Abstract
Although pressure is an important environmental parameter in microbial niches such as the deep sea and is furthermore used in food preservation to inactivate microorganisms, the fundamental understanding of its effects on bacteria remains fragmentary. Our group recently initiated differential fluorescence induction screening to search for pressure-induced Escherichia coli promoters and has already reported induction of the heat shock regulon. Here the screening was continued, and we report for the first time that pressure induces a bona fide SOS response in E. coli, characterized by the RecA and LexA-dependent expression of uvrA, recA, and sulA. Moreover, it was shown that pressure is capable of triggering lambda prophage induction in E. coli lysogens. The remnant lambdoid e14 element, however, could not be induced by pressure, as opposed to UV irradiation, indicating subtle differences between the pressure-induced and the classical SOS response. Furthermore, the pressure-induced SOS response seems not to be initiated by DNA damage, since ΔrecA and lexA1 (Ind−) mutants, which are intrinsically hypersensitive to DNA damage, were not sensitized or were only very slightly sensitized for pressure-mediated killing and since pressure treatment was not found to be mutagenic. In light of these findings, the current knowledge of pressure-mediated effects on bacteria is discussed.
Pressure is an environmental parameter that varies only between narrow limits and thus has little or no influence in most commonly studied microbial niches. However, in some specific niches and situations, the life and death of microorganisms are strongly affected by pressure. This is the case for piezophilic and piezotolerant microorganisms (respectively requiring or tolerating high pressure during growth) living in the deep sea and the deep subsurface (40, 54) and also for nonpiezophiles that are subjected to pascalization, an emerging process for preserving foods by treatment with ultrahigh pressure (100 to 1,000 MPa) (15). Although pressure, like temperature, is a thermodynamically well-known physical parameter, the effects of high pressure on microorganisms remain poorly characterized, unlike those of heat. Some effects of pressure on biomolecules and biological systems that have been well studied in vitro and explained on the basis of thermodynamic principles are protein denaturation and phase transition in membranes (5). Therefore, most pressure effects on microorganisms observed in vivo, such as inhibition of key enzymes (48) and processes (20, 41) and disruption of cellular structures (35) and membranes (22, 42), are believed to stem from these primary events.
Microorganisms that are adapted to normal atmospheric pressure (0.1 MPa), such as Escherichia coli, can often grow at pressures up to a few tens of megapascals, but only at a strongly reduced rate. The deep sea, on the other hand, with pressures ranging from 30 up to 100 MPa, constitutes a reservoir of piezophilic and piezotolerant microorganisms, which have become a model for studying piezophysiology and to gain insight into cellular adaptation strategies for coping with high-pressure stress. Although these organisms remain difficult to study, several research groups have revealed specific piezoprotective alterations in their membrane and protein components, such as an increased proportion of unsaturated fatty acids and a decreased occurrence of helix-destabilizing amino acid residues, respectively (reviewed in references 1 and 8).
Pressures in the range of 100 to 1,000 MPa kill most microorganisms and are being used in high-pressure food preservation, because they leave most of the sensorial and nutritional properties of the food intact, as opposed to thermal treatments (15). Detailed pressure inactivation studies, however, revealed significant differences in pressure sensitivities among vegetative bacterial species and even between strains within a single species (3, 9). Moreover, several groups have reported the selection of pressure-resistant mutants of E. coli (23, 28) and Listeria monocytogenes (31). Although not affecting pressure growth limits, the resistance in E. coli was extended by an extraordinary 500 MPa (28). Molecular characterization of such pressure-resistant mutants of E. coli and L. monocytogenes revealed in both cases the abundance of heat shock proteins (2, 32), stressing protein management as an important feature for withstanding extremely high pressures.
In order to understand the piezoprotective adaptations in dedicated deep-sea bacteria as well as in surface-dwelling bacteria that have mutated to become resistant to high-pressure inactivation, a better insight into cellular awareness of pressure is necessary. One way to achieve this is by characterizing the bacterial high-pressure response. To date, only a few groups have embarked on dissection of the genetic response of piezotolerant or piezosensitive microorganisms to pressure. For the deep-sea bacterium Photobacterium profundum SS9, Bartlett and colleagues discovered the expression of outer membrane proteins to be pressure dependent and to be regulated at the molecular level by homologues of the Vibrio cholerae ToxR and E. coli RpoE proteins (7, 16, 53). Using two-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis and gene arrays, two other studies revealed a link between the pressure response and the heat shock or freeze-thaw response of E. coli and Saccharomyces cerevisiae, respectively (30, 52). Using a differential fluorescence induction (DFI) screening (50), our group recently was able to demonstrate induction of several heat shock promoters after pressure treatment in E. coli, further substantiating the link between the genetic heat shock and pressure responses (2).
The characterization of the genetic response of microorganisms upon pressure treatment, however, remains fragmentary and is far from finished. In this paper, based on further DFI screening, we describe the unexpected discovery of a pressure-induced SOS response for E. coli. The SOS response is typically induced upon DNA damage, for example, after UV irradiation or mitomycin C treatment, resulting in stalling of the DNA replication fork and disassembly of the replication apparatus. This then results in the exposure of single-stranded DNA, which is rapidly sensed and stabilized by the RecA protein, generating a nucleoprotein filament that activates the autoproteolytic activity of LexA. Intact LexA acts as a repressor of the SOS response, controlling more than 40 genes involved in stabilization of single-stranded DNA, base or nucleotide excision repair, recombinational repair, translesion synthesis, and control of cell division (21, 36, 37). Pressure induction of the SOS response is surprising and has never been reported earlier, but it provides an explanation for some as yet unexplained pressure-related phenomena in bacteria.
MATERIALS AND METHODS
Strains and growth conditions.
The effects of high-pressure treatment were studied with E. coli strain MG1655 (13). Derivatives of MG1655 carrying either ΔrecA, lexA1 (Ind−), or e14::Tn10 were constructed by P1 transduction (46), using QC2411 (ΔrecA srl::Tn10) (19), AM121 (lexA1 [Ind−] malF3089::Tn10) (4), and CH1494 (e14-1272::Tn10) (14), respectively, as donor strains. Tetracycline-resistant transductants were isolated and, in the case of ΔrecA and lexA1 (Ind−) strains, checked for UV sensitivity. Lysogens of MG1655 wild-type and ΔrecA and lexA1 strains were constructed using λΦ[P3rpoH::lacZ] isolated from ADA600 (10) and selected on medium containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (20 μg/ml). Finally, C600 was used as a host for counting PFU.
Overnight cultures were obtained by growth in Luria Bertani broth (LB) (46) for 21 h at 37°C under well-aerated conditions. Antibiotics (Applichem, Darmstadt, Germany) were added when necessary to obtain the following concentrations: 100 μg of ampicillin/ml and 20 μg of tetracycline/ml.
Construction and screening of promoter trap library of MG1655.
A promoter trap library of MG1655, consisting of ca. 15,000 independent clones, each containing 1- to 3-kb fragments of a partial Sau3AI digest of MG1655 chromosomal DNA inserted upstream of the promoterless gfp gene of pFPV25 (50), was constructed earlier (2). Screening of the promoter trap library was based on the DFI technique described by Valdivia and Falkow (50) and slightly customized for isolating pressure-induced promoters as recently described (2).
Pressure treatment.
Overnight cultures were diluted 1/100 in fresh prewarmed LB with 100 μg of ampicillin/ml and further incubated until late exponential phase (optical density at 600 nm [OD600] = 0.6). Portions (4 ml) of this culture were then pelleted by centrifugation (5 min at 6,000 × g) and resuspended in the same volume of fresh prewarmed LB. For pressure induction, a 500-μl sample was sealed without air bubbles in a polyethylene bag and pressurized for 15 min in an 8-ml pressure vessel, maintained at 20°C with an external cooling circuit (Resato, Roden, The Netherlands). It should be noted that pressurization caused some adiabatic heating of the sample; however, this was less than 3°C at 100 MPa, which was the maximum inducing pressure used in this study. For inactivation experiments, pressures up to 300 MPa were used. After treatment, cultures were maintained at 37°C and used for the measurement of gfp induction, phage induction, or determination of viability.
Green fluorescent protein (GFP) fluorescence measurements.
After induction, 300-μl samples were transferred to microplate wells and placed in a fluorescence reader (Fluoroscan Ascent FL; Thermolabsystems, Brussels, Belgium). Fluorescence at 520 nm was then measured at 30-min intervals with intermittent shaking (every 5 min) at 37°C, using an excitation wavelength of 480 nm. At the same time, OD600 was measured and fluorescence was expressed per unit of OD600. Alternatively, 3 h after induction, cultures were analyzed by fluorescence-activated cell sorting (FACS) analysis using a FACSCalibur apparatus (Becton Dickinson, Erembodegem, Belgium) fitted with an argon laser emitting at 488 nm. Fluorescence data shown are representative results from at least four independent experiments. To determine the fold induction after pressure treatment, population means of fluorescence calculated by the FACSCalibur software were used.
Determination of viability.
Serial dilutions from pressurized and nonpressurized samples were plated on Tryptone Soy Agar (Oxoid, Basingstoke, United Kingdom) with a spiral plater (Spiral Systems Inc., Cincinnati, Ohio). Twenty-four hours later, colonies on the plates were counted, and reduction factors (RF) were determined as follows: RF = [no. of CFU/ml (nonpressurized sample)]/[no. of CFU/ml (pressurized sample)].
Construction of plasmids.
Specific GFP transcriptional fusions were constructed in pFPV25 with the promoter regions of recA (PrecA) and sulA (PsulA), which were obtained by PCR (Platinum Pfx DNA polymerase; Invitrogen, Merelbeke, Belgium), using the primer pairs 5′-TACGTCTAGATTATACTCCTGTCATGCCGGG-3′ and 5′-TAGCGGATCCTGTCTATTAGTGGTATCGCC-3′ for PrecA and 5′-GCATTCTAGATTAACGATGTGCATAGCCTC-3′ and 5′-GCATGGATCCCCCGAAGATACAACTCACC-3′ for PsulA. Both PCR products and pFPV25 were cut with BamHI and XbaI to allow directional cloning of the promoters upstream of gfp. Subsequently, these constructs were transformed to wild-type, ΔrecA and lexA1 (Ind−) backgrounds of MG1655 and confirmed by sequencing the promoter fragment and the gfp 5′ end. All restriction enzymes were purchased from Roche Diagnostics Belgium (Vilvoorde, Belgium).
Sequencing.
Inserts in the pFPV25 plasmid were sequenced by MWG Biotech AG (Ebersberg, Germany), using 5′-GACAAGTGTTGGCCATGGAACAGGTAG-3′ in the 5′ region of gfp as a sequencing primer.
Phage induction.
Late exponential cultures (OD600 = 0.6) of the λ lysogenized strains were pressure treated as described above and subsequently incubated at 37°C. At different time points after pressure treatment, 500-μl portions of pressure treated and untreated control cells were centrifuged (5 min at 24,000 × g), and the supernatant was sterilized by adding 30 μl of chloroform and vortexing. Afterwards, dilutions of this supernatant were added to 1 ml of stationary-phase C600 cells, which are a more efficient plating host than MG1655. Subsequently, 3 ml of TBMM top agar (34) was added to this culture, and the mixture was poured on a LB plate. After 24 h, the number of PFU was counted and recalculated to number of PFU/milliliter of the original culture.
UV treatment.
The UV oven used in this work (Bio-Link, Vilber Lourmat, France) was equipped with five fluorescent lamps of 8 W each, emitting from 180 to 280 nm with a peak at 254 nm. UV doses were programmed and are controlled by a radiometer that constantly monitors the UV light emission.
For measuring induction of promoters by UV, 1-ml portions of late-exponential-phase cultures (OD600 = 0.6) of strains containing promoter-gfp fusions or carrying a λ prophage were poured in a petri dish and irradiated in the UV oven (0.1 kJ/m2). Afterwards, the cultures were used for measurement of production of GFP or λ phage particles. For measuring UV inactivation or e14::Tn10 excision, a dilution series of cells was plated on LB agar and directly irradiated in the UV oven (0 to 0.1 kJ/m2). The distance between the lamps and the plates was 14 cm. After 24 h, colonies were counted. It should be noted that irradiating a 1-ml culture in a petri dish resulted in far less inactivation than direct irradiation of plated bacteria (about 1 log cycle for the wild type and the lexA1 mutant and 1.5 log cycles for the ΔrecA mutant).
RESULTS
uvrA induction by pressure, detected by DFI.
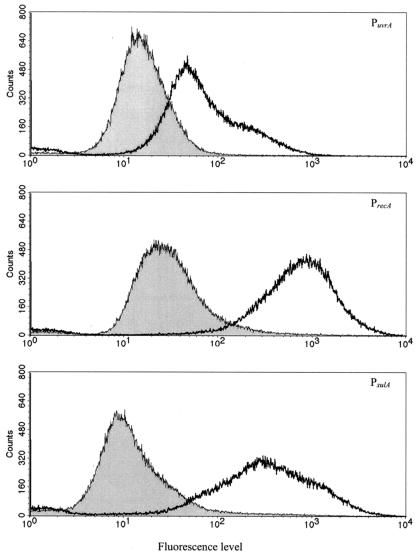
DFI screening of a random promoter probe library of E. coli MG1655, constructed in pFPV25, led to the isolation of several plasmids containing putative pressure-inducible promoters. Sequence analysis of the cloned fragments revealed in one particular clone the presence of the uvrA promoter (PuvrA) with its LexA binding box upstream of gfp. This clone showed a ca. fivefold increase in fluorescence 3 h after sublethal pressurization at 100 MPa for 15 min (Fig. 1), whereas the vast majority of clones containing random promoter fragments did not show pressure-induced GFP expression (data not shown). The functionality of PuvrA in the checked fragment was further confirmed by a dose-response curve showing UV induction (data not shown).
FIG. 1.
Flow-cytometry analysis of the high-pressure induction of PuvrA, PrecA, and PsulA fused to gfp in the wild-type MG1655 background, 3 h after treatment at 100 MPa for 15 min. The curves with the grey and transparent surfaces underneath represent populations of approximately 105 control cells and high-pressure-treated cells, respectively.
Induction of a bona fide SOS response by pressure.
Since uvrA is part of the SOS regulon, two additional gfp transcription fusions were constructed with the promoters of recA (PrecA) and sulA (PsulA). Like PuvrA, these promoters also harbor LexA binding boxes, but the LexA dissociation constant for each promoter is different (in order of increasing LexA affinity: PuvrA, PrecA, and PsulA). Both PrecA and PsulA proved to be responsive towards pressure treatment, showing ca. 18-fold and ca. 20-fold fluorescence induction at 100 MPa, respectively (Fig. 1).
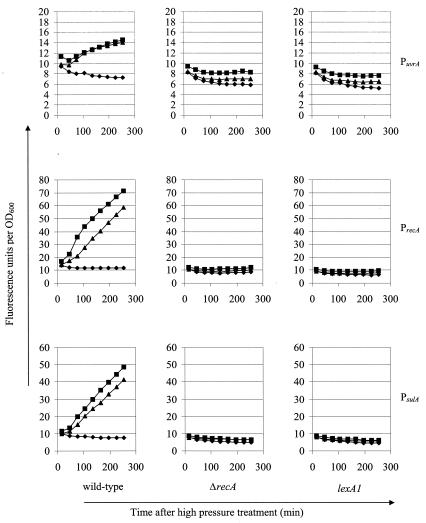
Since pressure can cause protein denaturation (5), one possible explanation for the observed induction of the SOS response was denaturation of the LexA repressor, resulting in clearance of the LexA boxes and in a short-cut mechanism of induction of the SOS response. Alternatively, the SOS response could be the result of a bona fide physiological response, dependent on the key regulators RecA and LexA. To distinguish between the two possibilities, we investigated the effect of a RecA deletion (ΔrecA) and a cleavage-resistant LexA protein (lexA1 Ind−) on pressure induction of PuvrA, PrecA, and PsulA. Figure 2 clearly shows that both defects abolish pressure induction of all three promoters at 100 MPa, similar to what they do for their induction by UV. Since ΔrecA strains were slightly more inactivated at 100 MPa than wild-type and lexA1 (Ind−) strains (see Fig. 4A), the same experiment was repeated at 75 MPa, where the differences in inactivation between all three strains were negligible and loss of responsiveness in the ΔrecA background could not be attributed to loss of viability. Although the induction at 75 MPa was slightly lower, a similar pattern was observed (data not shown). From this we can conclude that a genuine, RecA and LexA-dependent SOS response is elicited by sublethal pressures.
FIG. 2.
High-pressure (100 MPa, 15 min, ▪) and UV (0.1 kJ/m2, ▴) induction of PuvrA, PrecA, and PsulA fused to gfp in the wild-type MG1655, ΔrecA, and lexA1 (Ind−) backgrounds compared to results for untreated control cells (♦).
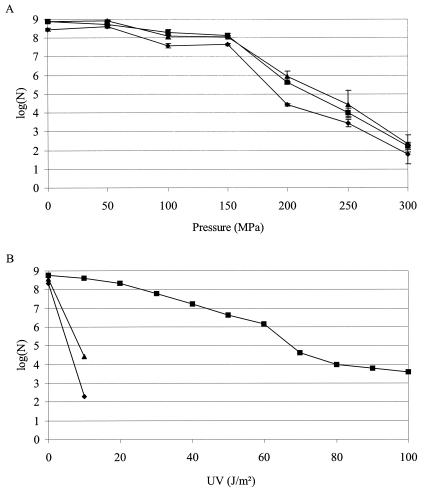
FIG. 4.
Number of survivors [log(N)] of wild-type (▪), ΔrecA (♦), and lexA1 (▴) cells of MG1655 after treatment at different pressures (A) or with different doses of UV (B). No survivors were recovered upon UV treatment of the ΔrecA and lexA1 cells at doses of ≥20 J/m2.
Induction of the lytic development of lysogenic λ by pressure.
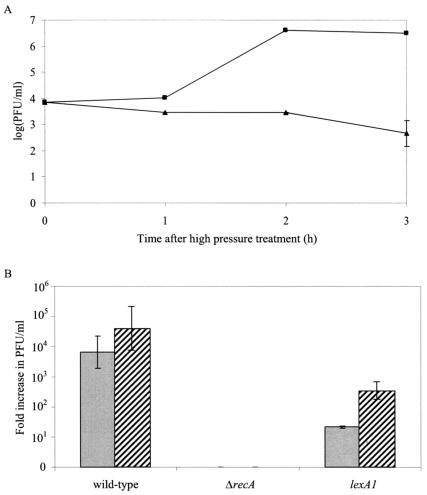
Since the activated RecA nucleoprotein fragment stimulates autocleavage not only of LexA but also of the phage λ CI repressor, responsible for maintaining lysogeny (36), we anticipated that pressure would cause induction of λ prophage from lysogenized MG1655. Figure 3A shows that an almost 104-fold induction of λ prophages indeed occurred 3 h after pressure treatment. Since this induction was not accompanied by visible lysis of the culture, we presume that only a subpopulation of cells was induced. In addition, pressure-mediated prophage induction was also shown to be LexA dependent (Fig. 3B), since lysogens displayed severely diminished induction in a lexA1 background. Finally, in a ΔrecA background, no phage induction could be observed (Fig. 3B). A similar pattern of prophage induction by UV irradiation (Fig. 3B) or mitomycin C treatment (data not shown) was observed.
FIG. 3.
(A) Induction of phage λ lysogens by high-pressure treatment (100 MPa, 15 min). Evolution of phage particle count [log(PFU/ml)] in untreated (▴) and high-pressure-treated (▪) cell suspensions of MG1655. (B) Fold increase in phage particle count (PFU/ml) 3 h after pressure treatment (100 MPa, grey bars) or UV treatment (0.1 kJ/m2, dashed bars) in cell suspensions of the MG1655 wild type or ΔrecA or lexA1 mutant. Titers of phage for untreated cell suspensions were ca. 1.2 × 103 PFU/ml for wild-type lysogens, 5 PFU/ml for ΔrecA lysogens, and 3.5 × 103 PFU/ml for lexA1 lysogens.
Stability of the e14 element is not affected by pressure treatment.
The e14 element is a 15.4-kb lambdoid bacteriophage remnant known to excise itself from the E. coli genome upon SOS induction by UV treatment (26). The use of an antibiotic resistance marker positioned in e14 (see “Strains and growth conditions”) allows direct demonstration of the presence or absence of this element (14). Loss of e14::Tn10 was assayed after pressure or UV treatment by streaking ca. 400 survivors on LB with 20 μg of tetracycline/ml. While ca. 60% of UV-treated (0.02 kJ/m2) cells showed loss of e14, no loss was observed for any of the pressures applied (Table 1).
TABLE 1.
Loss of the e14::Tn10 element by pressure or UV treatment
| Treatment | No. of survivors | Loss of tetracycline resistance in survivors (%) |
|---|---|---|
| None | 1.8 × 109 | 0 |
| Pressure | ||
| 50 MPa | 8.9 × 108 | 0 |
| 100 MPa | 7.1 × 108 | 0 |
| 150 MPa | 1.2 × 108 | 0 |
| 200 MPa | 1.7 × 106 | 0 |
| 250 MPa | 1.7 × 104 | 0 |
| UV (0.02 kJ/m2) | 1.8 × 107 | 60 |
ΔrecA and lexA1 mutants are not sensitized against pressure treatment.
MG1655 ΔrecA and lexA1 mutants displayed no sensitivity or only a very slightly increased sensitivity towards pressure treatments that are sublethal (0 to 150 MPa) or lethal (150 to 300 MPa) to MG1655 (Fig. 4A), although both mutants were, as expected, extremely sensitive towards UV irradiation (Fig. 4B). Together with the stability of the e14 element, unaffected by pressure, these observations provide a remarkable difference in the behavior of E. coli towards pressure versus other that toward SOS-inducing treatments described so far.
DISCUSSION
The bacterial SOS response is typically induced by DNA-damaging treatments, such as UV irradiation or exposure to mitomycin C (21, 36, 37); however, its induction by pressure as shown in this work (Fig. 1) was previously unreported. Although pressure is known to denature proteins, it has been shown to stabilize DNA helices in vitro by promoting hydrogen bonds and enhancing stacking of the hydrophobic bases, resulting in compact DNA (6). The induction of the heat shock response by high pressure, as has been reported previously (2), therefore seems more logical than activation of the SOS regulon. However, our results prove that the pressure-induced SOS response is not a consequence of LexA repressor denaturation by pressure but genuinely depends on the physiologically active RecA and LexA proteins, since mutant strains with a RecA deletion or producing the autocleavage-resistant LexA1 variant did not exhibit SOS induction (Fig. 2.).
In addition to the transcriptional activation of uvrA, recA, and sulA, λ prophage induction by pressure provided a second independent manifestation of the pressure-mediated SOS response that was also RecA and LexA dependent (Fig. 3A and B). Whereas lysogens in wild-type E. coli after pressure induction produced an almost 104-fold-increased level of phage particles, only about a 10-fold induction was observed in a lexA1 background. This residual 10-fold induction is probably due to activation of RecA protein that is formed by leaky expression of its LexA-dependent promoter. While pressure-induced expression of the gfp fusions is silenced in a lexA1 background because of the inability of LexA1 to cleave itself, the limited amount of RecA protein after activation by pressure could mediate some cleavage of the λ CI repressor. In wild-type lysogens, however, LexA also becomes cleaved, resulting in derepression of the recA promoter, production of additional RecA protein, and enhanced CI autocleavage stimulation. In lysogens in a ΔrecA background, the noninduced production of phage particles was strongly reduced (ca. 5 PFU/ml, compared to ca. 103 to 104 PFU/ml in the wild type), and no induction by pressure was observed (Fig. 3B) because the CI repressor is not cleaved in the absence of RecA. Similar patterns of prophage induction were obtained with UV irradiation, but UV induction resulted in higher titers of phage than pressure induction, indicating that the applied pressure treatments induced a smaller fraction of lysogens (Fig. 3B). Thus, although Fig. 1 shows a uniform induction of the SOS response throughout the population, cleavage of the CI repressor seems to occur only in a relatively small fraction of the pressurized cells. Interestingly, during the preparation of this report, our attention was drawn to a 40-year-old study of Rutberg (45) (hard-copy citations from 1953 to 1965 were added online to Medline on 30 September 2003), who demonstrated unexplainable similarities between UV and pressure induction of λ in E. coli K12. Therefore, in hindsight, our results provide a molecular description for this cryptic phenotype observed 40 years ago.
Some lambdoid prophages confer virulence properties to their host upon lysogenization (51). A well-known example is the Shiga toxin-converting (Stx) bacteriophages, which carry the genes for production of Shiga toxins in Stx-producing E. coli strains (47). In fact, we recently found Stx lysogens to be inducible by high-pressure treatment (A. Aertsen, D. Faster, and C. W. Michiels, submitted for publication). In food preservation, therefore, pressure treatment could potentially enhance the spread of these phages and lead to the emergence of new pathogens or pathogens with increased virulence. The probability of such a scenario is difficult to estimate, but recently, Toth et al. (49) demonstrated that Stx bacteriophages could indeed readily lysogenize enteropathogenic E. coli strains in porcine ligated ileal loops.
Interestingly, the e14 element, which is a lambdoid prophage remnant, remained unaffected by pressure treatment of differ-ent intensities, while an SOS response evoked by UV irradiation caused its excision in ca. 60% of the survivors (Table 1). Possibly, this difference can be attributed to the observation that the pressure-mediated SOS response induces the excision only in a smaller fraction of cells than with UV treatment, and we were unable to detect it accordingly. Indeed, it could be that none of the pressures used supports the sustained levels of activated RecA necessary to induce detectable e14 excision. A similar observation was made for λ prophage induction (see above). An additional distinguishing feature with classical SOS-inducing treatments is that ΔrecA and lexA1 mutants, which are hypersensitive towards DNA-damaging treatments, exhibited almost wild-type tolerance to pressure (Fig. 4A). Although ΔrecA strains were slightly more inactivated by pressure treatment than the wild type and lexA1 mutants, this difference was small compared to the difference in UV sensitivity between these strains (Fig. 4B). This feature may reflect a lack of DNA damage induction by high pressure. Indeed, MG1655 cultures showed no increased incidence of rifampin resistance after high-pressure treatment (data not shown). Moreover, in the literature, pressure has never been associated with mutagenesis. Although two early studies reported an increased occurrence of petite mutants in a pressurized S. cerevisiae culture (44) or pressure-induced color mutation in Euglena gracilis (27), both phenotypes might be related to loss of mitochondria or chloroplasts, respectively, rather than to mutagenesis. In the absence of DNA lesions after pressure treatment, the causal trigger of this SOS response remains enigmatic.
Nevertheless, the presence of an activated RecA nucleoprotein filament can reasonably be assumed, since cleavage of both LexA and CI seems to occur. This nucleoprotein complex originates when RecA binds to single-stranded DNA (ssDNA) in the cell (36), implicating the formation or exposure of ssDNA during or after pressure treatment. While the DNA helix itself is stabilized under pressure, the DNA replication machinery performs one of the most pressure-sensitive essential cellular processes in E. coli (8). In fact, all multisubunit protein complexes are highly susceptible to dissociation upon pressurization, and therefore, we believe pressure to cause disassembly of the replication fork. Without the requirement for DNA lesions to stall the replication fork, this direct denaturation of the replication complex can then result in exposure of ssDNA, which in turn can activate the RecA protein and trigger SOS induction.
Interestingly, in recent years several microbial responses to stresses seemingly unrelated to DNA damage, such as starvation, aging, and translational stress, have been shown to evoke or rely on members of the SOS regulon (12, 29). It has been hypothesized that the increased mutation and recombination rate, empowered by these SOS proteins, might generate genetic diversity in times of stress (43). When the specific physiological stress responses fall short, relaxation of the functions involved in safeguarding the integrity of the DNA might well be a last resort for a population to survive a high level of stress by increasing its genetic diversity. Future research should determine whether pressure can trigger adaptive mutations and assess its possible implications for microorganisms exposed to high pressure in the deep sea, the deep subsurface, or during high-pressure food preservation.
We have reason to believe that our findings present a new paradigm in explaining several pressure-related phenomena. The first pressure-responsive genes, discovered in Photobacterium profundum SS9 by the group of Bartlett (7), comprise ompH and ompL. Both encode outer membrane proteins, of which OmpL is predominant under ambient pressures and is replaced by OmpH under high pressures. Since the absence of these genes did not influence growth or survival of SS9 at high or ambient pressures, it was hypothesized that in the deep sea, characterized by high pressure and nutrient scarcity, OmpL was replaced by OmpH to facilitate nutrient uptake. Also in E. coli, reduced expression of OmpC, OmpF, and OmpX was observed upon growth at sublethal pressures (39). This repression seemed to occur independently from the normal EnvZ-OmpR signal transduction cascade. Interestingly, Garvey et al. (24) demonstrated earlier that OmpC, OmpF, and OmpA disappeared in E. coli upon the induction of the SOS response by nalidixic acid. Although this awaits further confirmation, we therefore propose that pressure might well influence outer membrane protein expression in the above-mentioned cases by inducing an SOS response.
Another typical pressure-related phenomenon is cell filamentation in piezosensitive bacteria growing at permissive high pressures (≤50 MPa) (38, 55). Interestingly, we found pressure-induced cell elongation also to occur in the first hours after sublethal high-pressure treatment (unpublished data). Moreover, during the preparation of this report, an independent study reported a similar effect in E. coli (33) after short high-pressure treatment. In many cases, cell elongation is typically the result of an SOS response and, more specifically, of SulA-mediated inhibition of FtsZ ring formation, which constitutes an early and crucial step in the cell division process. However, our observations were consistent with those presented by Kawarai et al. (33), showing no elimination of cell elongation in a sulA or recA mutant background after sublethal pressure shock. In addition, no effect of the absence of SulA or RecA on cell filamentation induced by growth at permissive pressures was observed (unpublished data). Although both Kawarai et al. (33) and Molina-Hoppner et al. (38) suggested that elongation results from direct high-pressure inhibition of FtsZ ring formation, we believe that high-pressure-induced cell elongation in this respect resembles the “transient filamentation” phenotype observed by Gottesman et al. (25) after UV treatment, which was also shown to be independent of SulA. Also the recent finding of Bidle and Bartlett (11) that recD of the piezotolerant deep-sea isolate SS9 allows E. coli to grow at high pressures without filamentation is more compatible with a model in which pressure-induced filamentation is mediated by the SOS response than with the model assuming a direct effect of pressure on FtsZ, because RecD is involved in DNA recombination and repair. The exact mechanism by which RecD affects pressure-induced filamentation in E. coli, however, awaits further clarification.
Finally, Chilukuri et al. (17) have identified piezoadaptive amino acid substitutions in the ssDNA binding proteins of marine Shewanella strains living at elevated pressures that are essential for the functionality of these proteins at high pressure, while Welch et al. (52) observed increased rates of synthesis of cold shock proteins, RecA, and some other proteins in cultures of E. coli growing at 55 MPa. The idea was then formulated that pressure might mimic cold shock and that elevated levels of DNA binding proteins would be necessary to compensate for decreased DNA binding at elevated pressures (18). Our data place these distinct observations in a broader framework of a pressure-induced SOS response.
In conclusion, we have demonstrated the high-pressure induction in E. coli of a genuine SOS response that nevertheless shows some peculiar differences from the typical SOS response as induced by DNA-damaging treatments. This SOS response provides a plausible explanation for several previous observations made with bacteria under high pressure and at the same time raises some intriguing new questions on how high pressure triggers a response that is normally initiated by DNA damage. In both ways, this work contributes to a better understanding of the effects of pressure on cells and cellular processes.
Acknowledgments
We acknowledge financial support by fellowships to authors A.A. and R.V.H. from the Flemish Institute for the Promotion of Scientific Technical Research (IWT) and from the Fund for Scientific Research Flanders (F.W.O. Vlaanderen), respectively, and by research grants OT/01/35 from the K.U.Leuven Research Fund and G.0195.02 from F.W.O. Vlaanderen.
We thank J. Vanderleyden (Centre for Microbial and Plant Genetics, K.U.Leuven) for providing access to his FACS facilities.
REFERENCES
- 1.Abe, F., C. Kato, and K. Horikoshi. 1999. Pressure-regulated metabolism in microorganisms. Trends Microbiol. 7:447-453. [DOI] [PubMed] [Google Scholar]
- 2.Aertsen, A., K. Vanoirbeek, P. De Spiegeleer, J. Sermon, K. Hauben, A. Farewell, T. Nyström, and C. W. Michiels. 2004. Heat shock protein-mediated resistance towards high hydrostatic pressure in Escherichia coli. Appl. Environ. Microbiol. 70:2660-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alpas, H., N. Kalchayanand, F. Bozoglu, A. Sikes, C. P. Dunne, and B. Ray. 1999. Variation in resistance to hydrostatic pressure among strains of food-borne pathogens. Appl. Environ. Microbiol. 65:4248-4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balashov, S., and M. Z. Humayun. 2002. Mistranslation induced by streptomycin provokes a RecABC/RuvABC-dependent mutator phenotype in Escherichia coli cells. J. Mol. Biol. 315:513-527. [DOI] [PubMed] [Google Scholar]
- 5.Balny, C., P. Masson, and K. Heremans. 2002. High pressure effects on biological macromolecules: from structural changes to alteration of cellular processes. Biochim. Biophys. Acta 1595:3-10. [DOI] [PubMed] [Google Scholar]
- 6.Barciszewski, J., J. Jurczak, S. Porowski, T. Specht, and V. A. Erdmann. 1999. The role of water structure in conformational changes of nucleic acids in ambient and high-pressure conditions. Eur. J. Biochem. 260:293-307. [DOI] [PubMed] [Google Scholar]
- 7.Bartlett, D., M. Wright, A. A. Yayanos, and M. Silverman. 1989. Isolation of a gene regulated by hydrostatic pressure in a deep-sea bacterium. Nature 342:572-574. [DOI] [PubMed] [Google Scholar]
- 8.Bartlett, D. H. 2002. Pressure effects on in vivo microbial processes. Biochim. Biophys. Acta 1595:367-381. [DOI] [PubMed] [Google Scholar]
- 9.Benito, A., G. Ventoura, M. Casadei, T. Robinson, and B. Mackey. 1999. Variation in resistance of natural isolates of Escherichia coli O157 to high hydrostatic pressure, mild heat, and other stresses. Appl. Environ. Microbiol. 65:1564-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bianchi, A. A., and F. Baneyx. 1999. Hyperosmotic shock induces the sigma32 and sigmaE stress regulons of Escherichia coli. Mol. Microbiol. 34:1029-1038. [DOI] [PubMed] [Google Scholar]
- 11.Bidle, K. A., and D. H. Bartlett. 1999. RecD function is required for high-pressure growth of a deep-sea bacterium. J. Bacteriol. 181:2330-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bjedov, I., O. Tenaillon, B. Gerard, V. Souza, E. Denamur, M. Radman, F. Taddei, and I. Matic. 2003. Stress-induced mutagenesis in bacteria. Science 300:1404-1409. [DOI] [PubMed] [Google Scholar]
- 13.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 14.Brody, H., A. Greener, and C. W. Hill. 1985. Excision and reintegration of the Escherichia coli K-12 chromosomal element e14. J. Bacteriol. 161:1112-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheftel, J. C. 1995. Review: high-pressure, microbial inactivation and food preservation. Food Sci. Technol. Int. 1:75-90. [Google Scholar]
- 16.Chi, E., and D. H. Bartlett. 1995. An rpoE-like locus controls outer membrane protein synthesis and growth at cold temperatures and high pressures in the deep-sea bacterium Photobacterium sp. strain SS9. Mol. Microbiol. 17:713-726. [DOI] [PubMed] [Google Scholar]
- 17.Chilukuri, L. N., D. H. Bartlett, and P. A. Fortes. 2002. Comparison of high pressure-induced dissociation of single-stranded DNA-binding protein (SSB) from high pressure-sensitive and high pressure-adapted marine Shewanella species. Extremophiles 6:377-383. [DOI] [PubMed] [Google Scholar]
- 18.Chilukuri, L. N., P. A. Fortes, and D. H. Bartlett. 1997. High pressure modulation of Escherichia coli DNA gyrase activity. Biochem. Biophys. Res. Commun. 239:552-556. [DOI] [PubMed] [Google Scholar]
- 19.Dukan, S., and T. Nystrom. 1999. Oxidative stress defense and deterioration of growth-arrested Escherichia coli cells. J. Biol. Chem. 274:26027-26032. [DOI] [PubMed] [Google Scholar]
- 20.Erijman, L., and R. M. Clegg. 1998. Reversible stalling of transcription elongation complexes by high pressure. Biophys. J. 75:453-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Friedberg, E. C., G. C. Walker, and W. Siede. 1995. DNA repair and mutagenesis. ASM Press, Washington, D.C.
- 22.Ganzle, M. G., and R. F. Vogel. 2001. On-line fluorescence determination of pressure mediated outer membrane damage in Escherichia coli. Syst. Appl. Microbiol. 24:477-485. [DOI] [PubMed] [Google Scholar]
- 23.Gao, X., J. Li, and K. C. Ruan. 2001. Barotolerant Escherichia coli induced by high hydrostatic pressure. Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao (Shanghai) 33:77-81. [PubMed] [Google Scholar]
- 24.Garvey, N., A. C. St John, and E. M. Witkin. 1985. Evidence for RecA protein association with the cell membrane and for changes in the levels of major outer membrane proteins in SOS-induced Escherichia coli cells. J. Bacteriol. 163:870-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gottesman, S., E. Halpern, and P. Trisler. 1981. Role of sulA and sulB in filamentation by Lon mutants of Escherichia coli K-12. J. Bacteriol. 148:265-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greener, A., and C. W. Hill. 1980. Identification of a novel genetic element in Escherichia coli K-12. J. Bacteriol. 144:312-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gross, J. A. 1965. Pressure-induced color mutation of Euglena gracilis. Science 147:741-742. [DOI] [PubMed] [Google Scholar]
- 28.Hauben, K. J., D. H. Bartlett, C. C. Soontjens, K. Cornelis, E. Y. Wuytack, and C. W. Michiels. 1997. Escherichia coli mutants resistant to inactivation by high hydrostatic pressure. Appl. Environ. Microbiol. 63:945-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Humayun, M. Z. 1998. SOS and Mayday: multiple inducible mutagenic pathways in Escherichia coli. Mol. Microbiol. 30:905-910. [DOI] [PubMed] [Google Scholar]
- 30.Iwahashi, H., H. Shimizu, M. Odani, and Y. Komatsu. 2003. Piezophysiology of genome wide gene expression levels in the yeast Saccharomyces cerevisiae. Extremophiles 7:291-298. [DOI] [PubMed] [Google Scholar]
- 31.Karatzas, K. A., and M. H. Bennik. 2002. Characterization of a Listeria monocytogenes Scott A isolate with high tolerance towards high hydrostatic pressure. Appl. Environ. Microbiol. 68:3183-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karatzas, K. A., J. A. Wouters, C. G. Gahan, C. Hill, T. Abee, and M. H. Bennik. 2003. The CtsR regulator of Listeria monocytogenes contains a variant glycine repeat region that affects piezotolerance, stress resistance, motility and virulence. Mol. Microbiol. 49:1227-1238. [DOI] [PubMed] [Google Scholar]
- 33.Kawarai, T., M. Wachi, H. Ogino, S. Furukawa, K. Suzuki, H. Ogihara, and M. Yamasaki. 2004. SulA-independent filamentation of Escherichia coli during growth after release from high hydrostatic pressure treatment. Appl. Microbiol. Biotechnol. 64:255-262. [DOI] [PubMed] [Google Scholar]
- 34.Kleckner, N., J. Bender, and S. Gottesman. 1991. Uses of transposons with emphasis on Tn10. Methods Enzymol. 204:139-180. [DOI] [PubMed] [Google Scholar]
- 35.Kobori, H., M. Sato, A. Tameike, K. Hamada, S. Shimada, and M. Osumi. 1995. Ultrastructural effects of pressure stress to the nucleus in Saccharomyces cerevisiae: a study by immunoelectron microscopy using frozen thin sections. FEMS Microbiol. Lett. 132:253-258. [DOI] [PubMed] [Google Scholar]
- 36.Kuzminov, A. 1999. Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol. Mol. Biol. Rev. 63:751-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Little, J. W. 1993. LexA cleavage and other self-processing reactions. J. Bacteriol. 175:4943-4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Molina-Hoppner, A., T. Sato, C. Kato, M. G. Ganzle, and R. F. Vogel. 2003. Effects of pressure on cell morphology and cell division of lactic acid bacteria. Extremophiles 7:511-516. [DOI] [PubMed] [Google Scholar]
- 39.Nakashima, K., K. Horikoshi, and T. Mizuno. 1995. Effect of hydrostatic pressure on the synthesis of outer membrane proteins in Escherichia coli. Biosci. Biotechnol. Biochem. 59:130-132. [DOI] [PubMed] [Google Scholar]
- 40.Newman, D. K., and J. F. Banfield. 2002. Geomicrobiology: how molecular-scale interactions underpin biogeochemical systems. Science 296:1071-1077. [DOI] [PubMed] [Google Scholar]
- 41.Niven, G. W., C. A. Miles, and B. M. Mackey. 1999. The effects of hydrostatic pressure on ribosome conformation in Escherichia coli: an in vivo study using differential scanning calorimetry. Microbiology 145:419-425. [DOI] [PubMed] [Google Scholar]
- 42.Pagan, R., and B. Mackey. 2000. Relationship between membrane damage and cell death in pressure-treated Escherichia coli cells: differences between exponential- and stationary-phase cells and variation among strains. Appl. Environ. Microbiol. 66:2829-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Radman, M. 2001. Fidelity and infidelity. Nature 413:115. [DOI] [PubMed] [Google Scholar]
- 44.Rosin, M. P., and A. M. Zimmerman. 1977. The induction of cytoplasmic petite mutants of Saccharomyces cerevisiae by hydrostatic pressure. J. Cell Sci. 26:373-385. [DOI] [PubMed] [Google Scholar]
- 45.Rutberg, L. 1964. On the effects of high hydrostatic pressure on bacteria and bacteriophage. 3. Induction with high hydrostatic pressure of Escherichia coli K lysogenic for bacteriophage lambda. Acta Pathol. Microbiol. Scand. 61:98-105. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 47.Schmidt, H. 2001. Shiga-toxin-converting bacteriophages. Res. Microbiol. 152:687-695. [DOI] [PubMed] [Google Scholar]
- 48.Simpson, R. K., and A. Gilmour. 1997. The effect of high hydrostatic pressure on the activity of intracellular enzymes of Listeria monocytogenes. Lett. Appl. Microbiol. 25:48-53. [DOI] [PubMed] [Google Scholar]
- 49.Toth, I., H. Schmidt, M. Dow, A. Malik, E. Oswald, and B. Nagy. 2003. Transduction of porcine enteropathogenic Escherichia coli with a derivative of a shiga toxin 2-encoding bacteriophage in a porcine ligated ileal loop system. Appl. Environ. Microbiol. 69:7242-7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Valdivia, R. H., and S. Falkow. 1996. Bacterial genetics by flow cytometry: rapid isolation of Salmonella typhimurium acid-inducible promoters by differential fluorescence induction. Mol. Microbiol. 22:367-378. [DOI] [PubMed] [Google Scholar]
- 51.Wagner, P. L., and M. K. Waldor. 2002. Bacteriophage control of bacterial virulence. Infect. Immun. 70:3985-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Welch, T. J., A. Farewell, F. C. Neidhardt, and D. H. Bartlett. 1993. Stress response of Escherichia coli to elevated hydrostatic pressure. J. Bacteriol. 175:7170-7177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Welch, T. J., and D. H. Bartlett. 1998. Identification of a regulatory protein required for pressure-responsive gene expression in the deep-sea bacterium Photobacterium species strain SS9. Mol. Microbiol. 27:977-985. [DOI] [PubMed] [Google Scholar]
- 54.Yayanos, A. A. 1995. Microbiology to 10,500 meters in the deep sea. Annu. Rev. Microbiol. 49:777-805. [DOI] [PubMed] [Google Scholar]
- 55.ZoBell, C. E., and A. B. Cobet. 1964. Filament formation by Escherichia coli at increased hydrostatic pressures. J. Bacteriol. 87:710-719. [DOI] [PMC free article] [PubMed] [Google Scholar]