Abstract
Burkholderia cenocepacia strain K56-2, a representative of the Burkholderia cepacia complex, is part of the epidemic and clinically problematic ET12 lineage. The strain produced plant tissue watersoaking (ptw) on onion tissue, which is a plant disease-associated trait. Using plasposon mutagenesis, mutants in the ptw phenotype were generated. The translated sequence of a disrupted gene (ptwD4) from a ptw-negative mutant showed homology to VirD4-like proteins. Analysis of the region proximal to the transfer gene homolog identified a gene cluster located on the 92-kb resident plasmid that showed homology to type IV secretion systems. The role of ptwD4, ptwC, ptwB4, and ptwB10 in the expression of ptw activity was determined by conducting site-directed mutagenesis. The ptw phenotype was not expressed by K56-2 derivatives with a disruption in ptwD4, ptwB4, or ptwB10 but was observed in a derivative with a disruption in ptwC. Complementation of ptw-negative K56-2 derivatives in trans resulted in complete restoration of the ptw phenotype. In addition, analysis of culture supernatants revealed that the putative ptw effector(s) was a secreted, heat-stable protein(s) that caused plasmolysis of plant protoplasts. A second chromosomally encoded type IV secretion system with complete homology to the VirB-VirD system was identified in K56-2. Site-directed mutagenesis of key secretory genes in the VirB-VirD system did not affect expression of the ptw phenotype. Our findings indicate that in strain K56-2, the plasmid-encoded Ptw type IV secretion system is responsible for the secretion of a plant cytotoxic protein(s).
The Burkholderia cepacia complex (Bcc) consists of nine genomovars recently elevated to species status (11, 65, 66, 68). Members of the Bcc include plant and animal pathogens as well as catabolically active soil saprophytes (64). Some Bcc members can cause life-threatening respiratory infections, particularly in persons with cystic fibrosis (CF) (43). Studies indicate that 85 to 90% of strains isolated from infected CF patients are B. cenocepacia or B. multivorans, with other Bcc species being infrequently isolated (41). Several epidemic clonal lineages of B. cenocepacia have been identified (12), including the ET12 lineage, which is responsible for infecting many CF patients in Canada and the United Kingdom (32, 39); the PHDC lineage, responsible for nearly all Bcc infections in the mid-Atlantic region of the United States (8); and the Midwest lineage, which is responsible for infecting numerous patients in CF centers in the midwestern region of the United States (42). Factors that account for the apparent enhanced capacity of epidemic clones for human infection are unknown.
Postulated clinically associated virulence determinants for B. cenocepacia include hemolysins (30, 67), siderophores (62), and cable pili (56). The hemolysin induces apoptosis and degranulation of phagocytes (30), and siderophore production, regulated by quorum sensing (40), plays a role in the primary colonization process of animal lung and spleen tissues (62). Cable pili are important in adherence to the respiratory mucosal blanket and epithelial cells (57).
In B. cepacia type strain ATCC 25416, a plant pathogenic representative of the Bcc, a plasmid-encoded pectate hydrolase (Peh) is a virulence factor necessary for maceration of onion tissue (24). Derivatives of ATCC 25416 that have been cured of the Peh-encoding plasmid do not macerate onion tissue. However, the Peh-negative derivatives remain capable of causing a plant tissue watersoaking (ptw) phenotype that results from loss of cell membrane integrity and the accumulation of fluids in the intracellular spaces of plant tissue (22). In a survey of isolates from the Bcc experimental strain panel (47), we found that all of the B. cepacia and B. cenocepacia strains tested and one B. vietnamiensis strain produced the ptw phenotype, suggesting that members of these genomic species may produce a cytotoxic factor(s) that affects plant cell integrity (50).
In recent years, a body of research has substantiated the concept that bacterial pathogens share common secretion mechanisms for the delivery of virulence determinants (9, 54). There are numerous examples of the importance of type III (54) and type IV secretion systems (10) in the infection process for both plant and animal pathogens, with evidence that elements of the secretion systems show an ancestral relationship to bacterial transport machinery. It has been hypothesized that type III secretion systems are derived from flagellar assembly constituents modified to function as a transport mechanism for virulence factors (46). The type IV secretion system from Agrobacterium tumefaciens was the first to be identified and is used to deliver oncogenic transfer DNA and effector proteins to plant cells during infection (10, 37). Presently, type IV secretion systems can be categorized as (i) conjugation systems that mediate DNA transfer to recipient cells, (ii) effector translocator systems that transfer molecules termed effectors to eukaryotic cells during infection, or (iii) DNA uptake or release systems mediating exchange of DNA with the milieu (18). The Legionella pneumophila Dot/Icm transporter is an example of a type IV secretion system that is capable of both conjugation and effector translocation (e.g., RalF) (52). Helicobacter pylori possesses two type IV secretion systems with differing roles. The Cag secretory apparatus functions in the translocation of CagA into host cells, and the Com system is used to uptake DNA to facilitate genetic variation and enhance cell survival (16, 18).
In this study we report on the presence of both a plasmid and a chromosomally encoded type IV secretion system in B. cenocepacia strain K56-2 and describe the involvement of the plasmid-encoded system in the export of a putative protein(s) responsible for the ptw phenotype.
MATERIALS AND METHODS
Media and growth conditions.
Descriptions of plasmids and bacterial strains used in this study are listed in Tables 1 and 2, respectively. Luria Bertani (LB) medium was used for routine maintenance of cultures. Minimal medium Vogel Bonner (69) amended with 1.0% glucose (VBG) was used in mating experiments. Minimal medium M9 (59) containing 1.0% glucose was used for culturing bacteria for supernatant analysis. B. cenocepacia and Escherichia coli strains were grown at 37°C. Media were supplemented with appropriate concentrations of antibiotics as needed for selection. Antibiotics were added to media at the following concentrations: 50 μg of tetracycline (Tc)/ml and 100 μg of trimethoprim (Tp)/ml for B. cenocepacia and 10 μg of Tc/ml, 10 μg of gentamicin (Gm)/ml, 40 μg of ampicillin/ml, and 100 μg of Tp/ml for E. coli.
TABLE 1.
Plasmids used in this study
| Plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| pK56-2 | 92-kb resident plasmid of B. cenocepacia strain K56-2 containing the ptw cluster | This study |
| pDrive | Cloning vector; Kmr, Apr, pUC origin, T7, SP6, lacZaα+ | Qiagen |
| pRK2013 | Kmr, Tra+, Mob+, ColE1 replicon | 20 |
| pBR325 | Tcr, Apr, Cmr, ColE1 replicon | 4 |
| pR388 | IncW, Sur, Tpr | 13 |
| pTnMod-RTp′ | Tn5, R6K oriR, Tpr | 15 |
| pScosBC1 | Cosmid with K56-2 genomic library | 62 |
| pSBC-9F5 | pScosBC1 with K56-2 genomic DNA containing ptwD4 and ptwC | This study |
| pSBC-3H3 | pScosBC1 with K56-2 genomic DNA containing ptwB4 and ptwB10 | This study |
| pJQ200SK | GmrsacB Mob+, P15A replicon | 55 |
| pURF047 | IncW, Ampr, Gmr, Mob+, lacZα+, Par+, derivative of pURF043 | 63 |
| pASE101 | pURF047 with 2.0-kb Tpr cassette inserted at ScaI site of Apr cassette | This study |
| pASE102 | pJQ200SK containing 10-kb BamHI fragment (pSBC-9F5) containing ptwD4 and ptwC inserted in the MCS | This study |
| pASE103 | pJQ200SK containing 11-kb BamHI fragment (pSBC-3H3) containing ptwB4 and ptwB10 inserted in the MCS | This study |
| pASE104 | pASE101 containing 10-kb BamHI fragment (pSBC-9F5) containing ptwD4 and ptwC inserted in the MCS | This study |
| pASE105 | pASE101 containing 11-kb BamHI fragment (pSBC1-3H3) containing ptwB4 and ptwB10 inserted in the MCS | This study |
| pASE106 | pASE102 with 1.9-kb Tcr cassette from pBR325 inserted in the ScaI site of ptwD4 | This study |
| pASE107 | pASE102 with 1.9-kb Tcr cassette from pBR325 inserted in the EcoRV site of ptwC | This study |
| pASE108 | pASE103 with 1.9-kb Tcr cassette from pBR325 inserted in the ApaI site of ptwB4 | This study |
| pASE109 | pASE103 with 1.9-kb Tcr cassette from pBR325 inserted in the SacI site of ptwB10 | This study |
| pASE110 | pDrive containing 1.5-kb PCR product containing virB6 | This study |
| pASE111 | pDrive containing 1.9-kb PCR product containing virB11 | This study |
| pASE112 | pJQ200SK containing virB6 from pASE110 | This study |
| pASE113 | pJQ200SK containing virB11 from pASE111 | This study |
| pASE114 | pASE112 with 1.9-kb Tcr cassette from pBR325 inserted into the ScaI site of virB6 | This study |
| pASE115 | pASE113 with 1.9 kb Tcr cassette from pBR325 inserted into the ScaI site of virB11 | This study |
Ap, ampicillin; Cm, chloramphenicol; Km, kanamyacin; Su, sulphonamide; Tc, tetracycline; Tp, trimethoprim; ptw, plant tissue watersoaking phenotype.
TABLE 2.
Bacterial strains used in this study
| Strain | Relevant characteristicsa | Reference or source |
|---|---|---|
| B. cenocepacia | ||
| K56-2 | CF respritory isolate, ptw+ | 62 |
| GM237 | K56-2 ptwD4::RTp′, ptw− | This study |
| GM238 | K56-2::RTp′, ptw+ | This study |
| GM241 | K56-2::RTp′, ptw− | This study |
| GM242 | K56-2 ptwD4::RTp′, ptw− | This study |
| GM243 | K56-2::RTp′, ptw− | This study |
| AE307 | K56-2 ptwD4::Tc, ptw− | This study |
| AE320 | K56-2 ptwC::Tc, ptw+ | This study |
| AE321 | K56-2 ptwB4::Tc, ptw− | This study |
| AE322 | K56-2 ptwB10::Tc, ptw− | This study |
| AE310 | AE307 (pASE104), ptw+ | This study |
| AE323 | AE321 (pASE105), ptw+ | This study |
| AE324 | AE322 (pASE105), ptw+ | This study |
| AE360 | K56-2 virB6::Tc, ptw+ | This study |
| AE361 | K56-2 virB11::Tc, ptw+ | This study |
| E. coli | ||
| DH10B | F−mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80dlacZΔM15 ΔlacX74 deoR recA1 endA1 araD139 Δ(ara, leu) 7697 galU galK1 rpsL nupG | Life Technologies, Inc. (Gaithersburg, Md.) |
| HB101 | F−recA13 | 6 |
| CC118λpir | ΔlacX74 galE galK thi-1 rpsE rpoB recA1, lysogenized with λ pir phage | 27 |
Tc, tetracycline; ptw, plant tissue watersoaking phenotype.
Plant tissue assay and growth study.
Plant tissue watersoaking activity was determined as previously described (24). Onion cultivar 1015Y was used for plant tissue assays. Onion bulb scales were inoculated, in triplicate, with 10 μl of an aqueous suspension of the isolate being tested as previously described (24). Aqueous bacterial suspensions were adjusted spectrophotometrically (A425 = 0.5) to yield a final concentration of ∼106 CFU/scale. Sterile double-distilled deionized water was used as a negative control and strain K56-2 served as the positive control in all experiments. Scales were placed on a sheet of aluminum foil that had been surface sterilized with 70% ethanol in containers layered with paper towels that were moistened with double-distilled deionized water, sealed, and incubated at 37°C. Plant tissue watersoaking activity was assessed at 24, 48, and 72 h postinoculation by measuring the vertical and horizontal diameters of the zones for triplicate samples in three independent experiments. Data were reported as the average area of tissue watersoaking ± standard deviations (SD) for the three independent experiments in triplicate.
To obtain a quantitative measure of growth in plant tissue for strains K56-2, AE307, and AE310, bulb scales were inoculated with an aqueous suspension of the strains as described above. At 0, 24, 48, and 72 h postinoculation watersoaked zones were measured and tissue was processed to determine the bacterial population. Scales were commuted and crushed in 0.0125 M phosphate buffer (pH 7.1) containing Triton X-100 (0.01%) by using a sterile mortar and pestle. A dilution series of each tissue slurry was plated to LB agar containing Gm (10 μg/ml) and Tc (10 μg/ml) and was incubated at 37°C for 48 h. Population data for each time interval was expressed as the average CFU/gram of tissue ± SD for three independent experiments in triplicate. Tissue watersoaking data were reported as the average area of watersoaking ± SD for three independent experiments in triplicate.
Activity testing of culture supernatants.
Cultures of strains K56-2, AE307, and AE310 were grown at 37°C and harvested at late exponential phase (A425 = 0.9) by centrifugation (16,000 × g, 15 min, at 5°C). Supernatants (1 liter) were filter sterilized using 0.22 μM filters (Pall) and were concentrated to 3 ml by using an Amicon ultrafiltration stirred cell (molecular weight cutoff, 10,000). Protein concentration of supernatant concentrates was determined by the method of Koch and Putnam (36). Concentrated uninoculated medium (1 liter) served as the negative control.
Plant protoplasts were used as the plant tissue system to obtain a quantitative measurement of activities of concentrated culture supernatants. Initial testing demonstrated that onion and carrot protoplasts were similar in sensitivity, and carrots were chosen because the presence of chromoplasts facilitated a more accurate evaluation of plasmolytic activity. Protoplasts were obtained using the following procedure (33). Young carrots were placed in a 10% sodium hypochlorite solution for 5 min and then were rinsed four times with sterile double-distilled deionized water. The epidermis was resected with a sterile scalpel and discarded. Cortical and phloem tissues were obtained, diced into fragments ∼2 mm in width, and placed in a sterile petri dish containing 20 ml of a filter-sterilized enzyme solution. The enzyme solution contained 10% mannitol (Fisher), 1.5% cellulase (CalBiochem), 0.25% macerase (CalBiochem), and 0.75% bovine serum albumin fraction V (Sigma) (33). Tissue was digested in the dark for 5 h at 28°C with gentle shaking (50 rpm). The carrot tissue was strained through nylon mesh and centrifuged (825 × g for 5 min at 25°C). The supernatant was removed using a pipette, avoiding disturbance of the protoplasts. The protoplasts were washed with 20 ml of 10% mannitol by resuspending with gentle swirling and centrifuged. The supernatant was removed and the pellet was gently resuspended in 10 ml of 10% mannitol, and a 5-ml 20% sucrose cushion was carefully layered in the bottom of the tube. The sample was centrifuged and the protoplasts were collected from the interface and counted with a hemacytometer (Hausser Scientific). On average, 1.8 × 106 protoplasts/ml were obtained.
The plasmolytic activity of supernatants was determined by using concentrates standardized for protein concentration and adjusted to 10% mannitol. The standardized concentrates, as well as dilutions (1:1, 1:5, and 1:10), were added to protoplasts. Assay mixtures and controls were incubated in 96-well microtiter plates (Corning) with an average concentration of 50 protoplasts per well. Assays, done in triplicate, were observed for changes in protoplast integrity at 2-h intervals for 8 h using a Zeiss inverted microscope. In addition, the effect of temperature on plasmolytic activity was tested by heating concentrates to 100°C for 10 min or at 80, 65, or 37°C for 1 h. Concentrates were also individually treated with 1-mg/ml final concentrations of proteinase K (Boehringer Mannheim) dissolved in 0.05 M Tris-HCl, pH 8; DNase I (3,200 U/mg; Sigma) in 50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, and 50 μg of bovine serum albumin/ml; or RNase A (100 U/mg; Sigma) in 0.25 M Tris-HCl, pH 6.8. All reaction mixtures were incubated at temperatures optimal for each enzyme's activity and were then adjusted to 10% mannitol before being mixed with protoplasts. Each incubation duration was performed in duplicate in three independent experiments with an average of 50 protoplasts examined at 0, 2, 4, 6, and 8 h. Data were expressed as percent plasmolysis ± SD for three independent experiments. Plasmolysis was analyzed as a function of time, dilution, treatment, and their interactions. Level of statistical significance of the different variables was determined with an analysis of variance. Statistical analysis was performed using SAS software and the general linear model procedure (SAS Institute Inc.).
Mating experiments.
For triparental matings, donor, mobilizer, and recipient suspensions were made in LB broth from cultures grown on solid media under selective conditions, as appropriate, for 18 h. Bacterial suspensions were adjusted spectrophotometrically (A600 = 0.5), mixed at an equal ratio (1:1:1), and transferred to a positively charged sterile membrane layered on a 100- by 15-mm LB agar petri dish. Following a 6-h incubation period, the cells from the mating and respective controls were washed twice in phosphate buffer (0.125 M, pH 7.1) by centrifugation (12,096 × g for 10 min at 5°C). The bacterial pellets were suspended in phosphate buffer and dilution plated to media with appropriate antibiotic selection. Single-colony isolates of individual transconjugants were obtained by streaking on selective media. Cultures were subjected to survey lysis followed by agarose gel electrophoresis (23) to confirm plasmid transfer.
Plasposon mutagenesis.
The plasposon pTnMod-RTp′ was employed to obtain initial ptw-negative mutants of strain K56-2 (15). Transconjugants were selected on VBG supplemented with Tp. After 48 h of incubation at 37°C, isolated colonies were transferred to homologous media to confirm selection. Trimethoprim-resistant transconjugants were evaluated for ptw activity by using the onion tissue assay. Transconjugants that no longer demonstrated the ptw phenotype were single-colony purified, retested, and retained for further analysis.
DNA isolation and sequence analysis.
Genomic DNA was extracted from plasposon-generated ptw-negative K56-2 mutants using a DNeasy kit (QIAGEN). Three micrograms of DNA were digested with PstI and self ligated by using T4 DNA ligase (New England Biolabs). The resulting plasmids were transformed into E. coli CC118λpir by CaCl2 transformation (59) and were selected on LB agar amended with Tp. Survey lysis and agarose gel electrophoresis were conducted to confirm the presence of plasmid DNA in the transformants. Plasmid DNA for sequencing was extracted using a HiSpeed Plasmid Midi kit (QIAGEN). Primers designed from the 5′ and 3′ termini of the plasposon insert were designated RTp1 and RTp2 (Table 3), respectively, and were used to sequence the cloned genomic DNA from B. cenocepacia strain K56-2. Sequencing was conducted by the Institute of Developmental and Molecular Biology, Gene Technologies Laboratory, Texas A&M University. MacVector 7.0 (Oxford Molecular, Ltd.) was used for DNA sequence analysis. DNA sequence for B. cenocepacia strain J2315 was produced by the Pathogen Sequencing Group at the Sanger Institute, Hinxton, Cambridge, and can be obtained from http://www.sanger.ac.uk/Projects/B_cenocepacia/.
TABLE 3.
Primers used in this study
| Primer | Sequence |
|---|---|
| RTp1 | 5′-GGTACCGTCGACATGCATGG-3′ |
| RTp2 | 5′-CAGTGCAAATTTATCCTGTG-3′ |
| PD4-1 | 5′-TGACTCAGCGAAGGAA-3′ |
| PD4-2 | 5′-ATCCGGTGGAAGCAA-3′ |
| PC-1 | 5′-CACTCCGACATCGAAT-3′ |
| PC-2 | 5′-AACTGTGTAGGACACT-3′ |
| PB10-1 | 5′-ATGCTTGTCGATGTTGCA-3′ |
| PB10-2 | 5′-TTCCGACGACTTCCAT-3′ |
| PB4-1 | 5′-AACTCAAGATGAATCAT-3′ |
| PB4-2 | 5′-AGAACGCATGGTGTTGA-3′ |
| VB1-1 | 5′-GACAGATCATGCGTGT-3′ |
| VB1-2 | 5′-AGGAGGAGACACAGGAGT-3′ |
| VB4-1 | 5′-ATCGCACTGTTGATCGTTG-3′ |
| VB4-2 | 5′-TCACGTTACGTGTGCATAC-3′ |
| VB6-1 | 5′-AAGGTGGTGTTCTATGAA-3′ |
| VB6-2 | 5′-ACAGTACGATATGCGTGAA-3′ |
| VB11-1 | 5′-TTCAAGTAACGATTGCTGA-3′ |
| VB11-2 | 5′-TGCAAATCGAGTTCTGGTA-3′ |
| VD4-1 | 5′-TGTTATGAGACAGATTGCA-3′ |
| VD4-2 | 5′-AGATAAGGATGGTCAGGT-3′ |
Site-directed mutagenesis.
Using PCR analysis, ptwD4, ptwC, ptwB4, and ptwB10 were found to be located on two cosmids from a previously constructed K56-2 library (62). Primer sets PD4 (PD4-1 and PD4-2), PC (PC-1 and PC-2), PB4 (PB4-1 and PB4-2), and PB10 (PB10-1 and PB10-2) (Table 3) were designed based on sequence data from the B. cenocepacia Genome Project. Each PCR (50 μl) was performed using a Taq PCR Core kit (QIAGEN) and a GeneAmp 2700 (Applied Biosystems). Reactions were run for 25 cycles, and parameters were as follows: denaturation at 95°C for 30 s; annealing at 55, 52, 56.5, and 54°C for 1 min for primer sets PD4, PC, PB4, and PB10, respectively; and extension at 72°C for 75 s. Resulting products were analyzed by agarose gel electrophoresis. It was determined that cosmid pSBC-9F5 contained ptwD4 and ptwC and that pSBC-3H3 harbored ptwB4 and ptwB10, respectively. Genes ptwD4, ptwC, ptwB4, and ptwB10, were disrupted using site-directed mutagenesis. A 10-kb BamHI fragment from pSBC-9F5 and an 11-kb BamHI fragment from pSBC-3H3 were individually inserted into the multiple cloning site (MCS) of pJQ200SK (55) to construct pASE102 and pASE103, respectively. Plasmids pASE102 and pASE103 were transformed into E. coli DH10B by electroporation (25 μF, 2.5 kV, 200 Ω) followed by selection on LB agar amended with Gm. Orientation of inserted DNA was determined by restriction enzyme digest analysis using SmaI. Only plasmids containing insertions that were in frame were used in the subsequent experiments. Using pASE102, plasmids pASE106 and pASE107 were constructed by inserting a Tc-resistance cassette, obtained as a 1.9-kb fragment from pBR325 (4), into the ScaI and EcoRV sites of ptwD4 and ptwC, respectively. Plasmids pASE108 and pASE109 were constructed by inserting the Tc cassette into the ApaI and SacI sites of ptwB4 and ptwB10 of pASE103, respectively. Constructs pASE106, pASE107, pASE108, and pASE109 were electroporated into E. coli DH10B, and transformants were selected on LB agar amended with Tc and Gm. Insertion of the Tc resistance cassette was confirmed by restriction enzyme digest analysis using BamHI and PCR using primer sets PD4, PC, PB4, and PB10 to confirm a 1.9-kb increase in plasmid size and PCR product, respectively. Plasmids pASE106, pASE107, pASE108, and pASE109 were individually mobilized into K56-2 by utilizing pRK2013 in triparental matings to yield genetic exchange mutants. Tetracycline-resistant transconjugants were single-colony purified and retested for Tcr. PCR using primer sets PD4, PC, PB4, and PB10 and plasmid survey lysis were used to confirm allelic exchange, reflected by the increase in PCR product and loss of the suicide vector. Single-colony isolates were tested for the ptw phenotype.
Genes virB6 and virB11, from the chromosomally located type IV secretion system, were disrupted by site-directed mutagenesis. Primer sets VB6 (VB6-1 and VB6-2) and VB11 (VB11-1 and VB11-2) (Table 3) were designed based on sequence data from the Sanger Centre. Each PCR was performed as described above, except that the cycle parameters were as follows: denaturation at 95°C for 30 s; annealing at 57.5 and 56°C for 1 min for primer sets VB6 and VB11, respectively; and extension at 72°C for 75 s. PCR products were individually ligated into vector pDrive (QIAGEN) and were transformed into E. coli DH10B by electroporation followed by selection on LB agar amended with Ap to yield pASE110 and pASE111, respectively. Genes virB6 and virB11 were excised from pASE110 and pASE111, respectively, by double digestion using restriction enzymes PstI and XhoI and were individually inserted into the MCS of pJQ200SK to construct pASE112 and pASE113, respectively. Plasmids pASE112 and pASE113 were transformed into E. coli DH10B by electroporation followed by selection on LB agar amended with Gm. Using pASE112 and pASE113, plasmids pASE114 and pASE115 were constructed by inserting a Tc resistance cassette, from pBR325, into the ScaI site of virB6 or virB11, respectively. Constructs pASE114 and pASE115 were individually electroporated into E. coli DH10B and transformants were selected on LB agar amended with Tc and Gm. Insertion of the Tc resistance cassette was confirmed by restriction enzyme digest analysis using BamHI and PCR primer sets VB6 and VB11 to confirm a 1.9-kb increase in plasmid size and PCR product, respectively. Plasmids pASE114 and pASE115 were individually mobilized into K56-2 by triparental matings to yield genetic exchange mutants. Tetracycline-resistant transconjugants were single-colony purified and retested for Tcr. PCR using primer sets VB6 and VB11 and plasmid survey lysis were used to confirm allelic exchange, reflected by the increase in PCR product and loss of the suicide vector. Single-colony isolates were tested for the ptw phenotype.
Complementation of disrupted genes.
Complementation of the disrupted ptwD4, ptwB4, or ptwB10 was accomplished in trans. Plasmid pURF047 (63) was amended by inserting a Tp resistance cassette, obtained from R388 as a BamHI fragment, into the ScaI site in the bla gene to obtain pASE101. A 10-kb BamHI fragment from pSBC-9F5 that contained ptwD4 and an 11-kb BamHI fragment from pSBC-3H3 that contained ptwB4 and ptwB11 were individually inserted into the MCS of pASE101, resulting in pASE104 and pASE105, respectively. Plasmids pASE104 and pASE105 were electrotransformed into E. coli DH10B, followed by selection on LB agar amended with Tp and Gm. Orientation of inserted DNA was determined by restriction enzyme digest analysis using ScaI. Only plasmids containing in-frame insertions were used in the subsequent experiments. Plasmid pASE104 was mobilized into AE307, and pASE105 was mobilized into AE321 and AE322 to complement insertionally disrupted ptw genes. Tetracycline-resistant and Tpr transconjugants were single-colony purified and retested for antibiotic resistance. Plasmid survey lysis and PCR were used to confirm plasmid transfer, which was reflected by the presence of both the resident and complement plasmid as well as inserted and uninserted PCR products.
RESULTS
Isolation and characterization of ptw-negative mutants.
A total of 5,000 transconjugants of strain K56-2 were obtained by using the plasposon mutagenesis system. Genomic DNA from 20 randomly selected transconjugants were digested with PstI, which does not cleave the TnMod-RTp′ insert. The DNA fragments were separated by agarose gel electrophoresis, and Southern blots were probed with the Tp cassette to determine the randomness of insertion. It was determined that the plasposon system was suitable for the generation of random mutations in the genome of B. cenocepacia strain K56-2 (data not shown).
From the pool of 5,000 transconjugants, 56 mutants were verified as ptw-negative using the plant tissue assay (Fig. 1) and demonstrated no other apparent phenotypic differences compared to phenotype of the parental strain. The DNA flanking the plasposon insertion site of 10 ptw-negative mutants was cloned and sequenced, and a BLAST search was performed. The translated gene products disrupted in clones GM237 and GM242 (Table 2) showed homology to VirD4-like proteins, whereas GM241 and GM243 (Table 2) showed homology to a putative regulator and promoter, respectively. Thus, our initial inquiry was focused on determining if strain K56-2 possessed and/or utilized a similar transfer system. Characterization of the remaining ptw-negative mutants will be addressed in a separate report.
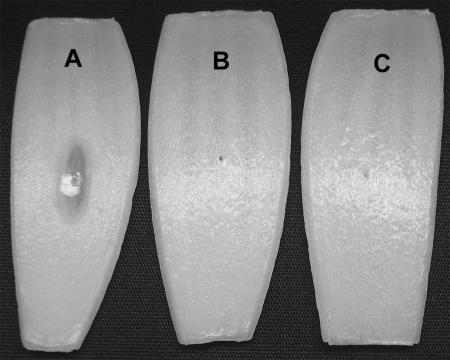
FIG. 1.
Plant tissue watersoaking assay. (A) Pierced onion bulb scale inoculated with B. cenocepacia strain K56-2 showing partial tissue collapse and translucence characteristic of the ptw phenotype. (B) Pierced onion bulb scale inoculated with sterile water (ptw negative). (C) Pierced onion bulb scale inoculated with ptw-negative mutant AE307. All ptw-negative mutants expressed no ptw activity on onion tissue, and inoculated scales showed the same symptoms as that in panel C.
Using sequence data from the B. cenocepacia Genome Project, a 60-kb region proximal to the identified virD4-like gene was analyzed. Strain J2315, the subject of the genome sequence project, is a member of the ET12 clonal lineage, as is strain K56-2 (47), and hence the arrangement of genes and their sequence was expected to be similar. A cluster of 11 genes potentially involved in the delivery of the ptw effector(s) was identified and designated the ptw cluster (Fig. 2). Southern analysis of K56-2 lysates (data not shown) determined that the ptw cluster was located on a 92-kb resident plasmid. Other isolates of the ET12 lineage (J2315, BC7, and C5424) were also shown to harbor the ptw cluster on their resident plasmids (data not shown). The resident plasmid, designated pK56-2, has a G-C ratio of 62.7%, and the ptw cluster has a G-C ratio of 61%, whereas the value for the whole genome is 66.9%.
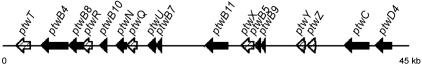
FIG. 2.
A 45-kb segment from plasmid pK56-2 containing the ptw cluster. Designation of genes was based on homology to gene products of transfer- and translocation-related proteins and are depicted by solid arrows. Open reading frames having no homologs are illustrated by patterned arrows.
DNA sequence and protein similarities of the ptw cluster.
The encoded products of the ptw cluster showed homology to various translocation and/or conjugation related proteins from other bacteria, including Agrobacterium tumefaciens, Salmonella enterica serovar Typhi, Vibrio cholerae, Novosphingobium aromaticivorans, and E. coli. Based on amino acid sequence similarity to TraD and other VirD4-like proteins, the predicted function of ptwD4 could involve nucleoside triphosphatase activity (NTPase) and may therefore serve as an active motor for secretion (60, 61). PtwB4 and its homolog VirB4 of the VirB-VirD system may also exhibit NTPase activity (2). Based on homology to R388 TrwC, PtwC may function as a component of a relaxosome (44). PtwB7, PtwB8, PtwB9, and PtwB10 were predicted to form complexes in the periplasm and/or membrane to create the pore (74). PtwB11 was also predicted to have NTPase activity and is part of the TraG subfamily, which belongs to the large superfamily of type II/IV secretion NTPases (53). PtwN, as well as PtwU, had no VirB-VirD homologs; however, they possessed homology to TraN and TraU of the F plasmid, respectively. In the F plasmid system TraN is an adhesin and TraU is involved in mating pair stabilization (38).
Based on analysis of gene products with sequence similarity and the probable involvement in the expression of the ptw phenotype, ptwD4, ptwC, ptwB4, and ptwB10 were chosen for further analysis. Translation of the ptwD4 sequence resulted in a protein that was 640 amino acids in length, with a predicted molecular size of 72 kDa and a pI of 6.6. Further analysis of the PtwD4 amino acid sequence, using reverse position specific BLAST (rpsBLAST) and the conserved domain database, revealed a predicted Walker A (P-loop) and B site for nucleotide binding (70) that is characteristic of type IV secretion ATP-binding proteins (74). The predicted P-loop nucleotide binding motif extends from 117 to 124 in the amino acid sequence, and the Walker B motif extends from 338 to 344. The translated sequence of ptwB4 was 1,013 amino acids in length, with a predicted molecular size of 108 kDa and a pI of 6.0. Further analysis of the PtwB4 amino acid sequence for hydrophobicity revealed four putative transmembrane domains, which is characteristic of type IV secretion NTPases (74). Translation of the ptwB10 sequence resulted in a protein that was 278 amino acids in length, with a predicted molecular size of 30 kDa and a pI of 5.2. Using rpsBLAST and the conserved domain database, PtwB10 was shown to contain a well-conserved C-terminal hydrophobic region (74). Translation of the ptwC sequence resulted in a protein that was 940 amino acids in length, with a predicted molecular size of 104 kDa and a pI of 6.0. A Pustell protein matrix analysis comparing PtwC and TrwC from R388 illustrated regions of similarity between the sequences. At the N terminus (amino acids 1 to 200), PtwC and TrwC showed 42% identity and signature motifs (2 and 3) that are conserved in proteins with DNA-nicking activities (44). An analysis of the PtwC C-terminal sequence indicated that PtwC contained 7 motifs (I, Ib, II, III, IV, V, and IV) characteristic of helicases (28).
Site-directed disruption of the ptw or virB-virD4 clusters and its effect on the ptw phenotype.
Translated products of the ptw cluster showed homology to proteins from known type IV secretion systems. Based on their predicted function, the role of ptwD4, ptwC, ptwB4, and ptwB10 in the expression of the ptw phenotype was determined. Watersoaking mutants of K56-2 with a disrupted ptwD4 were obtained in the initial screening of the plasposon generated mutants and by site-directed mutagenesis (Fig. 1 and Table 2). Site-directed mutagenesis of ptwD4, ptwC, ptwB4, and ptwB10 was accomplished by individual mobilization of pASE106, pASE107, pASE108, and pASE109, respectively, into K56-2. Tetracycline-resistant transconjugants showed a 1.9-kb increase in PCR product, which reflected the insertion of the Tc cassette (data not shown). Insertions in ptwD4, ptwB4, and ptwB10 resulted in loss of the ptw phenotype, whereas disruption of ptwC did not. Representative insertionally inactivated ptw-negative mutants of ptwD4, ptwB4, and ptwB10 were designated AE307, AE321, and AE322, respectively, whereas the inserted ptwC derivative was designated AE320.
Complementation of ptw-negative mutants was accomplished by mobilization of pASE104 into AE307 and pASE105 into AE321 or AE322 and selecting for Tcr and Tpr transconjugants. All transconjugants tested were ptw positive (Fig. 3). Plasmid pURF047 derivatives were used in complementation studies, because pURF047 is a derivative of pURF034 that contains the par locus and is maintained stably without selective pressure (>95% retention over 36 generations), which was a desirable parameter for plant tissue studies (14). A survey of transconjugant lysates confirmed plasmid transfer (data not shown). Representatives of ptw-positive complemented ptwD4, ptwB4, and ptwB10 mutants were designated AE310, AE323, and AE324, respectively (Fig. 3).
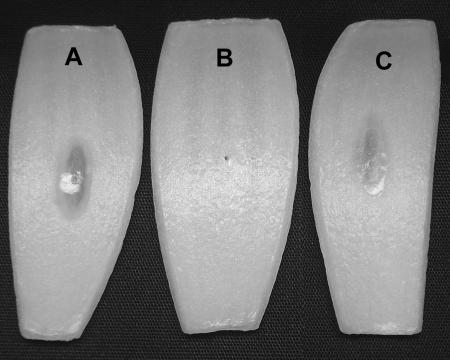
FIG. 3.
Complementation of ptw-negative mutants. (A) Pierced onion bulb scale inoculated with strain K56-2 showing characteristic ptw activity. (B) Pierced onion bulb scale inoculated with B. cenocepacia ptw-negative mutant AE307 showing no ptw activity. (C) Pierced onion bulb scale inoculated with complemented AE307 (AE310) showing restored ptw activity.
Genomic analysis of the J2315 sequence from the Sanger Centre identified a second cluster on chromosome II, which showed homology to the VirB-VirD type IV secretion system (Fig. 4). Using PCR analysis, the presence of virB1, virB4, virB6, virB11, and virD4 (Table 3), which span the entire virB-virD cluster, were confirmed in K56-2 (data not shown). In A. tumefaciens, virB6 is thought to mediate assembly of the pilus and a functional secretion machine through its effects on virB7 and virB9 multimerization (31), and virB11 is predicted to function as an NTPase, which provides the necessary energy for secretion (74). Based on their predicted function, the role of virB6 and virB11 in the expression of the ptw phenotype was assessed. Site-directed mutagenesis of virB6 and virB11 was accomplished by individual mobilization of pASE114 and pASE115 into K56-2, yielding AE360 and AE361, respectively. Tetracycline-resistant transconjugants showed a 1.9-kb increase in PCR product, which reflected the insertion of the Tc cassette (data not shown). Disruption of virB6 or virB11 did not result in loss of the ptw phenotype.
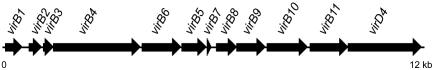
FIG. 4.
A 12-kb segment containing a VirB-VirD-like type IV secretion system cluster located on chromosome II of B. cenocepacia strain J2315. Designation of genes was based on homology to gene products of transfer- and translocation-related proteins and are depicted as solid arrows. Genes virB1, virB4, virB6, virB11, and virD4 have been identified in strain K56-2.
Partial characterization of watersoaking effector(s) and population dynamics.
Plant tissue assays indicated that the parental strain K56-2 and the complemented mutant (AE310) caused watersoaking, whereas the ptwD4 mutant (AE307) did not (Fig. 3). Growth and watersoaking were monitored over a 72-h period using the plant tissue assay to determine the contribution of ptw activity in survival and growth. Over the 72-h period both K56-2 and AE310 showed an approximate 100-fold increase in CFU/gram of tissue and an increase in the watersoaking area, whereas AE307 showed no watersoaking activity and an approximate 1,000-fold decrease in CFU/gram of tissue over the same period (Table 4).
TABLE 4.
Growth and watersoaking activity of B. cenocepacia strain K56-2 and its derivatives in onion tissue
| Time (h) | Strains
|
||
|---|---|---|---|
| K56-2 | AE307 | AE310 | |
| 0 | 6.5 × 105 ± 2.5 (0 ± 0)a | 4.2 × 105 ± 1.7 (0 ± 0) | 7.3 × 105 ± 2.0 (0 ± 0) |
| 24 | 6.1 × 104 ± 3.3 (90 ± 20) | 5.6 × 104 ± 1.0 (0 ± 0) | 4.4 × 104 ± 2.9 (120 ± 11) |
| 48 | 4.5 × 106 ± 1.7 (178 ± 42) | 6.7 × 103 ± 1.6 (0 ± 0) | 5.1 × 106 ± 3.5 (193 ± 14) |
| 72 | 4.4 × 107 ± 3.1 (304 ± 18) | 5.8 × 102 ± 3.1 (0 ± 0) | 8.6 × 107 ± 5.5 (347 ± 33) |
Average CFU/gram of tissue ± SD for three independent experiments in triplicate. Numbers in parentheses are the average area of tissue watersoaking (in square millimeters) ± SD for three independent experiments in triplicate.
Based on analysis of the ptw cluster and the putative role of a type IV secretion system in the export of an effector molecule(s), it was of interest to determine if culture supernatant concentrates showed effector activity. Over an 8-h duration, carrot protoplasts exposed to M9 medium concentrate did not experience significant plasmolysis compared to that of the protoplast controls (P > 0.05) (Table 5). However, significant (P ≤ 0.05) plasmolysis was observed for protoplasts exposed to the K56-2 or complemented mutant (AE310) concentrates as early as 2 h, and plasmolysis increased at a significant rate for dilutions tested (up to 1:5) over the 8-h time period compared to that of medium controls. Concentrated supernatant from the ptwD4 mutant (AE307) caused 6.7% plasmolysis after 8 h, which was not statistically different (P > 0.05) from plasmolysis resulting from treatment with medium controls. Dilution of the K56-2 and AE310 concentrates to a protein concentration of 0.02 μg/ml reduced activity to levels which approximated those of the control and AE307. Temperatures of 37, 65, and 80°C for 1 h or 100°C for 10 min did not affect the plasmolysis activity of the K56-2 concentrate. Complete inactivation of plasmolysis activity was observed when the K56-2 concentrate was treated with proteinase K; however, incubation with DNase I or RNase A showed no effect on activity.
TABLE 5.
Effect of supernatant concentrates from K56-2, ptwD4 mutant (AE307), and complemented ptwD4 mutant (AE310) on carrot protoplasts
| Supernatant concentrate | Concentrate dilutiona | % Plasmolysisb,c at time point (h):
|
|||
|---|---|---|---|---|---|
| 2 | 4 | 6 | 8 | ||
| M9 medium | 0 | 1.2 ± 1.1x | 2.7 ± 1.6x | 6.6 ± 1.7x | 8.2 ± 3.4x |
| 1:1 | 1.5 ± 1.7x | 3.4 ± 1.2x | 6.4 ± 2.0x | 7.5 ± 2.8x | |
| 1:5 | 2.6 ± 1.0x | 3.1 ± 1.6x | 4.3 ± 1.4x | 6.9 ± 2.7x | |
| 1:10 | 1.9 ± 1.5x | 2.9 ± 1.0x | 5.6 ± 2.3x | 7.3 ± 2.6x | |
| K56-2 | 0d | 18 ± 4.0y | 37 ± 4.1y | 57 ± 5.1y | 75 ± 5.8y |
| 1:1 | 18 ± 2.1y | 38 ± 5.0y | 56 ± 6.1y | 76 ± 7.6y | |
| 1:5 | 18 ± 4.3y | 36 ± 5.0y | 55 ± 5.6y | 73 ± 6.4y | |
| 1:10 | 1.6 ± 1.4x | 3.6 ± 1.0x | 5.9 ± 0.6x | 7.3 ± 1.4x | |
| AE307 | 0d | 1.8 ± 1.2x | 3.3 ± 0.7x | 4.9 ± 1.7x | 6.7 ± 1.7x |
| 1:1 | 2.2 ± 1.3x | 3.8 ± 2.1x | 5.4 ± 1.0x | 7.4 ± 1.9x | |
| 1:5 | 2.3 ± 1.4x | 3.5 ± 0.7x | 5.8 ± 1.5x | 8.4 ± 1.4x | |
| 1:10 | 2.3 ± 1.5x | 4.5 ± 1.1x | 6.1 ± 0.8x | 7.7 ± 0.6x | |
| AE310 | 0d | 19 ± 1.0y | 37 ± 3.0y | 52 ± 4.0y | 78 ± 7.7y |
| 1:1 | 19 ± 1.4y | 36 ± 4.9y | 52 ± 3.7y | 70 ± 6.6y | |
| 1:5 | 18 ± 2.0y | 39 ± 4.5y | 56 ± 4.2y | 77 ± 7.0y | |
| 1:10 | 1.5 ± 1.0x | 1.8 ± 1.0x | 6.0 ± 1.6x | 7.2 ± 1.0x | |
| None | 0 | 2.0 ± 1.6x | 3.1 ± 7.7x | 6.7 ± 1.6x | 11 ± 2.2x |
All dilutions were adjusted to 10% mannitol.
Data are mean values ± SD of two replicates for three independent experiments. At 0 h all plasmolysis values were 0x.
Values within the columns scored by the same letter were not statistically different at P = 0.05.
Undiluted concentrate adjusted to 0.2 μg of protein/ml in 10% mannitol.
DISCUSSION
Previously, the role of a polygalacturonase (PehA) as a virulence factor in B. cepacia strain ATCC 25416 was investigated (24). Derivatives cured of a resident plasmid that codes for the PehA no longer macerated onion tissue; however, the derivatives still produced a phenotype described as ptw. Of interest in the present study was the observation that all B. cepacia and B. cenocepacia isolates and a B. vietnamiensis strain from the Bcc experimental strain panel (47) produced the ptw phenotype in onion tissue (50). In this study K56-2, a B. cenocepacia strain that produced the plant disease-associated phenotype, was genetically analyzed and found to contain a plasmid-borne gene cluster, designated ptw, that encodes a type IV secretion system responsible for the translocation of the ptw effector protein(s).
The identified gene products in the ptw cluster had homology to proteins from known type IV secretion systems. Bacteria have evolved type IV secretion systems to transfer DNA or protein macromolecules to a wide array of target cell types (2, 10, 52). Originally, the nomenclature referred to the VirB-VirD-encoded translocation system of A. tumefaciens and two closely related systems encoded by the transfer region of the IncN plasmid pKM101 and the ptl operon of Bordetella pertussis (10). In the past decade, the type IV family has expanded considerably in number with the relaxation of defining criteria. Currently, type IV secretion systems are defined as translocation systems ancestrally related to any conjugation system of gram-negative or -positive bacteria (10, 25). However, it is important to distinguish functional translocation machines from mobile elements characteristically found in bacterial genomes. Ding et al. (18) have suggested that mutagenesis of a putative type IV system should yield a phenotype at least consistent with a translocation defect.
Christie (10) has suggested subclassification of type IV secretion systems based on ancestral lineage. Thus, the VirB-VirD type has been designated IVA, and the ColIb-P9 Tra and L. pneumophila Dot-Icm types have been designated IVB. This subclassification left open the possibility of further division of gram-negative and -positive secretion systems that differ from IVA and IVB. Therefore, an alternative grouping scheme has been suggested by Ding et al. (18) that separates systems based on function and does not replace the one previously described but rather expands its usefulness. This classification method groups type IV secretion systems as (i) conjugation systems, (ii) effector translocator systems, or (iii) DNA uptake or release systems (18). By definition, the conjugation systems deliver DNA substrates by establishing direct physical contact with target cells. Examples include the well-studied A. tumefaciens T-DNA transfer system and the F, RP4, and R388 plasmid transfer systems. Although the conjugation systems are known mainly for their role in distributing DNA among bacterial populations, they can also translocate protein substrates independently of DNA (73). There is also a subset of these systems that can transfer DNA and protein substrates to a range of eukaryotic cell types, including plant, fungal, and human (3, 71, 72). Most of the members of the type IV effector translocator group inject their substrates directly into the eukaryotic cytosol. This type of translocation is now recognized as the dominant virulence mechanism of the phytopathogen A. tumefaciens and of several medically important pathogens, including H. pylori, L. pneumophila, Brucella spp., and Bartonella spp. (7). Also included in this subfamily is the Ptl system of B. pertussis, even though this system exports its protein substrate independently of host-cell contact. Presently, at least 10 type IVA and several type IVB systems can be grouped as effector translocators and are thought to be essential for infection (18). The DNA uptake and release group, which presently contains three members, translocates DNA substrates across the cell envelope to or from the extracellular milieu (18). Neisseria gonorrhoeae uses a system encoded by the gonococcal genetic island to export DNA (17, 26). Recent studies have established that this system is very closely related to the F plasmid conjugation system of E. coli, even though it is not performing the same function (17). The two other members of this subfamily, Campylobacter jejuni and H. pylori, translocate DNA in the opposite direction, promoting genetic variation and cell survival (1, 29).
A notable feature of the type IV secretion systems is their extreme versatility. These systems can recognize a wide array of DNA and protein substrates, translocate substrates by both cell-contact-dependent and -independent mechanisms, and deliver substrates to an exceptionally wide range of prokaryotic and eukaryotic taxa (18). The ptw cluster contains homologs to components of multiple type IV secretion systems (Fig. 2). With respect to function, the plasmid-encoded type IV secretion system we have identified in B. cenocepacia strain K56-2 was involved in export of an effector(s) that was responsible for the ptw phenotype. The facts that the Ptw system was involved in the translocation of a protein(s), that it did not contain all of the necessary genetic components to support conjugation, and that no oriT homolog was identified indicate that functionally it is a member of the effector translocator subfamily. The difference in the G-C ratio between pK56-2 harboring the ptw cluster (62.7%) and the G-C ratio for the entire B. cenocepacia genome (66.9%) suggests acquisition by horizontal transfer (34).
The presence of a second type IV secretion system in B. cenocepacia is not unprecedented, because many bacteria, such as H. pylori and A. tumefaciens, have been found to harbor multiple secretion systems (18, 35). The type IV secretion system located on chromosome II showed most similarity to that of the A. tumefaciens VirB-VirD translocation system with respect to arrangement and gene product similarity (Fig. 4). The difference in the G-C ratio of the chromosomally located type IV secretion system (63%) and the G-C ratio of the entire genome (66.9%) suggests horizontal acquisition (34), as with the Ptw translocation system. Evidence suggests that the Ptw system is responsible for export of the protein(s) involved in expression of the ptw phenotype; however, the substrate(s) translocated by the VirB-D4 system is unknown.
Identification of a cluster of genes that encoded proteins with similarities to components of a type IV secretion system allowed for strategic generation of K56-2 mutants to support the hypothesis that such a system was involved in expression of the ptw phenotype. The genes selected for functional characterization were ptwD4, ptwC, ptwB4, and ptwB10. VirD4-like proteins (PtwD4 homologs) are cytoplasmic membrane NTP-binding proteins that are essential for coupling the relaxosome to the macromolecular transport system (51, 61). It is thought to interact with the oriT-bound relaxosome, which is made up of TrwC and TrwA, to facilitate DNA transfer (19, 45). In the Ptw system, however, there is no TrwA homolog. The ptwD4 product appears to have functions similar to those of Hp0524 of H. pylori. The hp0524 product is critical for the transfer of CagA from the bacterium to epithelial cells (21). Further analysis revealed that the PtwD4 amino acid sequence contained P-loop and Walker B sites, which are characteristic of type IV secretion ATP-binding proteins. The ptwC product had homology to TrwC, which is both a relaxase that cleaves at the nic site within the oriT in a strand- and sequence-specific manner, and a helicase, which is essential for transfer (48, 49). Analysis of PtwC revealed that the N terminus possessed signature motifs (2 and 3) conserved in proteins with DNA-nicking activities (44), and the C terminus possessed signature motifs (I, Ib, II, III, IV, V, and VI) conserved in DNA helicases (28); however, there is no identified oriT to facilitate the cleavage and thus linearization required for DNA transfer. VirB10, the PtwB10 homolog, is a protein located in the periplasmic space that is believed to form the core of the transfer machinery and is possibly part of the pore that spans the inner and outer membranes (74). PtwB10 was shown to contain the C-terminal hydrophobic region that is characteristic of VirB10-like proteins. PtwB4 showed similarity to VirB4 of A. tumefaciens, which is a putative nucleoside triphosphatase that may also serve as an active motor for secretion (74). The PtwB4 amino acid sequence was shown to possess four transmembrane regions that are characteristic of VirB4-like NTPases (74). The ptw phenotype was not expressed by K56-2 derivatives with a disruption in ptwD4, ptwB4, or ptwB10 but was expressed by a derivative with a disruption in ptwC. These results correlated with the predicted function of the gene products, because successful secretion of the ptw effector protein(s) would likely involve the following: a protein that potentially couples the moiety to the secretion system and supplies the necessary energy for export (PtwD4); a protein that acts in the assembly of the pore necessary for the passage of the substrate and provides energy necessary for the system to function (PtwB4); and a protein involved in structural formation of a pore (PtwB10), but not one involved in the generation of a relaxosome (PtwC).
By combining mutational analysis with cytotoxic assays employing both plant tissue and protoplasts, we have identified a type IV secretion system that appears to export an effector molecule(s) that is proteinaceous in nature and is responsible for the activity. This activity plays an important role in the ability of B. cenocepacia strain K56-2 to cause watersoaking and to grow in plant tissue (Fig. 2, Table 4). It appears that in the plant tissue assay the role of the effector(s) is to provide bacterial cells with needed nutrients by causing leakage of the plant cell cytosol. Presently, the role of the putative virulence effector protein(s) in contributing to growth and infection in a pulmonary environment is unknown; however, the ptw phenotype appears to be common among isolates belonging to B. cenocepacia that infect CF patients.
Protein DspE of Erwinia amylovora and WtsE of Pantoea stewartii subsp. stewartii are involved in elicitation of the watersoaking phenotype in their respective hosts. The dspE product is a 198-kDa protein that is required for the production of fireblight disease symptoms that include watersoaking of apple and pear tissue (5). A disruption in wtsE renders P. stewartii subsp. stewartii incapable of eliciting an observable watersoaking phenotype and causing a systemic corn leaf infection (22). In contrast to the ptw effector molecule(s), a type III secretion system is presumed to export DspE and WtsE from the phytopathogen to the plant hosts.
There are two types of substrates typically transported by both type III and type IV secretion systems (52, 58). The first group alters host processes by mimicking the function of a eukaryotic protein. Substrate proteins in this category, however, do not have significant sequence similarities with the eukaryotic factor they mimic, which makes identification difficult. The second class are genes that were “stolen” from eukaryotic host cells and then reshaped. The RalF protein of the L. pneumophila Dot/Icm system falls into the latter group (52). Substrates in this category are relatively easy to identify with genomic analysis. Secreted substrates from either group can vary in size from ∼22 kDa for A. tumefaciens VirF to ∼145 kDa for H. pylori CagA. They also vary in composition, from monomers to multisubunit structures that may include protein and/or DNA (18). Thus, there are no universally conserved primary sequence motifs or physical characteristics that are readily discernible for secreted effectors.
In conclusion, our study has identified a plasmid-encoded type IV secretion system in B. cenocepacia strain K56-2 that was functionally an effector translocator. The chromosomally located type IV secretion system showed modular similarity to the A. tumefaciens VirB-VirD translocation system; however, it was not responsible for the ptw phenotype in B. cenocepacia strain K56-2.
There are multiple examples for the role of type IV secretion systems in translocation of both plasmid DNA and proteins associated with plant and human pathogenesis (9, 10). The role of this putative virulence effector(s) in contributing to disease is being determined in studies using a murine model of pulmonary infection. Studies to identify the structural gene(s) for the effector protein(s) and regulatory subunits are in progress.
Acknowledgments
We are grateful to Pamela Sokol for providing K56-2 genomic library blots, to James Samuel for his critical reading of the manuscript, and James Starr for his statistical help. We also thank Michael B. Willett, Bianca S. Batista, Francisco Vielma, and Arwyn Hood for their technical assistance.
This work was supported by grant GONZAL03G0 from the Cystic Fibrosis Foundation.
REFERENCES
- 1.Bacon, D. J., R. A. Alm, D. H. Burr, L. Hu, D. J. Kopecko, D. P. Ewing, T. J. Trust, and P. Guerry. 2000. Involvement of a plasmid in virulence of Campylobacter jejuni 81-176. Infect. Immun. 68:4384-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baron, C., D. O'Callaghan, and E. Lanka. 2002. Bacterial secrets of secretion: Euro conference on the biology of type IV secretion processes. Mol. Microbiol. 43:1359-1365. [DOI] [PubMed] [Google Scholar]
- 3.Bates, S., A. M. Cashmore, and B. M. Wilkins. 1998. IncP plasmids are unusually effective in mediating conjugation of Escherichia coli and Saccharomyces cerevisiae. J. Bacteriol. 180:6538-6543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blank, R. D., and D. B. Wilson. 1982. Isolation and characterization of a 2000-bp derivative of pBR322. Plasmid 7:278-289. [DOI] [PubMed] [Google Scholar]
- 5.Bogdanove, A. J., J. F. Kim, Z. Wei, P. Kolchinsky, A. O. Charakowski, A. K. Conlin, A. Collmer, and S. V. Beer. 1998. Homology and functional similarity of an hrp-linked pathogenicity locus, dspEF, of Erwinia amylovora and the avirulence locus avrE of Pseudomonas syringae pathovar tomato. Proc. Natl. Acad. Sci. USA 95:1325-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyer, H., and D. Roulland-Dussoix. 1969. A complementation analysis of the restriction and modification DNA in Escherichia coli. J. Mol. Biol. 41:115-117. [DOI] [PubMed] [Google Scholar]
- 7.Burns, D. L. 2003. Type IV transporters of pathogenic bacteria. Curr. Opin. Microbiol. 6:29-34. [DOI] [PubMed] [Google Scholar]
- 8.Chen, J. S., K. A. Witzmann, T. Spiker, R. J. Fink, and J. J. LiPuma. 2001. Endemicity and inter-city spread of Burkholderia cepacia genomovar III in cystic fibrosis. J. Pediatr. 139:643-649. [DOI] [PubMed] [Google Scholar]
- 9.Christie, P. J., and J. P. Vogel. 2000. Bacterial type IV secretion: conjugation systems adapted to deliver effector molecules to host cells. Trends Microbiol. 8:354-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christie, P. J. 2001. Type IV secretion: intercellular transfer of macromolecules by systems ancestrally related to conjugation machines. Mol. Microbiol. 40:294-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coenye, T., P. Vandamme, J. R. W. Govan, and J. J. LiPuma. 2001. Taxonomy and identification of the Burkholderia cepacia complex. J. Clin. Microbiol. 39:3427-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coenye, T., and J. J. LiPuma. 2002. Multilocus restriction typing: a novel tool for studying global epidemiology of Burkholderia cepacia complex infection in cystic fibrosis. J. Infect. Dis. 185:1454-1462. [DOI] [PubMed] [Google Scholar]
- 13.Datta, N., and R. W. Hedges. 1972. Trimethoprim resistance conferred by W plasmids in Enterobacteriaceae. J. Gen. Microbiol. 72:349-355. [DOI] [PubMed] [Google Scholar]
- 14.DeFeyter, R., C. I. Kado, and D. W. Gabriel. 1990. Small, stable shuttle vectors for use in Xanthomonas. Gene 88:65-72. [DOI] [PubMed] [Google Scholar]
- 15.Dennis, J. J., and G. J. Zylstra. 1998. Plasposons: modular self-cloning minitransposon derivatives for rapid genetic analysis of gram-negative bacterial genomes. Appl. Environ. Microbiol. 64:2710-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dhar, S. K., R. K. Soni, B. K. Das, and G. Mukhopadhyay. 2003. Molecular mechanism of action of major Helicobacter pylori virulence factors. Mol. Cell. Biochem. 253:207-215. [DOI] [PubMed] [Google Scholar]
- 17.Dillard, J. P., and H. S. Seifert. 2001. A variable genetic island specific for Neisseria gonorrhoeae is involved in providing DNA for natural transformation and is found more often in disseminated infection isolates. Mol. Microbiol. 41:263-277. [DOI] [PubMed] [Google Scholar]
- 18.Ding, Z., K. Atmakuri, and P. J. Christie. 2003. The outs and ins of bacterial type IV secretion substrates. Trends Microbiol. 11:527-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fekete, R. A., and L. S. Frost. 2000. Mobilization of chimeric oriT plasmids by F and R100-1: role of relaxosome formation in defining plasmid specificity. J. Bacteriol. 182:4022-4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Figurski, D., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fischer, W., J. Puls, R. Buhrdorf, B. Gebert, S. Odenbreit, and R. Hass. 2001. Systemic mutagenesis of the Helicobacter pylori cag pathogenicity island: essential genes for CagA translocation in host cells and induction of interleukin-8. Mol. Microbiol. 42:1337-1348. [DOI] [PubMed] [Google Scholar]
- 22.Frederick, R. D., M. Ahmad, D. R. Majerczak, A. S. Arroyo-Rodriguez, S. Manulis, and D. L. Coplin. 2001. Genetic organization of the Pantoea stewartii subsp. stewartii hrp gene cluster and sequence analysis of the hrpA, hrpC, hrpN, and wtsE operons. Mol. Plant-Microbe Interact. 14:1213-1222. [DOI] [PubMed] [Google Scholar]
- 23.Gonzalez, C. F., and A. K. Vidaver. 1979. Bacteriocin, plasmid, and pectolytic diversity in Pseudomonas cepacia of clinical and plant origin. J. Gen. Microbiol. 110:161-170. [DOI] [PubMed] [Google Scholar]
- 24.Gonzalez, C. F., E. A. Pettit, V. A. Valadez, and E. Provin. 1997. Mobilization, cloning, and sequence determination of a plasmid-encoded polygalacturonase from a phytopathogenic Burkholderia (Pseudomonas) cepacia. Mol. Plant-Microbe Interact. 10:840-851. [DOI] [PubMed] [Google Scholar]
- 25.Grohmann, E., G. Muth, and M. Espinosa. 2003. Conjugative plasmid transfer in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67:277-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamilton, H. I., K. J. Schwartz, and J. P. Dillard. 2001. Insertion-duplication mutagenesis of Neisseria: use in characterization of DNA transfer genes in the gonococcal genetic island. J. Bacteriol. 186:4718-4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hodgman, T. C. 1988. A new superfamily of replicative proteins. Nature 333:22-23. [DOI] [PubMed] [Google Scholar]
- 29.Hofreuter, D., S. Odenbreit, and R. Haas. 2001. Natural transformation competence in Helicobacter pylori is mediated by the basic components of a type IV secretion system. Mol. Microbiol. 41:379-391. [DOI] [PubMed] [Google Scholar]
- 30.Hutchison M. L., I. R. Poxton, and J. R. W. Govan. 1998. Burkholderia cepacia produces a hemolysin that is capable of inducing apoptosis and degranulation of mammalian phagocytes. Infect. Immun. 66:2033-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jakubowski, S. J., V. Krishnamoorthy, and P. J. Christie. 2003. Agrobacterium tumefaciens VirB6 protein participates in formation of VirB7 and VirB9 complexes required for type IV secretion. J. Bacteriol. 185:2867-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson, W. M., S. D. Tyler, and K. R. Rozee. 1994. Linkage analysis of geographic and clinical clusters in Pseudomonas cepacia infections by multilocus enzyme electrophoresis and ribotyping. J. Clin. Microbiol. 32:924-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones, R. W., A. O. Jackson, and T. H. Morris. 1990. Defective-interfering RNAs and elevated temperatures inhibit replication of tomato bushy stunt virus in inoculated protoplasts. Virology 176:539-545. [DOI] [PubMed] [Google Scholar]
- 34.Karlin, S. 2001. Detecting anomalous gene clusters and pathogenicity islands in diverse bacterial genomes. Trends Microbiol. 9:335-343. [DOI] [PubMed] [Google Scholar]
- 35.Kersulyte, D., B. Velapatino, A. K. Mukhopadhyay, L. Cahuayne, A. Bussalleu, J. Combe, R. H. Gilman, and D. E. Berg. 2003. Cluster of type IV secretion genes in Helicobacter pylori's plasticity zone. J. Bacteriol. 185:3764-3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koch, A., and S. Putnam. 1971. Sensitive biuret method for determination of protein in an impure system such as whole bacteria. Anal. Biochem. 44:239-245. [DOI] [PubMed] [Google Scholar]
- 37.Kumar, R. B., and A. Das. 2002. Polar location and functional domains of the Agrobacterium tumefaciens DNA transfer protein VirD4. Mol. Microbiol. 43:1523-1532. [DOI] [PubMed] [Google Scholar]
- 38.Lawley, T. D., W. A. Klimke, M. J. Gubbins, and L. S. Frost. 2003. F factor conjugation is a true type IV secretion system. FEMS Microbiol. Lett. 224:1-15. [DOI] [PubMed] [Google Scholar]
- 39.Ledson, M. J., M. J. Gallagher, M. Jackson, C. A. Hart, and M. J. Walshaw. 2002. Outcome of Burkholderia cepacia colonization in an adult cystic fibrosis centre. Thorax 57:142-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lewenza, S., B. Conway, E. P. Greenberg, and P. A. Sokol. 1999. Quorum sensing in Burkholderia cepacia: identification of the LuxRI homologs CepRI. J. Bacteriol. 181:748-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.LiPuma, J. J. 1998. Burkholderia cepacia—management issues and new insights. Clin. Chest Med. 19:473-486. [DOI] [PubMed] [Google Scholar]
- 42.LiPuma, J. J., T. Spilker, L. Gill, P. W. Campbell, L. Liu, and E. Mahenthiralingam. 2001. Disproportionate distribution of Burkholderia cepacia complex species and transmissibility factors in cystic fibrosis. Am. J. Resp. Crit. Care Med. 164:92-96. [DOI] [PubMed] [Google Scholar]
- 43.LiPuma, J. J. 2003. Burkholderia cepacia complex as human pathogens. J. Nematol. 35:212-217. [PMC free article] [PubMed] [Google Scholar]
- 44.Llosa, M., S. Bolland, and F. de la Cruz. 1994. Genetic organization of the conjugal DNA processing region of the IncW plasmid R388. J. Mol. Biol. 233:448-464. [DOI] [PubMed] [Google Scholar]
- 45.Llosa, M., S. Zunzunegui, and F. de la Cruz. 2003. Conjugative coupling proteins interact with cognate and heterologous VirB10-like proteins while exhibiting specificity for cognate relaxosomes. Proc. Natl. Acad. Sci. USA 100:10465-10470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Macnab, R. M. 1999. The bacterial flagellum: reversible rotary propeller and type III export apparatus. J. Bacteriol. 181:7149-7153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mahenthiralingam, E., T. Coenye, J. W. Chung, D. P. Speert, J. R. W. Govan, P. Taylor, and P. Vandamme. 2000. Diagnostically and experimentally useful panel of strains from the Burkholderia cepacia complex. J. Clin. Microbiol. 38:910-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matson, S. W., W. C. Nelson, and B. S. Morton. 1993. Characterization of the reaction product of the ortT nicking reaction catalyzed by Escherichia coli DNA helicase I. J. Bacteriol. 175:2599-2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matson, S. W., J. K. Sampson, and D. R. N. Byrd. 2001. F plasmid conjugative DNA transfer. J. Biol. Chem. 276:2372-2379. [DOI] [PubMed] [Google Scholar]
- 50.Medrano, E. G. 2002. Ph.D. thesis. Texas A&M University, College Station, Tex.
- 51.Moncalián, G., E. CabeŸon, I. Alkorta, M. Valle, F. Moro, J. M. Valpuesta, F. M. Goñi, and F. de la Cruz. 1999. Characterization of ATP and DNA binding activities of TrwB, the coupling protein essential in plasmid R388 conjugation. J. Biol. Chem. 274:36117-36124. [DOI] [PubMed] [Google Scholar]
- 52.Nagai, H., and C. R. Roy. 2003. Show me the substrates: modulation of host cell function by type IV secretion systems. Cell. Microbiol. 5:373-383. [DOI] [PubMed] [Google Scholar]
- 53.Planet, P. J., S. C. Kachlany, R. DeSalle, and D. H. Figurski. 2001. Phylogeny of genes for secretion NTPases: identification of the widespread tadA subfamily and development of a diagnostic key for gene classification. Proc. Natl. Acad. Sci. USA 98:2503-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Plano, G. V., J. B. Day, and F. Ferracci. 2001. Type III export: new uses for an old pathway. Mol. Microbiol. 40:284-293. [DOI] [PubMed] [Google Scholar]
- 55.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 56.Sajjan, U. S., F. A. Sylvester, and J. Forstner. 2000. Cable-piliated Burkholderia cepacia binds to cytokeratin 13 of epithelial cells. Infect. Immun. 68:1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sajjan, U. S., Y. Wu, G. Kent, and J. Forstner. 2000. Preferential adherence of cable-piliated Burkholderia cepacia to respiratory epithelia of CF knockout mice and human CF lung explants. J. Med. Microbiol. 49:875-882. [DOI] [PubMed] [Google Scholar]
- 58.Salmond, G. P. C. 1994. Secretion of extracellular virulence factors by plant pathogenic bacteria. Annu. Rev. Phytopathol. 32:181-200. [Google Scholar]
- 59.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 60.Schröder, G., S. Krause, E. L. Zechner, B. Traxler, H. Yeo, R. Lurz, G. Waksman, and E. Lanka. 2002. TraG-like proteins of DNA transfer systems and of the Helicobacter pylori type IV secretion system: inner membrane gate for exported substrates? J. Bacteriol. 184:2767-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schröder, G., and E. Lanka. 2003. TraG-like proteins of type IV secretion systems: functional dissection of the multiple activities of TraG (RP4) and TrwB (R388). J. Bacteriol. 185:4371-4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sokol, P. A., P. Darling, D. E. Woods, E. Manhenthiralingam, and C. Kooi. 1999. Role of ornibactin biosynthesis in the virulence of Burkholderia cepacia: characterization of the pvdA gene encoding L-ornithine-N5-Oxygenase. Infect. Immun. 67:443-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Swarup, S., R. DeFeyter, R. H. Brlansky, and D. W. Gabriel. 1991. A pathogenicity locus from Xanthomonas citri enables strains from several pathovars of Xanthomonas campestris to elicit cankerlike lesions on citrus. Phytopathology 81:802-809. [Google Scholar]
- 64.Tomasek, P. H., B. Frantz, U. M. Sangodkar, R. A. Haugland, and A. M. Chakrabarty. 1989. Characterization and nucleotide sequence determination of a repeat element isolated from a 2, 4, 5-T degrading strain of Pseudomonas cepacia. Gene 76:227-238. [DOI] [PubMed] [Google Scholar]
- 65.Vandamme, P., D. Henry, T. Coenye, S. Nzula, M. Vancanneyt, J. J. LiPuma, D. P. Speert, J. R. W. Govan, and E. Mahenthiralingam. 2002. Burkholderia anthina sp. nov and Burkholderia pyrrocinia, two additional Burkholderia cepacia complex bacteria, may confound test results of new molecular diagnostic tools. FEMS Immunol. Med. Microbiol. 33:143-149. [DOI] [PubMed] [Google Scholar]
- 66.Vandamme, P., B. Holmes, T. Coenye, E. Mahenthiralingam, J. J. LiPuma, and J. R. W. Govan. 2003. Burkholderia cenocepacia sp. nov., a new twist on an old story. Res. Microbiol. 154:91-96. [DOI] [PubMed] [Google Scholar]
- 67.Vasil, M. L., D. P. Krieg, J. S. Kuhns, J. W. Ogle, V. D. Shortridge, R. M. Ostroff, and A. I. Vasil. 1990. Molecular analysis of hemolytic and phospholipase activities of Pseudomonas cepacia. Infect. Immun. 58:4020-4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vermis, K., T. Coenye, J. J. LiPuma, E. Mahenthiralingam, H. J. Nelis, and P. Vandamme. 2004. Identification of Burkholderia dolosa sp. nov. (formerly Burkholderia cepacia genomovar VI). Int. J. Syst. Evol. Microbiol. 54:689-691. [DOI] [PubMed]
- 69.Vogel, H. J., and D. M. Bonner. 1965. Acetylornithinase of Escherichia coli partial purification and some properties. J. Biol. Chem. 281:97-106. [PubMed] [Google Scholar]
- 70.Walker, J. E., M. Saraste, M. J. Runswick, and N. J. Gay. 1982. Distantly related sequence in the alpha- and beta-subunits of ATP sythase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1:945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ward, D. V., and P. C. Zambryski. 2001. The six functions of Agrobacterium VirE. Proc. Natl. Acad. Sci. USA 98:385-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Waters, V. L. 2001. Conjugation between bacterial and mammalian cells. Nat. Genet. 29:375-376. [DOI] [PubMed] [Google Scholar]
- 73.Wilkins, B. M., and A. T. Thomas. 2000. DNA-independent transport of plasmid primase protein between bacteria by the I1 conjugation system. Mol. Microbiol. 38:650-657. [DOI] [PubMed] [Google Scholar]
- 74.Yeo, H., and G. Waksman. 2004. Unveiling molecular scaffolds of the type IV secretion system. J. Bacteriol. 186:1919-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]