Abstract
The production of the Sinorhizobium meliloti exopolysaccharide, succinoglycan, is required for the formation of infection threads inside root hairs, a critical step during the nodulation of alfalfa (Medicago sativa) by S. meliloti. Two bacterial mutations, exoR95::Tn5 and exoS96::Tn5, resulted in the overproduction of succinoglycan and a reduction in symbiosis. Systematic analyses of the symbiotic phenotypes of the two mutants demonstrated their reduced efficiency of root hair colonization. In addition, both the exoR95 and exoS96 mutations caused a marked reduction in the biosynthesis of flagella and consequent loss of ability of the cells to swarm and swim. Succinoglycan overproduction did not appear to be the cause of the suppression of flagellum biosynthesis. Further analysis indicated that both the exoR95 and exoS96 mutations affected the expression of the flagellum biosynthesis genes. These findings suggest that both the ExoR protein and the ExoS/ChvI two-component regulatory system are involved in the regulation of both succinoglycan and flagellum biosynthesis. These findings provide new avenues of understanding of the physiological changes S. meliloti cells go through during the early stages of symbiosis and of the signal transduction pathways that mediate such changes.
Sinorhizobium meliloti and its legume host, alfalfa (Medicago sativa), establish an effective nitrogen-fixing symbiosis through a series of signal exchanges that starts with the exchange of Nod (nodulation) factors and flavonoids, which results in the formation of curled alfalfa root hairs that are colonized by S. meliloti cells (13, 20, 32). The colonized curled root hairs develop infection threads within the root hairs, which allow S. meliloti cells to invade the developing root nodules (14, 16, 31). A successful invasion of nodules by S. meliloti will result in the formation of pink nitrogen-fixing nodules. The pink color is due to the presence of leghemoglobin. Nodules that are not occupied by S. meliloti and/or not capable of fixing nitrogen are most often white due to the lack of leghemoglobin (20).
The formation of infection threads inside root hairs requires the presence of an S. meliloti exopolysaccharide, succinoglycan (9), in addition to the Nod factor (33). Succinoglycan is a polymer that consists of different numbers of a repeating unit consisting of one galactose and seven glucoses with three modification groups: acetyl, pyruvyl, and succinyl (17, 24). All three modifications must be present in order for the S. meliloti succinoglycan to be active in eliciting infection thread formation (9). Surprisingly, overproduction of succinoglycan appears to reduce efficiency of nodulation (12).
Two S. meliloti mutants, exoR95::Tn5 and exoS96::Tn5, were isolated based on their ability to overproduce succinoglycan (12). The exoR gene encodes a protein of 268 amino acids that shares no significant homology with any other protein in currently available databases (23). The exoS gene encodes the membrane-bound sensor of the ExoS/ChvI two-component regulatory system, and the exoS96 mutation might have resulted in the formation of a constitutively active version of the sensor (10). Two close homologs of the exoS gene, Agrobacterium tumefaciens chvG (8) and Brucella abortus bvrS (29), are essential to the pathogenicity of their respective hosts (8, 29). The A. tumefaciens chvG gene is involved in regulating acid-inducible genes (18), and B. abortus bvrS is involved in regulating membrane protein expression (15). Both the exoR95 and exoS96 mutations resulted in the upregulation of the 22 succinoglycan biosynthesis genes, with the exoYFQ operon as the primary target of the regulation (23). The exoY gene encodes a galactose transferase that carries out the first step of succinoglycan biosynthesis, and the exoY210::Tn5 mutation completely blocks succinoglycan biosynthesis (17, 25).
The exoR95 mutation resulted in a reduction of nodulation efficiency so that some of the plants inoculated with the exoR95 mutants were tall and green with pink nodules, while the others were short and yellow with white nodules (12). The exoR95 mutant also appeared to have a reduced efficiency in colonizing curled root hairs (9). The exoS96 mutation did not change the nodulation efficiency significantly, even though it appeared to cause a reduction in the efficiency in colonizing curled root hairs (9).
The exoR95 and exoS96 mutants have been linked to the reduction of cellular motility, based on the findings that the two mutants formed smaller colonies on swarming plates (35). Two S. meliloti mutations have been isolated that increase motility and the sizes of colonies on swarming plates and reduce succinoglycan production (35). These findings raised the possibility that the regulation of succinoglycan production and cellular motility could be coupled. The coupling of the regulation of bacterial exopolysaccharide production and the regulation of cell motility has been found in Ralstonia solanacearum (5), Vibrio cholerae (1, 34), and Salmonella enterica (7), which presumably increases the ability of the cells to interact with their prospective host cells.
S. meliloti cellular motility is supported by two to eight peritrichous semirigid flagella which allow the bacterium to move effectively in viscous liquid (6). These flagella rotate in one direction at different speeds, which allows the bacterial cells to alter direction (27). The S. meliloti cellular motility requires the function of the chemotaxis, flagellum, and motility genes, which are tightly regulated in a hierarchical order from class I to class III (30). Class I genes comprise the visN and visR genes (30). Class II genes, which comprise flagellar assembly and motor genes, are controlled by the class I genes (30). Class III genes include flagellin and chemotaxis genes (30). Any loss or change of gene function in the hierarchy could result in the loss of cellular motility, which would reduce nodulation efficiency (2-4, 6).
To further understand the roles of the ExoR protein and the ExoS/ChvI two-component signal transduction system in the S. meliloti-alfalfa symbiosis, systematic analysis of the symbiotic properties and cell motility of both mutants was conducted. Global gene expression profiles were analyzed by using oligonucleotide microarrays and confirmed by reverse transcription-PCR (RT-PCR). These results link the regulation of succinoglycan biosynthesis to the expression of the flagellum biosynthesis genes.
MATERIALS AND METHODS
Strains and bacterial media.
The S. meliloti strains used in this study are shown in Table 1. S. meliloti Rm1021 (Strr) was used as the wild-type strain (17), and MT616(pRK600) (Cmr) was used as a helper for conjugation (17). The S. meliloti phage used for general transduction was φM12 (19). The S. meliloti succinoglycan-deficient exoY210::Tn5 (Neor) mutant was used as a control (17). Two S. meliloti succinoglycan overproduction mutants, exoR95::Tn5 (Neor) and exoS96::Tn5 (Neor), were used to study the function of the exoR and exoS genes. To facilitate the construction of double mutants, the transposon Tn5 in the exoR95::Tn5 and exoS96::Tn5 mutants was replaced with the transposon Tn5-233 to generate exoR395::Tn5-233 (Gmr) and exoS396::Tn5-233 (Gmr) mutants, so that the exoR395::Tn5-233 and exoS396::Tn5-233 mutants were the same as the exoR95::Tn5 and exoS96::Tn5 mutants with the exception of the antibiotic markers (12). The exoR95 and exoR395 mutants have been used interchangeably in some of the experiments described in this paper. The same is true for the exoS96 and exoS396 mutants.
TABLE 1.
S. meliloti strains used in this study
| Strain | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Rm1021 | Wild type | 17 |
| Rm7210 | exoY210::Tn5 | 17 |
| Rm7095 | exoR95::Tn5 | 12 |
| Rm7096 | exoS96::Tn5 | 12 |
| Rm8395 | exoR395::Tn5-233 | G. C. Walker |
| Rm8396 | exoS396::Tn5-233 | G. C. Walker |
| RmHC1 | exoY210::Tn5 exoR395::Tn5-233 | This work |
| RmHC2 | exoY210::Tn5 exoR395::Tn5-233 | This work |
Luria-Bertani (LB) medium was used for the growth of Escherichia coli strains, and LB supplemented with 2.5 mM MgSO4 and 2.5 mM CaCl2 (LB/MC) was used for all S. meliloti strains (17). TY medium was used for growth of S. meliloti strains for microarray analysis. Agar (1.5%) was added to make solid medium. Antibiotics were used at the indicated concentrations: chloramphenicol (Cm), 10 μg/ml; gentamicin (Gm), 15 μg/ml; kanamycin, 25 μg/ml; neomycin (Neo), 200 μg/ml; streptomycin (Str), 500 μg/ml.
Nodulation efficiency.
To characterize the functions of ExoR and ExoS/ChvI proteins, exoR95 and exoS96 mutants and other strains were analyzed for their ability to establish nitrogen-fixing symbiosis with alfalfa by determining their nodulation efficiency, which was measured by the color and the number of nodules on roots of alfalfa plants inoculated with these strains. The symbiotic ability of the bacterial strains can also be measured by their ability to colonize curled alfalfa root hairs (root hair colonization efficiency), to invade alfalfa roots through root hairs (root hair invasion efficiency), and to invade root nodules (nodule invasion efficiency).
Nodulation efficiency assays were carried out as previously described (17). Alfalfa (M. sativa cv. Iroquois) seeds were surface sterilized and germinated in the dark. The bacterial cells (0.1 ml) were plated on the surface of solid plant growth medium, Jensen's agar (22), inside a square petri dish. For each bacterial strain, 0.1 ml of cell suspension was placed on top of solid plant growth medium, Jensen's agar. Ten 2-day-old alfalfa seedlings were spread out evenly across the plate. A stack of 10 plates was wrapped with aluminum foil and placed in a plant growth chamber on one side of the petri dishes so that the shoots of the seedlings were pointing up. To determine the effect of the size of the inoculum on nodulation efficiency, plants were inoculated with decreasing amounts of inoculum. One milliliter of cell suspensions at three different cell concentrations, 107, 105, and 103 cells/ml, was used to inoculate a set of 10 plants in one square petri dish. Ten plants were used for each strain at each cell concentration to minimize the influence of the individual alfalfa plant on the results. The alfalfa plants were checked for plant height, the numbers of nodules, and the percentage of nodules that were pink each week. The growth of the plants was documented using a Kodak digital camera.
The nodule invasion efficiency was measured as the percentage of the nodules that were pink. The formation of pink nodules is the result of bacterial colonization of the plant cells inside the nodule, which elicits the production of leghemoglobin (11).
Root hair invasion efficiency assays were carried out as previously described (9). Briefly, alfalfa seedlings growing on microscope slides covered with a layer of solid plant growth medium were inoculated with bacterial cell suspension and covered with a single layer of dialysis membrane. These slide assemblies were placed inside a 50-ml culture tube with beveled bottom. Culture tubes were filled with liquid Jensen's medium to cover the lower part of the slide to support growth. Plants were examined daily using an Olympus AX70TRF fluorescence microscope to score the numbers of colonized curled root hairs, root hairs with initiated infection threads, and extended infection threads. The combined number of all three events represents the total number of events of colonization of curled root hairs, because initiated infection threads and extended infection threads are the result of colonization of curled root hairs. The percentage of colonization of curled root hairs that further developed into extended root hairs represents the root hair invasion efficiency.
Cell motility and flagellum staining.
Cell motility was examined using swarming plates and phase-contrast microscopy as described previously (35). Briefly, bacterial strains were inoculated onto LB/MC soft agar medium and incubated for 4 days to determine colony size. Bacterial cells from mid-log phase were mixed with an equal volume of 0.4% high-viscosity carboxymethyl cellulose and observed under a phase-contrast microscope to determine their motility.
Bacterial flagella were stained using a method similar to that used in prior work (21). Slides were rinsed with 95% ethyl alcohol, dried with lintless paper tissue, and passed through a flame. A loopful of bacterial cell suspension was transferred to one end of the clean slide, and the slide was tilted to allow the drop of cell suspension to run to the other end. Cell suspensions were air dried. The slide was flooded with the flagellum mordant for 10 min, rinsed gently with distilled water, and flooded with Ziehl's carbol fuchsin for 5 min. The slide was rinsed off gently and air dried. Bacterial flagella were examined and photographed under a phase-contrast microscope.
Whole-genome analysis of gene expression. (i) Printing and layout of Sm6kOligo microarrays.
Each array contained 6.223 70-mer oligonucleotides and three PCR fragments printed in three replicates, except for a set of 12 control genes (sma1118, smb21183, smb21295, smc00323, smc00335, smc00363, smc00646, smc01106, smc02857, smc03859, smc03979, and smc04040), which were printed in 51 replicates. The 70-mer oligonucleotide set was designed and synthesized by QIAGEN (Hildesheim, Germany) based on the three NCBI refseqs: NC_003047 (chromosome), with the database file NC_003047.ffn; NC_003037 (pSymA), with the database file NC_003037.ffn; and NC_003078 (pSymB), with the database file NC_003078.ffn (updated in March 2002; ftp://ftp.ncbi.nih.gov/genomes/Bacteria/Sinorhizobium_meliloti/). As further controls, three alien DNA PCR fragments (spot report alien PCR product 1, Stratagene 252551; spot report alien PCR product 2, Stratagene 252552; spot report alien PCR product 3, Stratagene 252553 [Stratagene, La Jolla, Calif.]), four 70-mer oligonucleotides directed against transgenes (gusA, lacZ, nptII, and aacC1), and two 70-mer stringency control oligonucleotides directed against smc02725 and smc03990 (80% identity) were spotted in three replicates. As negative controls, 12 alien 70-mer oligonucleotides were spotted in 48 replicates.
PCR fragments (200 ng/μl) and oligonucleotides (40 μM) in 1.5 M betaine, 3× SSC (1× SSC is 0.15 M sodium chloride plus 0.015 M sodium citrate) (11a) were printed onto Creative Chip 3D slides (Eppendorf, Hamburg, Germany) using the MicroGrid II 610 spotter (BioRobotics, Cambridge, United Kingdom) equipped with 48 SMP3 stealth pins (TeleChem International, Sunnyvale, Calif.). DNA was cross-linked to the surface by incubation of the slides for 1 h at 80°C. Processing of the slides included the following washes: 0.1% Triton X-100 for 5 min at 20°C; 0.05% HCl, two times for 2 min at 20°C; 0.1 M KCl for 10 min at 20°C; H2O for 1 min at 20°C; 0.05% HCl-25% ethylene glycol for 15 min at 50°C; and H2O for 1 min at 20°C. Slides were dried by centrifugation (3 min, 185 × g, 20°C).
(ii) RNA purification and synthesis of labeled cDNA.
RNA was isolated as described previously (31). Cy3- and Cy5-labeled cDNA was prepared according to the method of deRisi et al. (12) from 10 to 15 μg of total RNA (http://www.microarrays.org/protocols.html). For each microarray experiment, five slide hybridizations were performed using the labeled cDNA synthesized from three independent RNA preparations obtained from three independent bacterial cultures.
Hybridizations, image acquisition, and data analysis.
For each comparison, hybridizations accounting for at least two biological and up to two technical replicates were conducted. Hybridizations, image acquisition, and data analysis were performed as described previously (31). Shortly, mean signal and mean local background intensities were obtained for each spot of the microarray images using the ImaGene 5.0 software for spot detection, image segmentation, and signal quantification (Biodiscovery Inc., Los Angeles, Calif.). The log2 value of the ratio of intensities was calculated for each spot by using the formula Mi = log2(Ri/Gi). Ri = Ich1i − Bgch1i, and Gi = Ich2i − Bgch2i, where Ich1i or Ich2i is the intensity of a spot in channel 1 or channel 2 and Bgch1i or Bgch2i is the background intensity of a spot in channel 1 or channel 2, respectively. A normalization method based on local regression that accounts for intensity and spatial dependence in dye biases was applied (37). Normalization and t-statistics were carried out using the EMMA 1.1 microarray data analysis software developed at the Bioinformatics Resource Facility (Center of Biotechnology, Bielefeld University; www.genetik.uni-bielefeld.de/EMMA/) (14). Genes were regarded as differentially expressed if P was ≤0.05 and M was ≥1.00 or ≤(−1.00) (at least a twofold difference).
Detection of gene expression using RT-PCR.
Total RNA was collected from the cells of S. meliloti Rm1021 and exoR95 and exoS96 mutants by using an RNA extraction kit (RNAex reagent; Huashun Biotechnology Co., Ltd., Shanghai). Briefly, bacterial cells were collected, mixed with RNAex reagent, and then extracted with chloroform. Total RNA was precipitated with isopropanol, washed with ethanol, dried, resuspended, and stored at −70°C. The detection of gene expression was carried out using an RT-PCR kit (TaKaRa Biotechnology, Dalian, China) following the instructions from the manufacturer. The set of primers for targeted genes (visR, visN, and flaA) and a set of primers for the control gene (rpsF) were used in the same RT-PCR. The RT-PCR products were resolved on agarose gel to determine whether a targeted gene was expressed.
RESULTS
Succinoglycan overproduction reduces nodulation efficiency of the exoR95 mutant but not of the exoS96 mutant.
To systematically and quantitatively determine the nodulation efficiency of exoR95 and exoS96 mutants, alfalfa plants growing inside petri dishes were inoculated with one of the two mutants or the wild-type strain Rm1021 as control. To better characterize the difference in nodulation efficiency, plants were inoculated with decreasing amounts of bacterial cells.
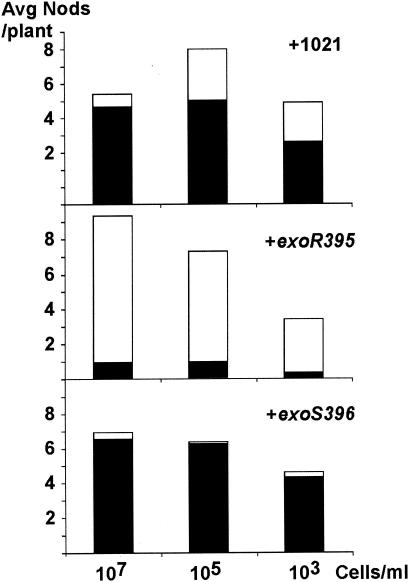
At the end of the fifth week, alfalfa plants that were not inoculated with any S. meliloti cells had already turned yellow and had started dying from lack of nitrogen. The alfalfa plants inoculated with all three concentrations, 107, 105, and 103 cells/ml, of the wild-type strain Rm1021 were tall and green (Fig. 1). The efficiency of the nodule invasion by the wild-type cells decreased 30% when the concentration of cells in the inoculum decreased to 103 cells/ml.
FIG. 1.
Nodulation efficiencies of the wild-type strain and the exoR95 and exoS96 mutants. The average numbers of nodules per plant on alfalfa plants inoculated with Rm1021 and the exoR395 and exoS396 mutants were plotted based on the size of inoculum. The numbers of nodules were further divided into pink nodules (black bars) and white nodules (white bars).
The plants inoculated with the exoR95 mutant were mostly yellow with a few light green leaves. Some of the plants were slightly greener than the others, which corresponded to a higher percentage of nodules that were pink on those plants. The nodule invasion efficiency was dramatically lower for the exoR95 mutant than for wild-type strain Rm1021, which suggested that the exoR95 mutant was unable to invade the nodules that it elicited on the alfalfa plants (Fig. 1).
The plants similarly inoculated with the exoS96 mutant were mostly green and tall, like those inoculated with the wild-type cells. The nodule invasion efficiency of the exoS96 mutant was not reduced but was higher than that of the wild-type cells (Fig. 1). The average number of pink nodules per plant was about the same for the plants inoculated with the wild-type cells or the exoS96 mutant, which is consistent with the notion that alfalfa plants can regulate the number of pink nodules based on their needs (11).
These results suggest that even though both the exoR95 and exoS96 mutations result in the overproduction of succinoglycan, only the exoR95 mutation causes a reduction in nodule invasion efficiency and, subsequently, a reduction in nodulation efficiency. The exoS96 mutation does not significantly alter the nodule invasion efficiency or nodulation efficiency.
Root hair invasion efficiency of the exoR95 and exoS96 mutants.
Root hair invasion is the key step in nodulation, and it can be quantitatively measured using fluorescently labeled S. meliloti cells as described in detail in Materials and Methods. Succinoglycan plays an important role in the initiation of infection thread formation during root hair invasion, and succinoglycan overproduction appears to interfere with root hair invasion (9). A large systematic analysis of the impact of succinoglycan overproduction on root hair invasion was needed to understand the role of the exoR and exoS/ChvI genes in symbiosis.
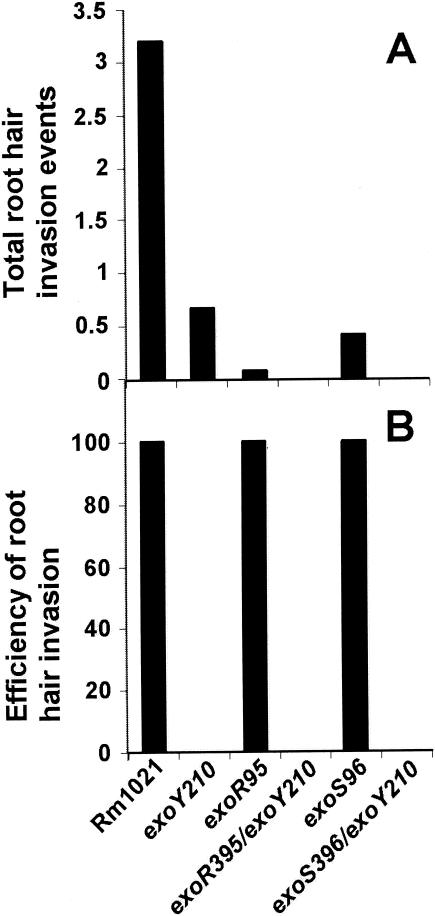
Root hair invasion can be divided into three stages: colonization of curled root hairs, initiation of infection thread formation, and extension of infection threads to the base of the root hairs. Root hair invasion can be blocked at the initiation of infection thread formation or the development of infection threads before they reach the base of the root hairs. On 12 alfalfa plants inoculated with wild-type strain Rm1021, the average number of total root hair invasion events per plant was 3.3 (Fig. 2A), and they were all in the form of long extended infection threads inside root hairs. This suggested that every root hair that was colonized by the bacterial cells developed long and extended infection threads. Root hair invasion by the wild-type strain was 100% efficient. On the 12 alfalfa plants inoculated with the exoY210 mutant, the average number of total root hair invasion events per plant was 0.67. None of them developed into infection threads, and so the root hair invasion efficiency of this mutant was 0% (Fig. 2B).
FIG. 2.
Root hair invasion efficiencies of the wild type and its succinoglycan-overproducing mutants. (A) Average numbers of total invasion events per plant on plants inoculated with wild-type strain Rm1021 and exoY210, exoR95, exoR395exoY210, exoS96, and exoS396exoY210 mutants. (B) Overall efficiencies of root hair invasion of the same set of strains.
On the 12 alfalfa plants inoculated with the exoR95 mutant, the average number of root hair invasion events per plant was 0.08 (Fig. 2A). Only one root hair containing an extended infection thread was found. No other forms of root hair invasion events were found, so that the one single curled root hair that was colonized by the exoR95 mutant developed an infection thread inside the colonized root hair. Similar results have been found in different sets of experiments (data not shown), suggesting that the root hair invasion by the exoR95 mutant was extremely efficient (Fig. 2B), but the number of root hairs that were colonized by the exoR95 mutant dropped dramatically compared to that of the wild-type strain. These findings suggest that nodulation of alfalfa by the exoR95 mutant might be blocked before or at the step of colonizing curled alfalfa root hairs.
On the 12 alfalfa plants inoculated with the exoS96 mutant, the average number of root hair invasion events per plant was 0.42 (Fig. 2A), which represented five root hairs with extended infection threads. No colonized root hairs or initiated infection threads were found, so that all of the curled colonized root hairs developed infection threads. The root hair invasion by the exoS96 mutant can be considered 100% efficient (Fig. 2B), but similar to that of the exoR95 mutant, the number of total root hair invasion events was dramatically lower for the exoS96 mutant. These findings suggest that nodulation of alfalfa by the exoS96 mutant is most likely blocked before or at the step of colonizing curled alfalfa root hairs.
If the reduction of colonization of the curled root hairs were the result of succinoglycan overproduction, blocking succinoglycan should allow these two mutants to become as efficient as the exoY210 mutant in colonizing curled root hairs. To test this hypothesis, the exoR395exoY210 and exoS396exoY210 double mutants were constructed by transducing the exoY210::Tn5 mutation into the exoR395::Tn5-233 or exoS396::Tn5-233 mutants. Both double mutants, exoR395exoY210 and exoS396exoY210, were even less efficient in colonizing curled root hairs. On the sets of 12 alfalfa plants inoculated with either one of the two double mutants, no curled root hair was found to be colonized. These findings raise the possibility that overproduction of succinoglycan might not be the only factor contributing to decreased efficiency in colonizing curled alfalfa root hairs.
Effects of the exoR95 and exoS96 mutations on cell motility.
The loss of cell motility has been shown to reduce the nodulation efficiency of S. meliloti (3), which could account for the lower nodulation efficiency of the exoR95 and exoS96 mutants. These findings and previous reports of the reduced cellular motility by the exoR95 and exoS96 mutations (35) made it essential to test the motility of these two mutants.
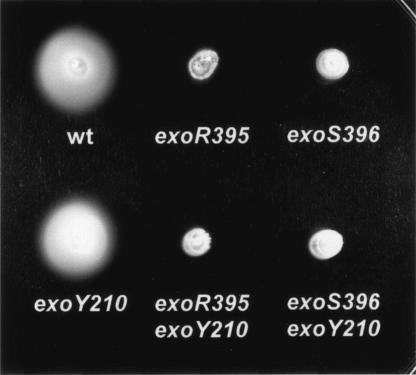
To examine the link between cellular motility and succinoglycan production, both wild-type strain Rm1021 and its succinoglycan-deficient mutant, exoY210, were tested on swarming plates as controls. Both Rm1021and the exoY210 mutant formed large diffuse colonies (Fig. 3), suggesting that blocking succinoglycan production does not affect cellular motility. When the exoR95 and exoS96 mutants were similarly examined on swarming plates, they formed smaller and smooth colonies, suggesting that both the exoR95 and exoS96 mutations decreased cellular motility. If overproduction of succinoglycan was the reason for the decrease in motility, blocking succinoglycan production by these two mutants should restore their cellular motility. Two double mutants, exoR395exoY210 and exoS396exoY210, were similarly tested on swarming plates, and they both formed small tightly packed colonies that were similar to those formed by single exoR95 and exoS96 mutants. These findings suggested that the suppression of cellular motility by the exoR95 and exoS96 mutation was not just the result of overproduction of succinoglycan.
FIG. 3.
The exoR395 and exoS396 mutants were nonmotile on swarming plates. Colonies formed by wild-type strain Rm1021 and exoY210, exoR395, exoS396, exoR395exoY210, and exoS396exoY210 mutants are shown.
To directly confirm that decreased cellular motility was related to the exoR95 and exoS96 mutations, bacterial cells of wild-type strain Rm1021 and the exoR95, exoS96, exoY210, exoR395exoY210, and exoS396exoY210 mutants were observed directly under a phase-contrast microscope. Bacterial cells of Rm1021 and the exoY210 mutant were motile, while the exoR95, exoS96, exoR395exoY210, and exoS396exoY210 mutant cells were nonmotile. These results were consistent with the findings of the swarming plate experiments and again suggested the overproduction of succinoglycan was not responsible for the decreased motility of the exoR95 and exoS96 mutants.
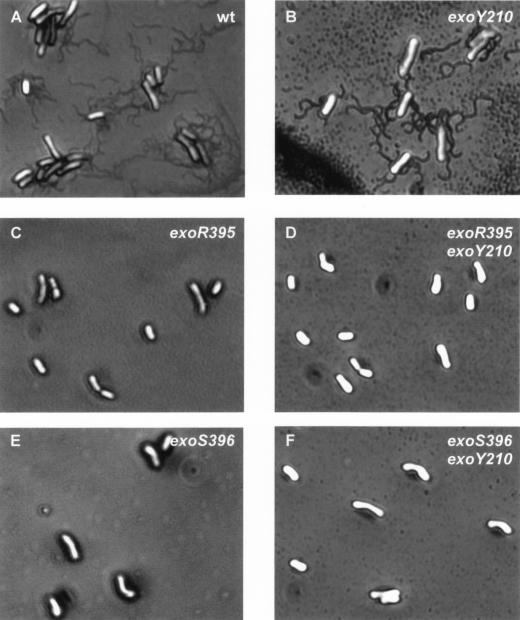
The exoR95 and exoS96 mutations result in the loss of flagella.
The production of flagella and the speed of flagellum rotation are highly regulated in S. meliloti (30). The loss of cell motility could be the result of the loss of flagella or interference in flagellum rotation. To determine whether the exoR95 and exoS96 mutations resulted in the loss of flagella, cells of the wild-type strain and the exoR95, exoS96, exoY210, exoR395exoY210, and exoS396exoY210 mutants were stained for flagella and examined under a phase-contrast microscope. The cells of wild-type Rm1021 and the exoY210 mutant possessed flagella (Fig. 4). The flagella of Rm1021 cells were examined in detail by using transmission electron microscopy, and the findings were in agreement with previous reports (data not shown). The cells of the exoR95, exoS96, exoR395exoY210, and exoS396exoY210 mutants did not have flagella. The fact that the exoY210 mutant produced flagella and that neither the exoR395exoY210 nor exoS396exoY210 double mutants produced flagella suggested that succinoglycan overproduction was not responsible for blocking flagellum biosynthesis. These findings also raised the possibility that ExoR and ExoS may play some roles in regulating flagellum biosynthesis in addition to regulating succinoglycan biosynthesis.
FIG. 4.
The exoR95 and exoS96 mutations blocked flagellum biosynthesis. The wild-type strain and the succinoglycan-deficient mutant exoY210 produced peritrichous flagella. The exoR95, exoS96, exoR395exoY210, and exoS396exoY210 mutants had no flagella.
The exoR and exoS96 mutations suppress the expression of the flagellin genes.
To confirm that the expression of S. meliloti flagellin biosynthesis genes is regulated by the ExoR protein or the ExoS/ChvI two-component regulatory system, the expression of all S. meliloti genes in the exoR95 and exoS96 mutant backgrounds was analyzed. Total RNA was isolated from free-living exoR95 and exoS96 mutants as well as from the wild-type strain, Rm1021. The total RNA was reverse transcribed into fluorescently labeled cDNA and hybridized to Sm6kOligo microarrays containing probes for all predicted protein-coding genes of S. meliloti Rm1021. The exoR95 mutation resulted in downregulation of the expression of 160 genes and putative open reading frames and in upregulation of 136 genes and putative open reading frames in the range of two- to eightfold. The exoS96 mutation caused downregulation of 129 genes and putative open reading frames and upregulation of 131 genes and putative open reading frames in the range of two- to eightfold. Both exoR95 and exoS96 mutations downregulated the expression of all five S. meliloti flagellum biosynthesis and regulatory genes 2.7- to 5.5-fold (Table 2). The expression of the fla genes was downregulated further in the exoR95 mutant background.
TABLE 2.
Changes in expression levels of S. meliloti flagellum genes resulting from the exoR95 and exoS96 mutations, as determined in microarray hybridizations
| S. meliloti flagellum gene | Function | Change of gene expression in specific genetic backgrounda
|
|
|---|---|---|---|
| exoR95 mutant | exoS96 mutant | ||
| flaA (smc03037)b | Flagellin subunit | −4.3c | −3.3 |
| flaB (smc03038) | Flagellin subunit | −5.5 | −4.1 |
| flaC (smc03040) | Flagellin subunit | −3.0 | −2.7 |
| flaD (smc03039) | Flagellin subunit | −4.6 | −3.9 |
| flaF (smc03050) | Regulator | −3.3 | −3.8 |
The levels of the changes in gene expression were determined by comparing a particular gene in the mutant background versus expression in the wild-type background.
The open reading frame smc03037 stands for the number 03037 open reading frame on the chromosome of S. meliloti.
The value −4.3, for example, indicates that the expression level of the flaA gene was decreased 4.3-fold in the exoR95 mutant compared to that in the wild-type strain.
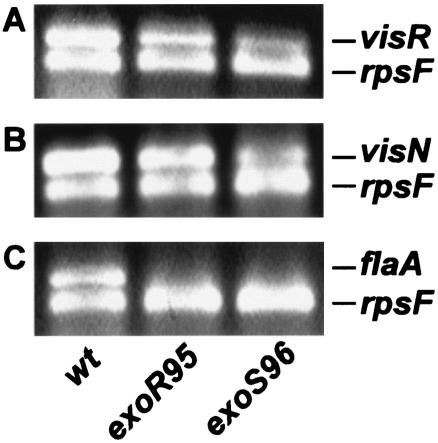
The effects of the exoR395 and exoS396 mutations, which were the same as the effects of the exoR95 and exoS96 mutations, on the expression of the flagellin gene were further analyzed using RT-PCR (Fig. 5). Expression of the rpsF gene, which encodes the S6 protein for the 30S ribosome subunit, was used as an internal control. Expression of the rpsF gene was detected in the wild type and the exoR395 and exoS396 mutants. Expression of the flaA gene, which encodes a flagellin subunit, was detected in the wild type but not in either of the mutants. Expression of the visN and visR genes, which serve as the primary regulators of flagellum biosynthesis, was detected in the wild type and the exoR395 and exoS396 mutants.
FIG. 5.
The exoR95 and exoS96 mutations suppressed expression of the flaA gene. The primers for both the rpsF and visR genes were mixed with total RNA from the wild type or the exoR95 and exoS96 mutants for RT-PCR. (A) The products of the RT-PCRs were resolved on an agarose gel and used to examine the expression of visR and rpsF. (B and C) Expression of the visN and rpsF genes (B) and of the flaA and rpsF genes (C) was similarly examined.
Together, these findings suggest that both ExoR and ExoS govern flagellum synthesis by regulating the expression of the fla genes, but they are not involved in regulating the expression the visN and visR genes, which play key roles in regulating S. meliloti flagellum biosynthesis.
DISCUSSION
The presence of succinoglycan is essential to successful root hair invasion and nodulation of alfalfa by S. meliloti (9, 17), and so it was puzzling that succinoglycan overproduction appeared to change nodulation efficiency of the exoR95 mutant but not of the exoS96 mutant (12). This suggests that there might be other factors related to the overproduction of succinoglycan. Since both the ExoR and ExoS proteins are involved in signal transduction, both of them could be involved in other symbiotic processes in addition to regulating succinoglycan production.
The systematic analysis of nodulation efficiency of the mutants showed that the exoR95 mutant was less efficient in nodule invasion than its wild-type parent, while the exoS96 mutant was as efficient as its wild-type parent. To determine at which point the exoR95 mutant becomes less efficient in nodule invasion, the efficiencies of colonizing curled root hair and root hair invasion were compared for the exoR95 and exoS96 mutants and other strains.
The efficiencies of root hair invasions of the two mutants were not affected, but the efficiencies of colonizing curled root hairs by these mutants were reduced dramatically. This lowered efficiency of colonizing curled root hairs could be compensated by a longer time of interaction between the mutants and alfalfa, which could explain why the drop in the efficiency of colonizing root hairs showed little impact on nodulation efficiency by the exoS96 mutant. These findings and the report of the exoR95 and exoS96 mutants showing reduced motility on swarming plates (35), as well as another report of the isolation of an exopolysaccharide-overproducing mutant with decreased motility (26), brought the cellular motility of these two mutants into focus.
Our analysis showed that neither the exoR95 nor exoS96 mutants were able to swarm or swim. When stained directly for flagella, neither exoR95 nor exoS96 mutants showed flagella, while the wild-type cells were flagellated. This reduction in flagellum production could be the result of energy stress on the cells or direct involvement of the ExoR and ExoS proteins in regulating flagellum biosynthesis. Further analyses showed that blocking succinoglycan production in the exoR395 and exoS396 mutant backgrounds did not restore flagellum production. These findings are consistent with the possibility that the ExoR protein and ExoS/ChvI two-component regulatory system are involved in the regulation of flagellum biosynthesis directly or indirectly.
To examine the possible involvement of the ExoR protein and the ExoS/ChvI two-component regulatory system in regulating expression of the flagellum biosynthesis genes, expression of the entire genome of the exoR95 and exoS96 mutants was examined using DNA microarray analysis technology. While the expression of numerous genes was changed by either the exoR95 mutation or the exoS96 mutation (the complete set of data will be published in the future), the expression of all five flagellum biosynthesis genes was consistently repressed by both mutations. These findings suggest that both the ExoR protein and the ExoS/ChvI two-component signal transduction system play key roles in regulating both succinoglycan production and flagellum biosynthesis, directly or indirectly.
S. meliloti fla genes are at the bottom of the chemotaxis regulatory hierarchy, and their expression is regulated by the VisR and VisN proteins at the first level and by the FliM protein and the protein encoded by open reading frame 38 at the second level (30). The regulatory mechanism(s) of the VisR and VisN proteins is not clear. It has been suggested that they might form heterodimers (30). Our findings that both exoR95 and exoS96 mutations downregulated the fla genes but not the visR and visN genes provided the first new set of links between succinoglycan and flagellum production through the ExoR protein and the ExoS/ChvI two-component regulatory system. The ExoR protein and the ExoS/ChvI system could regulate both succinoglycan and flagellum production via signal transduction pathways. Suppressor mutations have been isolated that alter the swarming and succinoglycan production phenotypes of the exoR95 mutant (data not shown), making it more likely that there are genes downstream from the ExoR protein and ExoS/ChvI system that are involved in regulating both succinoglycan and flagellum production. Alternatively, the expression of the genes regulated by the ExoR protein and ExoS/ChvI system could lead to physiological or structure changes that are sensed by the VisR and VisN proteins in regulating flagellum production. Efforts are currently under way to identify the genes that encode proteins that are downstream of ExoR or ExoS/ChvI and are involved in regulating succinoglycan and flagellum production. Other studies are being carried out to further study the clear differences in nodulation invasion efficiency between the exoR95 and exoS96 mutants.
All together, these findings suggested that flagellum biosynthesis and succinoglycan production might be coordinated at the level of gene expression. The multiple peritrichous flagella could increase the ability of S. meliloti cells to move towards alfalfa roots to bring themselves closer to the root hair surface (6), although flagella themselves might not be involved in attaching to the surfaces of root hairs (28). The flagellum production might be downregulated after the attachment. Once attached to and colonizing alfalfa roots, the expression of succinoglycan biosynthesis was induced (H.-P. Cheng, unpublished results), and succinoglycan elicits the formation of infection threads inside curled root hairs (9). Such coordinated production of flagella and succinoglycan could be necessary to ensure efficient nodulation.
The closely coordinated biosynthesis of flagella and exopolysaccharide has been found in many other bacterium-host interactions, since flagella play an important role in bacterial pathogenicity (36). The V. cholerae epsD and epsE genes appear to coordinate the biosynthesis of flagella and exopolysaccharide during biofilm formation (1, 34). The S. enterica igaA gene regulates the production of capsule polysaccharide and cell motility (7). The R. solanacearum phcA gene regulates polysaccharide production, endoglucanase activity, and motility (5). The studies of the coordinated biosynthesis of S. meliloti flagella and succinoglycan biosynthesis will contribute to the general understanding of the interactions between bacteria and their prospective hosts.
Acknowledgments
We thank Eva Schulte-Berndt for technical assistance.
This work was supported by grants from NIH (5S06GM08225) and PSC-CUNY (617320030 and 632140032), from Bundesministerium für Forschung und Technologie, Germany (national grants 0311752 and 031U213D) and the “Bioinformatik Initiative” by Deutsche Forschungsgemeinschaft (BIZ 7), and from The National High Technology (863) International Research Program (2001AA214211) and the National Key Program for Basic Research of China (2001CB108901).
REFERENCES
- 1.Ali, A., J. A. Johnson, A. A. Franco, D. J. Metzger, T. D. Connell, J. G. Morris, Jr., and S. Sozhamannan. 2000. Mutations in the extracellular protein secretion pathway genes (eps) interfere with rugose polysaccharide production in and motility of Vibrio cholerae. Infect. Immun. 68:1967-1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ames, P., and K. Bergman. 1981. Competitive advantage provided by bacterial motility in the formation of nodules by Rhizobium meliloti. J. Bacteriol. 148:728-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ames, P., S. A. Schluederberg, and K. Bergman. 1980. Behavioral mutants of Rhizobium meliloti. J. Bacteriol. 141:722-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergman, K., E. Nulty, and L. H. Su. 1991. Mutations in the two flagellin genes of Rhizobium meliloti. J. Bacteriol. 173:3716-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brumbley, S. M., and T. P. Denny. 1990. Cloning of wild-type Pseudomonas solanacearum phcA, a gene that when mutated alters expression of multiple traits that contribute to virulence. J. Bacteriol. 172:5677-5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caetano-Anolles, G., L. G. Wall, A. T. De Michell, E. M. Macchi, W. D. Bauer, and G. Favelukes. 1988. Role of motility and chemotaxis in efficiency of nodulation by Rhizobium meliloti. Plant Physiol. 86:1228-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cano, D. A., G. Dominguez-Bernal, A. Tierrez, F. G. Portillo, and J. Casadesus. 2002. Regulation of capsule synthesis and cell motility in Salmonella enterica by the essential gene igaA. Genetics 162:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Charles, T. C., and E. W. Nester. 1993. A chromosomally encoded two-component sensory transduction system is required for virulence of Agrobacterium tumefaciens. J. Bacteriol. 175:6614-6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng, H. P., and G. C. Walker. 1998. Succinoglycan is required for initiation and elongation of infection threads during nodulation of alfalfa by Rhizobium meliloti. J. Bacteriol. 180:5183-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng, H. P., and G. C. Walker. 1998. Succinoglycan production by Rhizobium meliloti is regulated through the ExoS-ChvI two-component regulatory system. J. Bacteriol. 180:20-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cullimore, J. V., R. Ranjeva, and J. J. Bono. 2001. Perception of lipo-chitooligosaccharidic Nod factors in legumes. Trends Plant Sci. 6:24-30. [DOI] [PubMed] [Google Scholar]
- 11a.Diehl, F., S. Grahlmann, M. Beier, and J. D. Hoheisel. 2001. Manufacturing DNA microarrays of high spot homogeneity and reduced background signal. Nucleic Acids Res. 29:E38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doherty, D., J. A. Leigh, J. Glazebrook, and G. C. Walker. 1988. Rhizobium meliloti mutants that overproduce the Rhizobium meliloti acidic calcofluor-binding exopolysaccharide. J. Bacteriol. 170:4249-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Downie, J. A., and S. A. Walker. 1999. Plant responses to nodulation factors. Curr. Opin. Plant Biol. 2:483-489. [DOI] [PubMed] [Google Scholar]
- 14.Gage, D. J., and W. Margolin. 2000. Hanging by a thread: invasion of legume plants by rhizobia. Curr. Opin. Microbiol. 3:613-617. [DOI] [PubMed] [Google Scholar]
- 15.Guzman-Verri, C., L. Manterola, A. Sola-Landa, A. Parra, A. Cloeckaert, J. Garin, J. P. Gorvel, I. Moriyon, E. Moreno, and I. Lopez-Goni. 2002. The two-component system BvrR/BvrS essential for Brucella abortus virulence regulates the expression of outer membrane proteins with counterparts in members of the Rhizobiaceae. Proc. Natl. Acad. Sci. USA 99:12375-12380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kijne, J. W. 1992. The Rhizobium infection process, p. 349-398. In G. Stacey, R. H. Burris, and H. J. Evans (ed.), Biological nitrogen fixation. Chapman & Hall, New York, N.Y.
- 17.Leigh, J. A., E. R. Signer, and G. C. Walker. 1985. Exopolysaccharide-deficient mutants of Rhizobium meliloti that form ineffective nodules. Proc. Natl. Acad. Sci. USA 82:6231-6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, L., Y. Jia, Q. Hou, T. C. Charles, E. W. Nester, and S. Q. Pan. 2002. A global pH sensor: Agrobacterium sensor protein ChvG regulates acid-inducible genes on its two chromosomes and Ti plasmid. Proc. Natl. Acad. Sci. USA 99:12369-12374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Long, S., J. W. Reed, J. Himawan, and G. C. Walker. 1988. Genetic analysis of a cluster of genes required for synthesis of the calcofluor-binding exopolysaccharide of Rhizobium meliloti. J. Bacteriol. 170:4239-4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Long, S. R. 1989. Rhizobium-legume nodulation: life together in the underground. Cell 56:203-214. [DOI] [PubMed] [Google Scholar]
- 21.Mayfield, C. I., and W. E. Inniss. 1977. A rapid, simple method for staining bacterial flagella. Can. J. Microbiol. 23:1311-1313. [DOI] [PubMed] [Google Scholar]
- 22.Meade, H. M., S. R. Long, G. B. Ruvkun, S. E. Brown, and F. M. Ausubel. 1982. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J. Bacteriol. 149:114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reed, J. W., J. Glazebrook, and G. C. Walker. 1991. The exoR gene of Rhizobium meliloti affects RNA levels of other exo genes but lacks homology to known transcriptional regulators. J. Bacteriol. 173:3789-3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reinhold, B. B., S. Y. Chan, T. L. Reuber, A. Marra, G. C. Walker, and V. N. Reinhold. 1994. Detailed structural characterization of succinoglycan, the major symbiotically important exopolysaccharide of Rhizobium meliloti strain Rm1021. J. Bacteriol. 176:1997-2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reuber, T. L., and G. C. Walker. 1993. Biosynthesis of succinoglycan, a symbiotically important exopolysaccharide of Rhizobium meliloti. Cell 74:269-280. [DOI] [PubMed] [Google Scholar]
- 26.Rodriguez-Navarro, D. N., A. J. Palomares, and J. Casadesus. 1991. Isolation and characterization of Rhizobium meliloti mutants affected in exopolysaccharide production. Microbiologia 7:13-22. [PubMed] [Google Scholar]
- 27.Scharf, B. 2002. Real-time imaging of fluorescent flagellar filaments of Rhizobium lupini H13-3: flagellar rotation and pH-induced polymorphic transitions. J. Bacteriol. 184:5979-5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smit, G., J. W. Kijne, and B. J. Lugtenberg. 1989. Roles of flagella, lipopolysaccharide, and a Ca2+-dependent cell surface protein in attachment of Rhizobium leguminosarum biovar viciae to pea root hair tips. J. Bacteriol. 171:569-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sola-Landa, A., J. Pizarro-Cerda, M. J. Grillo, E. Moreno, I. Moriyon, J. M. Blasco, J. P. Gorvel, and I. Lopez-Goni. 1998. A two-component regulatory system playing a critical role in plant pathogens and endosymbionts is present in Brucella abortus and controls cell invasion and virulence. Mol. Microbiol. 29:125-138. [DOI] [PubMed] [Google Scholar]
- 30.Sourjik, V., P. Muschler, B. Scharf, and R. Schmitt. 2000. VisN and VisR are global regulators of chemotaxis, flagellar, and motility genes in Sinorhizobium (Rhizobium) meliloti. J. Bacteriol. 182:782-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Timmers, A. C., M. C. Auriac, and G. Truchet. 1999. Refined analysis of early symbiotic steps of the Rhizobium-Medicago interaction in relationship with microtubular cytoskeleton rearrangements. Development 126:3617-3628. [DOI] [PubMed] [Google Scholar]
- 32.van Rhijn, P., and J. Vanderleyden. 1995. The Rhizobium-plant symbiosis. Microbiol. Rev. 59:124-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walker, S. A., and J. A. Downie. 2000. Entry of Rhizobium leguminosarum bv. viciae into root hairs requires minimal nod factor specificity, but subsequent infection thread growth requires nodO or nodE. Mol. Plant Microbe Interact. 13:754-762. [DOI] [PubMed] [Google Scholar]
- 34.Watnick, P. I., and R. Kolter. 1999. Steps in the development of a Vibrio cholerae El Tor biofilm. Mol. Microbiol. 34:586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei, X., and W. D. Bauer. 1999. Tn5-induced and spontaneous switching of Sinorhizobium meliloti to faster-swarming behavior. Appl. Environ. Microbiol. 65:1228-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson, J. W., M. J. Schurr, C. L. LeBlanc, R. Ramamurthy, K. L. Buchanan, and C. A. Nickerson. 2002. Mechanisms of bacterial pathogenicity. Postgrad. Med. J. 78:216-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Workman, C., L. J. Jensen, H. Jarmer, R. Berka, L. Gautier, H. B. Nielser, H. H. Saxild, C. Nielsen, S. Brunak, and S. Knudsen. 2002. A new non-linear normalization method for reducing variability in DNA microarray experiments. Genome Biol. 3:research0048 [DOI] [PMC free article] [PubMed]