Abstract
The genetic element flanking the Bacteroides fragilis pathogenicity island (BfPAI) in enterotoxigenic B. fragilis (ETBF) strain 86-5443-2-2 and a related genetic element in NCTC 9343 were characterized. The results suggested that these genetic elements are members of a new family of conjugative transposons (CTns) not described previously. These putative CTns, designated CTn86 and CTn9343 for ETBF 86-5443-2-2 and NCTC 9343, respectively, differ from previously described Bacteroides species CTns in a number of ways. These new transposons do not carry tetQ, and the excision from the chromosome to form a circular intermediate is not regulated by tetracycline; they are predicted to differ in their mechanism of transposition; and their sequences have very limited similarity with CTnDOT or other described CTns. CTn9343 is 64,229 bp in length, contains 61 potential open reading frames, and both ends contain IS21 transposases. Colony blot hybridization, PCR, and sequence analysis indicated that CTn86 has the same structure as CTn9343 except that CTn86 lacks a ∼7-kb region containing truncated integrase (int2) and rteA genes and it contains the BfPAI integrated between the mob region and the bfmC gene. If these putative CTns were to be demonstrated to be transmissible, this would suggest that the bft gene can be transferred from ETBF to nontoxigenic B. fragilis strains by a mechanism similar to that for the spread of antibiotic resistance genes.
Enterotoxigenic Bacteroides fragilis (ETBF) has been associated with diarrheal disease in livestock, young children, and adults (22, 23, 29, 30, 33, 41, 47). The only recognized virulence factor of these strains is a toxin termed B. fragilis toxin, or BFT. BFT has been characterized as a 20-kDa zinc-dependent metalloprotease (19) that mediates the cleavage of E-cadherin, resulting in an altered morphology of certain human intestinal carcinoma cell lines (particularly HT29/C1cells), fluid accumulation in ligated lamb ileal loops, and intestinal epithelial cell proliferation (4, 14, 21, 23, 25, 36, 41, 44, 45).
It has been reported that the bft gene is contained in a 6-kb pathogenicity island termed the B. fragilis pathogenicity island, or BfPAI (9, 20). In addition to the BfPAI, ∼12 kb of its flanking DNA has been sequenced (9; A. Franco and C. L. Sears, unpublished data). This work revealed that the BfPAI is flanked by genes encoding putative mobilization proteins (9). The left end of the BfPAI is flanked by a typical Bacteroides mobilizable region (11, 37), that is, the genes for DNA-processing enzymes (bfmA and bfmB) and the cis-acting oriT located adjacent to this genes. The region flanking the right end of the BfPAI contains a gene (bfmC) whose predicted protein shares significant identity to the TraG family proteins for mobilization and VirD4, a protein component of type IV secretion systems (9).
Colony blot analysis of a collection of ETBF and nontoxigenic B. fragilis (NTBF) strains indicated that there are three major populations of B. fragilis strains based on the presence of the BfPAI and its flanking regions: (i) pattern I strains, containing the BfPAI and its flanking region (all are ETBF strains); (ii) pattern II strains, lacking the BfPAI and its flanking regions (all are NTBF strains); and (iii) pattern III strains, containing the flanking region but lacking the BfPAI (all are NTBF strains) (9). It has also been reported that the region flanking the BfPAI and a ∼700-bp region upstream of bft are crucial to maximal BFT production by ETBF strains (10).
The G+C contents of the BfPAI (35%) and the flanking DNA (47 to 50%) differ greatly from that reported for the B. fragilis chromosome (43%) (http://www.sanger.ac.uk/Projects/B_fragilis), suggesting that the BfPAI and its flanking region are two distinct genetic elements originating from different organisms. Based on these results, we hypothesized that ETBF strains may have evolved by horizontal transfer of these two genetic elements into a pattern II NTBF strain (9).
Many strains of Bacteroides species carry large self-transmissible elements called conjugative transposons (CTns). CTns are genetic elements that move from the genome of a donor bacterium to the genome of a recipient bacterium by a process that requires intercellular contact. The CTn initiates conjugal transfer by excising from the chromosome to form a circular intermediate. This intermediate is nicked at the oriT, and a single-stranded copy is transferred to the recipient cell, recircularized, and integrated into the recipient (5, 7, 28, 35). Strains of Bacteroides species carry CTns that belong to at least two distinct families; the best described is CTnDOT and its relatives (31, 37). Many of these transposons confer resistance to tetracycline that is determined by tetQ. Excision of CTnDOT and formation of the circular intermediate are stimulated 1,000- to 10,000-fold by tetracycline (31, 38).
In this study, our analyses suggest that the genetic element flanking the BfPAI in ETBF strains and a related element present in pattern III NTBF strains are novel CTns not described previously in Bacteroides species.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The B. fragilis strains used in this study are described in Table 1. Bacteroides thetaiotaomicron strains BT4107 and BT400 were a gift from N. B. Shoemaker. Bacteroides strains were propagated anaerobically on BHC medium (37 g of brain heart infusion base [Difco Laboratories, Detroit, Mich.] per liter, with 0.1 mg of vitamin K per liter, 0.5 mg of hemin per liter, and 50 mg of l-cysteine per liter [all from Sigma, St. Louis, Mo.]).
TABLE 1.
B. fragilis strains used in this study
| Strain | Toxicity (pattern)a | Source or reference |
|---|---|---|
| 86-5443-2-2 | ETBF (I) | L. L. Myers (8) |
| VPI 13784 | ETBF (I) | T. D. Wilkins |
| DS233 | ETBF (I) | L. L. Myers |
| J-38-1 | ETBF (I) | L. L. Myers |
| 20793-3 | ETBF (I) | L. L. Myers |
| TM4000 | NTBF (II) | 11 |
| LM3 | NTBF (II) | 21 |
| LM9 | NTBF (II) | 21 |
| LM59 | NTBF (II) | 21 |
| LM46 | NTBF (II) | 21 |
| K518 | NTBF (II) | G.-T. Chung (9) |
| I-1345 | NTBF (III) | L. L. Myers (10) |
| NCTC 9343 | NTBF (III) | ATCC (12) |
ETBF indicates culture supernatants of the strains stimulate morphologic changes in HT29/C1 cells, whereas culture supernatants of NTBF strains do not. Pattern I, contains the BfPAI and flanking region; pattern II, lacks the BfPAI and flanking region; pattern III, lacks the BfPAI but contains its flanking region (9).
DNA isolation analysis.
Plasmid DNA was extracted by the alkali lysis method (32) or using QIAGEN columns (QIAGEN, Valencia, Calif.). Plasmids and cosmids were restriction mapped by Southern blot walking as described previously (8). Purification of DNA fragments and extraction from gel slices were performed with a QIAEX II gel extraction kit (QIAGEN). PCR products were cloned onto the pGEM-T Easy vector (Promega, Madison, Wis.) according to the instructions of the manufacturer.
Colony blot hybridizations.
Colony blots of B. fragilis strains were prepared by the technique described previously (8). Briefly, B. fragilis organisms grown overnight on BHC agar were transferred to Whatman 541 filters. The filters were microwave processed in alkali solution (0.5 M NaOH, 1.5 M NaCl) followed by neutralization in 2 M ammonium acetate. The probes were labeled with [α-32P]dCTP by random priming (Multiprime DNA labeling system; Amersham Pharmacia Biotech), hybridized at 37°C under high-stringency conditions in 50% formamide-5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate-1 mM EDTA-1× Denhardt's solution and washed with 5× SSC-0.1% sodium dodecyl sulfate at 65°C for 1 h. Finally, the filters were rinsed with 2× SSC at room temperature.
Construction and screening of the cosmid library.
Chromosomal DNA from ETBF strain 86-5443-2-2 was partially digested with Sau3A1 under conditions where the majority of fragments were 30 to 40 kb in size, and it was ligated into cosmid vector pHC79 digested with BamHI. Ligated DNA was packaged into lambda phage by using the Giga Pack II packing extract (Stratagene, La Jolla, Calif.) and transduced into Escherichia coli HB101. The cosmid library was screened to find the right end of CTn86 by using probe 11 (see Results). Screening of the cosmid library was performed by colony blot hybridization as described above.
PCR conditions.
The sequences of the primers and the parameters used for each PCR are shown in Table 2. PCRs were performed with Taq polymerase (1.5 U) in a 50-μl volume containing plasmid (5 to 10 ng) or chromosomal DNA (∼20 ng) as template, primers (25 pmol), deoxynucleoside triphosphates (200 μM), and MgCl2 (1.5 mM).
TABLE 2.
Primers and thermal cycler programs used for the PCRs in this study
| Probe | Primer | Primer sequence (5′-3′) | Primer 5′ positiona | Programb |
|---|---|---|---|---|
| 1 | Tn4R | GTCCTGTGGCAATGAAGACAATGC | 51 ORF 1 | 94°C, 1 min; 66°C, 1 min; 70°C, 2 min |
| Tn15 | TCCTCCATGTAATATTCCACCGCC | 171 ORF 3 | ||
| 2 | Tn15R | GGCGGTGGAATATTACATGGAGGA | 148 ORF 3 | 94°C, 1 min; 62°C, 1 min; 68°C, 2 min |
| Tn15A | GCTGCTACTGGAGCGAGAGTAAGT | 363 ORF 5 | ||
| 3 | Tn15AR | ACTTACTCTCGCTCCAGTAGCAGC | 387 ORF 5 | 94°C, 1 min; 62°C, 1 min; 68°C, 2 min |
| Tn24R | CGGTAGTATTCACATAGCTCGCC | 75 ORF 7 | ||
| 4 | Tn24 | GGCGAGCTATGTGAATACTACCG | 97 ORF 7 | 94°C, 1 min; 62°C, 1 min; 68°C, 2 min |
| Tn25 | CGCAGGTCAGACCCTGAGTCGA | 823 ORF 9 | ||
| 5 | Tn1 | GGTGTGCCATCGCAATGAAGCA | 1,169 ORF 14 (rteA) | 94°C, 1 min; 55°C, 1 min; 66°C, 2 min |
| Tn2 | CTCGGTATGGAGTATGCTCCAG | 111 ORF 16 | ||
| 6 | 15 | GGAACTTCGTGCTGCCCTCAAAG | 511 ORF 25 | 94°C, 1 min; 62°C, 1 min; 68°C, 2 min |
| Tn7 | GTAATTCGGTATCGTCCACCAGC | 628 ORF 27 | ||
| 7 | Tn11 | CAGCTACTCATGCCGTTCCAGGC | 242 ORF 28 | 94°C, 1 min; 66°C, 1 min; 70°C, 2 min |
| Tn12 | CGTTTGGCTCATTGTTCGGGCTG | 624 ORF 29 | ||
| 8 | Tn13 | CCACCGGTTGCTCCTTCGTTTC | 1,060 ORF 30 | 94°C, 1 min; 66°C, 1 min; 70°C, 2 min |
| Tn14 | GTGCAACCGTTGTCGCACGTAC | 101 ORF 30 | ||
| 9 | TranF | CGCAGAAGATCGAGGAACTCAC | 62 traN (CTn9343) | 94°C, 1 min; 62°C, 1 min; 68°C, 2 min |
| TranR | CGCTGTCAGCAGCGACTTGTTC | 1,056 traN (CTn9343) | ||
| 10 | Tn18 | CGAACAGCATGTCCGTGTTGCC | 2,125 ORF 42 (traG) | 94°C, 1 min; 66°C, 1 min; 70°C, 2 min |
| Tn19 | AGCCTTCAGGCACATGCCCGAC | 171 ORF 42 (traG) | ||
| 11 | Tn20 | GCGCGTCCCGATTGTTTCCAGG | 615 ORF 55 | 94°C, 1 min; 62°C, 1 min; 68°C, 2 min |
| Tn21 | GGGGATACGACATGTACCGCTC | 86 ORF 56 | ||
| 12 | Tn26 | CCGCAGAACGGCATATACCCTGG | 201 ORF 57 | 94°C, 1 min; 66°C, 1 min; 70°C, 2 min |
| Tn27 | CATGGGTGCGAGGTCCGTCCCTC | 127 ORF 59 | ||
| 13 | Tn22 | CGTGGACACGACTGCTGGAAGT | 259 ORF 61 | 94°C, 1 min; 66°C, 1 min; 70°C, 2 min |
| Tn23 | GGGGCACTTCTTCAACGATGGG | 198 ORF 63 | ||
| 14 | Tn5B | CGGTTGTACAGCTCCTCTTCTGTC | 728 ORF 65 | 94°C, 1 min; 62°C, 1 min; 68°C, 2 min |
| Tn5AR | GCTGCATGACATCGGTCGTATGAC | 1,244 ORF 66 | ||
| 15 | Tn5A | GTCATACGACCGATGTCATGCAGC | 1,267 ORF 66 | 94°C, 1 min; 62°C, 1 min; 68°C, 2 min |
| Tn5R | CTCTGTGAATAAGGTGGCTTGCC | 568 upstream ORF 66 | ||
| Tn2 | CTCGGTATGGAGTATGCTCCAG | 111 ORF 16 (bexA) | 94°C, 1 min; 55°C, 1 min; 66°C, 2 min | |
| Tn2A | GTATTCCACTGCCCAATATGCCGC | 625 ORF 16 | ||
| 86CTn8 | GGTTCTGGCAGGAGGAGTTTACC | 1,086 dprA (NCTC 9343) | 94°C, 1 min; 62°C, 1 min; 68°C, 2 min | |
| 86CTn2 | GTATCGTACAATAGAGAGGTCGCC | 1,249 tnpA1 (CTn86) | ||
| Tn22 | CGTGGACACGACTGCTGGAAGT | 259 tnpA2 (CTn86) | 94°C, 1 min; 62°C, 1 min; 68°C, 2 min | |
| 86CTn9 | CCAGCAACGACCTTCAACGCCTC | 693 ORF-C (NCTC 9343) | ||
| 86CTn2 | GTATCGTACAATAGAGAGGTCGCC | 1,249 tnpA1 (CTn86) | 94°C, 1 min; 51°C, 1 min; 72°C, 2 min | |
| Tn22 | CGTGGACACGACTGCTGGAAGT | 259 tnpA2 (CTn86) | ||
| Tn25A | CCTGTACGACTATGACAGGGCG | 240 tnpB (CTn9343) | 94°C, 1 min; 51°C, 1 min; 72°C, 2 min | |
| Tn22 | CGTGGACACGACTGCTGGAAGT | 259 tnpA2 (CTn9343) | ||
| Tn24 | GGCGAGCTATGTGAATACTACCG | 97 ORF-7 (NCTC 9343) | 94°C, 1 min; 62°C, 1 min; 68°C, 2 min | |
| Tn23 | GGGGCACTTCTTCAACGATGGG | 198 ORF-63 (NCTC 9343) | ||
| E11.2 | TGTGCGGGAACTCCTCTTGCTCTTCCTT | 257 upstream bfmA (CTn86) | 94°C, 1 min; 66°C, 30 s; 68°C, 4 min | |
| E13 | GCCGGATCGTGAAAAGGAATCG | 243 bfmA (CTn86) | ||
| 86CTn2 | GTATCGTACAATAGAGAGGTCGCC | 1,249 tnpA1 (CTn86) | 94°C, 60 s; 55°C, 60 s; 66°C, 2 min | |
| 86CTn1R | GCCTTCTGTGTCCTTCACCATAGA | 1,201 tnpA1 (CTn86) | ||
| Tn25D | GGGCAACTGTCCGTCCTGTCTG | 1,583 tnpA1 (CTn9343) | 94°C, 1 min; 66°C, 1 min; 70°C, 2 min | |
| Tn25 | CGCAGGTCAGACCCTGAGTCGA | 823 tnpA1 (CTn9343) | ||
| Tn25E | ATGTATCTACGGACCGACAGGC | 133 int2 (CTn9343) | 94°C, 1 min; 62°C, 1 min; 68°C, 2 min | |
| Tn25C | ATGTCTCCTCACATGCCACGTC | 669 int2 (CTn9343) | ||
| Tn1 | GGTGTGCCATCGCAATGAAGCA | 1,169 rteA (CTn9343) | 94°C, 1 min; 66°C, 1 min; 70°C, 2 min | |
| Tn1A | GGCGAGACTGTATTGGAGTGGG | 190 rteA (CTn9343) | ||
| Tn21A | CAGGCGTTCCGCCTCGTTCACC | 840 prmN1 (CTn9343) | 94°C, 1 min; 66°C, 1 min; 70°C, 2 min | |
| Tn21 | GGGGATACGACATGTACCGCTC | 86 prmN1 (CTn9343) |
In base pairs (primers used to construct the DNA probes were derived from CTn9343 and its flanking element).
All programs had an initial segment at 94°C for 1 min (except PCR with primers 86CTn2 and Tn22 and Tn25A and Tn22, which had an initial segment at 94°C for 5 min), were run for 30 cycles and had a 10-min final extension at the corresponding extension temperature. The programs to identify CTn86 and CTn9343 circular intermediate form (primers 86CTn2 and Tn22 and Tn25A and Tn22) were run for 20, 25, and 30 cycles to detect the PCR product in the linear range of the reaction.
Inverse PCR.
Chromosomal DNA from ETBF strain 86-5443-2-2 was restriction digested with an appropriate enzyme (PstI or HindIII) (see Fig. 3, below). The restriction fragments (final concentration, 2.5 ng/μl) were ligated to form circular fragments using 4 U of T4 DNA ligase/μl. A total of 40 ng of ligated DNA was used as a template for amplification using primers that specifically amplified the left end of CTn86 (see Fig. 2A and B, below). The sequence and relative location of the primers as well as the parameters used for each inverse PCR are shown in Table 2. PCRs were performed with elongase enzyme mix (Invitrogen, Carlsbad, Calif.) (1 μl) or Taq polymerase (1.5 U) when primers El1.2 and El3 or 86CTn1R and 86CTn2 were used, respectively. Reactions were in 50-μl volumes containing primers (25 pmol), deoxynucleoside triphosphates (200 μM), and MgCl2 (1.5 mM).
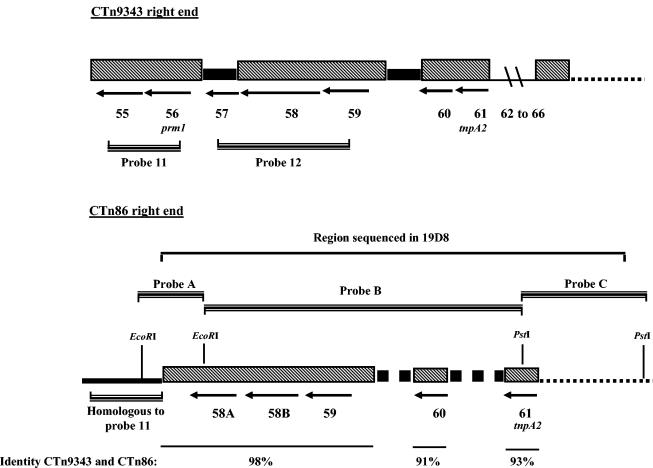
FIG. 3.
Comparison of the right ends of CTn9343 and CTn86. The thick black bars in CTn9343 and dotted thick bars in CT86 show DNA regions specific for CTn9343 and CTn86, respectively. Location of the region homologous to probe 11 used to screen the ETBF 86-5443-2-2 cosmid library is shown for CTn86. Arrows indicate the location of the ORFs and the direction of their transcription. The chromosomal DNA flanking the right ends of CTn86 and CTn9343 is shown as dotted lines.
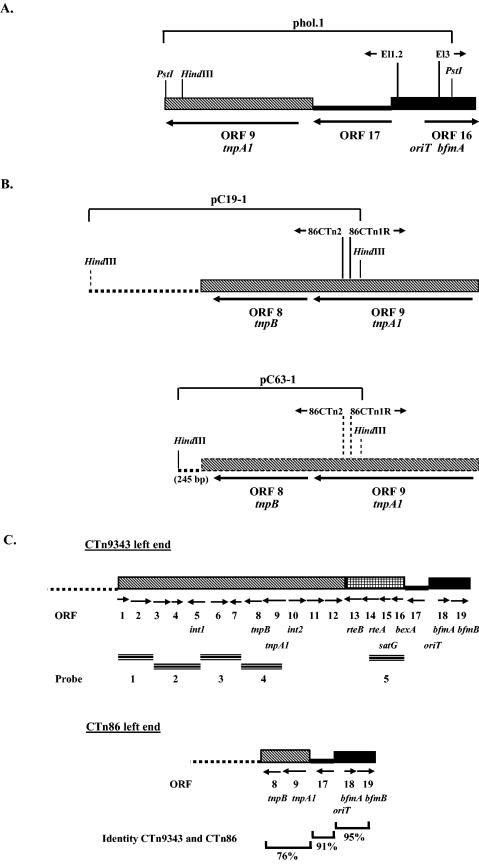
FIG.2.
Identification of the CTn86 left end by inverse PCR. (A and B) ORFs identified after sequencing the PCR products obtained using primers El1.2 and El3 (A) and 86CTn1R and 86CTn2 (B). Relative positions of primers El1.2 and El3 and 86CTn1R and 86CTn2 are shown. (C) Comparison of the left ends of CTn9343 and CTn86. Arrowheads show directions of the primers, and arrows indicate the locations of the ORFs and the direction of their transcription. The chromosomal DNA flanking the left ends of CTn86 and CTn9343 is shown as dotted lines.
Susceptibility to antibiotics.
Using serial twofold dilutions, MICs of tetracycline and virginiamycin M (streptogramin A), as well as of norfloxacin and moxifloxacin (fluoroquinolones), were determined for strains 86-5443-2-2 (pattern I), TM4000 (pattern II), and NCTC 9343 (pattern III) grown anaerobically in BHC medium. MICs were read after 24 h of incubation at 37°C.
Identification of CTn9343 and CTn86 intermediate circular forms.
To detect the joined ends of the excised CTn9343 and CTn86, primers were designed from the ends of the CTns (Tn25A and Tn22 for CTn9343, and 86CTn2 and Tn22 for CTn86) (Table 2). These primers are directed out from the ends of the CTns and cannot yield a PCR product unless the element is in circular form. ETBF 86-5443-2-2 and NTBF NCTC 9343 and I-1345 were grown overnight in BHC broth with and without virginiamycin M (2 μg/ml), moxifloxacin (0.08 μg/ml), or norfloxacin (4 μg/ml). ETBF 86-5443-2-2 and NTBF I-1345 were also grown in BHC broth containing tetracycline (1 μg/ml). Cell concentration was determined by densitometry after overnight growth. Similar concentrations of cells (optical density at 600 nm, 0.8) were spun down, and 10 μl of the pellet was used as a template for PCR (50-μl final volume). PCR conditions were selected to permit detection of the PCR products in the linear range of the reaction (Table 2).
RT-PCR.
Expression of CTn9343 genes was determined by reverse transcription-PCR (RT-PCR) (as previously described [10]) after growing strain NCTC 9343 in BHC broth and BHC broth containing virginiamycin M (2 μg/ml), moxifloxacin (0.08 μg/ml), or norfloxacin (4 μg/ml) and strain I-1345 in either BHC broth or BHC broth containing tetracycline (1 μg/ml). Briefly, total RNA of overnight cultures was obtained using TRIzol reagent (Gibco BRL) according to the manufacturer's protocol. The same amount of total RNA (4 μg) from NCTC 9343 and I-1345 grown in BHC and BHC with antibiotics was used to synthesize cDNA by using the SuperScript preamplification system for first-strand cDNA synthesis kit (Gibco BRL) following the instructions of the manufacturer. The cDNA samples were PCR amplified using primers Tn25D and Tn25, Tn25E and Tn25C, Tn1 and Tn1A, TranF and TranR, Tn18 and Tn19, and Tn21A and TnTn21 to identify expression of tnpA1, int2, rteA, traN, traG, and prmN1, respectively. Sequences and PCR conditions are described in Table 2.
Nucleotide sequence analysis.
Recombinant plasmids and PCR products were sequenced by the DNA Analysis Facility of Johns Hopkins University with an Applied Biosystems model 373A version 2.0.S dye terminator automated sequencer. DNA and amino acid sequences were analyzed using the NCBI BLAST server (1) and the Sequence Analysis software DNAMAN version 5.2.9 (Lynnon BioSoft, Quebec, Canada).
Nucleotide sequence accession number.
The accession number of the 25-kb region containing the left end of CTn86 and the flanking region is AY372755, and that for the 6-kb region containing the right end of CTn86 and the flanking region is AY375536.
RESULTS
Identification of CTn9343.
To identify the complete sequence of the genetic element flanking the BfPAI, we aligned the partial sequence (12 kb in total) of the genetic element flanking the BfPAI from ETBF 86-5443-2-2 with the sequence of B. fragilis strains NCTC 9343 (pattern III; lacks the BfPAI but contains its flanking region) and 638R (pattern II; lacks both the BfPAI and its flanking region) produced by the Wellcome Trust Sanger Institute (http://www.sanger.ac.uk/Projects/B_fragilis/). This identified that the 12-kb region sequenced in ETBF 86-5443-2-2 was 96% identical to the corresponding sequence in NCTC 9343. The ends of the flanking element then were determined by alignment of the appropriate sequences of strains NCTC 9343 and TM4000. These alignment results initially identified that the genetic element flanking the BfPAI was ∼80 kb in length and contained 66 putative open reading frames (ORFs) (Table 3). Sequence analysis using the NCBI BLAST server (1) suggested that this genetic element is a CTn (herein designated CTn9343).
TABLE 3.
Putative ORFs in CTn9343 and flanking regions
| ORF (gene name)a | No. of amino acids | Putative protein | % Identity/ % similarity (range)b | E valuec | Source | Accession no. |
|---|---|---|---|---|---|---|
| 1 | 120 | No significant identity | ||||
| 2 | 462 | No significant identity | ||||
| 3 | 211 | No significant identity | ||||
| 4 | 187 | No significant identity | ||||
| 5 (int1) | 268 | Integrase | 63/81 (263) | 6e-95 | S. pneumoniae | AAK99251.1 |
| 6 | 342 | Type I R-M system (chain S) | 32/43 (343) | 3e-34 | S. pneumoniae | AAK99252.1 |
| 7 | 102 | Type I R-M system (chain S) | 33/54 (62) | 0.90 | S. pneumoniae | AAK99252.1 |
| 8 (tnpB) | 263 | IS21 transposase | 100/100 (263) | e-145 | B. fragilis | AF303352.2 |
| IstB (IS21) | 34/56 (239) | 1e-36 | E. coli | BVECIT | ||
| 9 (tnpA1) | 548 | IS21 transposase | 100/100 (548) | 0.00 | B. fragilis | AF303352.1 |
| IstA (IS21) | 27/43 (382) | 3e-20 | E. coli | BVECIS | ||
| 10 (int2) | 240 | Integrase | 91/92 (205) | e-104 | B. fragilis | AF303352.5 |
| Integrase (CTnDOT) | 65/78 (223) | 1e-82 | B. thetaiotaomicron | CAC47921.1 | ||
| 11 | 349 | Hypothetical protein | 31/51 (357) | 4e-42 | T. erythraeum | ZP00072163.1 |
| 12 | 221 | Hypothetical protein | 23/41 (186) | 5e-14 | T. erythraeum | ZP00072164.1 |
| 13 (rteB) | 440 | RteB | 98/98 (440) | 0.00 | B. fragilis | AF303352.3 |
| RteB (CTnDOT) | 85/93 (449) | 0.00 | B. thetaiotaomicron | AAA22921 | ||
| 14 (rteA) | 423 | RteA | 96/96 (423) | 0.00 | B. fragilis | AF303352.4 |
| RteA (CTnDOT) | 64/78 (337) | e-117 | B. thetaiotaomicron | A41860 | ||
| 15 (satG) | 205 | Acetyltransferase | 72/83 (205) | 2e-88 | B. thetaiotaomicron | AA077491.1 |
| Acetyltransferase | 65/79 (204) | 3e-79 | C. acetobutylicum | AAK78753.1 | ||
| 16 (bexA) | 235 | Putative cation efflux pump | 58/73 (229) | 1e-69 | B. thetaiotaomicron | AA077007.1 |
| 17 | 314 | Hypothetical protein | 55/69 (310) | 1e-89 | B. thetaiotaomicron | AA077572.1 |
| 18 (bfmA) | 180 | Mobilization protein A | 97/100 (180) | 9e-97 | B. fragilis | AAD05337.1 |
| 19 (bfmB) | 316 | Mobilization protein B | 97/98 (316) | e-180 | B. fragilis | AAD05338.1 |
| MobB | 26/42 (257) | 2e-15 | B. vulgatus | CAA63435 | ||
| 20 (bfmC) | 534 | Mobilization protein C | 98/99 (534) | 0.00 | B. fragilis | AAD05339.1 |
| TraG family protein | 23/45 (453) | 6e-30 | P. gingivalis | AAQ66535.1 | ||
| Type IV secretion, VirD4 | 21/40 (369) | 4e-04 | V. vulnificus | AA007601.1 | ||
| 21 | 117 | No significant identity | ||||
| 22 | 1221 | Hypothetical protein | 21/41 (551) | 1e-17 | H. sommus | ZP00122678.1 |
| 23 | 438 | Hypothetical protein | 29/56 (168) | 1e-17 | C. perfringes | NP561571.1 |
| 24 | 134 | Hypothetical protein | 32/53 (80) | 4e-04 | C. perfringes | NP561571.1 |
| 25 | 195 | RNA polymerase ECF-type sigma factor | 40/67 (170) | 2e-34 | B. thetaiotaomicron | AA077570.1 |
| 26 | 318 | Putative anti-sigma factor | 38/56 (316) | 3e-57 | B. thetaiotaomicron | AA077569.1 |
| 27 | 1127 | Putative outer membrane protein involved in nutrient binding | 32/50 (1,168) | e-161 | B. thetaiotaomicron | AA077568.1 |
| 28 | 545 | Putative outer membrane protein involved in nutrient binding | 28/44 (577) | 8e-44 | B. thetaiotaomicron | AA077302.1 |
| 29 (xyl) | 519 | Putative xylanase | 35/54 (495) | 7e-74 | B. thetaiotaomicron | AA078149.1 |
| 30 (bsh) | 359 | Choloylglycine hydrolase | 36/56 (341) | 1e-53 | B. melitensis | AAL51724.1 |
| Penicillin acylase | 31/51 (340) | 1e-35 | L. monocytogenes | CAC98525.1 | ||
| 31 | 326 | Methylase HpyI | 23/37 (217) | 0.05 | H. pylori | AAB07029.1 |
| 32 | 265 | Chromosome-partitioning ATPase | 36/57 (260) | 1e-41 | B. thetaiotaomicron | AAO45329.1 |
| 33 | 614 | No significant identity | ||||
| 34 | 527 | Cell surface protein | 23/39 (312) | 2e-04 | M. acetivorans | AAM07636.1 |
| 35 | 183 | Putative peptidase | 40/53 (189) | 9e-28 | B. thetaiotaomicron | AAO79101.1 |
| 36 (traN) | 384 | TraN-like protein (CTnDOT) | 33/57 (365) | 2e-55 | B. thetaiotaomicron | AA045314.1 |
| 37 | 149 | No significant identity | ||||
| 38 (traM) | 326 | Tra-M-like protein (CTnDOT) | 30/46 (325) | 4e-28 | B. thetaiotaomicron | AA045313.1 |
| 39 | 213 | Hypothetical protein | 40/60 (204) | 2e-35 | B. thetaiotaomicron | AA045312.1 |
| 40 | 257 | Hypothetical protein | 25/44 (264) | 2e-15 | B. thetaiotaomicron | AA045311.1 |
| 41 (traI) | 173 | Tra-I-like protein (CTnDOT) | 28/49 (182) | 2e-09 | B. thetaiotaomicron | AA045310.1 |
| 42 (traG) | 806 | Tra-G-like protein (CTnDOT) | 43/64 (803) | 0.00 | B. thetaiotaomicron | AA045309.1 |
| 43 | 86 | No significant identity | ||||
| 44 | 102 | Hypothetical protein | 37/61 (77) | 2e-09 | B. thetaiotaomicron | AA045307.1 |
| 45 | 265 | No significant identity | ||||
| 46 | 534 | Platelet binding protein GspB | 20/38 (349) | 4e-05 | S. gordii | AAL13053.1 |
| 47 | 320 | No significant identity | ||||
| 48 | 293 | No significant identity | ||||
| 49 | 687 | Hypothetical protein | 27/45 (211) | 1e-11 | B. thetaiotaomicron | AA077766.1 |
| 50 | 180 | Hypothetical protein | 37/57 (189) | 7e-30 | B. thetaiotaomicron | AA076173.1 |
| 51 | 140 | No significant identity | ||||
| 52 | 172 | No significant identity | ||||
| 53 | 115 | No significant identity | ||||
| 54 | 210 | No significant identity | ||||
| 55 | 242 | No significant identity | ||||
| 56 (prmN1) | 303 | PrmN1 | 37/51 (300) | 3e-44 | B. uniformis | AF238307 |
| 57 | 185 | No significant identity | ||||
| 58 | 369 | Hypothetical protein | 32/47 (370) | 1e-35 | B. thetaiotaomicron | AA079131.1 |
| Replication protein | 35/53 (128) | 3e-13 | Bacteriophage bIL286 | AF3236669.16 | ||
| DnaD domain protein | 29/49 (127) | 9e-09 | E. faecalis | AA081863.1 | ||
| 59 | 165 | Hypothetical protein | 32/57 (101) | 3e-13 | B. thetaiotaomicron | AA079131.1 |
| DnaD domain protein | 31/49 (99) | 3e-06 | E. faecalis | AA081863.1 | ||
| 60 | 105 | No significant identity | ||||
| 61 (tnpA2) | 124 | IS21 transposase (TnpA) | 44/65 (121) | 8e-23 | B. fragilis | AA303352.1 |
| 62 | 447 | Type I R-M system (chain S) | 33/50 (420) | 4e-51 | S. pneumoniae | AAK99252.1 |
| 63 | 148 | ABC-type sugar transport system | 25/49 (136) | 6e-06 | F. nucleatum | EAA24799.1 |
| ATPase component | ||||||
| Doc protein | 30/52 (88) | 7e-04 | S. oneidensi | AAN53454.1 | ||
| 64 | 338 | Hypothetical protein | 44/64 (337) | 1e-74 | Magnetococcus sp. | ZP 00044096.1 |
| 65 (hsdM) | 506 | Type I R-M system (M subunit) | 55/73 (498) | e-161 | S. pneumoniae | AAK74667.1 |
| 66 (hsdR) | 771 | Type I R-M system (R subunit) | 49/67 (774) | 0.00 | S. pneumoniae | AAK99254.1 |
Name based on sequence homology.
Identity and similarity are presented as percent amino acid identity and similarity between the CTn9343 ORF and the best hit. The range is the number of amino acids over which this identity and similarity exist. Identity and similarity were determined by BLAST (1).
E value of <0.05 was considered statistically significant (1).
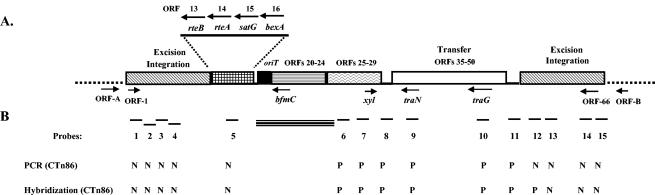
CTn9343 is organized in a modular fashion, with clusters of genes with related functions (Fig. 1A). Similar to CTnDOT, CTn9343 contains the regulatory genes rteA and rteB (ORFs 13 and 14, respectively); however, these genes are not adjacent to tetQ as in CTnDOT but rather are adjacent to ORFs that predict proteins with significant homology to proteins involved in resistance to virginiamycin M (satG) and fluoroquinolones (bexA) (Fig. 1A; Table 3). Putative genes involved in integration-excision, like integrases (int1 and int2), transposases (tnpA1, tnpA2, and tnpB), and primases (prmN1) (Table 3) are located at the left and right ends of CTn9343. The predicted protein encoded by int2 shares significant homology to the integrase of CTnDOT (Table 3). Adjacent to the right-end putative integration-excision region, there is a cluster of genes potentially involved in the transfer of the transposon. However, only four ORFs (ORFs 36, 38, 41, and 42) of this cluster of genes predict proteins with significant homology with transfer proteins encoded by CTnDOT (Table 3). Besides rteA, rteB, int2, and these four putative transfer genes, there is no additional sequence homology between CTn9343 and CTnDOT-related transposons. In contrast to most self-transmissible elements, the putative oriT-mob region (bfmA and bfmB) is not adjacent to the transfer region. In CTn9343, two clusters of genes (ORFs 20 to 24 and 25 to 29) and a region containing ORFs 30 to 34 separate the oriT-mob from the transfer region (Fig. 1A). The cluster containing ORFs 25 to 29, all predicted to be transcribed in the same direction, may be involved in the regulation of xylose utilization. The second cluster containing ORFs 20 to 24 is also predicted to be transcribed in the same direction, although no overall potential function for this region is evident. The last ORF of this cluster (ORF 20 [bfmC]) is a putative mobilization gene that encodes a protein predicted to share significant homology with a component of a type IV secretion system (Table 3). ORFs 30, 31, and 32 are predicted to encode proteins potentially involved in bile hydrolysis, protection from restriction endonucleases, and chromosome partitioning, respectively. Both putative ends of CTn9343 (ORFs 6, 62, 65, and 66) contain genes encoding proteins with significant homology to proteins conferring protection from restriction endonuclease cleavage.
FIG. 1.
(A) Initial schematic map of CTn9343. The chromosomal DNA flanking the integrated element is shown by dotted lines. The putative excision-integration, transfer, oriT-mob, and rteA-rteB-satG-bexA regions are shown. The thin line between ORF 29 and the transfer region contains genes that are predicted to encode proteins contributing to resistance to bile (ORF 30), protection from restriction endonuclease cleavage (ORF 31), and partitioning of DNA into bacterial cells (ORF 32) (Table 3). The thin line between the transfer and excision-integration regions contains genes (ORFs 51 to 55) that encode proteins without significant identity to any protein in the GenBank database (Table 3). Arrows show the locations of representative ORFs and the direction of their transcription. (B) Relative locations of the PCR products used as probes to characterize CTn86. The results of PCR and hybridization to these probes are indicated beneath each probe (P, positive; N, negative). The thick bar between probes 5 and 6 indicates the ∼12-kb region sequenced in ETBF 86-5443-2-2. The BfPAI in ETBF 86-5443-2-2 is located between oriT and bfmC.
The putative start codons of ORFs 1 and 66 are located in bp 1,804,281 and 1,727,162 of the NCTC 9343 chromosome, respectively. Alignment of the NCTC 9343 and 638R sequences indicated that the putative left end of CTn9343 is approximately 164 bp upstream of ORF 1, and the putative right end is 1,329 bp upstream of ORF 66. The closet ORF to the putative left end of CTn9343 is an ORF (designated ORF-A) of 915 bp located 471 bp upstream of ORF 1 (Fig. 1A). The predicted protein encoded by ORF-A shares significant identity (73%) with a protein of unknown function encoded by an ORF located upstream of a capsular biosynthesis locus (15). Like this locus, ∼1 kb upstream of ORF-A there is a cluster of genes that may be involved in the biosynthesis of capsular polysaccharide. The closest ORF upstream of the putative right end of CTn9343 is located 1,353 bp upstream of ORF 66 (Fig. 1A). This ORF, designated ORF-B, encodes a protein of 310 amino acids with no significant identity to any protein in the GenBank database.
Contribution of satG and bexA to antibiotic resistance in strain NCTC 9343.
To determine if satG and bexA confer resistance to virginiamycin M and fluoroquinolones, respectively, the MICs of virginiamycin M as well as of norfloxacin and moxifloxacin (fluoroquinolones) were determined. Colony blot hybridizations using a fragment containing satG and bexA as a probe (Fig. 1B, probe 5) determined that strains 86-5443-2-2 (pattern I; contains a genetic element related to CTn9343 flanking the BfPAI) and TM4000 (pattern II; lacks CTn9343 or a related genetic element) lack satG and bexA. Notably, the MICs for strains NCTC 9343, 86-5443-2-2, and TM4000 of virginiamycin and moxifloxacin were identical (10 and 0.15 μg/ml, respectively), whereas 86-5443-2-2 had a higher MIC of norfloxacin (16 μg/ml) than strains NCTC 9343 and TM4000 (8 μg/ml). These results indicate that the presence of satG and bexA in CTn9343 does not increase resistance to virginiamycin M and/or fluoroquinolones in strain NCTC 9343.
The BfPAI is contained in CTn86.
Our laboratory's previous studies demonstrated that in ETBF strains the BfPAI is integrated between bfmB and bfmC (9). We sequenced a ∼12-kb region flanking the BfPAI in ETBF strain 86-5443-2-2. An alignment of this sequence flanking the BfPAI with the appropriate sequence of CTn9343 showed that these two sequences were 96% identical (Fig. 1B). To determine whether the entire CTn9343 sequence flanks the BfPAI in strain 86-5443-2-2, 15 sets of primers spanning the entire CTn9343 sequence were designed (Table 2). Using these primers for PCR analysis and colony blot hybridizations (using the PCR products as probes), CTn9343 was characterized in ETBF 86-5443-2-2 (Fig. 1B). ETBF 86-5443-2-2 was PCR and probe positive to the central region of CTn9343 spanning probes 6 to 11; however, it was negative by PCR and hybridization to the regions spanning probes 1 to 5 and 13 to 15, suggesting that the ends of CTn9343 in ETBF 86-5443-2-2 were deleted. ETBF 86-5443-2-2 was probe positive but PCR negative to the region spanning probe 12, suggesting that the deletion of the end of CTn9343 occurred in this region. Because the CTn in ETBF 86-5443-2-2 differs from that of NCTC 9343, the genetic element flanking the BfPAI in ETBF 86-5443-2-2 was termed CTn86. None of the 15 probes derived from CTn9343 to characterize CTn86 (Fig. 1B) hybridized with B. thetaiotaomicron 4107, a strain that contains CTnDOT integrated into the chromosome (data not shown).
The left end of CTn86 is deleted.
Because CTn86 was probe negative to the region spanning probes 1 to 5, the left end of CTn86 was hypothesized to be close to the sequenced region oriT-mob (Fig. 1B). To identify the left end of CTn86, the sequence adjacent to the oriT-mob region was cloned by inverse PCR. Using primers El1.2 and El3 derived from the oriT-mob region, the next PstI fragment (4.3 kb) was cloned into pGEM-T to originate phol.1 (Fig. 2A). The 4.3-kb PstI fragment hybridized with all pattern I and III strains but not with any of the pattern II strains listed in Table 1, indicating that this fragment did not contain the end of CTn86 (data not shown). Sequence analysis of the 4.3-kb PstI fragment showed that in CTn86, ORF 17 is adjacent to ORF 9 (tnpA1), with a deletion of the region that encodes int1, ORF 11, ORF 12, rteB, rteA, satG, and bexA (Fig. 2A; see also Fig. 8, below).
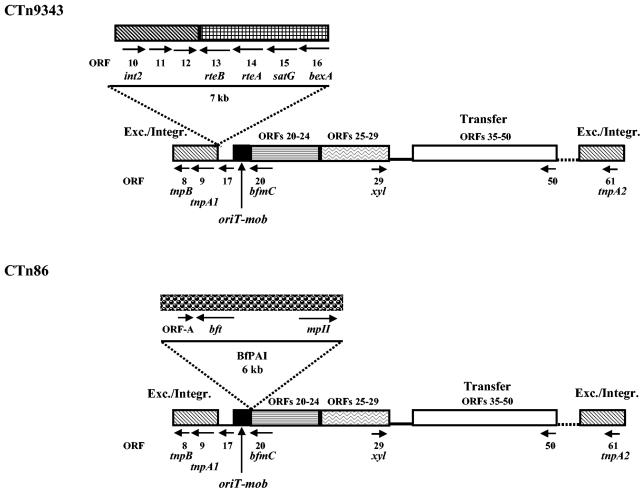
FIG. 8.
Comparison of the real schematic maps of CTn9343 and CTn86. Both CTns have the same basic structure, except that CTn9343 has an extra 7-kb region containing ORFs 10 to 16 and CTn86 contains the 6-kb BfPAI integrated between bfmB (oriT-mob) and bfmC.
To identify the next adjacent region of the left end of CTn86, a second inverse PCR was performed to clone the next HindIII fragment (Fig. 2B). Unexpectedly, two PCR products, 3.0 and 1.6 kb, were obtained. Both PCR products were cloned in pGEM-T to produce pC19-1 and pC63-1 for the 3.0- and 1.6-kb PCR products, respectively. Both PCR products hybridized by colony blotting with all pattern II and III B. fragilis strains tested (Table 1), suggesting that both PCR products contained the left end of CTn86 (data not shown). Sequence analysis of pC19-1 and pC63-1 revealed that ETBF 86-5443-2-2 contains two copies of CTn86 integrated in different regions of the chromosome. The sequence analyses also indicated that ORF 8 (tnpB) is the last gene in the left end of both CTn86 copies (Fig. 2B; see also Fig. 8). Comparison of the sequence of the left end of CTn86 with the appropriate sequence of CTn9343 revealed that these sequences shared only 76% identity in the region encoding ORF 9 (tnpA1) and ORF 8 (tnpB) (Fig. 2C).
Identification of the CTn86 right end.
The colony blotting and PCR results indicated that ETBF 86-5443-2-2 was positive to probe 12 but negative to PCR when primers for probe 12 were used (Fig. 1B). These results suggested that the right end of CTn86 is contained in the region spanning probe 12. To clone the right end of CTn86, probe 11 was used to screen a cosmid library of ETBF 86-5443-2-2. A positive cosmid (19D8) was restriction mapped, and three adjacent restriction fragments (probes A, B, and C) (Fig. 3) were used to probe a collection of B. fragilis strains (Table 1). Probe C, but not probes A and B, hybridized with all pattern II B. fragilis strains (data not shown), indicating that the region spanning probe C contains the right end of CTn86. Sequence analysis of the region spanning probe C revealed that ORF 61 (tnpA2) is the last ORF of the right end of CTn86. Since ORF 61 is downstream of the predicted right end of CTn86 (the region spanning probe 12), the DNA regions spanning probes A and B were also sequenced. Comparison of the sequences of probes A to C with the appropriate sequence of CTn9343 showed overall sequence identity of 95%; however, CTn86 has deletions and additions that altered its coding sequence (Fig. 3). CTn86 has a deletion of 446 bp containing the major part of ORF 57. This deleted region is part of probe 12 and explains the negative PCR, positive DNA hybridization results discussed above. Furthermore, ORF 58 of CTn9343 is not present in the CTn86 sequence. In this region, CTn86 encodes two ORFs (designed ORF 58A and 58B) of 444 and 612 bp, respectively. The predicted proteins encoded by ORF 58A and 58B do not share significant identity or similarity with any protein in the GenBank database. The DNA region downstream of ORF 60 (∼500 bp) also differs in CTn9343 and CTn86, and CTn86 contains an extra 730-bp sequence immediately downstream of ORF 61 (Fig. 3).
Identification of CTn86 integration sites.
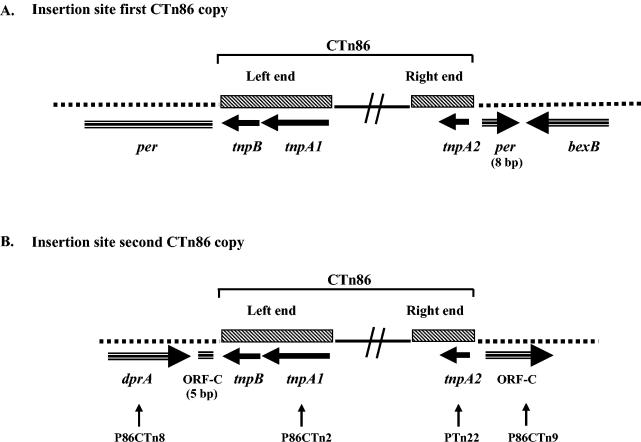
Sequence analysis of cosmid C19D8 (which contains the CTn86 right end and ∼1,700 bp flanking this region) (Fig. 3) showed that 173 bp downstream of the putative tnpA2 start codon there is an ORF of 1,365 bp that encodes a predicted protein sharing significant homology (52% identity and 72% similarity) with putative Na+-driven multidrug efflux pump proteins. Due to the predicted similar function of this ORF to the BexA protein described for B. thetaiotaomicron (18), this ORF was designated bexB (Fig. 4A). Sequence analysis of pC19-1 (which contains the CTn86 left end and ∼1,900 bp flanking this region) (Fig. 2B) revealed that the left end of one copy of CTn86 is flanked by an ORF that encodes a predicted protein with significant homology (38% identity and 59% similarity) to ABC transporter-permease proteins. This ORF was designated per (Fig. 4A). Alignment of the left and right sequences flanking CTn86 with the appropriate sequence of NCTC 9343 (http://www.sanger.ac.uk/Projects/B_fragilis/) revealed that in strain NCTC 9343 per is 2,295 bp in length (in pC19-1, only ∼1,900 bp were sequenced), and per and bexB are adjacent (99% identical to the region flanking CTn86). Alignment of the ends of CTn86 and flanking regions with the NCTC 9343 per and bexB genes showed also that integration of CTn86 interrupts the last 8 bp at the 3′ end of per (Fig. 4A). The per and bexB genes transcribe in opposite directions, and the start codon of per is located at bp 2,587,491 of the NCTC 9343 chromosome.
FIG. 4.
Insertion sites of the two copies of CTn86 into the ETBF 86-5443-2-2 chromosome. The dotted lines show the chromosomal DNA. Horizontal arrows indicate the locations of the ORFs and the direction of their transcription. The relative positions of the primers P86CTn8 and P86CTn2 and PTn22 and P86CTn9 used to identify by PCR the integration site of the second copy of CTn86 are shown. Integration of CTn86 interrupts the last 8 and first 5 bases of per and ORF-C, respectively.
To determine the integration site of the second copy of CTn86, the sequence of pC63-1 (which contains the CTn86 left end and ∼245 bp of its flanking region) (Fig. 2B) was analyzed. To determine the locus where CTn86 is integrated, the 245-bp region flanking CTn86 was aligned with the NCTC 9343 sequence (http://www.sanger.ac.uk/Projects/B_fragilis/). The alignment results indicated that the left end of CTn86 is flanked by an ORF of 1,122 bp encoding a protein with significant homology (73% identity and 83% similarity; score, 521) with SmF/DprA family proteins (accession no. AAO78176.1) (Fig. 4B). In Haemophilus influenzae and Helicobacter pylori, these proteins are required for natural chromosomal and plasmid transformation (2, 13). This ORF flanking CTn86 was designated dprA (Fig. 4B). The right end of the second copy of CTn86 is flanked by an ORF of 1,020 bp (designated ORF-C) that encodes a protein sharing significant homology (62% identity and 74% similarity) with a protein of B. thetaiotaomicron of unknown function (accession no. AAO78167.1) (Fig. 4B). ORF-C and dprA transcribe in the same direction, and the start codon of ORF-C is bp 5,173,855 of the NCTC 9343 genome. Integration of CTn86 between dprA and ORF-C was confirmed by PCR using primers derived from the left and right chromosome-CTn86 junction (primers 86CTn8 and 86CTn2 for the left end and Tn22 and 86CTn9 for the right end [Fig. 4B]). Sequencing of the PCR products revealed that the region flanking CTn86 in ETBF 86-5443-2-2 shares 99% identity with the appropriate sequence of NCTC 9343, and integration of CTn86 interrupts the first 5 bp at the N-terminal region of ORF-C (Fig. 4B).
CTn86 forms a circular intermediate.
CTns initiate conjugal transfer by excising from the chromosome to form a circular intermediate. CTn86 excision and formation of a circular intermediate were evaluated by PCR using primers designed from the end sequences of the integrated transposon and directed outward (primers 86CTn2 and Tn22). A 1.5-kb PCR product was detected intracellularly after growing ETBF 86-5443-2-2 overnight in BHC. Identification of the CTn86 circular form suggested that this genetic element can be transferred between B. fragilis strains. Strain 86-5443-2-2 is tetracycline resistant (MIC, 10 μg/ml); however, CTn86 does not contain the tetQ gene. To determine if tetracycline induces formation of the CTn86 circular intermediate, ETBF 86-5443-2-2 was grown overnight in BCH medium containing tetracycline (1 μg/ml) followed by semiquantitative PCR for the circular form. The PCR product quantity was similar after growth in the presence or absence of tetracycline (data not shown), indicating that excision and formation of the circular intermediate were not induced by tetracycline, like that of CTnDOT.
Proposed model for CTn86 integration.
Sequence analysis of the 1.5-kb PCR product revealed that the exact CTn86 left end is 38 bp downstream from the putative stop codon of ORF 8 (tnpB) and the right end is 74 bp upstream from the putative start codon of ORF 61 (tnpA2). In the circular form, these nucleotides are joined by a TA sequence (Fig. 5A). The ends of the transposon contain short inverted repeat (IR) sequences of 4 bp separated by the TA sequence (Fig. 5A). Sequence analysis of the CTn86 integrated form showed that the ends of both copies of CTn86 are flanked by direct repeat sequences of 7 bp (Fig. 5C), suggesting that these direct repeat sequences are the target integration sites of CTn86 and that integration of the transposon duplicates the target sites. The 7-bp target sites were identified at the carboxy terminus of per and the N terminus of ORF-C. However, due to duplication of the target site, the start codon of ORF-C was not affected (Fig. 5B and C). Comparison of the sequences of the ends of CTn86 with the 7-bp target sites and comparison of the sequences of the 7-bp target sites for both copies of CTn86 did not reveal any sequence similarity. These results suggest that integration of CTn86 is not site selective.
FIG. 5.
Model for integration of CTn86 into the B. fragilis chromosome. (A) Circular intermediate of CTn86 prior to integration. attCTn86 in the right-left junction is enlarged to show the IR sequence at the ends of CTn86 separated by TA. (B) Sequences of the regions of per and ORF-C where CTn86 integrates (B. fragilis aattB). The 7-bp target sites in per and ORF-C are indicated by boxes. There is not any sequence similarity between the ends of CTn86 and the target sites. (C) Sequences of the ends of CTn86 and flanking regions. Integration of CTn86 into the target site resulted in duplication of the target sites (shown in boxes).
The real ends of CTn9343 are tnpB and tnpA2.
Initial attempts to identify the joined ends of the circular form of CTn9343 by PCR using primers derived from ORF 1 (putative left end) and ORF 66 (putative right end) were unsuccessful. Colony blot hybridization of our collection of B. fragilis strains using the 15 probes spanning CTn9343 to characterize CTn86 (Fig. 1B) showed that only 8 of 80 NTBF pattern III strains (expected to contain CTn9343) were positives to probes 1 to 3 and 13 to 15 (Franco and Sears, unpublished) and, unexpectedly, 2 of 89 B. fragilis pattern II strains (expected to lack CTn9343 and CTn86 transposons) were positive to probes 1 to 3 and 13 to 15 (strains LM46 and K518 [Table 1]). BLASTP (1) analysis of the ORFs contained within the regions spanning probes 1 to 3 and 13 to 15 showed that ORF 5 encodes a protein with significant homology to integrases (designated int1), and ORFs 6, 7, 62, 65, and 66 encode proteins with homology to components of a type I restriction modification system (type I R-M system) (Table 3). Proteins encoded by ORFs 6 and 62 had significant homology with subunit S (hsdS) of the type I R-M system, and proteins encoded by ORFs 65 and 66 had significant homology with subunits M (hsdM) and R (hsdR), respectively (Table 3; Fig. 6). Because of these results and the results defining CTn86, the ends of CTn9343 were hypothesized to be adjacent to ORF 8 (tnpB) and ORF 61 (tnpA2), similar to CTn86, and the DNA regions flanking these ORFs (ORFs 1 to 7 at the left end and 62 to 66 at the right end, encompassed by probes 1 to 3 and 13 to 15) were hypothesized to represent another foreign genetic element.
FIG. 6.
Integration of CTn9343 in another foreign genetic element. The real ends of CTn9343 are tnpB (left) and tnpA2 (right). The regions flanking these ends (ORFs 1 to 7 and 62 to 66) are part of another genetic element integrated into the B. fragilis chromosome (dotted lines). Relative positions of primers Tn24 and Tn23 to generate hsdS4 are shown.
To address this latter hypothesis, primers Tn24 and Tn23 derived from ORF 7 and ORF 63 sequences (Fig. 6), respectively, were used for PCR with the two B. fragilis pattern II strains positive by hybridization to probes 1 to 3 and 13 to 15 (strains LM46 and K518 [Table 1]). The PCR yielded a product (ca. 1.5 kb) in both B. fragilis pattern II strains but not in strains NCTC 9343 or TM4000 (pattern II strains negative to probes 1 to 3 and 13 to 15), suggesting that the regions spanning probes 3 and 13 are adjacent in these two pattern II B. fragilis strains (Fig. 6). Sequence analysis of the PCR product identified an ORF (designated hsdS). The first 1,343 bp of this ORF had significant identity (93%) to ORF 62, and the last 129 bp shared significant identity (99%) with ORF 7. These results indicated that CTn9343 is contained in another foreign genetic element encoding a type I R-M system, and its integration in this genetic element interrupts the hsdS gene. Our results also indicate that, like CTn86, the left and right ends of CTn9343 are close to the stop codon of tnpB and start codon of tnpA2, respectively (Fig. 6; see also Fig. 8).
Identification of the CTn9343 intermediate circular form.
Once the likely real ends of CTn9343 were determined, primers derived from the left (Tn25A) and right (Tn22) ends were designed to detect the intermediate circular form. Similar to ETBF 86-5443-2-2, a ∼0.8-kb PCR product was detected intracellularly after growing NCTC 9343 overnight in BHC broth, suggesting that CTn9343 may be transferable between B. fragilis strains.
To determine if virginiamycin M and/or fluoroquinolones induce formation of the CTn9343 circular intermediate, strain NCTC 9343 was grown overnight in the presence of virginiamycin M as well as norfloxacin and moxifloxacin (fluoroquinolones). No induction of NCTC 9343 was detected by PCR in the presence of these antibiotics (data not shown). Furthermore, RT-PCR analysis showed similar levels of expression of rteA, putative integration-excision genes int2, tnpA1, and prmN1, and putative transfer genes traN and traG after growth of strain NCTC 9343 in the presence or absence of these antibiotics (data not shown). These results suggested that virginiamycin M and/or fluoroquinolones do not induce formation of the CTn9343 circular intermediate. Strain NCTC 9343 is tetracycline sensitive, and so induction of the CTn9343 circular form by tetracycline was determined using the tetracycline-resistant strain I-1345 (10). Similar to NCTC 9343, strain I-1345 is pattern III and hybridizes with the 15 probes derived from different regions spanning CTn9343 (Fig. 1B). These results suggest that strain I-1345 contains a genetic element similar to CTn9343. The CTn9343 circular form was detected in strain I-1345, and the circular form was not induced by tetracycline (data not shown). Similarly, RT-PCR analysis showed that tetracycline did not induce expression of rteA, int2, tnpA1, prmN1, traN, or traG in strain I-1345. These results suggest that tetracycline does not induce CTn9343 excision and formation of circular intermediates.
Proposed model for CTn9343 integration.
Sequence analysis of the 0.8-kb PCR product to identify the circular form revealed that the exact ends of CTn9343 are 89 bp downstream from the putative stop codon of ORF 8 (tnpB) and 74 bp upstream from the putative start codon of ORF 61 (tnpA2). These bases are joined by an AT sequence and, like CTn86, the ends of CTn9343 have a short IR sequence of 4 bp separated by the AT sequence (Fig. 7A). Similar to CTn86, the target site of CTn9343 is a 7-bp sequence that is duplicated after the integration of the transposon (Fig. 7B and C). The 7-bp target site of CTn9343 does not share any similarity with the target sites of CTn86 or with the ends of CTn9343, suggesting that integration of CTn9343 is not site specific.
FIG. 7.
Model for integration of CTn9343 into the B. fragilis chromosome. (A) Circular intermediate of CTn9343 prior to integration. attCTn9343 in the right-left junction is enlarged to show the IR sequence at the ends of CTn9343 separated by AT. (B) Sequence of the region of hsdS where CTn86 integrates (B. fragilis aattB). The 7-bp target site in hsdS is indicated by the box. The arrow shows the direction of hsdS transcription. Integration of CTn9343 interrupts HsdS in tyrosine (Y) 398. Like CTn86, there is no sequence similarity between the ends of CTn9343 and the target site. (C) Sequences of the ends of CTn9343 and flanking regions. Like CTn86, integration of CTn9343 into the target site resulted in duplication of the target sites (shown in boxes).
DISCUSSION
In this study, the genetic element flanking the BfPAI in ETBF 86-5443-2-2 and a related genetic element in NCTC 9343 were identified and characterized. The results suggest that these genetic elements are members of a new family of CTns not described previously. These putative CTns, designated CTn86 and CTn9343, for ETBF 86-5443-2-2 and NCTC 9343, respectively, differ from previously described Bacteroides species CTns in a number of ways. These new transposons do not carry tetQ, and the excision and formation of circular intermediates are not regulated by tetracycline; they are predicted to have a different mechanism of transposition; and their sequences have very limited sequence homology with CTnDOT or other described CTns.
Initial alignment results with NCTC 9343 (pattern III) and 638R (pattern II) sequences indicated that CTn9343 was ∼80 kb in length; however, further colony blot hybridizations, PCR, and sequence analysis determined that the real ends of CTn9343 define a genetic element of 64,229 bp. The results indicated that the ∼16-kb region flanking CTn9343 is another foreign genetic element that encodes a type I R-M system (43) and a lambda family integrase (int1) (24). The highest homologies of the putative proteins comprising this type I R-M system (HsdS, HsdR, and HsdM), as well as Int1, are with those of Streptococcus pneumoniae (Table 3), suggesting that B. fragilis strains may have acquired this genetic element by horizontal transfer from a gram-positive organism.
CTn9343 may also have arisen from gram-positive bacteria. CTn9343 harbors antibiotic resistance genes (fluoroquinolones and virginiamycin M) that are similar to those from gram-positive organisms. The predicted protein encoded by the virginiamycin M resistance gene (satG) is 65% identical and 79% similar to an analog protein of Clostridium acetobutylicum (Table 3). Virginiamycin M is usually used in animal feed; this factor may have selected for the evolution and transfer of CTn9343. Recently, a new Bacteroides CTn, CTnGERM1, has been reported that also contains genes found in gram-positive bacteria (39). These results support the idea that CTns can be acquired by Bacteroides species from other genera. B. fragilis strains may have acquired CTn9343 and its flanking genetic element together during the transposition of CTn9343 or, alternatively, B. fragilis strains may have first acquired the genetic element encoding a type I R-M system and then it was interrupted by integration of CTn9343.
The MIC results suggested that the B. fragilis strains tested are intrinsically resistant to virginiamycin M, and the presence of satG did not increase the resistance to this antibiotic in strain NCTC 9343. Similarly, the presence of bexA did not increase resistance to the tested fluoroquinolones. The predicted protein encoded by bexA shares significant homology with a multidrug efflux transporter involved in fluoroquinolone resistance in B. thetaiotaomicron (18). A portion of the norfloxacin resistance of B. fragilis and B. thetaiotaomicron is attributed to active efflux of the antibiotic by BexA (17, 18). However, BexA may not be functional in CTn9343, since the protein lacks 200 N-terminal amino acids in comparison to the analog protein encoded by B. thetaiotaomicron.
The structure of CTn9343 appears to be modular, containing genes derived from different bacteria, bacteriophages, viruses, plasmids, and unknown sources (Table 3). The G+C content of the entire CTn9343 sequence (46.5%) is different from that reported for the B. fragilis chromosome (43% [http://www.sanger.ac.uk/Projects/B_fragilis/]). However, there are several CTn9343 regions whose G+C contents differ significantly from the rest of the transposon. For example, the putative transfer region (see below) has a G+C content of 53%, and the region containing the rteB, rteA, satG, and bexA genes has a G+C content of 45%. These results suggest that CTn9343 originated from different genetic sources. In contrast, the G+C content (43%) of the cluster of genes (ORFs 25 to 29) encoding the enzyme for utilization of xylose together with an ECF-type sigma factor and two-component systems (Table 3) is characteristic of the genome of Bacteroides species (46), suggesting that CTn9343 also contains genes of Bacteroides species origin.
There is limited homology between CTn9343, CTnDOT, and related CTns. None of the DNA probes used to characterize CTn86 (Fig. 1B) hybridized with CTnDOT under the conditions tested (see Materials and Methods). Only the region of CTn9343 containing int2-rteB-rteA and four putative transfer genes (ORFs 36, 38, 41, and 42) have homology with CTnDOT. This may indicate that these regions were acquired by CTn9343 and CTnDOT from a common ancestor. The putative integrase (Int2) encoded by CTn9343 has significant homology with the integrase of CTnDOT (Table 3); however, the integrase of CT9343 lacks 128 amino acids from the N terminus and 188 amino acids from the carboxy terminus (this includes the region termed box 2, which is conserved in the lambda integrase family and is considered important to integrase function) of CTnDot. In the lambda integrase family, the carboxy terminus, missing in the CTn9343 Int2, contains the catalytic site that carries out the DNA breaking and joining reactions that mediate recombination (24).
In the case of CTnDOT, the integrase gene and regulatory rteA, rteB, and rteC genes together with the exc gene are essential for excision and integration (5, 6, 42). The CTnDOT rteA and rteB genes together with the tetracycline resistance gene tetQ form an operon. RteA and RteB are members of a two-component regulatory system; however, recently it has been reported that neither RteA nor RteB affects expression of the tetQ-rteA-rteB operon (40). This operon may be regulated by TetQ by a translational attenuation mechanism in response to tetracycline (40). In contrast to CTnDOT, CTn9343 lacks tetQ, and rteA and rteB are in a putative operon that also includes satG and bexA. Exposure of NCTC 9343 cells to virginiamycin M or fluoroquinolones (antibiotic resistance encoded by satG and bexA, respectively) as well as tetracycline did not increase formation of the CTn9343 circular intermediate or expression of rteA or putative genes involved in the excision-integration or transfer of CTn9343. These results suggested that the rteB/rteA/satG/bexA operon does not regulate excision and transfer of CTn9343. The rteB/rteA/satG/bexA operon may not be functional in CTn9343, because rteA and bexA encode truncated proteins. RteA lacks 349 amino acid residues from the carboxy-terminal end in comparison to RteA of CTnDOT, and BexA lacks 200 amino acids from the N terminus.
The homology of ORFs 36, 38, 41, and 42 to CTnDOT transfer proteins TraN, TraM, TraI, and TraG and the location of these ORFs in a large cluster of genes (ORF 35 to 50), all of which (except ORF 37) are predicted to be transcribed in the same direction, suggest that this region is the transfer region of CTn9343. Beside these four transfer proteins, there is no additional sequence similarity between the CTn9343 putative transfer region and other conjugative elements. It has been reported that the transfer region of CTnDOT is totally different from that of gram-positive and gram-negative organisms (3). Only TraG shares sequence similarity with proteins encoded by other transmissible elements (3). The presence in CTn9343 of genes with similarity to traN, traM, traI, and traG genes may indicate that the proteins encoded by these genes are absolutely necessary for conjugal transfer in Bacteroides species.
Colony blot hybridization, PCR, and sequence analysis indicated that CTn86 has a structure similar to CTn9343, except that CTn86 lacks an ∼7-kb region identified in CTn9343 containing int2, ORF 11, ORF 12, rteB, rteA, satG, and bexA and it contains the BfPAI (Fig. 8). The G+C content of the BfPAI (35%) also differs greatly from that of other regions of CTn86, suggesting acquisition from a different organism.
Even though the region encoding int2 and the regulatory genes, rteA and rteB, has been deleted in CTn86, this transposon forms circular intermediates. These results further suggest that the truncated Int2 and RteA proteins are not functional in CTn9343, and other genes in the transposon regulate the transposition. Analysis of the ends and flanking regions of CTn9343 and CTn86 in the circular and integrate forms (Fig. 5 and 7), respectively, suggests that these transposons integrate in a similar way to IS21. IS21 is bordered by IRs of different lengths and contains two contiguous genes, istA and istB, forming an operon (16). The hallmark of the IS21 transposition mechanism is the spontaneous formation of IS21 tandem repeats, designated (IS21)2 (27). In the (IS21)2 configuration, the two insertion sequences are typically separated by 2 or 3 bp, termed a junction sequence (26). The tandemly repeated copies of IS21 promote insertion of entire plasmids carrying (IS21)2 in a transposition event involving the abutted terminal IR sequences (26). During this cut-and-paste process, the junction sequence of (IS21)2 is lost, and the outer ends of IS21 are dispensable. The cointegrates formed have a single IS21 copy at each junction, and the IS21 elements are bordered by direct repeats of 4 bp, i.e., the target duplication (26).
The left ends of CTn9343 and CTn86 have two consecutive genes, tnpA1 and tnpB, that predict encoded proteins with significant homology to IS21 IstA and IstB, respectively. IstA and IstB are required for integration (34). The right end has a gene that encodes a truncated TnpA1 (tnpA2). In CTn9343 and CTn86, joining of the transposon ends to form the circular intermediates yields a structure where one copy contains tpnA1 and tnpB and the second contains tnpA2. The ends of both copies contain IR sequences separated by a 2-bp junction sequence (Fig. 5A and 7A). Similar to the structure of (IS21)2, where the ends contain short IR sequences separated by 2 nucleotides, transposition of the circular forms into the B. fragilis chromosome duplicates the 7-bp target site with loss of the 2-bp junction sequence (Fig. 5B and C and 7B and C). The tnpA2 gene encodes a truncated protein of 124 amino acid residues in CTn9343 and CTn86. It is unknown if TnpA2 is functional in CTn9343 and CTn86 or only serves to join with the right end to form an active circular form.
Together, this predicted mechanism of excision and integration described for CTn9343 and CTn86 differs from that of CTnDOT or other reported CTns (5, 7, 28). The absence of homology between the 7-bp target sites with the ends of CTn9343 and CTn86, as well as the absence of a consensus sequence in the 7-bp target sites of CTn9343 and CTn86, suggest that, in contrast to CTnDOT, insertion of CTn9343 and CTn86 is not site specific.
In conclusion, based on sequence analysis and identification of the circular intermediates, the genetic element flanking the BfPAI in ETBF strain 86-5443-2-2 and a related genetic element in strain NCTC 9343 are predicted to be CTns (CTn86 and CTn9343, respectively). These putative CTns may be members of a new family of CTns. These putative CTns are distinct in that CTn86 lacks a 7-kb region containing a truncated integrase (int2) and rteA genes and possesses the BfPAI. If CTn86 were demonstrated to be transmissible, this would suggest that the bft gene can be transferred from ETBF to NTBF strains by a mechanism similar to that for the spread of antibiotic resistance genes. Further studies are necessary to determine if the transfer genes of CTn86 and CTn9343 are functional.
Acknowledgments
I thank Cynthia L. Sears for review of the manuscript and helpful discussions and Janeth P. Castillo for excellent technical assistance.
This work was supported by grant number RO1A148708 (A.A.F.) from the National Institutes of Health.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ando, T., D. A. Israel, K. Kusugami, and M. J. Blaser. 1999. HP0333, a member of the dprA family, is involved in natural transformation in Helicobacter pylori. J. Bacteriol. 181:5572-5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonheyo, G., D. Graham, N. B. Shoemaker, and A. A. Salyers. 2001. Transfer region of a Bacteroides conjugative transposon, CTnDOT. Plasmid 45:41-51. [DOI] [PubMed] [Google Scholar]
- 4.Chambers, F. G., S. S. Koshy, R. F. Saidi, D. P. Clark, R. D. Moore, and C. L. Sears. 1997. Bacteroides fragilis toxin exhibits polar activity on monolayers of human intestinal epithelial cells (T84 cells) in vitro. Infect. Immun. 65:3561-3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng, Q., B. J. Paszkiet, N. B. Shoemaker, J. F. Gardner, and A. A. Salyers. 2000. Integration and excision of a Bacteroides conjugative transposon, CTnDOT. J. Bacteriol. 182:4035-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng, Q., Y. Sutanto, N. B. Shoemaker, J. F. Gardner, and A. A. Salyers. 2001. Identification of genes required for excision of CTnDOT, a Bacteroides conjugative transposon. Mol. Microbiol. 41:625-632. [DOI] [PubMed] [Google Scholar]
- 7.Churchward, G. 2002. Conjugative transposons and related mobile elements, p. 177-191. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, D.C.
- 8.Franco, A. A., L. M. Mundy, M. Trucksis, S. Wu, J. B. Kaper, and C. L. Sears. 1997. Cloning and characterization of the Bacteroides fragilis metalloprotease toxin gene. Infect. Immun. 65:1007-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franco, A. A., R. K. Cheng, G.-T. Chung, S. Wu, H.-B. Oh, and C. L. Sears. 1999. Molecular evolution of the pathogenicity island of enterotoxigenic Bacteroides fragilis. J. Bacteriol. 181:6623-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franco, A. A., R. K. Cheng, A. Goodman, and C. L. Sears. 2002. Modulation of bft expression by the Bacteroides fragilis pathogenicity island and its flanking region. Mol. Microbiol. 45:1067-1077. [DOI] [PubMed] [Google Scholar]
- 11.Hecht, D. W., T. J. Jagielo, and M. H. Malamy. 1991. Conjugal transfer of antibiotic resistance factors in Bacteroides fragilis: the btgA and btgB genes of plasmid pBFTM10 are required for its transfer from B. fragilis and for its mobilization by IncP beta plasmid R751 in Escherichia coli. J. Bacteriol. 173:7471-7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson, J. L. 1978. Taxonomy of the Bacteroides. I. Deoxyribonucleic acid homologies among Bacteroides fragilis and other saccharolytic Bacteroides species. Int. J. Syst. Bacteriol. 28:245-268. [Google Scholar]
- 13.Karudapuram, S., X. Zhao, and G. J. Barcak. 1995. DNA sequence and characterization of Haemophilus influenzae dprA+, a gene required for chromosomal but not plasmid DNA transformation. J. Bacteriol. 177:3235-3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koshy, S. S., M. H. Montrose, and C. L. Sears. 1996. Human intestinal epithelial cells swell and demonstrate actin rearrangement in response to the metalloprotease toxin of Bacteroides fragilis. Infect. Immun. 64:5022-5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krinos, C. M, M. J. Coyne, K. G. Weinacht, A. O. Tzianabos, D. L. Kasper, and L. E. Comstock. 2001. Extensive surface diversity of a commensal microorganism by multiple DNA inversions. Nature 414:555-558. [DOI] [PubMed] [Google Scholar]
- 16.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyamae, S., H. Nikaido, Y. Tanaka, and F. Yoshimura. 2000. Active efflux of norfloxacin by Bacteroides fragilis. Antimicrob. Agents Chemother. 42:2119-2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyamae, S., O. Ueda, F. Yoshimura, J. Hwang, Y. Tanaka, and H. Nikaido. 2001. A MATE family multidrug efflux transporter pumps out fluoroquinilones in Bacteroides thetaiotaomicron. Antimicrob. Agents Chemother. 45:3341-3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moncrief, J. S., R. Obiso, Jr., L. A. Barroso, J. J. Kling, R. L. Wright, R. L. Van Tassell, D. M. Lyerly, and T. D. Wilkins. 1995. The enterotoxin of Bacteroides fragilis is a metalloprotease. Infect. Immun. 63:175-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moncrief, J. S., A. J. Duncan, R. L. Wright, L. A. Barroso, and T. D. Wilkins. 1998. Molecular characterization of the fragilysin pathogenicity islet of enterotoxigenic Bacteroides fragilis. Infect. Immun. 66:1735-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mundy, L. M., and C. L. Sears. 1996. Detection of toxin production by Bacteroides fragilis: assay development and screening of extraintestinal clinical isolates. Clin. Infect. Dis. 23:269-276. [DOI] [PubMed] [Google Scholar]
- 22.Myers, L. L., B. D. Firehammer, D. S. Shoop, and M. M. Border. 1984. Bacteroides fragilis: a possible cause of acute diarrheal disease in newborn lambs. Infect. Immun. 44:241-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Myers, L. L., D. S. Shoop, L. L. Stackhouse, F. S. Newman, R. J. Flaherty, G. W. Letson, and R. B. Sack. 1987. Isolation of enterotoxigenic Bacteroides fragilis from humans with diarrhea. J. Clin. Microbiol. 25:2330-2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nunes-Duby, S. E., H. J. Kwon, R. S. Tirumalai, T. Ellenberger, and A. Landy. 1998. Similarities and differences among 105 members of the Int family of site-specific recombinases. Nucleic Acids Res. 26:391-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Obiso, R. J., Jr., A. O. Azghani, and T. D. Wilkins. 1997. The Bacteroides fragilis toxin fragilysin disrupts the paracellular barrier of epithelial cells. Infect. Immun. 65:1431-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reimmann, C., R. Moore, S. Little, A. Savioz, N. S. Willets, and D. Hass. 1989. Genetic structure, function and regulation of the transposable element IS21. Mol. Gen. Genet. 215:416-424. [DOI] [PubMed] [Google Scholar]
- 27.Riess, G., B. W. Holloway, and A. Puhler. 1980. R68.45, a plasmid with chromosome mobilizing ability (Cma) carries a tandem duplication. Genet. Res. 36:99-109. [DOI] [PubMed] [Google Scholar]
- 28.Rudy, C., K. L. Taylor, D. Hinerfeld, J. R. Scott, and G. Churchward. 1997. Excision of a conjugative transposon in vitro by the Int and Xis proteins of Tn916. Nucleic Acids Res. 25:4061-4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sack, R. B., L. L. Myers, J. Almeido-Hill, D. S. Shoop, W. C. Bradbury, R. Reid, and M. Santosham. 1992. Enterotoxigenic Bacteroides fragilis: epidemiological studies of its role as a human diarrhoeal pathogen. J. Diarrhoeal Dis. Res. 10:4-9. [PubMed] [Google Scholar]
- 30.Sack, R. B., M. J. Albert, K. Alam, P. K. Neogi, and M. S. Akbar. 1994. Isolation of enterotoxigenic Bacteroides fragilis from Bangladeshi children with diarrhea: a controlled study. J. Clin. Microbiol. 32:960-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salyers, A. A., N. B. Shoemaker, A. M. Stevens, and L.-Y. Li. 1995. Conjugative transposons: an unusual and diverse set of integrated gene transfer elements. Microbiol. Rev. 59:579-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed., p. 1.25-1.27. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.San Joaquin, V. H., J. C. Griffis, C. Lee, and C. L. Sears. 1995. Association of Bacteroides fragilis with childhood diarrhea. Scand. J. Infect. Dis. 27:211-215. [DOI] [PubMed] [Google Scholar]
- 34.Schmid, S., T. Seitz, and D. Haas. 1998. Cointegrase, a naturally occurring, truncated form of IS21 transposases, catalyzes replicon fusion rather than simple insertion of IS21. J. Mol. Biol. 282:571-583. [DOI] [PubMed] [Google Scholar]
- 35.Scott, J. R., F. Bringel, D. Marra, G. Van Alstine, and C. K. Rudy. 1994. Conjugative transposition of Tn916: preferred targets and evidence for conjugative transfer of a single strand and for a double-stranded circular intermediate. Mol. Microbiol. 11:1099-1108. [DOI] [PubMed] [Google Scholar]
- 36.Sears, C. L., L. L. Myers, A. Lazenby, and R. L. Van Tassell. 1995. Enterotoxigenic Bacteroides fragilis. Clin. Infect. Dis. 20(Suppl. 2):S142-S148. [DOI] [PubMed] [Google Scholar]
- 37.Smith, C. J., G. D. Tribble, and D. P. Bayley. 1998. Genetic elements of Bacteroides species: a moving story. Plasmid 40:12-29. [DOI] [PubMed] [Google Scholar]
- 38.Stevens, A. M., N. B. Shoemaker, L. Y. Li, and A. A. Salyers. 1993. Tetracycline regulation of genes on Bacteroides conjugative transposons. J. Bacteriol. 175:6134-6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, Y., G.-R. Wang, A. Shelby, N. B. Shoemaker, and A. A. Salyers. 2003. A newly discovered Bacteroides conjugative transposon, CTnGERM1, contains genes also found in gram-positive bacteria. Appl. Environ. Microbiol. 69:4595-4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, Y., N. B. Shoemaker, and A. A. Salyers. 2004. Regulation of a Bacteroides operon that controls excision and transfer of the conjugative transposon CTnDOT. J. Bacteriol. 186:2548-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weikel, C. S., F. D. Grieco, J. Reuben, L. L. Myers, and R. B. Sack. 1992. Human colonic epithelial cells, HT29/C1, treated with crude Bacteroides fragilis enterotoxin dramatically alter their morphology. Infect. Immun. 60:321-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whittle, G., N. B. Shoemaker, and A. A. Salyers. 2002. Characterization of genes involved in modulation of conjugal transfer of the Bacteroides conjugative transposon CTnDOT. J. Bacteriol. 184:3839-3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson, G. G. 1981. Organization of restriction-modification systems. Nucleic Acids Res. 19:2539-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu, S., K.-C. Lim, J. Huang, R. F. Saidi, and C. L. Sears. 1998. Bacteroides fragilis enterotoxin cleaves the zonula adherens protein, E-cadherin. Proc. Natl. Acad. Sci. USA 95:14979-14984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu, S., P. J. Morin, D. Maouyo, and C. L. Sears. 2003. Bacteroides fragilis enterotoxin induces c-Myc expression and cellular proliferation. Gastroenterology 124:392-400. [DOI] [PubMed] [Google Scholar]
- 46.Xu, J., M. K. Bjursell, J. Himrod, S. Deng, L. K. Carmichael, H. C. Chiang, L. V. Hooper, and J. I. Gordon. 2003. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299:2074-2076. [DOI] [PubMed] [Google Scholar]
- 47.Zhang, G., B. Svenungsson, A. Karnell, and A. Weintraub. 1999. Prevalence of enterotoxigenic Bacteroides fragilis in adult patients with diarrhea and healthy controls. Clin. Infect. Dis. 29:590-594. [DOI] [PubMed] [Google Scholar]