Bacteria belonging to the family Deinococcaceae survive exposure to >1.5 megarads of ionizing irradiation or to extreme desiccation without lethality or mutagenesis (2, 31, 35). This tolerance derives from the ability of these species to accurately mend numerous double-strand DNA breaks (DSBs), thus reassembling an intact genome from hundreds of fragments in a manner that restores chromosomal continuity. The only known mechanism that enables accurate repair of DSBs in bacteria is RecA-dependent homologous recombination, whereby information lost at a lesion is restored by a homologous DNA sequence that acts as a template (22-24). As such, DNA repair via homologous recombination strictly depends upon the ability of cellular systems to perform a rapid and efficient genomewide search for homologous DNA sites (27). However, following extensive DNA fragmentation, no intact template remains. Homologous search conducted under such circumstances would necessarily entail repetitive reinspection of multiple randomly dispersed DNA fragments, rendering the process inherently futile (9a, 36). Indeed, the first phase of DNA repair in Deinococcus radiodurans was shown to be RecA independent (9), implying that this phase does not rely on homologous recombination.
The high resistance of bacterial spores to irradiation and desiccation indicates that DSBs inflicted by these assaults on dormant spores are efficiently and accurately mended upon germination. However, DNA repair involving homologous search processes cannot occur in germinating spores, because bacterial spores regularly carry only one copy of their genomes (5). Consequently, germinating spores lack the template required for accurate homologous-recombination-mediated repair of DSBs.
STRUCTURAL SOLUTIONS: HOLLIDAY JUNCTIONS AND DNA TOROIDS
Biochemical and genetic studies, including the complete sequencing of the D. radiodurans genome, indicated that this organism possesses a typical bacterial complement of DNA repair enzymes (50) and that these proteins are, by and large, similar to those found in other bacteria (3, 4). A recent analysis of the effects exerted by acute irradiation upon D. radiodurans gene expression did not elucidate a genetic basis of DNA repair (29). These observations, which imply that the complement of DNA repair proteins in D. radiodurans is not sufficient to confer resistance, led to the suggestion that repair of DSBs in this organism is promoted by a continuous alignment of genome copies (36; Daly and Minton, Science 270:1318, 1995). Such an alignment, presumably maintained by multiple four-stranded Holliday junctions, would provide a means for error-free DNA repair by supplying an ever-present nearby template, hence eliminating the need for a logistically impractical homologous search. Multiple Holliday junctions between DNA molecules would, however, represent a major obstacle to DNA transactions, and indeed, they were shown by optical mapping analysis to be absent in the D. radiodurans genome (28).
An alternative to genome alignment by Holliday junctions was implied by structural studies of D. radiodurans, which demonstrated that chromatin in the organism adopts a toroidal shape. It was suggested that within this tightly packed shape, ends of DNA fragments generated by DSBs are continuously held in close physical proximity, thus enabling their accurate repair in a template-independent pathway (26, 33). Such repair processes may proceed through nonhomologous end joining (NHEJ), as well as via homologous annealing of protruding single strands that are present at the ends of DNA fragments (9).
In this commentary, we survey recently reported findings, which indicate that toroidal DNA conformations represent a common feature in highly resistant life forms, such as members of the family Deinococcaceae, as well as dormant and germinating bacterial spores. We discuss other observations that imply that the complement of DNA repair enzymes in these life forms evolved specifically to enable accurate repair of DSBs through end joining within a tightly packed DNA organization. Taken together, these findings support the notion that a toroidal DNA conformation is used in bacteria to facilitate the mending of DSBs when RecA-dependent homologous repair cannot be effectively employed.
DNA TOROIDS: STRUCTURAL AND PHYSIOLOGICAL ASPECTS
Cryoelectron microscopy of DNA toroids obtained in vitro indicated that within these structures, DNA molecules attain a high degree of order and compactness (17). The inherent lateral order and high packaging density make DNA toroids rigid matrices in which diffusion of DNA fragments generated by irradiation or desiccation is substantially restricted (33). Indeed, in vitro studies demonstrated that within toroidal DNA structures, annealing of cohesive DNA ends, as well as enzymatic ligation of cohesive and blunt DNA ends, is enhanced by 5 to 6 orders of magnitude relative to its rate and efficiency in dispersed DNA structures (18, 51). As such, DNA toroids, in which free DNA ends are kept close together and their local concentration is substantially enhanced, provide uniquely suitable scaffolds for DNA repair through high-fidelity DNA end-joining processes.
DNA toroids in vivo.
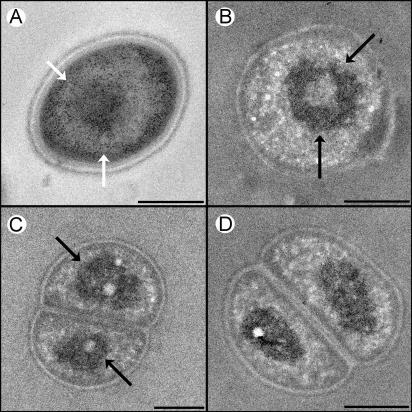
DNA molecules in D. radiodurans cells adopt a distinct toroidal shape that sets the species apart from most other bacteria, in which a dispersed and amorphous morphology of the genome is regularly discerned (26). Studies conducted in our laboratory demonstrated that the genomes of two additional members of the family Deinococcaceae, Deinococcus radiopugnans and Deinococcus radiophilus, are also assembled as toroids (Fig. 1). A similar toroidal DNA shape was detected in dormant spores of Bacillus subtilis (13). Notably, the toroidal structure was shown to persist in germinating spores of both B. subtilis and Bacillus megaterium (43).
FIG. 1.
Transmission electron micrographs of cryofixed D. radiopugnans (A and C) and D. radiophilus (B and D) cells. (A) Regular staining. The darkly stained particles are ribosomes, while the lightly stained space contains chromatin. (B, C, and D) Cells stained with the DNA-specific reagent osmium-ammine-SO2 (27). DNA toroids (indicated by arrows) are evident in panels A, B, and C, whereas in panel D the toroids are detected edge on. Because thin sections are used, some (cross-sectioned) specimens reveal only one compartment. Scale bars, 0.5 μm.
The observation that members of the family Deinococcaceae, as well as bacterial spores, adopt a toroidal DNA conformation is significant, because both life forms cannot promote conventional high-fidelity DNA repair pathways. Following extensive DNA fragmentation in deinococcal species, homologous search processes, and hence repair of DSBs through homologous recombination, become ineffective (36). Analogously, DNA repair through homologous search cannot occur in germinating spores, because bacterial spores regularly carry only one copy of their genomes (5). Thus, species belonging to the family Deinococcaceae and bacterial spores share three conspicuous features. Both life forms survive irradiation and desiccation in doses that are lethal to other species (2, 39, 47), both forms are incapable of repairing DSBs through homologous recombination, and most significantly, both species belonging to the family Deinococcaceae and spores maintain their DNA complements in a toroidal conformation, within which accurate DNA repair by NHEJ processes may occur.
DNA toroids and cellular morphology.
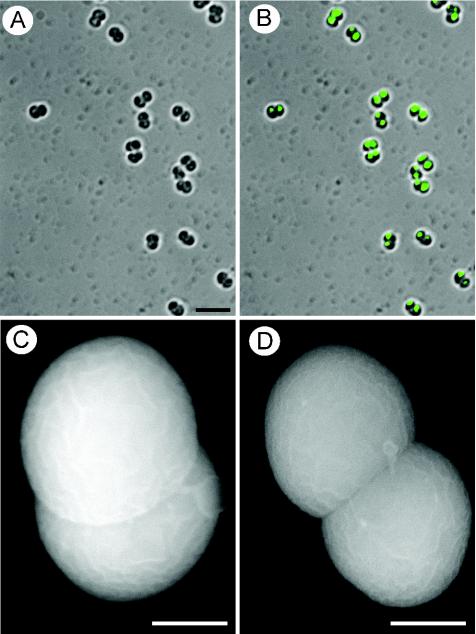
D. radiodurans cells reveal a tetrad morphology (26) and carry 4 to 10 genome copies (16), which are segregated within the four compartments (26). Further studies have indicated that D. radiopugnans and D. radiophilus are diplococcal, composed of two compartments within which genome copies are segregated (Fig. 2). The multicoccal morphology is significant because of the notion that when bacteria contain several segregated genome copies, these copies reveal different levels of transcriptional activity and hence different extents of packaging (41). In species of the family Deinococcaceae, in which individual chromosomes are segregated into two or four compartments, such differential packaging would explain how a single vegetative cell could have both metabolically active chromosomes that allow growth and condensed toroidal nucleoids that promote resistance. This notion is supported by the finding that D. radiodurans cells carry several genome copies (16), whereas dormant spores, which do not need an active decondensed genome, maintain only one copy (5). The notion is further buttressed by the existence of orifices in the membranes that separate the compartments in species of the family Deinococcaceae, indicating that the compartments are not fully separated (26). Indeed, the nucleoid in one or two compartments of vegetative D. radiodurans cells exhibits a dispersed morphology, whereas the chromatin in the other compartments adopts a toroidal structure (26). Notably, the metabolically dormant spores do not contain active chromosomes and hence do not require multicoccal morphology.
FIG. 2.
Morphology and DNA segregation in D. radiopugnans cells from 4-day-old cultures. (A and B) Shown are light (A) and fluorescence (B) microscopy of cells labeled with DAPI (4′,6′-diamidino-2-phenylindole). DNA segregation in both compartments of each diplococcal unit is evident. A diplococcal morphology is demonstrated by all cells, as indicated by both light (A and B) and scanning electron (C and D) microscopy. Scale bars, 5 (A and B) and 0.5 (C and D) μm.
FACTORS THAT STABILIZE TOROIDAL STRUCTURES
In vitro studies have indicated that a toroidal DNA shape represents a particularly stable mode of DNA condensation (6, 40). Several factors combine to further enhance the intrinsic stability of this particular shape in Deinococcaceae and in bacterial spores.
Temperature.
The toroidal DNA shape in species of the family Deinococcaceae becomes substantially more pronounced at low temperature yet is hardly discernible as the temperature is raised to 42°C (J. Englander and A. Minsky, unpublished results). Consistent with this finding is the observation that the radioresistance of deinococcal species is decreased by 2 orders of magnitude at elevated temperatures (21).
Mn2+ ions.
In vitro studies have demonstrated that the divalent ion Mn2+ is uniquely efficient in promoting ordered, toroidal DNA condensation (7, 30, 44). This observation is significant, because the genome of D. radiodurans maintains an exceptionally large concentration of Mn2+ ions (25). The ability of Mn2+ ions to specifically stabilize condensed DNA morphologies under dehydrating conditions (44) is particularly notable, as D. radiodurans DNA damage tolerance has been proposed to reflect an evolutionary adaptation to dehydration (32).
However, it has been demonstrated that when the concentration of DNA-condensing factors is increased beyond a given threshold, DNA decondensation and resolubilization are effected, possibly due to DNA charge reversal (10, 38, 42). Notably, relatively high (>2.5 μM) concentrations of Mn2+ ions sensitize D. radiodurans cells to irradiation without affecting their viability or growth under unstressed conditions (8). Indeed, when exposed to large concentrations of Mn2+, the D. radiodurans genome reveals an amorphous, nontoroidal morphology (26). It thus appears that factors that modulate the formation and stability of DNA toroids, such as temperature and divalent ions, correspondingly affect damage tolerance.
DNA-binding proteins.
Small DNA-binding, acid-soluble proteins (SASPs), which are ubiquitous in bacterial spores, specifically stabilize toroidal DNA packaging in vitro (15), as well as within spores (13, 43). In vitro studies have indicated that the DNA-SASP toroidal complex is ordered and highly condensed (13). Indeed, spores that lack SASPs do not form a ringlike DNA structure and are substantially more sensitive to UV light and desiccation than wild-type spores (47). Similarly, the absence of toroidal DNA structures in D. radiodurans renders the organism susceptible to irradiation, as mentioned above.
The D. radiodurans DNA-binding protein HU has recently been shown to reveal a particularly high affinity for prebent DNA sequences, thus specifically stabilizing these structural motifs (14). Apparently, in addition to the factors mentioned above and in analogy to the sporal SASP, the ubiquitous HU protein acts to promote toroidal DNA packaging in the species of the family Deinococcaceae by stabilizing a highly curved DNA trajectory.
Growth phase.
Starved stationary-state D. radiodurans cells are threefold more resistant to ionizing irradiation than actively growing cells (35). This observation is consistent with the finding that the toroidal DNA organization is substantially more pronounced in stationary-state D. radiodurans cells than in actively growing bacteria (26). This finding is, however, inconsistent with the premise that DNA repair in D. radiodurans is promoted solely by induced enzymatic pathways, because these pathways become increasingly inefficient during prolonged starvation (34).
DNA REPAIR ENZYMES IN DEINOCOCCACEAE AND IN SPORE-FORMING BACTERIA
Whereas the sequencing and analysis of the D. radiodurans genome indicated that the complement of DNA repair enzymes in this resistant species is similar to that found in nonresistant bacteria (50), several intriguing differences were discerned.
RecA and RecBCD.
RecA and RecA-like proteins play critical roles in homologous recombination (22, 45). Studies conducted with a recA-defective mutant indicated that the initial DSB repair phase in D. radiodurans is, however, RecA independent (9). This phase, which is initiated immediately following acute irradiation and which proceeds for several hours, is highly efficient, resulting in error-free mending of more than one-third of the multiple DSBs. This finding has been taken to imply the presence of RecA-independent annealing between complementary single-stranded DNA segments created at the ends of the fragments (9). We claim that while such annealing may indeed assist DNA repair, its contribution would be limited relative to NHEJ processes because single strands generated at DSB sites are unlikely to become long enough to allow significant annealing in the absence of RecBCD exonuclease in D. radiodurans (31) (see below). We note that, regardless of the relative contributions of NHEJ and single-strand annealing to DNA repair, both processes would be substantially facilitated and accelerated within the scaffold of tightly packed DNA toroids, in which the continuity of DNA fragments is physically preserved (33).
The RecA protein in D. radiodurans, which exhibits 53% sequence identity with Escherichia coli RecA, is constitutively expressed at low levels but is transiently induced to higher levels following extensive DNA damage (19, 20). Significantly, in contrast to the RecA proteins in other bacterial strains, D. radiodurans RecA binds preferentially to double-stranded rather than to single-stranded DNA and hydrolyzes ATP more rapidly upon binding to double-stranded DNA than to single-stranded molecules (19, 20). These unique traits, as well as recent observations which imply that the recombination activity of RecA in D. radiodurans does not represent a critical factor in DNA repair processes (37, 46), highlight the notion that the actual modes through which RecA exerts its functions in D. radiodurans remain poorly understood.
The heterotrimeric helicase-nuclease RecBCD plays an essential role in homologous repair of DSBs in bacteria by producing single-stranded DNA tails (24) and stimulating the loading of RecA onto these tails (1). As such, the RecBCD complex can formally be considered an enzyme that extends DNA damage at a DSB site through its nuclease activity. The unexpected absence of RecBCD in D. radiodurans (31, 50), along with the unique preferential binding of D. radiodurans RecA to double-stranded DNA (19), supports the notion that repair of DSBs in this organism relies on repair enzymes that evolved to exert their activities on double-stranded DNA species, presumably by promoting NHEJ within a rigid toroidal matrix.
DNA ligases and NHEJ.
In eukaryotic cells, DSBs are repaired by homologous recombination or by NHEJ (12). NHEJ is specifically promoted by ATP-dependent DNA ligases that are ubiquitous in eukaryotes but are considered to be absent in bacterial cells, which regularly encode NAD-dependent ligases involved in DNA replication (12). Until recently, it was assumed that a NHEJ system is not present in prokaryotes, and bacterial high-fidelity repair of DSBs was thought to rely solely on homologous recombination.
Recent studies have revealed, however, that a unique family of ATP-dependent DNA ligases is present in several bacterial species, including Mycobacterium tuberculosis, B. subtilis, and Bacillus halodurans, in addition to typical NAD-dependent ligases (11, 48, 49). These bacterial species were also found to contain Ku-like proteins, which are homologous to the eukaryotic Ku protein that acts to recruit ATP-dependent DNA ligases onto DSB sites. The presence of ATP-dependent ligases and Ku homologues in bacteria that spend long periods of their life cycles in stationary phase or that are regularly exposed to harsh environments was proposed to imply that an NHEJ system might represent an important mode of repair of DSBs in these species (49).
In addition to an NAD-dependent ligase, an ATP-dependent DNA ligase was identified in D. radiodurans (50). The ATP-dependent ligase is induced by irradiation, whereas the typical NAD-dependent ligase is down-regulated (29), implying that the ATP-dependent ligase might be involved in postirradiation repair in D. radiodurans. Significantly, the small mass (22 kDa) of D. radiodurans ATP-dependent ligase, which sets it apart from the typically much larger DNA ligases, is likely to facilitate access of the enzyme to DSBs within the tightly packed toroids.
In contrast to spore-forming bacteria, a Ku homologue was not identified in D. radiodurans. However, because Ku-dependent stimulation of ligation is partially attributed to the ability of the protein to juxtapose two DNA ends (11), its activity in Deinococcaceae might not be required, as DNA ends are kept together within the toroidal DNA matrix.
DNA TOROIDS AND DNA REPAIR
The observations summarized here imply that a tight toroidal DNA organization is uniquely adjusted to promote the repair of multiple DSBs by both NHEJ and RecA-independent annealing in a manner that drastically minimizes errors. They further indicate that this structurally dependent strategy is specifically adopted by prokaryotes that are inactive during significant periods of their life cycles, e.g., dormant spores, or that are regularly exposed to harsh environments, such as Deinococcaceae. These highly resistant life forms, in which repair of DSBs by homologous recombination is impossible or ineffective, evolved mechanisms (proteins such as SASPs and HU or high concentrations of Mn2+ ions) that promote the formation and stability of DNA toroids. These organisms also evolved a complement of DNA repair enzymes that enables NHEJ within toroids. In some cases (e.g., Deinococcaceae), they adopted polycoccal morphologies that allow cellular growth while preserving tight DNA packaging. Finally, the considerations presented here corroborate the notion that particular genome structures represent crucial factors in the maintenance of DNA integrity in living systems exposed to harsh environmental conditions (13, 27, 33, 34).
The views expressed in this Commentary do not necessarily reflect the views of the journal or of ASM.
REFERENCES
- 1.Anderson, D. G., and S. C. Kowalczykowski. 1997. The translocating RecBCD enzyme stimulates recombination by directing RecA protein onto ssDNA in a chi-regulated manner. Cell 90:77-86. [DOI] [PubMed] [Google Scholar]
- 2.Battista, J. R. 1997. Against all odds: the survival strategies of Deinococcus radiodurans. Annu. Rev. Microbiol. 51:203-224. [DOI] [PubMed] [Google Scholar]
- 3.Battista, J. R. 2000. Radiation resistance: the fragments that remain. Curr. Biol. 10:R204-R205. [DOI] [PubMed] [Google Scholar]
- 4.Battista, J. R., A. M. Earl, and M. J. Park. 1999. Why is Deinococcus radiodurans so resistant to ionizing radiation? Trends Microbiol. 7:362-365. [DOI] [PubMed] [Google Scholar]
- 5.Belliveau, B. H., T. C. Beaman, and P. Gerhardt. 1990. Heat resistance correlated with DNA content in Bacillus megaterium spores. Appl. Environ. Microbiol. 56:2919-2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bloomfield, V. A. 1996. DNA condensation. Curr. Opin. Struct. Biol. 6:334-341. [DOI] [PubMed] [Google Scholar]
- 7.Cherstvy, A. G., A. A. Kornyshev, and S. Leikin. 2002. Temperature-dependent DNA condensation triggered by rearrangement of adsorbed cations. J. Phys. Chem. B 106:13362-13369. [Google Scholar]
- 8.Chou, F. I., and S. T. Tan. 1990. Manganese(II) induces cell division and increases in superoxide dismutase and catalase activities in an aging deinococcal culture. J. Bacteriol. 172:2029-2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daly, M. J., and K. W. Minton. 1996. An alternative pathway of recombination of chromosomal fragments precedes recA-dependent recombination in the radioresistant bacterium Deinococcus radiodurans. J. Bacteriol. 178:4461-4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Daly, M. J., and K. W. Minton. 1995. Resistance to radiation. Science 270:1318. [DOI] [PubMed] [Google Scholar]
- 10.de la Cruz, M. O., L. Belloni, M. Delsanti, J. P. Dalbiez, O. Spalla, and M. Drifford. 1995. Precipitation of highly-charged polyelectrolyte solutions in the presence of multivalent salts. J. Chem. Phys. 103:5781-5791. [Google Scholar]
- 11.Doherty, A. J., S. P. Jackson, and G. R. Weller. 2001. Identification of bacterial homologues of the Ku DNA repair proteins. FEBS Lett. 500:186-188. [DOI] [PubMed] [Google Scholar]
- 12.Eisen, J. A., and P. C. Hanawalt. 1999. A phylogenomic study of DNA repair genes, proteins, and processes. Mutat. Res. 435:171-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frenkiel-Krispin, D., R. Sack, J. Englander, E. Shimoni, M. Eisenstein, E. Bullitt, R. Horowitz-Scherer, C. S. Hayes, P. Setlow, A. Minsky, and S. G. Wolf. 2004. Structure of the DNA-SspC complex: implications for DNA packaging, protection, and repair in bacterial spores. J. Bacteriol. 186:3525-3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghosh, S., and A. Grove. 2004. Histone-like protein HU from Deinococcus radiodurans binds preferentially to four-way DNA junctions. J. Mol. Biol. 337:561-571. [DOI] [PubMed] [Google Scholar]
- 15.Griffith, J., A. Makhov, L. Santiago-Lara, and P. Setlow. 1994. Electron microscopic studies of the interaction between a Bacillus subtilis alpha/beta-type small, acid-soluble spore protein with DNA: protein binding is cooperative, stiffens the DNA, and induces negative supercoiling. Proc. Natl. Acad. Sci. USA 91:8224-8228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen, M. T. 1978. Multiplicity of genome equivalents in the radiation-resistant bacterium Micrococcus radiodurans. J. Bacteriol. 134:81-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hud, N. V., and K. H. Downing. 2001. Cryoelectron microscopy of lambda phage DNA condensates in vitreous ice: the fine structure of DNA toroids. Proc. Natl. Acad. Sci. USA 98:14925-14930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jary, D., and J. L. Sikorav. 1999. Cyclization of globular DNA. Implications for DNA-DNA interactions in vivo. Biochemistry 38:3223-3227. [DOI] [PubMed] [Google Scholar]
- 19.Kim, J. I., and M. M. Cox. 2002. The RecA proteins of Deinococcus radiodurans and Escherichia coli promote DNA strand exchange via inverse pathways. Proc. Natl. Acad. Sci. USA 99:7917-7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim, J. I., A. K. Sharma, S. N. Abbott, E. A. Wood, D. W. Dwyer, A. Jambura, K. W. Minton, R. B. Inman, M. J. Daly, and M. M. Cox. 2002. RecA protein from the extremely radioresistant bacterium Deinococcus radiodurans: expression, purification, and characterization. J. Bacteriol. 184:1649-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kitayama, S., Harsojo, and A. Matsuyama. 1980. Sensitization of Micrococcus radiophilus to gamma-rays by post-irradiation incubation with chloramphenicol or at non-permissive temperature. J. Radiat. Res. 21:257-262. [DOI] [PubMed] [Google Scholar]
- 22.Kowalczykowski, S. C. 1991. Biochemical and biological functions of Escherichia coli RecA protein: behavior of mutant RecA proteins. Biochimie 73:289-304. [DOI] [PubMed] [Google Scholar]
- 23.Kowalczykowski, S. C. 1991. Biochemistry of genetic recombination: energetics and mechanism of DNA strand exchange. Annu. Rev. Biophys. Biophys. Chem. 20:539-575. [DOI] [PubMed] [Google Scholar]
- 24.Kowalczykowski, S. C., D. A. Dixon, A. K. Eggleston, S. D. Lauder, and W. M. Rehrauer. 1994. Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev. 58:401-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leibowitz, P. J., L. S. Schwartzberg, and A. K. Bruce. 1976. The in vivo association of manganese with the chromosome of Micrococcus radiodurans. Photochem. Photobiol. 23:45-50. [DOI] [PubMed] [Google Scholar]
- 26.Levin-Zaidman, S., J. Englander, E. Shimoni, A. K. Sharma, K. W. Minton, and A. Minsky. 2003. Ringlike structure of the Deinococcus radiodurans genome: a key to radioresistance? Science 299:254-256. [DOI] [PubMed] [Google Scholar]
- 27.Levin-Zaidman, S., D. Frenkiel-Krispin, E. Shimoni, I. Sabanay, S. G. Wolf, and A. Minsky. 2000. Ordered intracellular RecA-DNA assemblies: a potential site of RecA-mediated activities. Proc. Natl. Acad. Sci. USA 97:6791-6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin, J. Y., R. Qi, C. Aston, J. P. Jing, T. S. Anantharaman, B. Mishra, O. White, M. J. Daly, K. W. Minton, J. C. Venter, and D. C. Schwartz. 1999. Whole-genome shotgun optical mapping of Deinococcus radiodurans. Science 285:1558-1562. [DOI] [PubMed] [Google Scholar]
- 29.Liu, Y. Q., J. Z. Zhou, M. V. Omelchenko, A. S. Beliaev, A. Venkateswaran, J. Stair, L. Y. Wu, D. K. Thompson, D. Xu, I. B. Rogozin, E. K. Gaidamakova, M. Zhai, K. S. Makarova, E. V. Koonin, and M. J. Daly. 2003. Transcriptome dynamics of Deinococcus radiodurans recovering from ionizing radiation. Proc. Natl. Acad. Sci. USA 100:4191-4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma, C. L., and V. A. Bloomfield. 1994. Condensation of supercoiled DNA induced by MnCl2. Biophys. J. 67:1678-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Makarova, K. S., L. Aravind, Y. I. Wolf, R. L. Tatusov, K. W. Minton, E. V. Koonin, and M. J. Daly. 2001. Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiol. Mol. Biol. Rev. 65:44-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mattimore, V., and J. R. Battista. 1996. Radioresistance of Deinococcus radiodurans: functions necessary to survive ionizing radiation are also necessary to survive prolonged desiccation. J. Bacteriol. 178:633-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Minsky, A. 2003. Structural aspects of DNA repair: the role of restricted diffusion. Mol. Microbiol. 50:367-376. [DOI] [PubMed] [Google Scholar]
- 34.Minsky, A., E. Shimoni, and D. Frenkiel-Krispin. 2002. Stress, order and survival. Nat. Rev. Mol. Cell Biol. 3:50-60. [DOI] [PubMed] [Google Scholar]
- 35.Minton, K. W. 1994. DNA repair in the extremely radioresistant bacterium Deinococcus radiodurans. Mol. Microbiol. 13:9-15. [DOI] [PubMed] [Google Scholar]
- 36.Minton, K. W., and M. J. Daly. 1995. A model for repair of radiation-induced DNA double-strand breaks in the extreme radiophile Deinococcus radiodurans. Bioessays 17:457-464. [DOI] [PubMed] [Google Scholar]
- 37.Narumi, I. 2003. Unlocking radiation resistance mechanisms: still a long way to go. Trends Microbiol. 11:422-425. [DOI] [PubMed] [Google Scholar]
- 38.Nguyen, T. T., I. Rouzina, and B. I. Shklovskii. 2000. Reentrant condensation of DNA induced by multivalent counterions. J. Chem. Phys. 112:2562-2568. [Google Scholar]
- 39.Nicholson, W. L., N. Munakata, G. Horneck, H. J. Melosh, and P. Setlow. 2000. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol. Mol. Biol. Rev. 64:548-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Noguchi, H., S. Saito, S. Kidoaki, and K. Yoshikawa. 1996. Self-organized nanostructures constructed with a single polymer chain. Chem. Phys. Lett. 261:527-533. [Google Scholar]
- 41.Norris, V. 1995. Hypothesis: chromosome separation in Escherichia coli involves autocatalytic gene expression, transertion and membrane-domain formation. Mol. Microbiol. 16:1051-1057. [DOI] [PubMed] [Google Scholar]
- 42.Pelta, J., F. Livolant, and J. L. Sikorav. 1996. DNA aggregation induced by polyamines and cobalthexamine. J. Biol. Chem. 271:5656-5662. [DOI] [PubMed] [Google Scholar]
- 43.Ragkousi, K., A. E. Cowan, M. A. Ross, and P. Setlow. 2000. Analysis of nucleoid morphology during germination and outgrowth of spores of Bacillus species. J. Bacteriol. 182:5556-5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rau, D. C., and V. A. Parsegian. 1992. Direct measurement of temperature-dependent solvation forces between DNA double helices. Biophys. J. 61:260-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roca, A. I., and M. M. Cox. 1997. RecA protein: structure, function, and role in recombinational DNA repair. Prog. Nucleic Acid Res. Mol. Biol. 56:129-223. [DOI] [PubMed] [Google Scholar]
- 46.Satoh, K., I. Narumi, M. Kikuchi, S. Kitayama, T. Yanagisawa, K. Yamamoto, and H. Watanabe. 2002. Characterization of RecA424 and RecA670 proteins from Deinococcus radiodurans. J. Biochem. 131:121-129. [DOI] [PubMed] [Google Scholar]
- 47.Setlow, P. 1995. Mechanisms for the prevention of damage to DNA in spores of Bacillus species. Annu. Rev. Microbiol. 49:29-54. [DOI] [PubMed] [Google Scholar]
- 48.Weller, G. R., and A. J. Doherty. 2001. A family of DNA repair ligases in bacteria? FEBS Lett. 505:340-342. [DOI] [PubMed] [Google Scholar]
- 49.Weller, G. R., B. Kysela, R. Roy, L. M. Tonkin, E. Scanlan, M. Della, S. K. Devine, J. P. Day, A. Wilkinson, F. D. di Fagagna, K. M. Devine, R. P. Bowater, P. A. Jeggo, S. P. Jackson, and A. J. Doherty. 2002. Identification of a DNA nonhomologous end-joining complex in bacteria. Science 297:1686-1689. [DOI] [PubMed] [Google Scholar]
- 50.White, O., et. al. 1999. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 286:1571-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zimmerman, S. B. 1993. Macromolecular crowding effects on macromolecular interactions: some implications for genome structure and function. Biochim. Biophys. Acta 1216:175-185. [DOI] [PubMed] [Google Scholar]