hdeAB and glutamate-dependent acid resistance (gad) genes are induced by DsrA.
To confirm that changes in transcript levels resulted from DsrA function, we measured acid resistance gene transcripts after induction of a DsrA variant, DsrA*H, which is compromised in its ability to regulate hns or rpoS (Fig. 1, all panels, lane M) (19). Variant plasmid pBRdsrA*H generated levels of DsrA*H mutant RNA comparable to those generated by the wild-type DsrA plasmid (Fig. 1, panel 6, compare lanes D and M). The plasmids that produce DsrA and DsrA*H are isogenic except for five point mutations within the dsrA gene. DsrA*H is unable to base pair with hns mRNA and cannot significantly activate the translation of rpoS mRNA, although DsrA*H can pair with an altered hns allele (19). These data indicate that DsrA and not other plasmid products or sequences induced these acid resistance genes.
Acid resistance phenotype test.
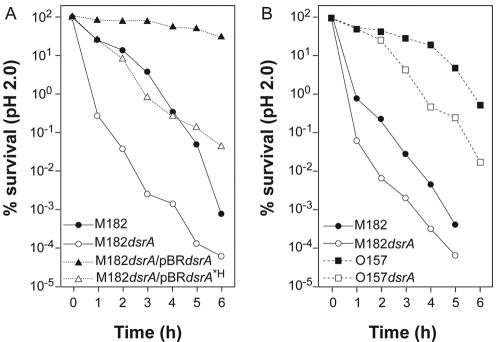
The pattern of gene induction described above suggests a role for DsrA in acid resistance. Consistent with these observations, in preliminary experiments E. coli K-12 strain M182, overexpressing DsrA from one of several plasmids, displayed increased acid tolerance at pH 3.8 by a factor of 12- to 5,000-fold relative to that of control M182 cells (data not shown). To confirm a physiological role for DsrA in acid resistance, we compared E. coli M182 to an otherwise isogenic dsrA-null mutant (M182 dsrA::cat) for the ability to survive immersion in pH 2 medium. Growth and acid treatment were at 37°C. The K-12 strains with dsrA deleted were killed more readily at pH 2 than was the wild type (Fig. 2A and B, compare filled and open circles). The downward trend indicates a 102- to 103-fold disadvantage in survival at low pH between 1 and 5 h for the dsrA mutant, supporting a physiological role for DsrA.
FIG. 2.
Plating assay for acid resistance. The percent survival of E. coli and isogenic dsrA mutants and overexpressers is plotted against time spent at pH 2.0. (A) Comparison of K-12 strain M182 with different levels of DsrA and DsrA*H. Filled circles represent wild-type M182, and open circles represent dsrA::cat, a null variant of M182. Filled triangles represent dsrA::cat complemented with wild-type DsrA from a plasmid; open triangles represent dsrA::cat complemented with the plasmid-encoded dsrA*H variant. Results are the average of three trials. The average standard deviation is <11% and never greater than 23%. (B) Comparison of K-12 strain M182 and EHEC strain O157:H7. Circles represent nonvirulent K-12 strain M182; squares represent pathogenic E. coli O157:H7. Filled symbols represent dsrA+ strains; open symbols represent dsrA mutant strains. Representative data are shown from at least three independent trials; all trials gave similar trends, but with sufficient variability that the data were not superimposable.
To verify that DsrA is responsible for the acid resistance seen, we complemented the dsrA-null strain with wild-type or mutant DsrA produced in trans from a plasmid (Fig. 2A, filled and open triangles). Wild-type DsrA overproduction in the dsrA-null mutant rendered M182 strongly acid resistant even up to 6 h of treatment at pH 2. The strain overproducing DsrA was up to 106-fold more acid resistant than the dsrA mutant.
Curiously, we saw a partial restoration of acid resistance in the dsrA-null mutant when we overproduced the altered DsrA*H variant in trans from a plasmid (Fig. 2A, compare open circles and open triangles). The DsrA*H-producing plasmid restored a level of acid resistance comparable to that of the wild-type strain (Fig. 2A, cf. filled circles and open triangles). However, wild-type DsrA was considerably more effective than DsrA*H for promoting acid resistance survival when overproduced (103-fold difference at 6 h) (Fig. 2A, cf. open and filled triangles). As DsrA*H cannot act directly on hns or rpoS RNA, an independent mechanism of acid resistance is implied, possibly via direct DsrA base pairing with other RNAs, such as, for example, putative mRNA targets argR, ilvI, and rbsD (19). Nevertheless, the strong and persistent acid resistance phenotype of the DsrA-overproducing strain, coupled with compromised acid resistance in dsrA-null mutants relative to the wild type, substantiates the physiological relevance of DsrA in acid resistance.
Acid resistance in a pathogen.
The ability to survive at a low pH in the stomach is considered to be a factor that permits pathogenic bacteria to establish an infection (9 and references therein). We theorized that the enterohemorrhagic pathogen E. coli O157:H7 might use DsrA to induce acid resistance. Accordingly, we assayed E. coli O157:H7 and its dsrA null mutant for acid resistance. An E. coli O157 clinical isolate displayed patterns of acid resistance different from those of K-12 laboratory strain M182 (Fig. 2B, cf. circles and squares), with the pathogenic strain E. coli O157 being considerably more acid resistant than K-12. Again we found a clear trend of compromised acid resistance in the dsrA::cat mutant (Fig. 2B, cf. filled and open squares), with a 10- to 75-fold reduction in acid resistance of E. coli O157 in the absence of DsrA.
DISCUSSION
DsrA global effects.
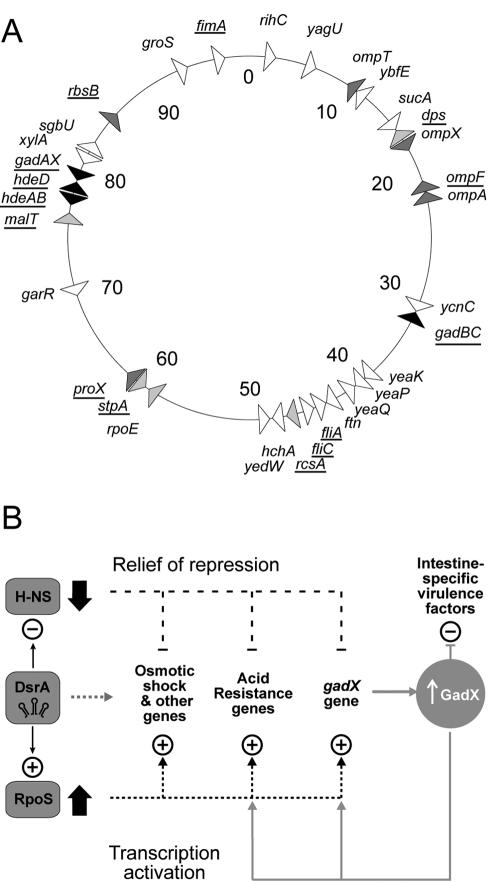
The mRNA levels of many genes increase, while levels of several mRNAs decrease in response to DsrA overproduction (Table 1). The affected genes are distributed around the chromosome and perform multiple functions (Table 1 and Fig. 3A). Most notably, acid resistance genes are induced by DsrA overexpression (Table 1 and Fig. 1). Apart from acid resistance, no single unifying theme emerged from our analysis of these data, although regulated genes include membrane proteins and sugar metabolism operons (Fig. 3A). These array data, in combination with studies of specific genes, serve as a departure point for generation of testable hypotheses. The role of condition-specific regulatory factors may predominate; for example, only two of the flagellar regulatory and synthesis genes were induced (Table 1, lines 22 and 23), and not a multigene cascade. Thus, hypotheses generated from these data could be individually tested under appropriate conditions and strains, as was done for acid resistance (Fig. 2). Not all genes were detected by arrays (e.g., stpA, Table 1, line 18), possibly because of suboptimal annealing of cDNA primers, mRNA secondary structures, or degradation at the 3′ ends.
FIG. 3.
Control by DsrA. (A) Map of DsrA-responsive loci. Triangles indicate the direction of gene transcription. Black, acid resistance genes; light grey, osmotic-shock genes; dark grey, regulatory genes; white, other genes. Genes known to be H-NS/RpoS regulated are underlined (see the supplemental material for references). (B) DsrA regulatory circuits. +, activation; −, repression. Different environmental signals lead to increased DsrA, which binds hns mRNA to block translation and binds rpoS mRNA to increase translation (left) (17). A decrease in H-NS protein (↓) relieves the repression of genes, as an increase in RpoS protein (↑) coordinately activates transcription of genes, resulting in increased acid resistance and virulence. A grey dashed line to the right of DsrA shows putative direct DsrA binding to other target mRNAs. A secondary GadX circuit (solid gray lines) maintains acid resistance and blocks the production of strain-specific virulence factors, such as Per and LEE, as described in the text.
DsrA enhances acid resistance.
In comparisons of wild-type and dsrA-null mutants, both E. coli K-12 and pathogenic O157 strains are compromised in the ability to survive a low-pH challenge if dsrA is knocked out (Fig. 2). Also, dsrA mutants were rendered strongly acid resistant by overproduction of DsrA in trans. Taken together, these results imply a physiological role for DsrA in acid resistance. Multiple acid resistance responses (gadAX, gadBC, and hdeAB operons) are depicted as part of a DsrA regulatory circuit (Fig. 3B). DsrA presumably acts indirectly, via H-NS and RpoS. However, in some cases DsrA may act directly (Fig. 3B, gray dotted arrow), by RNA-RNA interactions with specific target mRNAs, as it does with hns and rpoS. One such example is the periplasmic ribose-binding protein operon (rbs) transcript, which was identified as a potential DsrA target by virtue of complementarity (19), and which is depressed about threefold when DsrA is induced (Table 1, line 14).
The greater effectiveness of wild-type DsrA than DsrA*H in complementing the dsrA-null mutant for survival at low pH should be considered (Fig. 2A). Clearly, DsrA*H cannot induce hde or gad acid resistance genes (Fig. 1). It is feasible that either DsrA*H or plasmid sequences titrate out a regulatory molecule such as Hfq or LeuO, respectively (16, 36). LeuO and Hfq have both been shown to complement an hns mutant for repression of certain acid-inducible genes (29, 30). Another possibility is that an additional, H-NS-independent pathway is responsible for DsrA*H partially complementing the DsrA null mutant. DsrA interacts with hns and rpoS mRNAs by different base pairing via discrete portions of the DsrA molecule (17). The hypothesis that DsrA might interact with another target via different nucleotides than those mutated in DsrA*H is therefore a viable option. DsrA contains regions of antisense complementarity to at least another three genes, namely, argR, ilvI, and rbsD (19), although direct base pairing between DsrA and these mRNAs has not been demonstrated.
Other acid resistance gene networks that are independent of DsrA (e.g., those regulated by EvgA-YdeO-YhiE or GadW) undoubtedly combine with these networks to impart acid resistance (23, 41). It is noteworthy that E. coli O157 possesses the same three acid resistance systems found in K-12 (9). Since E. coli O157 contains >1.4 Mb of DNA absent from K-12 (reviewed in reference 45), it is likely that the pathogen utilizes additional and divergent regulation of gene expression that significantly enhances acid resistance.
Concerted acid and osmotic shock stress responses.
Surprisingly, while low temperature induces DsrA (24), low temperature does not induce gadA in wild-type or dsrA-null strains (R.A.L., unpublished data), suggesting specific integration of appropriate environmental signals in acid resistance regulation. Also, besides its role in acid resistance, DsrA protects E. coli from osmotic shock. DsrA induces the proU hyperosmotic shock operon proVWX by more than threefold (Table 1, line 9) (19) and the hypo-osmotic shock gene ompF by 2- to 2.5-fold (Table 1, line 12). Under hyperosmotic conditions, dsrA knockout mutants are compromised for survival (21). Thus, osmotic and acid stress responses may be integrated via DsrA (Fig. 3B). Interestingly, both hyper- and hypo-osmotic shock conditions induce expression of gad acid resistance genes (10), consistent with interdependence of acid and osmotic shock protective mechanisms. A unifying theme is that maintenance of membrane chemiosmotic potentials and control of cell permeability would be common to these and other stress responses.
DsrA as a virulence factor coordinator.
The mRNA for the acid resistance and virulence factor regulatory protein GadX is induced by DsrA overexpression (Table 1, line 5; Fig. 1, panel 1, and 3B), repressed by H-NS, and activated by RpoS (Fig. 3B) (40). GadX (formerly YhiX) is an AraC-like regulator produced from the gadX promoter, as well as cotranscriptionally from the gadAX operon. GadX functions as a master activator (and autoactivator) for gadAX, gadBC, hdeAB, and hdeD, and other acid resistance-related genes (Fig. 3B) (23, 41). GadX also represses Per, a regulatory protein of enteropathogenic E. coli that is absent from enterohemorrhagic E. coli (EHEC) strains such as O157. In these enteropathogenic E. coli strains, Per activates virulence factors produced from a bacterial pathogenicity island (LEE) (31 and references therein). In EHEC strains, LEE virulence factors are induced by quorum sensing and a regulatory cascade that involves RpoS induction, as well as factors that antagonize H-NS silencing of LEE (7, 37). Coordination of acid resistance and adherence phenotypes by DsrA modulation of RpoS and H-NS levels could benefit bacteria that pass from the low-pH environment of the stomach to sites of potential attachment and effacement in the intestine.
Supplementary Material
Acknowledgments
We thank Jonathan Hibbs and Susan Gottesman for generously providing bacterial strains; Kelynne Reed for strains and advice; Lori Conlan, Colin Coros, Coby Slagter-Jäger, and David Edgell for comments on the manuscript; and M. Carl and J. Dansereau for help with the manuscript and figures, respectively. We appreciate the services of the Wadsworth Center Molecular Genetics and Genomics Core Facilities. Sarah Woodson provided laboratory resources, as well as valuable comments on the manuscript.
This work was supported by NIH grants GM39422 and GM44844 to M.B. and GM46686 to Sarah Woodson.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org.
REFERENCES
- 1.Altuvia, S., and E. G. H. Wagner. 2000. Switching on and off with RNA. Proc. Natl. Acad. Sci. USA 97:9824-9826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnqvist, A., A. Olsen, and S. Normark. 1994. Sigma S-dependent growth-phase induction of the csgBA promoter in Escherichia coli can be achieved in vivo by sigma 70 in the absence of the nucleoid-associated protein H-NS. Mol. Microbiol. 13:1021-1032. [DOI] [PubMed] [Google Scholar]
- 3.Atlung, T., and H. Ingmer. 1997. H-NS: a modulator of environmentally regulated gene expression. Mol. Microbiol. 24:7-17. [DOI] [PubMed] [Google Scholar]
- 4.Barbosa, T. M., and S. B. Levy. 2002. Activation of the Escherichia coli nfnB gene by MarA through a highly divergent marbox in a class II promoter. Mol. Microbiol. 45:191-202. [DOI] [PubMed] [Google Scholar]
- 5.Belfort, M., K. Ehrenman, and P. S. Chandry. 1990. Genetic and molecular analysis of RNA splicing in Escherichia coli. Methods Enzymol. 181:521-539. [DOI] [PubMed] [Google Scholar]
- 6.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 7.Bustamante, V. H., F. J. Santana, E. Calva, and J. L. Puente. 2001. Transcriptional regulation of type III secretion genes in enteropathogenic Escherichia coli: Ler antagonizes H-NS-dependent repression. Mol. Microbiol. 39:664-678. [DOI] [PubMed] [Google Scholar]
- 8.Casadaban, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138:179-207. [DOI] [PubMed] [Google Scholar]
- 9.Castanie-Cornet, M., T. A. Penfound, D. Smith, J. F. Elliott, and J. W. Foster. 1999. Control of acid resistance in Escherichia coli. J. Bacteriol. 181:3525-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Biase, D., A. Tramonti, F. Bossa, and P. Visca. 1999. The response to stationary-phase stress conditions in Escherichia coli: role and regulation of the glutamic acid decarboxylase system. Mol. Microbiol. 32:1198-1211. [DOI] [PubMed] [Google Scholar]
- 11.Donnenberg, M. S., and J. B. Kaper. 1991. Construction of an eae deletion mutation of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59:4310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gajiwala, K. S., and S. K. Burley. 2000. HDEA, a periplasmic protein that supports acid resistance in pathogenic enteric bacteria. J. Mol. Biol. 295:605-612. [DOI] [PubMed] [Google Scholar]
- 13.Gorden, J., and P. L. Small. 1993. Acid resistance in enteric bacteria. Infect. Immun. 61:364-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gottesman, S. 2002. Stealth regulation: biological circuits with small RNA switches. Genes Dev. 16:2829-2842. [DOI] [PubMed] [Google Scholar]
- 15.Hengge-Aronis, R. 1999. Interplay of global regulators and cell physiology in the general stress response of Escherichia coli. Curr. Opin. Microbiol. 2:148-152. [DOI] [PubMed] [Google Scholar]
- 16.Klauck, E., J. Bohringer, and R. Hengge-Aronis. 1997. The LysR-like regulator LeuO in Escherichia coli is involved in the translational regulation of rpoS by affecting the expression of the small regulatory DsrA-RNA. Mol. Microbiol. 25:559-569. [DOI] [PubMed] [Google Scholar]
- 17.Lease, R. A., and M. Belfort. 2000. Riboregulation by DsrA RNA: trans-actions for global economy. Mol. Microbiol. 38:667-672. [DOI] [PubMed] [Google Scholar]
- 18.Lease, R. A., and M. Belfort. 2000. A trans-acting RNA as a control switch in Escherichia coli: DsrA RNA modulates function by forming alternative structures. Proc. Natl. Acad. Sci. USA 97:9919-9924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lease, R. A., M. E. Cusick, and M. Belfort. 1998. Riboregulation in Escherichia coli: DsrA RNA acts by RNA:RNA interactions at multiple loci. Proc. Natl. Acad. Sci. USA 95:12456-12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Majdalani, M., C. Cunning, D. Sledjeski, R. Elliot, and S. Gottesman. 1998. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl. Acad. Sci. USA 95:12462-12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Majdalani, N., S. Chen, J. Murrow, K. St. John, and S. Gottesman. 2001. Regulation of RpoS by a novel small RNA: the characterization of RprA. Mol. Microbiol. 39:1382-1394. [DOI] [PubMed] [Google Scholar]
- 22.Massé, E., F. E. Escorcia, and S. Gottesman. 2003. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 17:2374-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masuda, N., and G. M. Church. 2003. Regulatory network of acid resistance genes in Escherichia coli. Mol. Microbiol. 48:699-712. [DOI] [PubMed] [Google Scholar]
- 24.Repoila, F., and S. Gottesman. 2001. Signal transduction cascade for regulation of RpoS: temperature regulation of DsrA. J. Bacteriol. 183:4012-4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richmond, C. S., J. D. Glasner, R. Mau, H. Jin, and F. R. Blattner. 1999. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 27:3821-3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Romeo, T. 1998. Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol. Microbiol. 29:1321-1330. [DOI] [PubMed] [Google Scholar]
- 27.Rudd, K. E. 2000. EcoGene, a genomic sequence database for Escherichia coli K-12. Nucleic Acids Res. 28:60-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Shi, X., and G. N. Bennett. 1995. Effects of multicopy LeuO on the expression of the acid-inducible lysine decarboxylase gene in Escherichia coli. J. Bacteriol. 177:810-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi, X., and G. N. Bennett. 1994. Plasmids bearing hfq and the hns-like gene stpA complement hns mutants in modulating arginine decarboxylase gene expression in Escherichia coli. J. Bacteriol. 176:6769-6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shin, S., M. Castanie-Cornet, J. W. Foster, J. A. Crawford, C. Brinkley, and J. B. Kaper. 2001. An activator of glutamate decarboxylase genes regulates the expression of enteropathogenic Escherichia coli virulence genes through control of the plasmid-encoded regulator, Per. Mol. Microbiol. 41:1133-1150. [DOI] [PubMed] [Google Scholar]
- 32.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 33.Simons, R. W., and N. Kleckner. 1988. Biological regulation by antisense RNA in prokaryotes. Annu. Rev. Genet. 22:567-600. [DOI] [PubMed] [Google Scholar]
- 34.Sledjeski, D., and S. Gottesman. 1995. A small RNA acts as an antisilencer of the H-NS-silenced rcsA gene of Escherichia coli. Proc. Natl. Acad. Sci. USA 92:2003-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sledjeski, D. D., A. Gupta, and S. Gottesman. 1996. The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J. 15:3993-4000. [PMC free article] [PubMed] [Google Scholar]
- 36.Sledjeski, D. D., C. Whitman, and A. Zhang. 2001. Hfq is necessary for regulation by the untranslated RNA DsrA. J. Bacteriol. 183:1997-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sperandio, V., J. L. Mellies, W. Nguyen, S. Shin, and J. B. Kaper. 1999. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 96:15196-15201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stim-Herndon, K. P., T. M. Flores, and G. N. Bennett. 1996. Molecular characterization of adiY, a regulatory gene which affects expression of the biodegradative acid-induced arginine decarboxylase gene (adiA) of Escherichia coli. Microbiology 142:1311-1320. [DOI] [PubMed] [Google Scholar]
- 39.Tao, H., C. Bausch, C. Richmond, F. R. Blattner, and T. Conway. 1999. Functional genomics: expression analysis of Escherichia coli growing on minimal and rich media. J. Bacteriol. 181:6425-6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tramonti, A., P. Visca, M. De Canio, M. Falconi, and D. De Biase. 2002. Functional characterization and regulation of gadX, a gene encoding an AraC/XylS-like transcriptional activator of the Escherichia coli glutamic acid decarboxylase system. J. Bacteriol. 184:2603-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tucker, D. L., N. Tucker, Z. Ma, J. W. Foster, R. L. Miranda, P. S. Cohen, and T. Conway. 2003. Genes of the GadX-GadW regulon in Escherichia coli. J. Bacteriol. 185:3190-3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wagner, E. G. H., S. Altuvia, and P. Romby. 2002. Antisense RNAs in bacteria and their genetic elements. Adv. Genet. 46:361-398. [DOI] [PubMed] [Google Scholar]
- 43.Wassarman, K. M. 2002. Small RNAs in bacteria: diverse regulators of gene expression in response to environmental changes. Cell 109:141-144. [DOI] [PubMed] [Google Scholar]
- 44.Wassarman, K. M., A. Zhang, and G. Storz. 1999. Small RNAs in Escherichia coli. Trends Microbiol. 7:37-45. [DOI] [PubMed] [Google Scholar]
- 45.Whittam, T. S., and A. C. Bumbaugh. 2002. Inferences from whole-genome sequences of bacterial pathogens. Curr. Opin. Genet. Dev. 12:719-725. [DOI] [PubMed] [Google Scholar]
- 46.Yoshida, T., C. Ueguchi, and T. Mizuno. 1993. Physical map location of a set of Escherichia coli genes (hde) whose expression is affected by the nucleoid protein H-NS. J. Bacteriol. 175:7747-7748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoshida, T., C. Ueguchi, H. Yamada, and T. Mizuno. 1993. Function of the Escherichia coli nucleoid protein, H-NS: molecular analysis of a subset of proteins whose expression is enhanced in a hns deletion mutant. Mol. Gen. Genet. 237:113-122. [DOI] [PubMed] [Google Scholar]
- 48.Zhang, A., S. Rimsky, M. E. Reaban, H. Buc, and M. Belfort. 1996. Escherichia coli protein analogs StpA and H-NS: regulatory networks, similar and disparate effects on nucleic acid dynamics. EMBO J. 15:1340-1349. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.