Abstract
DNA microarrays encompassing the entire genome of Yersinia pestis were used to characterize global regulatory changes during steady-state vegetative growth occurring after shift from 26 to 37°C in the presence and absence of Ca2+. Transcriptional profiles revealed that 51, 4, and 13 respective genes and open reading frames (ORFs) on pCD, pPCP, and pMT were thermoinduced and that the majority of these genes carried by pCD were downregulated by Ca2+. In contrast, Ca2+ had little effect on chromosomal genes and ORFs, of which 235 were thermally upregulated and 274 were thermally downregulated. The primary consequence of these regulatory events is profligate catabolism of numerous metabolites available in the mammalian host.
Bubonic plague caused by Yersinia pestis is generally recognized as the most devastating acute infectious disease experienced by mankind. It is therefore of interest that this organism has evolved within the last 10,000 years from Yersinia pseudotuberculosis (1), known to cause chronic enteropathogenic disease. Despite their very close resemblance, plague bacilli have both lost central genes of intermediary metabolism retained in its predecessor and acquired unique genes by lateral transfer (7). For example, even though early studies showed that Y. pestis possesses functional Embden-Meyerhof (28) and Entner-Doudoroff (20) pathways plus a complete tricarboxylic acid (TCA) cycle (13, 27), the species-specific absence of detectable glucose 6-phosphate dehydrogenase (Zwf) prevents use of hexose via the pentose-phosphate pathway (21). Similarly, loss of aspartase (AspA) activity in Y. pestis but not Y. pseudotuberculosis prevents complete catabolism of l-glutamic acid, which undergoes conversion and excretion as l-aspartate (12). In addition, Y. pestis possesses additional species-specific mutations that cause nutritional requirements at 26°C, prevent utilization of potential metabolites, and eliminate host cell invasins and adhesins (7); these events are now characterized by genomic sequencing (11, 23). The nature of nutritional requirements at 37°C is more complex and, as noted below, dependent upon plasmid profile, the presence or absence of Ca2+, Na+, dicarboxylic amino acids, and regulatory functions addressed in this report.
Established functions unique to Y. pestis are encoded by species-specific ∼10-kb pPCP and ∼100-kb pMT. The former encodes plasminogen activator (Pla) required for tissue invasion from dermal sites infected by fleabite whereas the structural genes for anti-phagocytic capsular fraction 1 (Caf1) and murine toxin (MT), required for survival in the flea, reside on pMT (7, 25). Plague bacilli and the enteropathogenic yersiniae share a ∼70-kb plasmid (pCD in Y. pestis) encoding a type III protein secretion system (TTSS) that delivers cytotoxins termed Yops to the cytosol of professional and nonprofessional phagocytes (8) and excretes soluble LcrV (V antigen), which inhibits generation of proinflammatory cytokines by upregulating interleukin-10 (6). These functions provide the basis for the acute symptoms of plague as well as chronic afflictions caused by enteropathogenic Y. pseudotuberculosis and Yersinia enterocolitica. Expression of pCD-mediated activities are upregulated at 37°C via thermoinduction of the pCD-encoded transcriptional activator LcrF (18). Nevertheless, the organisms undergo bacteriostasis at this temperature in vitro unless either 2.5 mM Ca2+ is present (16) or Na+ and dicarboxylic amino acids are eliminated (5, 14; R. R. Brubaker, unpublished data). The addition of Ca2+ to culture media (but not removal of Na+ or dicarboxylic amino acids) downregulates LcrV, Yops, and the TTSS (7, 25; Brubaker, unpublished). Cure of pCD causes outright avirulence, emphasizing the importance of this low calcium response (LCR) in promoting disease. Little is known about the extent or nature of the regulatory cascade initiated by Ca2+ or the ability of this cation or temperature to regulate chromosomal genes.
The genome of the Y. pestis bv. Medievalis strain KIM5, used in the present study, and that of the Y. pestis bv. Orientalis strain CO92 have been sequenced (11, 23). To identify functions regulated by Ca2+ and temperature, we defined the genome-wide expression of transcripts displaying coordinate LCR-mediated regulation by using a high-density PCR fragment-based microarray containing all ∼4,500 chromosomal and plasmid genes of Y. pestis KIM5. The isogenic substrain D27 lacking the deletable ∼100-kb chromosomal Pgm sequence was used as a source of mRNA; temperature-mediated changes within this region have been previously defined in detail (25).
Bacterial growth, RNA isolation, microarray analyses, and real-time reverse transcription (RT)-PCR.
Sequences used to design the microarray containing the entire Y. pestis genome were obtained from plasmids pMT1 (accession no. AF074611), pCD1 (AF074612), pPCP1 (AL109969), and chromosome (AL590842) provided by GenBank. The ORF-specific primers were designed using the PRIMER3 program (http://www.genome.wi.mit.edu) and synthesized with a 5′-C6 amino-modification (MWG Biotech, Inc., High Point, N.C.). After PCR amplification, the purified fragments were spotted in triplicate on SuperAldehyde substrate by TeleChem International, Inc., Sunnyvale, California. The exact composition of the microarray primers, amplified fragments, and other supplemental information noted in the text is provided at http://bbrp.llnl.gov/microbial/Ypestis. Chemically defined BCS medium (14) was used to cultivate conditionally virulent (nonpigmented) Y. pestis KIM5 substrain D27 for two transfers at 26°C without added Ca2+. The organisms were then inoculated at an optical density of 0.25 (620 nm) into parallel subcultures incubated without added Ca2+ at 26 and 37°C or at 37°C with 4.0 mM Ca2+; all three of these environments permits essentially full-scale growth (14). RNA samples were isolated at different time points after the temperature shift; samples containing about 2 × 109 cells were immediately mixed with an equal volume of cold RNA stabilization solution RNAlater (Ambion, Austin, Tex.) and harvested by centrifugation. Further RNA purification was performed using a RNeasy Midi kit (QIAGEN, Valencia, Calif.) followed by DNase I treatment on RNeasy Mini spin columns (QIAGEN). Total RNA concentration was determined using a RiboGreen RNA quantitation kit (Molecular Probes, Inc., Eugene, Oreg.); 20 μg of RNA was labeled with either Alexa Fluor 488 or Alexa Fluor 546 using ARES DNA labeling kits (Molecular Probes, Inc.). The labeled cDNAs were purified using a QIAquick PCR purification kit (QIAGEN), dried in a SpeedVac concentrator (ThermoSavant, Holbrook, N.Y.), and resuspended in 90 μl of formamide containing hybridization buffer (Schleicher & Schuell, Inc., Keene, N.H.). Hybridizations were performed in a Hybri-Well chamber, 21 by 41 by 0.15 mm (Sigma, St. Louis, Mo.), for 12 h at 42°C, slides were washed with microarray wash buffers (TeleChem International, Inc.), and images were obtained with a ScanArray Lite 4000 confocal laser scanner (Packard BioScience, Billerica, Mass.). RNA preparations isolated at each time point from cultures grown under three distinct conditions were labeled separately with both red and green fluorescent dyes and hybridized with each other. The image for each array was captured using the software package Gleams v. 3.0 (NuTec Sciences, Stafford, Tex.), which produced a separate estimated intensity for the red and green signals that were subtracted for background. The resulting intensity estimates were then normalized on each array separately using intensity-dependent normalization techniques (36). Briefly, the red and green intensities from each spot (denoted R and G) were transformed into a log ratio M (derived from log2 R/G) and a log geometric mean intensity A [derived from (log2 M + log2 A)/2]. A scatter plot was then used to estimate a function f(A) representing the average value of M over the chip, as a function of A (essentially zero). The log ratio M for each spot was then normalized by subtracting the corresponding f(A). The experimental approach consisted of separate loop designs at 1-, 4-, and 10-h time points after inoculation of the third transfer. Each loop contained vertices for three treatments at a single time point (26°C without calcium added, 37°C without calcium added, and 37°C with calcium added). Each pair of treatments in a loop was hybridized twice with dye-swapping, resulting in six hybridization intensity ratios for each time point. Eighteen degrees of freedom were available for estimating in-gene 2-log ratios, or contrasts, between treatments. A linear model was used to estimate two contrasts at each of the three time points in the experiment. The first contrast estimated the difference in log intensity between expression at 26 and 37°C with no calcium added at either temperature. The second estimated the difference in log intensity at 37°C due to the addition of calcium. Because of the design, each contrast was a weighted linear combination of all six hybridization intensities at that time point. Twelve degrees of freedom remained to estimate in-gene variability (additional statistical treatment at http://bbrp.llnl.gov/microbial/Ypestis). TaqMan assay was used to validate the microarray results (http://bbrp.llnl.gov/microbial/Ypestis).
Array quality and genome-wide transcript perspective.
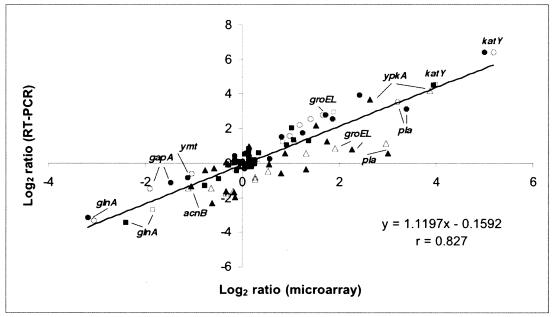
The average value of the log ratio of each gene plotted as a function of intensity was essentially horizontal and had a value of zero, indicating no residual intensity-dependent normalization. A plot of the F ratio for each gene against the estimate of the signal for each gene provided acceptable P value thresholds (http://bbrp.llnl.gov/microbial/Ypestis). This approach demonstrated that only genes carried by pCD1 mediating the TTSS were regulated by temperature as well as Ca2+ (see Table 1 at http://bbrp.llnl.gov/microbial/Ypestis). In contrast, neither chromosomally carried genes nor those located on pPCP1 or pMT1 were affected by Ca2+ (P < 0.05). Approximately 10% of chromosomal genes found to be up- or downregulated by temperature had P values of less than 0.05 and differed in expression by at least 1.5-fold. An additional, but minor, group of genes differed in expression by more than twofold (at P values in the range 0.05 to 0.1) (see Table 2 at http://bbrp.llnl.gov/microbial/Ypestis). In total, 235 chromosomal genes were upregulated after the temperature shift to 37°C, while 274 were downregulated. Clustering analysis (data not shown) revealed that nearly 80% of the chromosomal thermoregulated genes had altered transcriptional levels at 1-h postshift (early response genes) while the remainder were differentially expressed at 4 or at 10 h (late response genes). Most of the genes that responded early to temperature shift did not remain differentially regulated at the later determinations, suggesting immediate roles in adapting to the change in growth conditions (Fig. 1). The microarray expression data were evaluated by TaqMan analysis of 12 genes representing different functional categories. The average n-fold changes in quantity of cDNA molecules present between the three growth conditions for each time point were determined by this real-time RT-PCR procedure and compared to those derived from the microarray analysis. There was strong positive correlation (r = 0.827) between results obtained by the two techniques (Fig. 2).
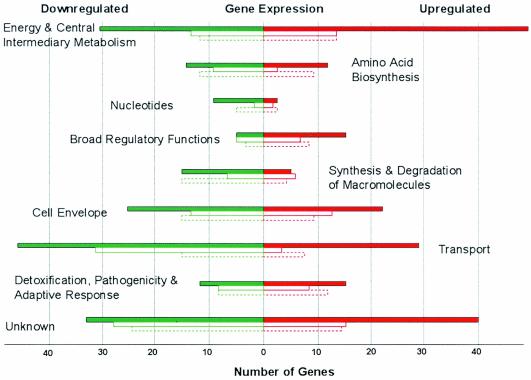
FIG. 1.
Functional classification of chromosomal thermoregulated genes of Y. pestis. Colors represent upregulated (red) and downregulated (green) genes, and bars correspond to the number of differentially expressed genes at the 1- (closed bar), 4- (open solid line bar), and 10-h (open dashed line bar) time points. The functional category assignment was based on the annotated genome of Y. pestis CO92 (23).
FIG. 2.
Correlation of DNA microarray and TaqMan RT-PCR assays. The n-fold difference in transcript levels obtained by both methods for three growth conditions at each of three time points were log-transformed and the values were plotted against each other. Closed and open markers represent samples grown with and without calcium, respectively. The time points correspond to circles (1 h), squares (4 h), and triangles (10 h). The line of best fit for all values is shown (r = 0.827). Select genes are shown for illustrative purposes.
Plasmid genes.
As already noted, components of the TTSS displayed a wide range of thermoregulation; genes of the lcrGVHyopBD operon and the effectors yopH, yopE, yopJ, ypkA, yopT, and yopM consistently showed the highest levels of expression (see Table 1 at http://bbrp.llnl.gov/microbial/Ypestis). The genes yopK, lcrE (yopN), tyeA, sycN, yscX, and the pseudogene ylpA were also strongly upregulated while genes of the secretion apparatus per se, including the negative regulator lcrQ (yscM), the operons yscNOPQRSTU and yscBCDEFGHIJKL, plus the individual chaperones sycE and sycT, were upregulated to a lesser extent. Thermoinduction levels for all of these genes increased progressively over time but were significantly reduced at postshift times of 4 and 10 h in the presence of Ca2+. The genes yscY, lcrD, lcrR, and sycH displayed the lowest levels of thermoinduction among all genes carried by pCD, and Ca2+ exerted little effect on their transcription. A chaperone-like protein (orf7) and a hypothetical protein (orf84) qualified as novel genes carried by pCD1 capable of undergoing upregulation by temperature and downregulation by calcium. In addition, genes encoding LcrF, the lipoprotein YscW (VirG), and a hypothetical protein (orf60) were regulated by temperature but not calcium. The latter were characterized by early induction followed by a decline in expression as opposed to orf73, orf74, and orf75 (all of unknown function), which exhibited late thermal induction. This analysis of genes carried by pCD1 is consistent with previously published results (7, 25) and now provides global, quantitative, and temporal values for activities required for formation and function of the LCR. The determination also uncovered six novel thermoregulated genes in pCD1 that might contribute to yersiniae virulence.
While neither pPCP1 nor pMT1 possessed genes regulated by Ca2+, these plasmids encoded a number of thermoregulated functions. The pPCP1 genes pla, pst, pimi, and YPPCP_08c encoding the plasminogen activator, pesticin, pesticin immunity, and a putative transcriptional regulator, respectively, were induced at all time points after shift to 37°C. In contrast, the genes located on pMT1 displayed a temporal pattern of upregulation. Both the caf operon encoding capsule components and its transcriptional regulator (caf1R) became upregulated over time, achieving a 100-fold increase in caf1 at 10 h (the strongest upregulation value encountered in the entire genome). This thermoinducible phenotype is in full agreement with findings established decades previously (7, 25). The ymt gene of pMT1 encoding MT was downregulated in agreement with previously published observations (25). Six of the nine newly identified thermoinduced ORFs of pMT1 were located in the vicinity of the capsular operon or ymt (putative genes orf12, orf13, orf108, orf110, orf111, and orf112). Of the remainder, orf55 and orf54 were hypothetical genes while orf38 was a putative periplasmic solute-binding protein.
Genes of carbon and energy metabolism.
Numerous enzymes that directly or indirectly facilitate substrate phosphorylation in cells growing in the steady state at 26°C on d-gluconate as a source of energy underwent downregulation upon shift to 37°C (Fig. 3; also see Table 2 at http://bbrp.llnl.gov/microbial/Ypestis). Examples are 6-phosphogluconate dehydrogenase (gnd) of the pentose-phosphate pathway, 2-keto 3-deoxy 6-phosphogluconate aldolase (eda) of the Entner-Doudoroff pathway, numerous glycolytic enzymes (pfkA, fda, tpi, gapA, gmpA, eno, and pykF), carbohydrate phosphotransferase system components (ccr, fru, manXYZ, nagE), the six-gene cluster encoding the maltose transport system (malMBKEFG), and the related genes malZPQ. The components of the pentose-phosphate pathway were unchanged after 6-phosphogluconate dehydrogenase promoted interconversion of 3- to 7-carbon intermediates and glucose 6-phosphate isomerase (pgi), which interconverts fructose 6-phosphate and glucose 6-phosphate. Gene zwf encoding glucose 6-phosphate dehydrogenase is evidently intact in Y. pestis (11, 23) and we observed its constitutive expression (data not shown) despite the absence of detectable enzymatic activity in plague bacilli (21). In contrast to the majority of the genes involved in the utilization of carbohydrates, shift to 37°C upregulated members of the d-gluconate transport system including a putative gluconokinase (YPO3953), gluconate permease (gntT), and a transcription factor for the latter (gntR). The ribose uptake genes rbsK encoding ribokinase and rbsD for ribose permease also underwent early induction at 37°C. The ABC galactose transporter operon mglBAC was similarly induced as was the entire galETKM operon of galactose metabolism. Furthermore, the genes glpF, glpK, and glpD encoding the glycerol uptake facilitator, glycerol kinase, and aerobic glycerol 3-phosphate dehydrogenase, respectively, underwent strong (10- to 25-fold) early upregulation at 37°C.
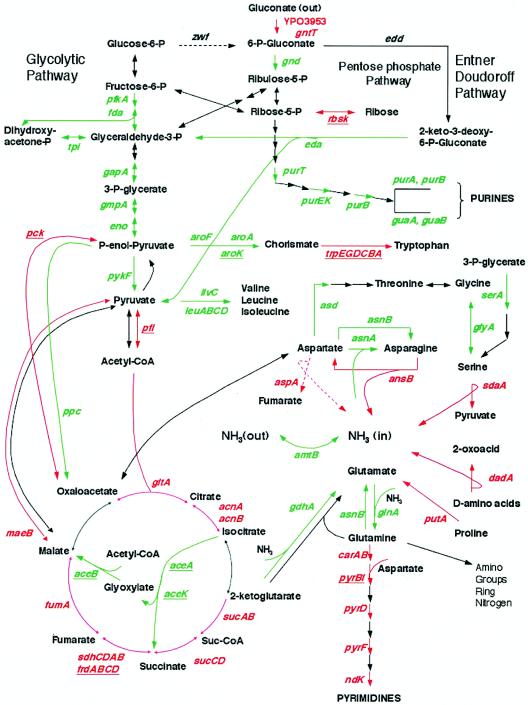
FIG. 3.
Y. pestis primary metabolic pathways affected by shift of growth temperature. The colors show the expression changes upon shift in growth temperature from 26 to 37°C. The red arrows and the green arrows represent the genes induced and repressed at 37°C, respectively. The gene names are colored in the same way as the arrows with the late-regulated gene names further underlined. The dashed lines indicate enzymes known to be inactive in Y. pestis (i.e., genes zwf and aspA). The gene names are designated according to the annotated Y. pestis CO92 genome (23). The amount of change is listed in Table 2 at http://bbrp.llnl.gov/microbial/Ypestis.
Concomitant changes in oxidative catabolism favoring reliance on a full TCA cycle occurred after shift to 37°C and are shown in Fig. 3. Phosphotransacetylase (pta) and enzymes of the glyoxylate bypass (aceBAK) required for generation of acetyl-coenzyme A (CoA) and its utilization during gluconeogenesis were downregulated after temperature shift. In contrast, some but not all enzymes of the TCA cycle underwent at least initial induction, including citrate synthase (gltA), aconitases (acnA and acnB), the 2-oxyglutarate dehydrogenase complex (sucABCD), the succinate dehydrogenase complex (sdhCDAB), and fumarate hydratase (fumA). Furthermore, upregulation of cytochromes (cybB and cybC) and an attendant terminal electron acceptor (katY) was dramatic (see Table 2 at http://bbrp.llnl.gov/microbial/Ypestis). These results indicate that shift to host temperature results in reliance on oxidative phosphorylation mediated by the TCA cycle and an enhanced ability to utilize a variety of carbohydrates present in mammalian tissue. The latter are all converted to the level of pyruvate where they can enter the TCA cycle either after reductive decarboxylation as acetyl-CoA or as oxaloacetate or malate following carboxylation via upregulated phosphoenolpyruvate carboxykinase (pck) or NADP+-dependent malic enzyme (maeB), respectively (Fig. 3). As noted below, at 37°C the TCA cycle accommodates terminal oxidation of numerous additional sources of carbon and energy in addition to carbohydrates.
Nitrogen and amino acid metabolism.
Many genes involved in nitrogen assimilation (including NTR-regulated genes) were strongly downregulated upon shift to 37°C, including the ammonium transport facilitator encoded by amtB and the neighboring nitrogen regulator glnK, glutamine synthetase (glnA), and both glutamine- (asnB) and ammonia-dependent (asnA) asparagine synthetases. Also repressed were the nitrogen regulators ntrB and ntrC (Fig. 3; also see Table 2 at http://bbrp.llnl.gov/microbial/Ypestis). In contrast, at least 11 genes required for efficient catabolism of amino acids were rapidly induced upon temperature shift, including those encoding d-amino-acid dehydrogenase (dadA), l-serine deaminase (sdaA), l-asparaginase (ansB), carbamoylphosphate synthetase (carAB), proline/delta 1-pyrroline 5-carboxylate dehydrogenase (putA), proline permease (putP), and a putative aminotransferase (YPO0623). Some of these enzymes account for the bulk of released metabolic ammonia via reactions that directly or indirectly promote deamination during formation of α-keto acids entering the TCA cycle (Fig. 3). As previously noted, aspartase activity is cryptic in Y. pestis (12); nevertheless, aspA transcription was upregulated following temperature shift even though the medium lacked added l-glutamate. The medium also lacked l-histidine and l-serine, thus, not surprisingly, the histidine biosynthetic (hisGDCBHAFI) and transport (hisJQMP) operons underwent early postshift induction at 1 h and then became repressed as the culture approached stationary phase. l-Serine was previously shown to undergo uncontrolled reductive deamination in Y. pestis at 37°C (12). The biosynthesis of l-serine was downregulated at the branch point from glycolysis (gene serA) as was conversion of glycine to l-serine via serine hydroxymethyltransferase (glyA); in addition, the oxidative cleavage of glycine via the gcvP and gcsH products was repressed. These changes serve to isolate the l-serine precursors 3-phosphoglycerate and glycine (obtained in minimal medium from threonine aldolase encoded by ltaA), thereby minimizing loss of metabolic carbon otherwise destined for glycolysis or biosynthesis of aliphatic amino acids through l-threonine. With the exception of l-tryptophan, synthesis of other amino acids (present in abundance) was downregulated after shift to 37°C often at branch points or early within specific pathways (Fig. 3). Examples are asd (encoding aspartate semialdehyde dehydrogenase), aroF, aroA, and aroK (facilitating aromatic amino acid biosynthesis to chorismate), ilvC-encoded ketol-acid reductoisomerase (initiating synthesis of l-isoleucine and l-valine), and the biosynthetic l-leucine operon (leuABCD).
Lipid metabolism.
Shift to 37°C caused modest downregulation of fabB and fabC encoding the 3-ketoacyl synthase complex involved in lipid biosynthesis (see Table 2 at http://bbrp.llnl.gov/microbial/Ypestis). However, this change prompted upregulation of 3-ketoacyl thiolase (fadA and probably YPO2746), the fatty acid oxidation complex (fadB and faoA), and yafH (probable acyl-CoA dehydrogenase), as well as fadL encoding long-chain fatty acid transport. These results would be expected if shift to host temperature promotes β-oxidation of fatty acids.
Nucleotide metabolism.
Although the culture medium lacked added purines and pyrimidines, marked differences occurred between their patterns of regulation after shift to 37°C. Purine biosynthesis was downregulated before IMP at the level of GAR (phosphoribosyl glycinamide formyltransferase) transformylase (purT), phosphoribosylaminoimidazole carboxylase (purEK), and adenylosuccinate lyase (purB); subsequent conversion of IMP to both GMP and AMP was also repressed (Fig. 3). Nucleoside permease (nupC) mediating high-affinity transport of adenine and pyrimidine nucleosides was similarly downregulated. Transcription of all other genes in the de novo pathway, the coordinate purine repressor (purR), and genes required for purine interconversion was not influenced by temperature shift. In contrast, carbamoylphosphate synthase (carAB), aspartate carbamoyltransferase (pyrB), and its regulator (pyrI), dihydroorotate dehydrogenase (pyrD), and orotidine 5′-phosphate decarboxylase (pyrF) of the de novo pyrimidine biosynthetic pathway were upregulated after temperature shift. Furthermore, nucleoside diphosphate kinase (ndk) that performs the last reaction in the synthesis of nucleoside triphosphates was induced although cytidylate kinase (cmk) was downregulated. Other than ndk, the entire set of genes encoding pyrimidine salvage pathways and deoxyribonucleotide interconversions were not thermoregulated.
Macromolecular synthesis.
As expected, a modest number of genes mediating macromolecular synthesis (e.g., DNA replication and ribosomal proteins) were downregulated after 10 h at 37°C as the bacteria approached early stationary phase (see Table 2 at http://bbrp.llnl.gov/microbial/Ypestis). Several genes involved in lipopolysaccharide biosynthesis were also downregulated, including nagB and nagE concerned with glucosamine metabolism. Similarly, fabZ, lpxA, and lpxB encoding hydroxymyristoyl-dehydratase, UDP-GlcNAc acyltransferase, and lipid A-disaccharide synthase, respectively, were downregulated; these activities are involved in the initial steps of lipid A biosynthesis.
Unknown ORFs.
A significant number of genes encoding putative exported, membrane, or unknown proteins were thermoregulated (see Table 2 at http://bbrp.llnl.gov/microbial/Ypestis). The possible operon YPO1996-1994, which lacks significant homology to any entry in the current version of GenBank, displayed the strongest level of temperature-dependent upregulation within this category.
Pathogenicity.
Determinants considered in this category constitute products of chromosomal genes that were found by annotation to map within putative pathogenicity islands (11, 23), result in avirulence when lost by mutation (7), or exist as homologues of virulence effectors of other species. The Mn-cofactored superoxide dismutase encoded by sodA was reported to facilitate survival and multiplication of Y. enterocolitica in mice (26). However, sodA was downregulated at all time points in the present study although sodC encoding the Cu-Zn-cofactored enzyme displayed a pattern of prompt induction with later downregulation at 10 h (see Table 2 at http://bbrp.llnl.gov/microbial/Ypestis). As noted above, the gene katY encoding Y. pestis/Y. pseudotuberculosis-specific catalase-peroxidase was strongly induced (more than 30-fold at 1 h) during the shift to 37°C in agreement with classical (9) and recent (15) observations. Although katA remained upregulated at 10 h, thiol peroxidase (tpx) was downregulated at this time. Genes encoding the putative copper resistance proteins YPO1784 and YPO1785 were induced early. Accordingly, out of 10 genes annotated as belonging to the detoxification functional group, 7 were differentially regulated, while the remainder (sodB, ahpC, and cutF) were expressed constitutively (data not shown). Also induced was the serine protease encoded by htrA (gsrA) known to facilitate resistance to oxidative stress, growth at elevated temperature, and survival of many pathogens in macrophages including Y. pestis (35). The two-component system PhoPQ was similarly upregulated: this function is a known global regulator of virulence in yersiniae (22) and other bacterial species.
Of the chromosomally encoded fimbrial-type adhesins previously described by Parkhill et al. (23), only the psaABC locus encoding antigen 4/pH 6 antigen (4, 9) underwent strong initial upregulation (>50-fold for psaA). This pattern is in accord with that defined by the initial study of thermoregulation (9) and in more recent observations (33). The pseudogene inv (YPO1793) encoding invasin in Y. pestis is inactivated by IS1541 (31) but underwent downregulation as reported for the ClpB protease-dependent inv of Y. enterocolitica (3). However, ClpB (YPO2946) was not downregulated after temperature shift in Y. pestis although the close homologue YPO0506 (one of six paralogues in the genome) was repressed. YPO0506 is located within a large cluster of 18 genes (YPO0499 to YPO0516) that were strongly (up to 10-fold) downregulated at all postshift time points; most ORFs comprising this cluster are hypothetical proteins. In contrast to inv, the gene ail (YPO2905) for attachment-invasion locus (modulated by the distinct protease ClpP) was upregulated 10 h after shift. The clpP gene was expressed constitutively in our experiments in accord with the finding that ail is transcribed in Y. enterocolitica during stationary phase at 37°C (24). Of the additional three Ail-like proteins found in the genome of Y. pestis CO92 (23), only YPO2505 was downregulated at 37°C although the putative invasin gene (YPO3944) containing 22 large degenerate repeats was induced. Urease activity in Y. pestis is also cryptic due to a single base addition in ureD (30). Nevertheless, the structural genes of the urease locus (operon ureAB) as well as the putative urea transporter (YPO2672) were downregulated at 37°C.
Although not involved in mammalian pathogenesis, the series of genes encoding homologues of insecticidal toxin complexes (23, 34) underwent thermoregulation and are therefore considered here. The cluster of genes YPO3675-3682 was severely downregulated at 37°C. This cluster, assigned by Parkhill et al. (23) as a potential pathogenicity island, includes genes encoding putative insecticidal toxins tcaABC as well as transcriptional regulator and hypothetical ORFs of phage-related origin. Two copies of another cryptic insecticidal toxin encoded by tccC (YPO3673 and YPO3674) are located next to this cluster but were not thermoregulated. Genes located in other putative pathogenicity islands displaying differential expression included YPO0590, YPO0595-0597, YPO0623-0628, YPO0881, YPO1091, and YPO1242-1252. These genes are typically putative ORFs with unknown function or are associated with degenerate bacteriophage.
Adaptive response.
Genes in this category modulate responses to distinct environments within the host that present unique challenges to invading pathogens. The chromosomal psp locus of Y. enterocolitica encoding phage shock proteins plays a role in virulence and was induced concomitantly with the pYV-encoded TTSS (10). The pspA gene of this locus was upregulated early at 37°C as was the universal stress protein A encoded by uspA (see Table 2 at http://bbrp.llnl.gov/microbial/Ypestis). The latter provides coupling of glucose and acetate metabolism, thus helping bacteria to effectively utilize both of these carbon sources (32). The carbon starvation protein encoded by cstA and regulated in Escherichia coli by the cyclic AMP (cAMP) and cAMP receptor protein complex (29) was also induced following temperature shift. Also upregulated was arcB encoding the ArcAB aerobic respiration control sensor protein, cold shock proteins (genes cspH and cspE), and heat shock proteins (genes ibpA and ibpB). The later are associated with the high-level production of certain heterologous proteins in E. coli (2). The universal heat shock chaperonins GroEL and GroES were robustly upregulated (three- to fourfold) at all time points tested. In contrast, ORF YPO3784 (annotated as an additional carbon starvation protein) was downregulated at 37°C as was the cAMP receptor protein complex-regulated gene osmY encoding an osmotically inducible protein (19). The oxidative stress sigma factor encoded by rpoE, known to modulate virulence in Salmonella (17), underwent notable upregulation at 37°C. Also induced was gene yfiA, a putative modulator for the nitrogen assimilation sigma factor RpoN (sigma 54). In addition to the categories of genes described above, 8% of the Y. pestis pseudogenes (25 out of 312) are thermoregulated under the experimental conditions employed in this work (see Table 3 at http://bbrp.llnl.gov/microbial/Ypestis).
Concluding comments.
Plague bacilli exist in nature at ambient temperature within the flea vector or in the mammalian host at 37°C in either Ca2+-sufficient plasma, lymph, and interstitial fluid or in Ca2+-deficient cytoplasm released into focal lesions. DNA microarray technology was used to characterize thermoregulated changes in global regulation that occur in chemically defined medium reflecting introduction into these niches. This approach avoided the nutritional stepdown inherent in the LCR (37), thereby maintaining the bacteria in a steady state and avoiding premature expression of functions associated with entry into stationary phase. Accordingly, the observed early changes in gene expression reflect real adaptation to the host rather than changes associated with the onset of bacteriostasis. Transcriptional profiles following shift from 26 to 37°C revealed 15 novel thermoregulated genes on the three Y. pestis plasmids. Only genes carried by pCD1, including components of type III secretion (including two genes of unknown function), were downregulated by Ca2+. In addition, approximately 10% of all chromosomal genes were influenced by temperature (but not Ca2+). Of these, 235 were upregulated and 274 were downregulated upon shift from 26 to 37°C, thereby inhibiting glycolysis while favoring terminal oxidation of a variety of substrates (including carbohydrates, amino acids, and fatty acids known to exist within the host). Shift to 37°C also repressed Ntr-controlled genes involved in nitrogen assimilation and biosynthesis of amino acids and purines (but not pyrimidines). Numerous global regulators and transport systems were also thermoregulated as was expression of known and putative genes associated with general adaptive responses, oxidative stress, and modulators of innate immunity (e.g., invasins, adhesins, cytotoxins, and inhibitors of proinflammatory cytokines). These studies indicate that, in nature, plague bacilli favor fermentative patterns of metabolism during slow growth within the flea but exhibit pronounced oxidative catabolism during rapid proliferation in the host. Differential transcription during temperature shift also identified a useful list of putative virulence-associated genes to target as novel candidates for future research on the control of this pathogen.
Acknowledgments
This work was performed under the auspices of the U.S. Department of Energy by the University of California, Lawrence Livermore National Laboratory, under contract no. W-7405-Eng-48. Partial support was also provided by National Institutes of Health grant R21 AI53508-01 and the Region V ‘Great Lakes' RCE (NIH award 1-U54-AI-057153).
We thank Arthur Kobayashi for designing the website for this project.
REFERENCES
- 1.Achtman, M., K. Zurth, G. Morelli, G. Torrea, A. Guiyoule, and E. Carniel. 1999. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. USA 96:14043-14048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen, S. P., J. O. Polazzi, J. K. Gierse, and A. M. Easton. 1992. Two novel heat shock genes encoding proteins produced in response to heterologous protein expression in Escherichia coli. J. Bacteriol. 174:6938-6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Badger, J. L., B. M. Young, A. J. Darwin, and V. L. Miller. 2000. Yersinia enterocolitica ClpB affects levels of invasin and motility. J. Bacteriol. 182:5563-5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ben-Efraim, S., M. Aronson, and L. Bichowsky-Slomnicki. 1961. New antigenic component of Pasteurella pestis formed under specific conditions of pH and temperature. J. Bacteriol. 81:704-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brubaker, R. R. 1967. Growth of Pasteurella pseudotuberculosis in simulated intracellular and extracellular environments. J. Infect. Dis. 117:403-417. [DOI] [PubMed] [Google Scholar]
- 6.Brubaker, R. R. 2003. Interleukin-10 and inhibition of innate immunity to yersiniae: roles of Yops and LcrV (V antigen). Infect Immun. 71:3673-3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brubaker, R. R. 2000. Yersinia pestis and bubonic plague. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackelbrandt (ed.), The prokaryotes, an evolving electronic resource for the microbiological community. Springer Verlag, New York, N.Y. [Online.] http://link.springer-nv.com/link/service/books/10125/index.htm.
- 8.Cornelis, G. R. 2002. The Yersinia Ysc-Yop ‘Type III' weaponry. Nat. Rev. Mol. Cell Biol. 3:742-752. [DOI] [PubMed] [Google Scholar]
- 9.Crumpton, M. Y., and D. A. L. Davies. 1956. An antigenic analysis of Pasteurella pestis by diffusion of antigens and antibodies in agar. Proc. R. Soc. Lond. Ser. B 145:109-134. [DOI] [PubMed] [Google Scholar]
- 10.Darwin, A. J., and V. L. Miller. 2001. The psp locus of Yersinia enterocolitica is required for virulence and for growth in vitro when the Ysc type III secretion system is produced. Mol. Microbiol. 39:429-444. [DOI] [PubMed] [Google Scholar]
- 11.Deng, W., V. Burland, G. I. Plunkett, A. Boutin, G. F. Mayhew, P. Liss, N. T. Perna, D. J. Rose, B. Mau, D. C. Schwartz, S. Zhou, J. D. Fetherston, L. E. Lindler, R. R. Brubaker, G. V. Plano, S. C. Straley, K. A. McDonough, M. L. Nilles, J. L. Matson, F. R. Blattner, and R. D. Perry. 2002. Genome sequence of Yersinia pestis KIM. J. Bacteriol. 184:4601-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dreyfus, L. A., and R. R. Brubaker. 1978. Consequences of aspartase deficiency in Yersinia pestis. J. Bacteriol. 136:757-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Englesberg, E., and J. B. Levy. 1955. Induced synthesis of tricarboxylic acid cycle enzymes as correlated with the oxidation of acetate and glucose by Pasteurella pestis. J. Bacteriol. 69:418-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fowler, J. M., and R. R. Brubaker. 1994. Physiological basis of the low calcium response in Yersinia pestis. Infect. Immun. 62:5234-5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia, E., Y. A. Nedialkov, J. Elliott, V. L. Motin, and R. R. Brubaker. 1999. Molecular characterization of KatY (antigen 5), a thermoregulated chromosomally encoded catalase-peroxidase of Yersinia pestis. J. Bacteriol. 181:3114-3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Higuchi, K., L. L. Kupferberg, and J. L. Smith. 1959. Studies on the nutrition and physiology of Pasteurella pestis. III. Effects of calcium ions on the growth of virulent and avirulent strains of Pasteurella pestis. J. Bacteriol. 77:317-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Humphreys, S., A. Stevenson, A. Bacon, A. B. Weinhardt, and M. Roberts. 1999. The alternative sigma factor, σE, is critically important for the virulence of Salmonella typhimurium. Infect. Immun. 67:1560-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lambert de Rouvroit, C., C. Sluiters, and G. R. Cornelis. 1992. Role of the transcriptional activator, VirF, and temperature in the expression of the pYV plasmid genes of Yersinia enterocolitica. Mol. Microbiol. 6:395-409. [PubMed] [Google Scholar]
- 19.Lange, R., M. Barth, and R. Hengge-Aronis. 1993. Complex transcriptional control of the σS-dependent stationary-phase-induced and osmotically regulated osmY (csi-5) gene suggests novel roles for Lrp, cyclic AMP (cAMP) receptor protein-cAMP complex, and integration host factor in the stationary-phase response of Escherichia coli. J. Bacteriol. 175:7910-7917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mortlock, R. P. 1962. Gluconate metabolism of Pasteurella pestis. J. Bacteriol. 84:53-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mortlock, R. P., and R. R. Brubaker. 1962. Glucose-6-phosphate dehydrogenase and 6-phosphogluconate dehydrogenase activities of Pasteurella pestis and Pasteurella pseudotuberculosis. J. Bacteriol. 84:1122-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oyston, P. C., N. Dorrell, K. Williams, S. R. Li, M. Green, R. W. Titball, and B. W. Wren. 2000. The response regulator PhoP is important for survival under conditions of macrophage-induced stress and virulence in Yersinia pestis. Infect. Immun. 68:3419-3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. G. Holden, M. B. Prentice, M. Sebaihia, K. D. James, C. Chercher, K. L. Mungall, S. Baker, D. Basham, S. D. Bentley, K. Brooks, A. M. Cerdeño-Tárraga, T. Chillingworth, A. Cronin, R. M. Davies, P. Davis, G. Dougan, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Leather, S. Moule, P. C. F. Oyston, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523-527. [DOI] [PubMed] [Google Scholar]
- 24.Pederson, K. J., S. Carlson, and D. E. Pierson. 1997. The ClpP protein, a subunit of the Clp protease, modulates ail gene expression in Yersinia enterocolitica. Mol. Microbiol. 26:99-107. [DOI] [PubMed] [Google Scholar]
- 25.Perry, R. D., and J. D. Fetherston. 1997. Yersinia pestis—etiologic agent of plague. Clin. Microbiol. Rev. 10:35-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roggenkamp, A., T. Bittner, L. Leitritz, A. Sing, and J. Heesemann. 1997. Contribution of the Mn-cofactored superoxide dismutase (SodA) to the virulence of Yersinia enterocolitica serotype O8. Infect. Immun. 65:4705-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Santer, M., and S. Ajl. 1954. Metabolic reactions of Pasteurella pestis. I. Terminal oxidation. J. Bacteriol. 67:379-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santer, M., and S. Ajl. 1955. Metabolic reactions of Pasteurella pestis. II. The fermentation of glucose. J. Bacteriol. 69:298-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schultz, J. E., and A. Matin. 1991. Molecular and functional characterization of a carbon starvation gene of Escherichia coli. J. Mol. Biol. 218:129-140. [DOI] [PubMed] [Google Scholar]
- 30.Sebbane, F., A. Devalckenaere, J. Foulon, E. Carniel, and M. Simonet. 2001. Silencing and reactivation of urease in Yersinia pestis is determined by one G residue at a specific position in the ureD gene. Infect. Immun. 69:170-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simonet, M., B. Riot, N. Fortineau, and P. Berche. 1996. Invasin production by Yersinia pestis is abolished by insertion of an IS200-like element within the inv gene. Infect. Immun. 64:375-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tao, H., C. Bausch, C. Richmond, F. R. Blattner, and T. Conway. 1999. Functional genomics: expression analysis of Escherichia coli growing on minimal and rich media. J. Bacteriol. 181:6425-6440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thulasiraman, V., S. L. McCutchen-Maloney, V. L. Motin, and E. Garcia. 2001. Detection and identification of virulence factors in Yersinia pestis using SELDI ProteinChip system. BioTechniques 30:428-432. [DOI] [PubMed] [Google Scholar]
- 34.Waterfield, N. R., D. J. Bowen, J. D. Fetherston, R. D. Perry, and R. H. ffrench-Constant. 2001. The tc genes of Photorhabdus: a growing family. Trends Microbiol. 9:185-191. [DOI] [PubMed] [Google Scholar]
- 35.Williams, K., P. C. Oyston, N. Dorrell, S. Li, R. W. Titball, and B. W. Wren. 2000. Investigation into the role of the serine protease HtrA in Yersinia pestis pathogenesis. FEMS Microbiol. Lett. 186:281-286. [DOI] [PubMed] [Google Scholar]
- 36.Yang, Y. H., S. Dudoit, P. Luu, D. M. Lin, V. Peng, J. Ngai, and T. P. Speed. 2002. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zahorchak, R. J., W. T. Charnetzky, R. V. Little, and R. R. Brubaker. 1979. Consequences of Ca2+ deficiency on macromolecular synthesis and adenylate energy charge in Yersinia pestis. J. Bacteriol. 39:792-799. [DOI] [PMC free article] [PubMed] [Google Scholar]