Abstract
Vibrio anguillarum can utilize hemin and hemoglobin as sole iron sources. In previous work we identified HuvA, the V. anguillarum outer membrane heme receptor by complementation of a heme utilization mutant with a cosmid clone (pML1) isolated from a genomic library of V. anguillarum. In the present study, we describe a gene cluster contained in cosmid pML1, coding for nine potential heme uptake and utilization proteins: HuvA, the heme receptor; HuvZ and HuvX; TonB, ExbB, and ExbD; HuvB, the putative periplasmic binding protein; HuvC, the putative inner membrane permease; and HuvD, the putative ABC transporter ATPase. A V. anguillarum strain with an in-frame chromosomal deletion of the nine-gene cluster was impaired for growth with heme or hemoglobin as the sole iron source. Single-gene in-frame deletions were constructed, demonstrating that each of the huvAZBCD genes are essential for utilization of heme as an iron source in V. anguillarum, whereas huvX is not. When expressed in Escherichia coli hemA (strain EB53), a plasmid carrying the gene for the heme receptor, HuvA, was sufficient to allow the use of heme as the porphyrin source. For utilization of heme as an iron source in E. coli ent (strain 101ESD), the tonB exbBD and huvBCD genes were required in addition to huvA. The V. anguillarum heme uptake cluster shows some differences in gene arrangement when compared to homologous clusters described for other Vibrio species.
Iron is an essential element for most bacteria, serving as a cofactor in key metabolic processes such as nucleotide biosynthesis, electron transfer, and energy transduction. Most bacterial pathogens require iron for growth and to establish an infection, and thus they have developed efficient mechanisms to obtain iron from the host (32). The small amounts of extracellular iron are quickly bound by high-affinity carrier proteins such as transferrin in serum and lactoferrin in secretions. Vibrio anguillarum is the etiological agent of a septicemic disease known as vibriosis, which affects a large number of marine fish species. Within the 10 O serogroups described by Sørensen and Larsen (35), only serotypes O1 and O2 and, to a lesser extent, serotype O3 are considered important fish pathogens. Although V. anguillarum is the best known fish pathogen of the genus Vibrio, the nature of its virulence mechanisms is not thoroughly understood. Strains of pathogenic V. anguillarum serotypes can acquire iron by the production and secretion of siderophores (2, 4, 16, 19, 38). Heme and some heme-containing proteins, including hemoglobin and hemoglobin-haptoglobin, can also be used as iron sources by a siderophore-independent mechanism (23, 24).
In this way, many gram-negative pathogens have the ability to obtain iron through utilization of free heme or heme proteins from the host tissues (7, 18), and heme utilization genes have been identified in numerous species, including Yersinia enterocolitica (36, 37), Vibrio cholerae (13, 26, 29), Escherichia coli O157 (39), Vibrio vulnificus (20), Plesiomonas shigelloides (14), and Shigella dysenteriae (28) among others. Specific receptors are involved in heme binding and transport. Receptor-mediated uptake of heme includes translocation of the ligands into the periplasm by an energy-dependent process that requires a functional TonB system (30, 36). The TonB protein, which is anchored in the cytoplasmic membrane and associated with two accessory proteins, ExbB and ExbD, spans the periplasm and interacts with the ligand-loaded receptor. The TonB-ExbBD system is believed to be involved in transducing the energy of the proton motive force of the cytoplasmic membrane into transport energy required by the receptor. The pernicious oxidative effects of free heme dictate the presence of a periplasmic binding protein to transport heme across the periplasmic compartment. Transport of heme or iron across the cytoplasmic membrane is driven by ATP hydrolysis, and an ATP-binding cassette (ABC) transporter is commonly involved in transport through the cytoplasmic membrane (17).
In previous studies we identified HuvA, the outer membrane receptor involved in heme uptake in V. anguillarum. The huvA gene was isolated from a V. anguillarum H-775-3 cosmid library by its ability to restore heme utilization in 101ESD, an E. coli mutant strain that fails to grow under iron-limiting conditions and cannot use heme as an iron source (25). A TonB-ExbB-ExbD system has also been recently found in V. anguillarum. It was observed that a tonB mutant strain was still able to take up heme, suggesting that V. anguillarum may harbor two TonB systems (M. Stork, M. Di Lorenzo, S. Mouriño, C. R. Osorio, M. L. Lemos, and J. H. Crosa, unpublished data).
The goal of the present study was to identify and characterize the genes involved in heme transport in V. anguillarum. Sequences of the heme uptake cluster genes were determined and analyzed, and deletion mutants were constructed and tested for the ability to grow with hemin or hemoglobin as the sole iron source. Complementation of E. coli mutants with V. anguillarum genes for restoration of heme utilization as an iron and porphyrin source was also evaluated.
MATERIALS AND METHODS
Plasmids, bacteria, and media.
Plasmids and bacterial strains used in this study are listed in Table 1. V. anguillarum cells were routinely grown at 25°C in tryptic soy agar (Difco) supplemented with 1% NaCl (TSA-1), as well as in M9 minimal medium (27) supplemented with 0.2% Casamino Acids (Difco) (CM9). E. coli strains were grown at 37°C in Luria-Bertani (LB) broth, LB agar, or CM9 supplemented with antibiotics when appropriate. Strain EB53 aroB hemA and its derivatives were routinely grown in LB medium supplemented with 2 μg of 5-aminolevulinic acid (ALA; Sigma) ml−1. All strains were stored frozen at −80°C in LB broth with 20% glycerol. Antibiotics were used at the following final concentrations: tetracycline hydrochloride at 15 μg ml−1, kanamycin at 25 μg ml−1, and ampicillin sodium salt at 50 μg ml−1. 2,2′-Dipyridyl (Sigma), used to chelate nonheme iron, was prepared at 10 mM in ultrapure water (milli-Q; Millipore). Bovine hemin (Sigma) was dissolved at 5 mM in 10 mM NaOH. Bovine hemoglobin (Sigma) was dissolved at 1 mM in ultrapure water.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| V. anguillarum H-775-3 | Serotype O1 plasmidless avirulent strain, deficient in anguibactin biosynthesis, derived from strain 775 by curing of pJM1 | J. H. Crosa |
| E. coli | ||
| DH5α | Cloning strain | Laboratory stock |
| SM17-1-λpir | Cloning strain | Laboratory stock |
| EB53 | aroB hemA | 6 |
| 101ESD | HB101 derivative, deficient in enterobactin biosynthesis Δ(entC-entA) | J. H. Crosa |
| Plasmids | ||
| pGEMT-Easy | PCR cloning vector, Apr | Promega |
| pWKS30 | Low-copy-number plasmid vector, Apr | 41 |
| pACYC177 | Cloning vector, Apr Kmr | 33 |
| pACYC184 | Cloning vector, Apr Cmr | 33 |
| pVK100 | Mobilizable cloning vector, Tcr Kmr | 15 |
| pRK2013 | Mobilizing plasmid for triparental mating, Kmr | 5 |
| pKEK229 | Suicide vector derived from pCVD442, Apr | 3 |
| pNidKan | pKEK229 with a 1.5-kb PstI-PstI fragment containing the Km cassette from pKANπ | This study |
| pML1 | 21-kb fragment from a gene library of strain H775-3 cloned into pVK100, Tcr | 25 |
| pCAR121 | ApaL1 fragment of pML1 containing huvA gene cloned into pACYC177; Kmr | This study |
| pCAR126 | ApaL1 fragment of pML1 containing huvX, tonB-exbBD, and huvB genes and partial huvZ and huvC genes cloned into pACYC177, Kmr | This study |
| pCAR115 | PCR-amplified huvA gene cloned into pWKS30, Apr | This study |
| pCAR179 | huvZ gene cloned in pWKS30, Apr | This study |
| pCAR181 | huvX gene cloned in pWKS30, Apr | This study |
| pSML11 | EcoRV fragment of pML1 containing complete huvZX genes cloned into pWKS30 | This study |
| pSML23 | 9-kb PCR fragment encoding the heme uptake system of V. anguillarum H-775-3 cloned into pWKS30, Apr | This study |
| pSML32 | PstI-EcoRI fragment of pSML23 containing huvBCD genes cloned into pWKS30, Apr | This study |
| pSML33 | SacI-ApaI fragment of pSML23 containing tonB-exbBD and huvBCD genes cloned into pWKS30, Apr | This study |
| pSML34 | SacI-HindIII fragment of pML1 containing tonB-exbBD genes cloned into pWKS30, Apr | This study |
Recombinant DNA techniques.
Recombinant DNA methods including restriction-enzyme digestions, ligation reactions, agarose gel electrophoresis, and plasmid analysis were performed following standard protocols (34). Chromosomal DNA was isolated by using the Easy-DNA kit (Invitrogen). Plasmid purification and elution of DNA fragments from agarose gels were performed with kits from QIAGEN. Southern blot analyses were performed with Hybond-N+ membranes (Amersham Biosciences), using the ECL Direct Nucleic Acid Labeling and Detection system (Amersham Biosciences), following the manufacturer's instructions. E. coli strains were transformed by a standard calcium chloride method (34). Triparental mating for transfer of cosmid pML1 into E. coli EB53 was performed as described previously (8). E. coli strain HB101 harboring the helper plasmid pRK2013 was used as a mobilizing strain (5).
DNA sequencing and data analysis.
DNA sequences were determined by the dideoxy-chain termination method by using the Big Dye Terminator v3.0 DNA sequencing kit (Applied Biosystems) on an automated sequencer, ABI 377 (Applied Biosystems). Restriction maps were generated and DNA translation was performed with the BioEdit Sequence Alignment Editor (11). Homologies of the deduced amino acid sequences were determined by consulting the EMBL and SWALL databases, using the FASTA3 and BLAST algorithms, at the European Bioinformatics Institute website.
Construction of chromosomal mutations.
Gene deletions in V. anguillarum H-775-3 were constructed by using PCR amplifications of two fragments of each gene, which when ligated together would result in an in-frame (nonpolar) deletion. Amplification was carried out using the Expand High Fidelity PCR system (Roche). The oligonucleotides used to amplify the carboxy- and amino-terminal fragments of each gene are listed in Table 2. Construction of an in-frame huvAZXBCD tonB exbBD mutant was accomplished by ligating the PCR products obtained with primer pairs HuvA.A-HuvA.B and HuvD.C2-HuvD.D. Allelic exchange was carried out using the suicide vector pNidKan. Plasmid pKEK229 (3) was cut with PstI, and a kanamycin resistance cassette (Genblock; Pharmacia) was inserted, producing pNidKan. As a pCVD442 derivative, pNidKan contains R6K ori, requiring the pir gene product for replication, and the sacB gene, conferring sucrose sensitivity. Construction of in-frame deletions of huvX, huvZ, huvC, huvD, huvB, huvA and deletion of the complete gene cluster occurred in several steps. The PCR-amplified carboxy-terminal gene fragments were ligated into pWKS30, and resulting plasmids were cut with appropriate enzymes and ligated to the amino-terminal PCR fragments of each corresponding gene. This process resulted in the formation of mutant alleles ΔhuvZ (removes coding sequences for amino acids 76 to 126), ΔhuvX (removes coding sequences for amino acids 90 to 144), ΔhuvC (removes coding sequences for amino acids 19 to 291), ΔhuvB (removes coding sequences for amino acids 18 to 259), ΔhuvD (removes coding sequences for amino acids 183 to 345), and ΔhuvA (removes coding sequences for amino acids 178 to 698) and the complete gene cluster deletion mutant Δhuv (removes coding sequences encompassing amino acid 178 encoded by huvA and amino acid 345 encoded by huvD). Each deleted allele cloned in pWKS30 was digested with NotI and SalI and ligated into the NotI/SalI sites of the suicide vector pNidKan. The resulting plasmids were mated from E. coli S17-1-λpir into V. anguillarum H-775-3, and transformants with the plasmid integrated into the chromosome by homologous recombination were selected on agar medium containing kanamycin and ampicillin. A second recombination event was obtained by selecting for sucrose resistance (10% wt/vol) and resistance to the specific antibiotic for the recipient strain (ampicillin). This led to obtention of V. anguillarum ΔhuvZ, ΔhuvX, ΔhuvC, ΔhuvB, ΔhuvD, ΔhuvA, and Δhuv mutant strains. Southern blot hybridization analysis was used to verify allelic exchange of the parental gene. In addition, for each mutant strain, the region involved in the deletion construction was PCR amplified and sequenced to ensure that the constructs were nonpolar (data not shown).
TABLE 2.
Oligonucleotides used in this study
| Amplified region | Primer | Nucleotide sequencea |
|---|---|---|
| Mutant constructions | ||
| huvA, amino terminal | HuvA.A | 5′-GCTCTAGAAAGCTTTATCAACAATTGATT-3′ |
| HuvA.B | 5′-TCCCCCGGGGGAAAGCTTTGCTTGTCCGCC-3′ | |
| huvA, carboxy terminal | HuvA.C | 5′-TCCCCCGGGAACGACTACTCTCAAGCTGAG-3′ |
| HuvA.D | 5′-GCGAATTCTGGGTAAAGAGGGCGAGTTAC-3′ | |
| huvZ, amino terminal | HuvZ.C | 5′-GCGGATCCGACGTGCGGGTTCACCTCCAA-3′ |
| HuvZ.D | 5′-GCGAATTCTATGATTTTACGAATTTAAAA-3′ | |
| huvZ, carboxy terminal | HuvZ.A | 5′-GCTCTAGAGCTTCCGTGCCCCTAACTTTA-3′ |
| HuvZ.B | 5′-GCGGATCCGACGGCTTAAGCCAGCTGCAA-3′ | |
| huvX, amino terminal | HuvX.A | 5′-GCTCTAGATGTCGATATCACCGCTTACTT-3′ |
| HuvX.B | 5′-GCGGATCCTTTATGGGACGTGAAAGTCAT-3′ | |
| huvX, carboxy terminal | HuvX.C | 5′-GCGGATCCCATTCCCCAATCCGCTAATGA-3′ |
| HuvX.D | 5′-GCAAGCTTGCATCGACTAATTTCGGTTGA-3′ | |
| huvB, amino terminal | HuvB.A | 5′-GCTCTAGACAAGCGCCCATTGCTTTATAA-3′ |
| HuvB.B | 5′-GCGGATCCGTCATTGGAGTGCAGCACGAG-3′ | |
| huvB, carboxy terminal | HuvB.C | 5′-GCGGATCCGCTCTCGTCGGCGGGTTAGGC-3′ |
| HuvB.D | 5′-GCGAATTCGTGAGTAAGAGTGCACCAAGT-3′ | |
| huvC, amino terminal | HuvC.A | 5′-GCTCTAGACACGCTTTTAAAAACCGTATG-3′ |
| HuvC.B | 5′-GCGGATCCGTCATTGGAGTGCAGCACGAG-3′ | |
| huvC, carboxy terminal | HuvC.C | 5′-GCGGATCCATTGTGACTGCGCTGGTGGGT-3′ |
| HuvC.D | 5′-GCGAATTCGCATTATAGCGAAGGGACGCG-3′ | |
| huvD, amino terminal | HuvD.A | 5′-GCTCTAGACTTGTTGGTCGGCGCAATTTT-3′ |
| HuvD.B | 5′-GCGGATCCCTTTGCTAACTCATTCGGTTG-3′ | |
| huvD, carboxy terminal | HuvD.C | 5′-GCGGATCCTCGTCGCTTGTCGCCGCACAT-3′ |
| HuvD.D | 5′-GCGAATTCGCTCGATCAGTGATGCGGTAT-3′ | |
| huvAZXBCD tonB exbBD, carboxy terminal | HuvD.C2 | 5′-TCCCCCGGGTCGTCGCTTGTCGCCGCACAT-3′ |
| Gene amplification | ||
| huvA | huvA-5′ | 5′-GCGAATTCTGGTGACAGCAGCGGCATCC-3′ |
| huvA-Eco-3′ | 5′-GCGAATTCAGGAGTTCATGGAACAACAAAGCC-3′ | |
| huvX | VA-huvZ-D | 5′-GCGAATTCTATGATTTTACGAATTTAAAA-3′ |
| VA-HutZ-1 | 5′-CTCACATTCGGACGGCCTTCA-3′ | |
| huvZ ΔhuvX | PX3 | 5′-GCCTTTTCTTACAATGTCC-3′ |
| VA-TonB-4 | 5′-TGGTTTGGGCTTGTTGGCTC-3′ |
Restriction sites for Xbal, BamHI, HindIII, EcoRI, and Sma1 are underlined.
Subcloning of pML1 and construction of plasmids carrying V. anguillarum genes.
Oligonucleotides used to amplify genes of the V. anguillarum heme uptake system are listed in Table 2. Cosmid pML1 was cut with ApaLI, and two fragments were subcloned in pACYC177, yielding pCAR121 (containing huvA) and pCAR126 (containing huvX, tonB-exbBD, huvB, and partial huvZC genes). An EcoRV fragment containing complete huvZX genes was cloned in pWKS30 to yield pSML11. A SacI-HindIII fragment including complete tonB-exbBD genes was cloned in pWKS30 to yield pSML34.
The complete nine-gene heme uptake cluster was PCR amplified with primers huvA-5′ and HuvC.D. The PCR product was cut with EcoRI and cloned in pWKS30 to produce pSML23. A PstI-EcoRI fragment of pSML23 containing huvBCD genes, and a SacI-ApaI fragment, including the tonB system plus huvBCD genes, were subcloned in pWKS30 to yield pSML32 and pSML33, respectively (in cases where a unique promoter exists upstream of tonB from which the six downstream genes are transcribed, huvBCD genes cloned in pSML32 will not be expressed).
The complete huvA gene including upstream and downstream DNA was amplified with primers huvA-5′ and huvA-Eco-3′. The PCR product was cut with EcoRI and cloned in pWKS30 to yield pCAR115. The complete huvX gene including upstream DNA was amplified with primers VA-huvZ-D and VA-HutZ-1. The PCR product was cloned in pGEMT-Easy, and the insert was cut with EcoRI and subcloned in pWKS30 to yield pCAR181. The complete huvZ gene including the putative huvX promoter and in-frame-deleted huvX open reading frame (ORF) was amplified from V. anguillarum ΔhuvX with primers PX3 and VA-TonB-4 (this allows the cloning of huvZ independently of huvX while still maintaining the putative huvX promoter, from which it is feasible that transcription of huvZ may occur). The PCR product was cloned in pGEMT-Easy, and the insert was cut with EcoRI and cloned in pWKS30 to yield pCAR179. All the PCR-amplified V. anguillarum genes to be used in complementation assays were obtained with the Expand High Fidelity PCR system (Roche) and further DNA sequenced to ensure that no PCR errors were artificially introduced.
Hemin and hemoglobin utilization assays.
To test the ability of V. anguillarum mutants to utilize hemin or hemoglobin as an iron source, overnight cultures of the parental strain, V. anguillarum H-775-3, and the mutant strains were adjusted to the same optical density and diluted 1:100 in fresh CM9 broth containing the iron source (hemin, 10 μM; or hemoglobin, 1 μM) with or without the iron chelator 2,2′-dipyridyl at 100 μM. Cultures were shaken at 25°C, and absorbance at 600 nm was monitored at 1-h intervals over 12 h.
Complementation experiments.
To test which genes of the cluster were essential for the utilization of hemin and hemoglobin as iron and porphyrin sources, E. coli strain EB53 aroB hemA and strain 101ESD Δ(entC-entA) were transformed with several plasmids. One-hundred-microliter portions of overnight cultures were added to 3 ml of molten soft nutrient broth (NB) or CM9 and plated onto appropriately prepared NB, CM9, or CM9 supplemented with 100 μM 2,2′-dipyridyl plates. Sterile filter paper disks were loaded with 20 μl of 5 and 0.05 mM hemin and 1 mM hemoglobin. Disks spotted with 20 μl of 2 mg-ml−1 ALA or 5 mM FeSO4 were included as positive controls for utilization of porphyrin and iron sources, respectively. Results were annotated as positive or negative after 24 h of incubation.
Nucleotide sequence accession number.
The EMBL accession number for the sequence described in this article is AJ496544.
RESULTS
Nucleotide sequence analysis of V. anguillarum heme utilization genes.
The genetic organization of the V. anguillarum heme utilization gene cluster was determined by partial DNA sequence analysis of pML1, a cosmid which enabled E. coli 101ESD to utilize heme as an iron source and which contains the heme receptor gene, huvA (25). The two strands of a DNA region downstream of huvA spanning ca. 6,000 bp were sequenced, and eight closely linked ORFs were identified (Fig. 1). The predicted products display significant similarity to components of other heme uptake systems in Vibrio and Plesiomonas species (Table 3).
FIG. 1.

(A) Physical map of the heme uptake gene cluster of V. anguillarum and mutant allele construction. ORFs are depicted as arrows, which indicate the direction of transcription, and the vertical numbers show the start and the end points of each gene. Deleted regions within each gene are shown as stippled boxes. (B) Relevant plasmid derivatives of the heme uptake cluster and restriction sites used in subcloning procedures.
TABLE 3.
Proteins with homology to products of V. anguillarum huvZ, huvX, and huvBCD genes
| V. anguillarum protein | Homologue | EMBL accession no. | Amino acid identity (%) | Amino acid similarity (%) |
|---|---|---|---|---|
| HuvZ | V. cholerae hypothetical protein | Q9KL41 | 82 | 93 |
| V. parahaemolyticus hypothetical protein | Q87J24 | 82 | 92 | |
| V. vulnificus hypothetical protein | Q8D3S0 | 81 | 90 | |
| P. shigelloides HugZ | Q93ST0 | 64 | 78 | |
| HuvX | V. cholerae hypothetical protein | Q9KL40 | 78 | 91 |
| V. parahaemolyticus hypothetical protein | Q87J25 | 74 | 88 | |
| V. vulnificus hypothetical protein | Q8D3S1 | 71 | 87 | |
| P. shigelloides HugX | Q93SS9 | 51 | 70 | |
| HuvB | V. parahaemolyticus HutB | Q87J30 | 68 | 82 |
| V. cholerae HutB | Q9KL36 | 66 | 81 | |
| V. vulnificus heme transporter | Q8D3S6 | 64 | 80 | |
| P. shigelloides HugB | Q93SS3 | 44 | 64 | |
| HuvC | V. cholerae HutC | Q9KL35 | 75 | 90 |
| V. parahaemolyticus HutC | Q87J31 | 74 | 87 | |
| V. vulnificus heme transporter | Q8D3S7 | 75 | 85 | |
| P. shigelloides HugC | Q93SS2 | 61 | 78 | |
| HuvD | V. cholerae HutD | O52047 | 70 | 83 |
| V. parahaemolyticus HutD | Q87J32 | 71 | 86 | |
| V. vulnificus heme transporter | Q8D3S8 | 60 | 77 | |
| P. shigelloides HugD | Q93SS1 | 48 | 66 |
A gene which we termed huvZ was found 125 bp downstream of the huvA stop codon and is transcribed from the opposite strand. huvZ encodes a 176-amino-acid protein with homology to proteins linked to heme transport systems (Table 3). The function of HuvZ homologues remains uncharacterized, though some observations suggest that P. shigelloides HugZ could be involved in preventing heme toxicity (14). Interestingly, database comparisons evidence that HuvZ has homology to proteins containing a flavin mononucleotide-binding split barrel (data not shown). This homology suggests that this protein may play a role in processes of electron transfer, with the heme group being involved.
The next ORF in the cluster corresponds to the huvX gene, located 55 nucleotides upstream of the huvZ start codon, and codes for a predicted protein of 171 amino acids. The highest similarities were to proteins linked to heme transport systems of gram-negative bacteria (Table 3). None of the HuvX homologues has a known function, but it has been suggested that P. shigelloides HugX could be involved in preventing heme toxicity (14).
Six additional genes are transcribed from the same strand as huvA. The three ORFs located adjacent to huvX encode the three proteins TonB, ExbB, and ExbD. The remaining three ORFs located downstream of exbD in this cluster code for proteins which show characteristics of heme transport proteins (Table 3). The predicted start codon for the seventh ORF of the cluster, huvB, is 51 nucleotides downstream from the stop codon of exbD. It encodes a predicted 282-amino-acid protein with homology to putative periplasmic heme-binding proteins. These proteins are believed to be involved in the transport of heme across the periplasm from the receptor to the ABC transporter located in the inner membrane.
The eighth ORF of the cluster encodes a 314-amino-acid protein which we termed HuvC, which has homology to members of a family of ABC-type permease proteins involved in the uptake of iron (Table 3). The last ORF of the cluster encoded a protein termed HuvD, which showed homology to the ATP-binding protein component of permeases involved in heme transport (Table 3). The first putative ATG of the V. anguillarum HuvD ORF determines a protein of 199 amino acids, which is shorter than homologues such as V. cholerae HutD or P. shigelloides HugD. Two TTG triplets, which is an unusual start codon in eubacteria (9), are located in frame upstream of the candidate ATG start codon and can be considered putative start codons for V. anguillarum HuvD. The amino acid sequence translated from any of these TTG triplets show high similarity to the N-terminal amino acids of V. cholerae HutD. Considering one of these two in-frame TTG triplets as the start codon, the larger V. anguillarum predicted HuvD ORF encodes a protein with features common to ABC transporter ATPases: the walkerA nucleotide-binding consensus motif GPNGAGKS (located 42 bp upstream of the first ATG codon) and the walkerB motif LMLDE at positions 103 to 107 downstream of the putative ATG start. These two sites are part of a highly conserved ATP-binding motif that constitute an ATP-binding pocket (40). In addition, an ABC transporter signature motif, LSGGE, is found at positions 77 to 81 of HuvD (Fig. 2). The presence of these features suggests that V. anguillarum HuvD is the ATPase component of the heme ABC transporter.
FIG. 2.
Alignment of the V. anguillarum HuvD ORF with homologues of other Vibrio species. Predicted amino acid sequence starting from a putative TTG start codon is shown in italics. Conserved GPNGAGKS, LMLDE, and LSGGE domains are shown in bold. Asterisks denote amino acids conserved in all the compared sequences.
Similarities with other heme uptake gene clusters.
The spatial organization of heme uptake genes in the chromosome of V. anguillarum is similar to that described for other Vibrio species and for P. shigelloides (Fig. 3). This fact suggests that the genes of the heme transport cluster were acquired by horizontal gene transfer, simultaneously with the TonB genes. This may have represented a selective advantage, allowing efficient heme transport by the recipient strain. However, some differences in gene order are observed. Among the Vibrio species, V. anguillarum is unique in that the outer membrane receptor gene is linked to the rest of the heme transport genes. Similarly, upstream of the huvX homologue, all other Vibrio species, as well as P. shigelloides, contain a gene which is transcribed in the same direction as huvX and huvZ homologues and is absent in the V. anguillarum heme uptake cluster. This gene codes for a putative coproporphyrinogen oxidase (it has been named HutW, PhuW, or HugW), an enzyme that converts coproporphyrinogen III into protoporphyrin IX, one of the steps in the heme biosynthesis pathway (31).
FIG. 3.
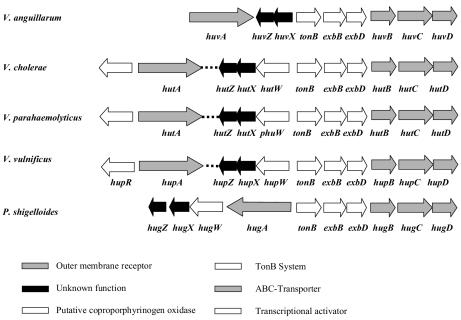
Comparative chromosomal arrangement of heme uptake cluster genes in Vibrio species and P. shigelloides.
Finally, a gene (hupR) coding for a member of the LysR family of transcriptional activators, which regulates expression of the heme receptor, has been found in V. vulnificus (21). Homologues of this gene are encountered upstream of V. parahaemolyticus hutA (22) and V. cholerae hutA (12). However, the nucleotide sequence immediately upstream of V. anguillarum huvA does not code for an HupR homologue (data not shown).
Phenotypic analysis of heme uptake system chromosomal mutants with hemin or hemoglobin as the only iron source.
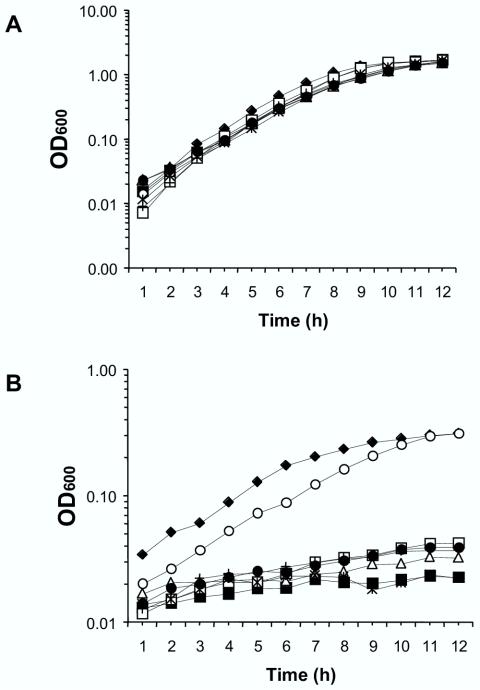
In order to evaluate the importance of individual genes of the heme uptake system in the utilization of hemin or hemoglobin, individual in-frame deletions of huvA, huvZ, huvX, huvB, huvC, and huvD and an in-frame deletion of the complete nine-gene cluster were constructed. The H-775-3 strain, deficient in siderophore anguibactin biosynthesis, was chosen in order to eliminate background growth resulting from anguibactin-mediated iron uptake in iron-restricted media. Growth assays were carried out in triplicate, and results shown here are the means of three independent experiments. Strains were first assayed in an iron-sufficient medium with hemin (10 μM). No significant differences in growth rates were seen in iron-sufficient medium between mutant strains and V. anguillarum H-775-3 (Fig. 4A). The same mutants were then grown in an iron-restricted medium with the iron chelator 2,2′-dipyridyl added at a concentration of 100 μM and containing 10 μM hemin as the sole iron source. Under these conditions, no significant differences in growth were observed between the ΔhuvX strain and the parental strain, V. anguillarum H-775-3, with the slight differences observed likely due to a lower initial inoculum. This demonstrates that this gene is not essential for the utilization of hemin as the sole iron source in V. anguillarum. By contrast, the rest of the assayed mutants were drastically affected in their ability to use hemin. Mutation of any huvAZBCD gene decreased bacterial growth to minimal levels (Fig. 4B). The Δhuv strain showed a reduction in growth with hemin as the sole iron source comparable to that observed in the single ΔhuvA, -Z, -B, -C, and -D mutants (Fig. 4B). The same results were obtained for all the assayed strains when hemoglobin was used as the iron source instead of hemin (data not shown).
FIG. 4.
Growth of V. anguillarum H-775-3 (⧫) and ΔhuvA (▪), ΔhuvZ (▵), ΔhuvX (○), ΔhuvB (□), ΔhuvC (•), ΔhuvD (+), and Δhuv (*) mutants in CM9 minimal medium. Growth was with hemin (10 μM) as the iron source without (A) and with (B) the free-iron chelator 2,2′-dipyridyl (100 μM). Results are expressed as the averages of three independent experiments. OD600, optical density at 600 nm.
Utilization of heme compounds by E. coli EB53 and 101ESD complemented with V. anguillarum genes.
The ability of V. anguillarum heme transport genes to allow the use of hemin or hemoglobin as porphyrin and iron sources was evaluated in E. coli EB53 (aroB hemA) and in E. coli 101 ESD [Δ(entC-entA)], respectively. The aroB and entC-entA mutations render E. coli unable to produce its own siderophore, enterochelin, and the hemA mutation disrupts synthesis of heme. Therefore, E. coli EB53 cannot grow unless supplied with the heme biosynthetic precursor ALA. EB53 could satisfy the porphyrin deficiency by utilizing exogenously supplied hemin, as long as a genetic system for hemin uptake is provided. Similarly, 101ESD cannot grow in the presence of iron chelators unless supplied with a utilizable source of iron.
Growth around hemin (0.05 and 5 mM) or hemoglobin (1 mM) disks on nutrient agar plates (NB) was used to assay the use of porphyrin sources by E. coli EB53. To test whether the introduction of the entire heme transport gene cluster would promote hemin and hemoglobin utilization as porphyrin sources, cosmid pML1, containing the entire heme transport region within a ca. 15-kb V. anguillarum H-775-3 genome fragment, was introduced into EB53 by triparental mating. This transformant utilized hemin and hemoglobin as porphyrin sources. A subclone of pML1 (pCAR121) containing only huvA proved to be sufficient to confer the utilization of hemin and hemoglobin as porphyrin sources upon E. coli EB53. This indicates that E. coli EB53 (a K-12 derivative) encodes all the additional functions necessary for transporting and utilizing heme as a porphyrin when an outer membrane heme receptor is provided. None of other plasmid combinations assayed, in which huvA was absent, could complement EB53 (data not shown).
To determine which genes are necessary for the utilization of hemin and hemoglobin as iron sources, E. coli 101ESD was complemented with V. anguillarum genes and tested on CM9 minimal medium plates supplemented with 100 μM 2,2′-dipyridyl and with hemin and hemoglobin supplied on paper disks. Results are summarized in Table 4. E. coli 101ESD transformed with plasmids containing huvA alone did not grow in this medium, indicating that other genes in addition to huvA are needed for the utilization of hemin or hemoglobin as an iron source. However, E. coli 101ESD transformed with cosmid pML1 utilized both compounds as iron sources. In order to determine the minimum genetic background necessary for utilization of heme as an iron source, different gene combinations were assayed.
TABLE 4.
Utilization of hemin and hemoglobin as iron sources by E. coli 101ESD complemented with V. anguillarum heme uptake genesa
| Strain | Gene(s) present | Utilization of:
|
||
|---|---|---|---|---|
| Hemoglobin (0.1 mM) | Hemin (5 mM)c | FeSO4 | ||
| 101ESD | None | − | − | + |
| 101ESD/pCAR121 | huvA | − | − | + |
| 101ESD/pCAR126 | huvX tonB-exbBD-huvB | − | − | + |
| 101ESD/pML1 | huvAZX tonB-exbBD-huvBCD | + | + | + |
| 101ESD/pCAR115/pCAR126 | huvAX tonB-exbBD-huvB | − | − | + |
| 101ESD/pCAR121/pCAR179 | huvAZ | − | − | + |
| 101ESD/pSML23 | huvAZX tonB-exbBD-huvBCD | + | + | + |
| 101ESD/pCAR121/pCAR181 | huvAX | − | − | + |
| 101ESD/pCAR121/pSML11 | huvAZX | − | − | + |
| 101ESD/pCAR121/pSML32 | huvA-exbD-huvBCDb | − | − | + |
| 101ESD/pCAR121/pSML34 | huvA tonB-exbBD | − | − | + |
| 101ESD/pCAR121/pSML33 | huvA tonB-exbBD-huvBCD | + | + | + |
Assay was conducted on CM9 plates supplemented with 100 μM 2,2′-dipyridyl.
Genes exbD-huvBCD in pSML32 will not be expressed if they are transcribed from a promoter upstream of tonB.
The same results were obtained when 0.05 mM hemin was used.
Heme iron utilization did not occur when strain 101ESD/pCAR121 was transformed with pSML11 (huvZX) or with pCAR126 (huvX tonB exbBD huvB). The complete nine-gene cluster was PCR amplified, cloned in pWKS30 to yield pSML23, and transformed into 101ESD. This strain utilized hemin and hemoglobin as iron sources, indicating that this capacity is encoded within the nine-gene cluster described here and that this utilization is not due to extra genes present in pML1.
Complete tonB exbBD huvBCD genes cloned in pSML33 and transformed into strain 101ESD/pCAR121 allowed this strain to utilize hemin and hemoglobin as iron sources. However, 101ESD/pCAR121/pSML32 and 101ESD/pCAR121/pSML34 failed to grow with hemin or hemoglobin. These results together demonstrate that tonB exbBD genes in combination with huvA are not sufficient for heme iron utilization unless huvBCD genes are also provided. It is feasible that huvBCD genes are the crucial ones in combination with huvA to allow transport of heme compounds into the cytoplasm of E. coli and further utilization as iron sources. The finding that tonB exbBD genes need to be provided in the same plasmid together with downstream huvBCD genes in order to allow complementation may be explained by the existence of a single promoter upstream of tonB, as has been reported for V. cholerae (29).
DISCUSSION
Utilization of hemin and heme-containing proteins as iron sources has been reported for V. anguillarum (23, 24), but the molecular mechanism supporting heme uptake is unknown. In a recent work, the V. anguillarum outer membrane heme receptor gene huvA was characterized, and a huvA mutant obtained by chemical mutagenesis showed a reduction in virulence for fish (25). In the present study, we characterized a gene cluster in V. anguillarum that has similarities with heme iron assimilation systems found in other Vibrio species and P. shigelloides, both at the amino acid level and gene organization.
Homologues of V. anguillarum HuvZX proteins have been described, associated with the heme utilization systems of V. cholerae, V. vulnificus, Vibrio parahaemolyticus, and P. shigelloides, but to date their roles remain unascertained. In the present study we report the mutational analysis of V. anguillarum huvZX genes. Our results have shown that a huvX deletion mutant is able to use heme nearly as efficiently as the parental strain. This suggests that either huvX is not directly involved in the use of heme as an iron source or additional V. anguillarum genes may substitute for the function of huvX. However, deletion of huvZ drastically reduces the ability of the bacterium to grow with heme as the sole iron source, indicating that this gene is essential for heme iron utilization. A HuvZ-related activity may be present in E. coli 101ESD, since the heme uptake system of V. anguillarum can be easily reconstituted in 101ESD without HuvZ. However, the actual function of HuvZ remains unknown. A possibility would be that HuvZ is involved in removing iron from heme. It has been proposed that the oxidative cleavage of heme mediated by heme oxygenases is a mechanism for iron acquisition for some bacteria. Heme oxygenase genes have been described recently for Neisseria meningitidis (44), and Corynebacterium diphtheriae (42), but no significant homology exists between these described heme oxygenase genes and HuvZ. Recently, Wyckoff et al. (43) demonstrated that hutZ (homologous to huvZ) is also essential for heme iron utilization in V. cholerae. These authors could not demonstrate a heme oxygenase activity for HutZ, suggesting that it may act as a heme carrier or storage protein. Further studies are needed in order to ascertain the role played by HuvZ in the utilization of heme as an iron source.
V. anguillarum huvBCD genes are essential for heme uptake. Nonpolar deletions in any of these three genes drastically reduce the growth of V. anguillarum with hemin or hemoglobin as the sole iron source. In contrast, it has been reported that V. cholerae hutB, hutC, or hutD mutants still retain significant growth when hemin is used as the only iron source (29). Thus, it is possible that proteins of other transport systems of V. cholerae can substitute for the physiological role of either HutB, -C, or -D, while in V. anguillarum each one of these proteins is essential for heme uptake.
Complementation studies carried out with E. coli EB53 showed that the V. anguillarum outer membrane heme receptor HuvA is sufficient for utilization of heme compounds as porphyrin sources. Complementation of E. coli heme-deficient mutants with an outer membrane heme receptor provided in trans has been previously reported (36). Interestingly, the P. shigelloides heme receptor gene hugA could not complement an E. coli DHE-1 hemA mutant for the use of heme as a porphyrin source unless P. shigelloides tonB-exbB-exbD genes were also provided in trans (14).
In the use of heme compounds as an iron source, huvA plus tonB exbBD huvBCD are needed to reconstitute the V. anguillarum heme transport system in E. coli 101ESD (an HB101 derivative). Similarly, Occhino et al. (29) reported that utilization of hemin as an iron source can be reconstituted in E. coli 1017 (HB101 derivative, ent) with V. cholerae hutA, tonB exbBD, and hutBCD genes. In contrast, Henderson et al. (14) observed that a plasmid containing hugWXZ was necessary in addition to hugA and tonB-exbBD to reconstitute the P. shigelloides heme iron utilization system in E. coli 1017.
We hypothesize that the V. anguillarum huvBCD genes are crucial for heme iron utilization in E. coli 101ESD but the tonB and exbBD genes are not. Complementation of EB53 for heme utilization as a porphyrin source did not depend on the presence of the V. anguillarum TonB system in trans, which means that huvA alone is active in an E. coli background. We propose that hutBCD genes are transcribed from a promoter located upstream of the tonB gene, and thus actual complementation for heme iron utilization is achieved only when the six genes tonB, exbBD, and huvBCD, are provided together on a plasmid. It remains to be explained why heme utilization as a porphyrin source in E. coli EB53 does not depend on huvBCD genes while utilization as an iron source in E. coli 101ESD does require huvBCD. This could be due to a difference in strain background, as for example, the presence of an inner membrane transporter in EB53.
As reported by Stojiljkovic and Hantke (36), the Y. enterocolitica outer membrane heme receptor HemR alone is sufficient to complement E. coli for utilization of heme as a porphyrin source, and it is possible that hemin, once in the periplasm, can be incorporated into cytochromes located in the cytoplasmic membrane (10). However, utilization of heme compounds as iron sources would be feasible only as long as the heme compound is transported from the periplasmic space into the cytoplasm, where it is expected that additional proteins are implicated in degradation of the heme molecule and in the release of iron. As proposed by Stojiljkovic and Hantke (36), the difference between porphyrin and iron utilization from heme may be merely quantitative, being the amount of heme needed to satisfy the cell's requirements for iron, much larger than the amount necessary as a porphyrin source. This being the case, E. coli EB53 complemented with V. anguillarum HuvA could transport trace amounts of heme as a porphyrin source into the cytoplasm via nonspecific E. coli ABC transporters. However, heme iron utilization could be effective only as long as a specific ABC transporter (HuvBCD) is provided.
The gene coding for the V. anguillarum outer membrane heme receptor is linked to the rest of heme transport genes. Such a spatial organization is unusual in other species of the family Vibrionaceae, where the outer membrane receptor gene is separated from the rest of the transport genes by hundreds of kilobases (1, 12, 22). Other differences in the V. anguillarum heme uptake cluster include the absence of a putative coproporphyrinogen oxidase gene and a gene for a LysR transcriptional activator homologue, which are present in V. vulnificus, V. cholerae, and V. parahaemolyticus (1, 12, 22). It is possible that the heme transport cluster originally included homologues of these genes, which eventually underwent a spatial reorganization in the genome of V. anguillarum.
In conclusion, we have shown in this study that the heme uptake cluster of V. anguillarum H-775-3 includes nine genes, five of which proved to be essential for utilization of heme as an iron source. The gene arrangement of the V. anguillarum heme uptake cluster has unique features which differentiate it from homologous clusters found in other gram-negative bacteria.
Acknowledgments
We gratefully acknowledge J. H. Crosa, K. E. Klose, and V. Braun for providing strains and plasmids.
This work was supported by grants AGL2000-0492 and AGL-2003-00086 from the Ministry of Science and Technology of Spain (cofunded by the FEDER Programme from the European Union) and grant PGIDT01PXI26202PN from Xunta de Galicia to M.L.L.
REFERENCES
- 1.Chen, C.-Y., K.-M. Wu, Y.-C. Chang, C.-H. Chang, H.-C. Tsai, T.-L. Liao, Y.-M. Liu, H.-J. Chen, A. B.-T. Shen, J.-C. Li, T.-L. Su, C.-P. Shao, C.-T. Lee, L.-I. Hor, and S.-F. Tsai. 2003. Comparative genome analysis of Vibrio vulnificus, a marine pathogen. Genome Res. 13:2577-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Conchas, R. F., M. L. Lemos, J. L. Barja, and A. E. Toranzo. 1991. Distribution of plasmid- and chromosome-mediated iron uptake systems in Vibrio anguillarum strains of different origins. Appl. Environ. Microbiol. 57:2956-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Correa, N., C. M. Lauriano, R. McGee, and K. E. Klose. 2000. Phosphorylation of the flagellar regulatory protein FlrC is necessary for Vibrio cholerae motility and enhanced colonization. Mol. Microbiol. 35:743-755. [DOI] [PubMed] [Google Scholar]
- 4.Crosa, J. H. 1989. Genetics and molecular biology of siderophore-mediated iron transport in bacteria. Microbiol. Rev. 53:517-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ditta, G., S. Stanfield, D. Corbim, and D. R. Helinski. 1980. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. USA 77:7347-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eberspächer, B., and V. Braun. 1980. The involvement of cytochromes in the uptake of ferrichrome by Escherichia coli K-12. FEMS Microbiol. Lett. 7:61-64. [Google Scholar]
- 7.Genco, C. A., and D. W. Dixon. 2001. Emerging strategies in microbial haem capture. Mol. Microbiol. 39:1-11. [DOI] [PubMed] [Google Scholar]
- 8.Goldberg, J. B., and D. E. Ohman. 1984. Cloning and expression in Pseudomonas aeruginosa of a gene involved in the production of alginate. J. Bacteriol. 158:1115-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gualerzi, C. O., and C. L. Pon. 1990. Initiation of mRNA translation in prokaryotes. Biochemistry 29:5881-5889. [DOI] [PubMed] [Google Scholar]
- 10.Haddock, B. A., and H. U. Schairer. 1973. Electron-transport chains of Escherichia coli. Reconstitution of respiration in a 5-aminolevulinic acid-requiring mutant. Eur. J. Biochem. 35:34-45. [DOI] [PubMed] [Google Scholar]
- 11.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 12.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henderson, D. P., and S. M. Payne. 1993. Cloning and characterization of the Vibrio cholerae genes encoding the utilization of iron from haemin and haemoglobin. Mol. Microbiol. 7:461-469. [DOI] [PubMed] [Google Scholar]
- 14.Henderson, D. P., E. E. Wyckoff, C. E. Rashidi, and A. L. Oldham. 2001. Characterization of the Plesiomonas shigelloides genes encoding the heme iron utilization system. J. Bacteriol. 183:2715-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knauf, V. C., and E. W. Nester. 1982. Wide host range cloning vectors: a cosmid clone bank of an Agrobacterium Ti plasmid. Plasmid 8:45-54. [DOI] [PubMed] [Google Scholar]
- 16.Köster, W. L., L. A. Actis, L. S. Waldbeser, M. E. Tolmasky, and J. H. Crosa. 1991. Molecular characterization of the iron transport system mediated by the pJM1 plasmid in Vibrio anguillarum 775. J. Biol. Chem. 266:23829-23833. [PubMed] [Google Scholar]
- 17.Köster, W. 2001. ABC transporter-mediated uptake of iron, siderophores, heme and vitamin B12. Res. Microbiol. 152:291-301. [DOI] [PubMed] [Google Scholar]
- 18.Lee, B. C. 1995. Quelling the red menace: haem capture by bacteria. Mol. Microbiol. 18:383-390. [DOI] [PubMed] [Google Scholar]
- 19.Lemos, M. L., P. Salinas, A. E. Toranzo, J. L. Barja, and J. H. Crosa. 1988. Chromosome-mediated iron uptake system in pathogenic strains of Vibrio anguillarum. J. Bacteriol. 170:1920-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Litwin, C. M., and B. L. Byrne. 1998. Cloning and characterization of an outer membrane protein of Vibrio vulnificus required for heme utilization: regulation of expression and determination of the gene sequence. Infect. Immun. 66:3134-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Litwin, C. M., and J. Quackenbush. 2001. Characterization of a Vibrio vulnificus LysR homologue, HupR, which regulates expression of the haem uptake outer membrane protein, HupA. Microb. Pathog. 31:295-307. [DOI] [PubMed] [Google Scholar]
- 22.Makino, K., K. Oshima, K. Kurokawa, K. Yokoyama, T. Uda, K. Tagomori, Y. Iijima, M. Najima, M. Nakano, A. Yamashita, Y. Kubota, S. Kimura, T. Yasunaga, T. Honda, H. Shinagawa, M. Hattori, and T. Iida. 2003. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V. cholerae. Lancet 361:743-749. [DOI] [PubMed] [Google Scholar]
- 23.Mazoy, R., and M. L. Lemos. 1991. Iron-binding proteins and heme compounds as iron sources for Vibrio anguillarum. Curr. Microbiol. 23:221-226. [Google Scholar]
- 24.Mazoy, R., and M. L. Lemos. 1996. Identification of heme-binding proteins in the cell membranes of Vibrio anguillarum. FEMS Microbiol. Lett. 135:265-270. [DOI] [PubMed] [Google Scholar]
- 25.Mazoy, R., C. R. Osorio, A. E. Toranzo, and M. L. Lemos. 2003. Isolation of mutants of Vibrio anguillarum defective in haeme utilisation and cloning of huvA, a gene coding for an outer membrane protein involved in the use of haeme as iron source. Arch. Microbiol. 179:329-338. [DOI] [PubMed] [Google Scholar]
- 26.Mey, A. R., and S. M. Payne. 2001. Haem utilization in Vibrio cholerae involves multiple TonB-dependent haem receptors. Mol. Microbiol. 42:835-849. [DOI] [PubMed] [Google Scholar]
- 27.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Mills, M., and S. M. Payne. 1997. Identification of shuA, the gene encoding the heme receptor of Shigella dysenteriae, and analysis of invasion and intracellular multiplication of a shuA mutant. Infect. Immun. 65:5358-5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Occhino, D. A., E. E. Wyckoff, D. P. Henderson, T. J. Wrona, and S. M. Payne. 1998. Vibrio cholerae iron transport: haem transport genes are linked to one of two sets of tonB, exbB, exbD genes. Mol. Microbiol. 29:1493-1507. [DOI] [PubMed] [Google Scholar]
- 30.O′Malley, S. M., S. L. Mouton, D. A. Occhino, M. T. Deanda, J. R. Rashidi, K. L. Fuson, C. E. Rashidi, M. Y. Mora, S. M. Payne, and D. P. Henderson. 1999. Comparison of the heme iron utilization systems of pathogenic vibrios. J. Bacteriol. 181:3594-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Panek, H., and M. R. O'Brian. 2002. A whole genome view of prokaryotic biosynthesis. Microbiology 148:2273-2282. [DOI] [PubMed] [Google Scholar]
- 32.Ratledge, C., and L. G. Dover. 2000. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54:881-941. [DOI] [PubMed] [Google Scholar]
- 33.Rose, R. E. 1988. The nucleotide sequence of pACYC177. Nucleic Acids Res. 16:356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook, J., and D.W. Rusell. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Sørensen, U. S. B., and J. L. Larsen. 1986. Serotyping of Vibrio anguillarum. Appl. Environ. Microbiol. 51:593-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stojiljkovic, I., and K. Hantke. 1992. Hemin uptake system of Yersinia enterocolitica: similarities with the other TonB-dependent systems in Gram-negative bacteria. EMBO J. 11:4359-4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stojiljkovic, I., and K. Hantke. 1994. Transport of haemin across the cytoplasmic membrane through a haemin-specific periplasmic binding-protein-dependent transport system in Yersinia enterocolitica. Mol. Microbiol. 13:719-732. [DOI] [PubMed] [Google Scholar]
- 38.Stork, M., M. Di Lorenzo, T. J. Welch, L. M. Crosa, and J. H. Crosa. 2002. Plasmid-mediated iron uptake and virulence in Vibrio anguillarum. Plasmid 48:222-228. [DOI] [PubMed] [Google Scholar]
- 39.Torres, A. G., and S. M. Payne. 1997. Haem iron-transport system in enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 23:825-833. [DOI] [PubMed] [Google Scholar]
- 40.Walker, J. E., M. Saraste, M. J. Runswick, and N. J. Gay. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1:945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing, and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]
- 42.Wilks, A., and M. P. Schmitt. 1998. Expression and characterization of a heme oxygenase (HmuO) from Corynebacterium diphtheriae. J. Biol. Chem. 273:837-841. [DOI] [PubMed] [Google Scholar]
- 43.Wyckoff, E. E., M. Schmitt, A. Wilks, and S. M. Payne. 2004. HutZ is required for efficient heme utilization in Vibrio cholerae. J. Bacteriol. 186:4142-4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu, W., A. Wilks, and I. Stojiljkovic. 2000. Degradation of heme in gram-negative bacteria: the product of the hemO gene of Neisseriae is a heme oxygenase. J. Bacteriol. 182:6783-6790. [DOI] [PMC free article] [PubMed] [Google Scholar]