Abstract
Exposure of bacteria to diverse growth-limiting stresses induces the synthesis of a common set of proteins which provide broad protection against future, potentially lethal stresses. Among Bacillus subtilis and its relatives, this general stress response is controlled by the σB transcription factor. Signals of environmental and energy stress activate σB through a multicomponent network that functions via a partner switching mechanism, in which protein-protein interactions are governed by serine and threonine phosphorylation. Here, we tested a central prediction of the current model for the environmental signaling branch of this network. We used isoelectric focusing and immunoblotting experiments to determine the in vivo phosphorylation states of the RsbRA and RsbS regulators, which act in concert to negatively control the RsbU environmental signaling phosphatase. As predicted by the model, the ratio of the phosphorylated to unphosphorylated forms of both RsbRA and RsbS increased in response to salt or ethanol stress. However, these two regulators differed substantially with regard to the extent of their phosphorylation under both steady-state and stress conditions, with RsbRA always the more highly modified. Mutant analysis showed that the RsbT kinase, which is required for environmental signaling, was also required for the in vivo phosphorylation of RsbRA and RsbS. Moreover, the T171A alteration of RsbRA, which blocks environmental signaling, also blocked in vivo phosphorylation of RsbRA and impeded phosphorylation of RsbS. These in vivo results corroborate previous genetic analyses and link the phosphorylated forms of RsbRA and RsbS to the active transmission of environmental stress signals.
In Bacillus subtilis the general stress response triggers the synthesis of more than 150 proteins which confer resistance to diverse lethal stresses (reviewed in references 14 and 23). Transcription of the general stress regulon is controlled by σB, whose activity is regulated by a signal transduction network in which key protein-protein interactions are determined by serine and threonine phosphorylation. This mechanism has been dubbed partner switching (4) and is found, wholly or in part, among evolutionarily diverse eubacteria (19, 22, 23). Study of this mechanism in B. subtilis should therefore help explain the principles which govern a broad array of bacterial signaling pathways.
A model for the B. subtilis signaling network is shown in Fig. 1. In this model, separate environmental and energy signaling pathways converge on the RsbV and RsbW regulators, which directly control σB by means of the partner switching mechanism (Fig. 1A). RsbW has two activities in unstressed cells. First, it acts as an anti-σ factor which binds σB and prevents its association with RNA polymerase (3, 6). Second, it acts as a serine kinase which specifically phosphorylates and inactivates the RsbV anti-anti-σ (3, 10). Following an activating stress, the phosphate is removed from RsbV-P by either the RsbP energy phosphatase or the RsbU environmental phosphatase (26, 28, 30), leading RsbV to complex with RsbW and force the release of σB. In this scheme, RsbW switches its binding partner in response to stress.
FIG. 1.
Model of the σB signal transduction network. (A) Two independent signaling pathways converge on the RsbV anti-anti-σ and the RsbW anti-σ, the direct regulators of σB activity. The energy pathway terminates with the RsbP phosphatase (Energy PP2C), which contains a PAS domain implicated in energy sensing; the environmental pathway terminates with the RsbU phosphatase (Environmental PP2C), which is activated by upstream signaling elements. Phosphorylated RsbV (RsbV-P) is the antagonist form found in unstressed cells. When activated by stress, either RsbP or RsbU can dephosphorylate RsbV-P, allowing it to bind and inactivate the RsbW anti-σ. (B) In the environmental signaling pathway, RsbS and RsbT are paralogs of RsbV and RsbW, respectively. RsbS is the antagonist form in unstressed cells, and RsbRA, RsbRB, RsbRC, and RsbRD are redundant coantagonists that function with RsbS to bind the RsbT kinase in an inactive complex. Following environmental stress, RsbT phosphorylates RsbRA and RsbS, releasing RsbT to bind and activate the RsbU phosphatase. The RsbX feedback phosphatase returns the system to its prestress condition. Phosphorylation of RsbRB, RsbRC, and RsbRD is not shown but is thought to resemble that of RsbRA. (C) RsbR coantagonist proteins share a carboxyl-terminal domain (shaded) with the smaller RsbS antagonist (1, 25). In the RsbR family, this domain contains two conserved threonine (T) residues, and RsbT is known to phosphorylate RsbRA on T171 and T205 in vitro (12). In contrast, RsbS bears an aspartate (D) and a serine (S) at these corresponding positions. Genetic evidence suggests that phosphorylation of S59 is required to relieve RsbS antagonist function (16, 30).
The subject of this communication is a second partner switch found in the environmental signaling branch (Fig. 1B). In this switch, RsbS and RsbT are paralogs of RsbV and RsbW, respectively (16). However, there are three significant differences between the two switches. First, the target of the RsbS-RsbT switch is an enzyme, namely, the RsbU environmental signaling phosphatase (30). Second, RsbT positively regulates RsbU activity, essentially serving as a regulatory subunit of the phosphatase (16, 17, 30). And third, RsbS alone is insufficient to sequester RsbT in an inactive complex. Also required is at least one of a family of paralogous proteins, the RsbR coantagonists (1, 9, 18), which possess carboxyl-terminal domains that resemble the entire length of the smaller RsbS antagonist (Fig. 1C). The first member of the RsbR family to be discovered was RsbRA, whose structural gene lies in the sigB operon, immediately upstream from that encoding RsbS (29). The genes encoding the other family members—RsbRB, RsbRC, and RsbRD—are unlinked to the sigB operon and to each other (1, 18, 20). Notably, RsbRA, RsbRB, and RsbS copurify from cell extracts in a large complex (18), and RsbRC and RsbRD have recently been identified as constituents of the same complex (A. L. Weigel, T. J. Kim, S. Neissen, J. R. Yates, and C. W. Price, unpublished data). The properties of this complex are thought to facilitate the sensing or transmission of environmental stress signals (18).
According to the model shown in Fig. 1, in the absence of environmental stress the RsbR coantagonists and the RsbS antagonist jointly bind RsbT, preventing its association with RsbU (9, 18). Following an environmental signal, such as acid, ethanol, heat, or salt stress, the RsbR family members and RsbS are specifically phosphorylated by RsbT (1). These phosphorylation events release RsbT to bind and activate RsbU by direct protein-protein interaction (9, 17, 30). RsbU then communicates the stress signal to the downstream partner switch by dephosphorylating RsbV-P, resulting in the activation of σB (28, 30). This model further holds that the environmental signal is damped by the RsbX feedback phosphatase (24, 27, 30), whose expression is under σB control (15). The biochemical and genetic evidence supporting this model includes a consideration of the phenotypes caused by alteration of the residues upon which RsbS and RsbRA are phosphorylated (2, 12, 16, 18, 30). However, it has yet to be established whether the in vivo phosphorylation state of the RsbR family members and RsbS do in fact change as a result of environmental stress.
Here we test key predictions of the model and show that both RsbRA and RsbS are indeed phosphorylated in vivo as part of the response to ethanol or salt stress and that this modification requires the RsbT kinase. However, we also find that the phosphorylation states of RsbRA and RsbS differ substantially in both stressed and unstressed cells, with RsbRA the more highly phosphorylated. Based on these results, we propose a refinement of the model for environmental stress signaling.
MATERIALS AND METHODS
Growth of bacterial strains.
All B. subtilis strains were derivatives of the wild-type Marburg strain (Table 1). Cells were grown at 37°C in shake flasks containing buffered Luria broth lacking salt (7). Early logarithmic cells were either used directly for unstressed controls (time zero) or stressed by the addition of salt or ethanol to a final concentration of 0.3 M or 4% (vol/vol), respectively. We used two different methods to harvest cells for isoelectric focusing (IEF) analysis, depending upon the purpose of the experiment.
TABLE 1.
B. subtilis strains
| Strain | Genotype | Reference or construction |
|---|---|---|
| PB2 | trpC2 | 168 Marburg strain |
| PB198 | amyE::ctc-lacZ trpC2 | 8 |
| PB421 | rsbTΔ1 trpC2 | pCK1(16) → PB2a |
| PB422 | rsbSΔ1 trpC2 | 16 |
| PB427 | rsbRAΔ1 trpC2 | 2 |
| PB465 | rsbSS59A trpC2 | 16 |
| PB477 | rsbSS59D Pspac(rsbV+rsbW+sigB+rsbX+) trpC2 | 16 |
| PB502 | rsbRAT205D trpC2 | 2 |
| PB505 | rsbRAT205A trpC2 | 2 |
| PB556 | rsbRAT171A-T205A trpC2 | 18 |
| PB557 | rsbRAT171D trpC2 | 12 |
| PB558 | rsbRAT171D-T205D trpC2 | 12 |
| PB829 | rsbRAT171A trpC2 | 18 |
Arrow indicates transformation of donor plasmid into recipient.
When preservation of the phosphorylation state of RsbS and RsbRA was not an issue, as was the case for the experiments shown below in Fig. 2, we harvested 10 ml of culture by centrifugation at 4°C in a Sorvall SS34 rotor, run at 8,000 rpm for 5 min. The cell pellets were washed by resuspension in 1 ml of sonication buffer (50 mM Tris-HCl [pH 6.8], 150 mM NaCl, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride), after which they were transferred to a 1.5-ml Eppendorf tube. Cells were collected by centrifugation in a microcentrifuge (1 min at 4°C), resuspended in 0.5 ml of sonication buffer, and then broken by sonication with four 30-s treatments separated by 20 s on ice, using a Sonic Dismembrator (model 300; Fisher Scientific, Pittsburgh, Pa.) with a microtip on the 35% setting. Cell debris was removed by centrifugation in a microcentrifuge (10 min at 4°C), and the supernatants were analyzed by IEF.
FIG. 2.
Identification of the RsbS and RsbRA isoforms separated by IEF. Wild-type and mutant cell extracts were analyzed by IEF, and the RsbS or RsbRA signals were detected with specific antibodies, as described in Materials and Methods. In all panels, gel images are oriented with their alkaline regions uppermost, toward the cathode, and the numbered lines to the right indicate the approximate positions of unmodified, singly modified, and doubly modified isoforms. (A) Lane 1, wild-type strain (PB2) (wt); lane 2, the rsbS deletion mutant (PB422) (ΔS). Wild-type extracts were also used for the λPP assays and were incubated for 18 h at 30°C (lanes 3 to 5). Lane 3, cell extract (CE) alone; lane 4, addition of λ reaction buffer and MnCl2 (+λB); lane 5, further addition of λPP (+λPP). (B) Wild-type extracts are in lanes 1 and 4; mutant extracts are in lanes 2 and 3. Lane 2, strain with the RsbS S59A alteration (PB465); lane 3, RsbS S59D (PB477). (C) Lane 1, the wild-type strain (PB2) (wt); lane 2, the rsbRA deletion mutant (PB427) (ΔRA). Wild-type extracts were also used for the λPP assays shown in lanes 3 to 5, labeled as described for panel A. (D) Wild-type extracts are in lanes 1 and 8, and mutant extracts are in lanes 2 to 7. Lane 2, strain with the RsbRA T171A alteration (PB829); lane 3, RsbRA T171D (PB557); lane 4, RsbRA T205A (PB505); lane 5, RsbRA T205D (PB502); lane 6, RsbRA T171A-T205A (PB556); lane 7, RsbRA T171D-T205D (PB558).
When harvesting by centrifugation could significantly influence the results, as in the experiments shown below in Fig. 3 and 4, we employed a rapid harvesting procedure. Here we filtered 10 ml of cell culture through 25-mm-diameter MF-Millipore membrane filters with a pore size of 0.22 μm (Millipore, Billerica, Mass.). The cells and the MF membranes were transferred to 1.5-ml Eppendorf tubes containing 1 ml of cold Z-buffer (100 mM sodium phosphate [pH 7.0], 10 mM KCl, 1 mM MgSO4) supplemented with phosphatase inhibitors (50 mM NaF, 0.1 mM sodium orthovanadate, 60 mM β-glycerophosphate, and 15 mM p-nitrophenyl phosphate). Following a brief mix to wash the cells from the filters, the samples were collected by centrifugation in a microcentrifuge (30 s at 4°C), after which the membrane and supernatant were removed. The cell pellet was then quickly frozen on dry ice-ethanol and stored at −80°C until all samples were collected. This entire sampling procedure was accomplished in less than 60 s. Prior to assay, cells were resuspended in 0.5 ml of sonication buffer containing the phosphatase inhibitors and broken by sonication, as described above for the standard harvesting procedure.
FIG. 3.
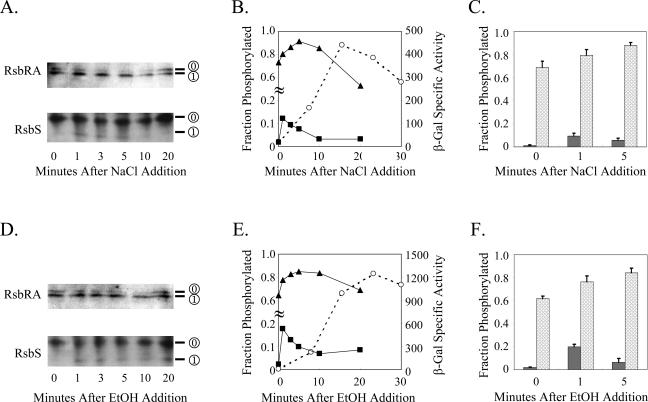
Isoform balance of RsbS and RsbRA changes after stress. Wild-type (PB2) cells were stressed by either salt or ethanol addition and then rapidly harvested by filtration at the indicated times (minutes). Samples were analyzed by IEF and immunoblotting as described in Materials and Methods. (A and D) Gel images, oriented with their alkaline regions uppermost and with isoform positions indicated on the right. (B and E) Quantification of the digitized images from panels A and D. Note the discontinuity (≈) in the y axis, designating the fraction of phosphorylated RsbS and RsbRA. ▪, RsbS-P/total RsbS; ▴, RsbRA-P/total RsbRA; ○, β-galactosidase activity of a σB-dependent lacZ reporter fusion carried by strain PB198, cultured in parallel with the cells used for the IEF assay. (C and F) RsbS-P/total RsbS (dark grey bars) and RsbRA-P/total RsbRA (light grey bars) found in three independent stress experiments (average ± standard deviation).
FIG. 4.
Dependence of RsbS and RsbRA phosphorylation on the RsbT environmental signaling kinase and the T171 residue of RsbRA. Wild-type and mutant cells were stressed by ethanol addition and then rapidly harvested by filtration at the indicated times (minutes). Gel images are oriented with their alkaline regions uppermost and with isoform positions indicated on the right. For both panels A (RsbS) and B (RsbRA), lanes 1 to 3 show extracts from the wild type (strain PB2), lanes 4 to 6 show extracts from the rsbT deletion mutant (PB421), lanes 7 to 9 show extracts from the rsbRAT171A substitution mutant (PB829), and lane 10 shows an additional extract from unstressed wild-type cells.
IEF.
IEF of 20-μl cell extract samples was done in a Mini-Protean II cell with a 15-well comb (Bio-Rad Laboratories, Hercules, Calif.), with the lower chamber containing 10 mM phosphoric acid as anolyte and the upper chamber containing 20 mM sodium hydroxide as catholyte. All samples were adjusted for equal amounts of protein and then run at 200 V for the first 0.5 h and at 300 V for another 2.5 h. However, to optimize isoform separation, gel and sample preparations were different for RsbS and RsbRA.
For RsbS (calculated pI, 4.14), a 4% acrylamide gel was made using a stock 30% acrylamide-bis (29:1) solution (Bio-Rad Laboratories), 9.37 M urea, 1% Triton X-100, 22 mM 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 3.85% Pharmalyte, pH range 4.2 to 4.9 (Sigma, St. Louis, Mo.), 1.15% Pharmalyte pH 3.0 to 10.0 (Sigma), 0.027% ammonium persulfate, and 0.107% N,N,N′,N′-tetramethylethylenediamine (TEMED). After removing the residual liquid from the sample wells, the gel was assembled in the Mini-Protean II cell. Thereafter, the wells were filled with RsbS loading buffer (4 M urea, 1.2% Pharmalyte pH 4.2 to 4.9, 0.36% Pharmalyte pH 3.0 to 10.0) to receive the samples. Protein concentration in each sample was measured with the Bio-Rad protein assay reagent (Bio-Rad Laboratories), and equal amounts of protein were dried completely under vacuum. These dried samples were solubilized with 25 μl of RsbS sample buffer (5 M urea, 40 mM CHAPS, 0.77% Pharmalyte pH 4.2 to 4.9, 0.23% Pharmalyte pH 3.0 to 10.0, 0.25 M dithiothreitol, and 0.075% sodium dodecyl sulfate [SDS]). Twenty-microliter aliquots of these sample solutions were loaded into the bottom of the wells, using a long-tipped pipette to minimize mixing with loading buffer. IEF was then conducted as described above.
For RsbRA (calculated pI, 4.73), a 5% acrylamide gel was made using a 30% acrylamide-bis solution (29:1), 8 M urea, 1% Triton X-100, 2% Pharmalyte pH 4.2 to 4.9, 0.6% Pharmalyte pH 3.0 to 10.0, 0.021% ammonium persulfate, and 0.167% TEMED. After removing the residual liquid from the wells, the gel was assembled and the wells were filled with RsbRA sample buffer (2 M urea, 0.5% Pharmalyte pH 4.2 to 4.9, 0.15% Pharmalyte pH 3.0 to 10.0, 0.5% Triton X-100, 0.5% β-mercaptoethanol, 0.01% bromphenol blue). Sample protein concentration was measured, and the amounts were equalized. Each adjusted sample was then mixed with an equal volume of 4× RsbRA sample buffer (8 M urea, 2% Pharmalyte pH 4.2 to 4.9, 0.6% Pharmalyte pH 3.0 to 10.0, 2% Triton X-100, 1% β-mercaptoethanol, 0.04% bromphenol blue) and loaded into the bottom of the wells.
Immunological methods.
Anti-RsbRA and anti-RsbS antibodies (11) were kindly provided by William Haldenwang. Antibody specificity was confirmed by Western blot analysis of wild-type and mutant cell extracts separated on SDS-polyacrylamide gel electrophoresis. For analysis of the IEF gels, the separated proteins were transferred to polyvinylidene difluoride membranes (Bio-Rad Laboratories). These membranes were blocked by immersion in 5% nonfat dried milk-TBS-T for 1 h (TBS-T is 10 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.5% Tween 20). Membrane blots were exposed to primary antibody in 5% nonfat dried milk-TBS-T for 1 h at 24°C, washed, and then incubated with peroxide-conjugated secondary anti-mouse immunoglobulin G antibody (Santa Cruz Biotechnology Inc., Santa Cruz, Calif.). The blots were washed, and bound antibody was visualized using the ECL Plus Western blotting detection kit (Amersham Pharmacia Biotech, Piscataway, N.J.), according to the manufacturer's instructions, with the image captured on Kodak BioMax light film (Eastman Kodak Company, Rochester, N.Y.). For quantitative analysis, the exposed film was scanned and digitized using an Epson Perfection 636 (Epson America, Inc., Long Beach, Calif.), and the band intensities were determined using Quantity One version 4.1.1 software (Bio-Rad Laboratories).
λPP assays.
To determine if the RsbS and RsbRA isoforms were phosphorylated on serine, threonine, or tyrosine residues, we added 40 U of λ protein phosphatase (λPP; New England BioLabs, Beverly, Mass.) per μl of cell extract, adjusted to 50 mM Tris-HCl (pH 7.5), 0.1 mM EDTA, 5 mM dithiothreitol, 0.01% Brij 35, and 2 mM MnCl2 by the addition of 10× λPP reaction buffer and 20 mM MnCl2. Extracts were incubated for 18 h at 30°C and then analyzed by IEF and immunoblotting. Controls included (i) cell extracts alone and (ii) cell extracts with λPP reaction buffer but no λPP. During the course of these experiments, we discovered that one of the two RsbS isoforms was unstable in control ii, suggesting that it was the substrate of an endogenous activity in the cell extract (data not shown). To clearly establish the effect of λPP, extracts for RsbS analysis were heated for 3 min at 100°C before adding λPP and λPP reaction buffer. By contrast, the RsbRA isoforms appeared stable in both control assays, and so these extracts were not subjected to heat treatment.
β-Galactosidase accumulation assays.
Strain PB198 carries a σB-dependent ctc-lacZ reporter fusion at its amyE locus. Samples were collected at the indicated times and assayed as described by Miller (21). Cells were washed with Z-buffer and permeabilized using SDS and chloroform and then incubated at 28°C in 1-ml reaction mixes containing ο-nitrophenyl-β-d-galactopyranoside. Reactions were stopped with 0.5 ml of 1 M Na2CO3 and centrifuged to remove cellular interference, and the A420 was recorded. Protein levels were determined with the Bio-Rad protein assay reagent (Bio-Rad Laboratories). β-Galactosidase activity was defined as ΔA420 × 1,000 per min per mg of protein.
RESULTS
Separation and identification of RsbRA and RsbS isoforms.
The RsbRA coantagonist and RsbS antagonist are known to be phosphorylated by the RsbT serine-threonine kinase in vitro (1, 12, 30). Furthermore, phenotypes elicited by substitutions at the known or presumed sites of phosphorylation—threonines 171 and 205 of RsbRA or serine 59 of RsbS—support the hypothesis that modification of these proteins influences the transmission of environmental stress signals (2, 12, 16, 18, 30). However, the phenotypes of some RsbRA substitutions are significantly altered by the presence or absence of the other members of the RsbR coantagonist family (18). In addition, any of these substitutions has the potential to affect protein structure or function independently of phosphorylation.
We therefore sought to directly test the hypothesis by developing IEF protocols to separate the isoforms of RsbRA and RsbS in extracts of wild-type cells. These protocols are described in Materials and Methods together with the two sampling methods we used. For the work described in this section, cells were harvested by a 5-min centrifugation at 4°C before being washed, chilled on ice, and frozen at −80°C. This sampling procedure itself was expected to activate the general stress response, yielding a mix of modified and unmodified regulators that allowed us to test the method and identify the various isoforms.
Turning first to RsbS, Fig. 2A shows that anti-RsbS antibody detected two signals in wild-type extracts subjected to IEF, as would be expected for a regulator that is modified on a single residue (lane 1). Neither of these signals was visible in extracts of mutant cells lacking RsbS (lane 2). Two lines of evidence led us to conclude that the more acidic of these isoforms was phosphorylated RsbS.
First, λPP is known to remove the phosphate group from serine, threonine, or tyrosine residues (31). Significantly, incubation of wild-type extracts with λPP decreased the signal of the more acidic isoform of RsbS and increased that of the more basic isoform (Fig. 2A, lanes 3 to 5). During the course of this experiment, we discovered that this conversion also occurred in the absence of λPP, presumably due to an endogenous enzyme which became active in the presence of λPP buffer and MnCl2. Therefore, in order to directly test the effect of the λPP, we heated the cell extracts for 3 min at 100°C before proceeding with the incubations shown in Fig. 2A.
Second, we analyzed mutant extracts that contained versions of RsbS in which serine 59 (S59) had been changed to either an alanine residue, which produces a form of RsbS that cannot be phosphorylated in vitro (30), or to a negatively charged aspartate residue, which is thought to mimic a phosphoserine (16, 30). Consistent with the results of the λPP assay, the extract containing the S59A variant had only one signal (Fig. 2B, lane 2), and this was near the position of the more basic isoform found in wild-type extracts (lanes 1 and 4). By contrast, the extract containing the S59D variant had no signal at this basic position but did manifest a more acidic signal (lane 3). We noted, however, the lack of close correspondence in the positions of the more acidic isoform found in wild-type cells and the form found in mutant cells bearing the S59D variant. This is consistent with the acidity difference of phosphoserine and an aspartate residue and indicates that the aspartate substitution is not an exact mimic for phosphoserine in RsbS.
For simplicity, we will refer to the more basic RsbS signal found in wild-type cells as the unmodified form (RsbS) and the more acidic signal as the modified form (RsbS-P). These same terms can additionally apply to RsbRA, which also appears to be present in two forms in wild-type cells.
Turning now to RsbRA, we had anticipated that signal interpretation would be complicated as a result of the two potential phosphorylation sites on this regulator. However, our IEF procedure readily separated three RsbRA isoforms, and their identities could be deduced from appropriate controls. As shown in Fig. 2C, the anti-RsbRA antibody first detected two signals in wild-type extracts (lane 1), neither of which was visible in extracts of mutant cells lacking RsbRA (lane 2). As was the case for RsbS, incubation with λPP decreased the signal of the more acidic RsbRA isoform and increased that of the more basic isoform (lanes 3 to 5). In contrast to RsbS, here the more acidic RsbRA isoform was relatively stable in cell extracts, and so no heat treatment was necessary prior to the incubations shown in Fig. 2C. Because RsbRA can be phosphorylated on two threonine residues (12), these results did not allow us to infer whether the more acidic signal reflected phosphorylation on threonine 171 (T171), on threonine 205 (T205), or both.
We addressed this issue by analyzing the mutant extracts shown in Fig. 2D, employing strains in which either or both threonine residues were changed to alanine or aspartate. The extracts bearing the T171 variants each had only a single signal: the signal in the T171A variant of RsbRA (Fig. 2D, lane 2) was near the location of the more basic isoform found in wild-type cells (lanes 1 and 8), and the signal in the T171D variant (lane 3) was near the location of the more acidic isoform. By contrast, the extracts containing the T205 variants each had two signals: the T205A variant (lane 4) had signals which focused to similar positions as those in wild-type extracts, whereas the T205D variant (lane 5) had one signal at about the same position as the more acidic isoform in wild-type cells and a second, new signal that was even more acidic. We interpret these results to indicate that in wild-type cells RsbRA is stably phosphorylated on T171 but not on T205. In this view, and given these growth and harvest conditions, the only circumstance under which the most acidic isoform appeared was when the T205 residue was altered to the charged aspartate.
This interpretation was supported by the analysis of strains bearing double alterations at both T171 and T205. The extract containing the T171A-T205A variant (lane 6) had only a single signal resembling the basic isoform in wild type, whereas the extract containing the T171D-T205D variant (lane 7) had a single signal near the position of the most acidic isoform found in the T205D variant (lane 5). As was the case for the aspartate substitution of RsbS, we note here that the aspartate substitutions of RsbRA did not focus to the same positions as the presumed phosphorylated forms. This is consistent with the acidity difference between an aspartate residue and the phosphothreonine.
For simplicity, we will refer to the more basic RsbRA signal found in wild-type cells as the unmodified form (RsbRA) and to the more acidic signal as the modified form (RsbRA-P). Our conclusion that RsbRA is normally phosphorylated on one and not both threonine residues is further supported by our analysis of salt- and ethanol-stressed cells, which is reported in the following section.
Shift of isoform balance during salt and ethanol stress.
A rapid sampling method was needed to reproducibly detect the changes in RsbS and RsbRA modification resulting from environmental stress. For the experiments described here, we harvested the cells by filtration, washed them with a buffer containing a mixture of phosphatase inhibitors, and then quickly froze them in dry ice-ethanol. The entire process was completed in 60 s or less. Using this approach, we found that the relative proportions of RsbS-P and RsbRA-P increased following salt or ethanol stress. However, the two regulators manifested striking differences both in the balance of isoforms found in unstressed cells and in the kinetics of their phosphorylation after stress.
Figure 3A shows the data for a representative salt stress experiment, Fig. 3B graphically indicates the change in the phosphorylated fraction of RsbS and RsbRA during the course of this experiment, and Fig. 3C summarizes the results of three independent salt stress experiments. We obtained similar results for the ethanol stress experiments shown in Fig. 3D to F. Notably, only the unmodified form of RsbS was found in unstressed cells. Immediately following salt or ethanol stress, we detected a small amount of the modified form, with its peak level recorded at the 1-min time point. In salt-stressed cells the modified form represented about 10% of total RsbS (Fig. 3C), whereas during the stronger ethanol stress the modified form represented about 20% of the total (Fig. 3F). For each stress, the amount of this modified form then decreased, reverting to a new steady-state level that was somewhat higher than that found in prestress cells. This decrease was presumably due to the action of the RsbX feedback phosphatase (24, 27, 30). Because the peak level of RsbS-P was detected at the first time point, the amount produced may have been underestimated. Nonetheless, the observed increase in RsbS-P correlated with the onset of the environmental stress response, measured indirectly by means of β-galactosidase accumulation from a reporter fusion (Fig. 3B and E).
In sharp contrast to the case with RsbS, unstressed cells manifested a balance between the modified and unmodified forms of RsbRA, with about 60 to 70% of the total found in the modified form (Fig. 3). Following salt or ethanol stress, the ratio of modified to unmodified RsbRA further increased, but more slowly than observed for RsbS, with the peak of the modified form recorded at the 5-min time point. Paralleling the observations for RsbS, this increase in the amount of RsbRA-P correlated with the onset of the environmental stress response (Fig. 3B and E). Based on the IEF patterns observed for the substituted forms of RsbRA (Fig. 2D), our interpretation of the results shown in Fig. 3 is that in unstressed cells RsbRA is already largely phosphorylated on T171, and environmental stress further increases the level of this isoform. Neither of the stress conditions shown here produced a detectable signal representative of the isoform phosphorylated on both T171 and T205. Experiments described in the following section underscore the importance of the T171 residue for the in vivo phosphorylation of RsbRA.
Isoform balance is dependent on the RsbT kinase and the T171 residue of RsbRA.
RsbT is known to specifically phosphorylate both RsbS and RsbRA in vitro, and direct biochemical analysis has identified T171 and T205 as the sites on which RsbRA is modified (12, 30). Complementing and extending these in vitro results, genetic analysis has shown that loss of RsbT kinase activity eliminates the environmental stress response (16, 17) and that the T171A alteration of RsbRA substantially blocks environmental signaling (18). In order to test the observed correlation between the environmental stress response and the increased phosphorylation of RsbS and RsbRA (Fig. 3), we next compared the isoforms present in wild-type and mutant cell extracts.
For these experiments, shown in Fig. 4, we examined ethanol-stressed cells over the 0-to-5-min interval found to embody the largest change in isoform balance (Fig. 3). In wild-type cells we observed two forms of RsbS and RsbRA (Fig. 4, wt lanes), and these manifested essentially the same pre- and poststress balance noted in Fig. 3. By contrast, for the mutant lacking the RsbT kinase, only the unmodified forms of RsbS and RsbRA were detected (Fig. 4, ΔrsbT lanes). Yet another pattern was found for the mutant bearing the T171A alteration of RsbRA. Here, the RsbS-P fraction was reduced by a factor of three, calculated by scanning the relevant images (Fig. 4A, compare wt and rsbRAT171A lanes). This result was consistent with the known role of RsbRA in stimulating the in vitro phosphorylation of RsbS by RsbT (9, 12) and suggests that a similar in vivo activity is negatively affected by the T171A substitution. This substitution had a more profound effect on RsbRA phosphorylation, with the RsbRA-P fraction reduced below the limits of detection in both stressed and unstressed cells (Fig. 4B, compare wt and rsbRAT171A lanes). From the results shown in Fig. 4, we conclude that phosphorylation of RsbRA and RsbS is dependent on RsbT in vivo and that phosphorylation of RsbRA is dependent on the integrity of its T171 residue. Moreover, given the deleterious effects that loss of RsbT function or the RsbRAT171A substitution have on environmental signaling in vivo (16-18), these results strengthen the correlation between RsbS and RsbRA phosphorylation and the transmission of environmental stress signals.
DISCUSSION
We have tested a key prediction of the model shown in Fig. 1, which holds that the transmission of environmental stress signals is correlated with the phosphorylation of the RsbS antagonist and RsbRA coantagonist proteins. This correlation had been inferred from the phenotypes elicited by substitutions at the RsbS and RsbRA residues that are the known (or presumed) substrates of the RsbT serine-threonine kinase in vitro (2, 12, 16, 18, 30). Here, we analyzed cell extracts by IEF and confirmed that the proportion of modified RsbS and RsbRA did indeed increase following salt and ethanol stress (Fig. 3). In further confirmation of the model, we also showed that the appearance of the modified forms of both RsbS and RsbRA was dependent on the RsbT kinase (Fig. 4).
Our data suggest that RsbS is unmodified in unstressed cells and that a surprisingly small amount of RsbS is rapidly phosphorylated during the stress response. Based on the limited range of stresses used in this study—a relatively weak salt stress and a relatively strong ethanol stress—it also appears that both the peak levels and the poststress, steady-state levels of RsbS-P correlate with the strength of the stress. This relatively low phosphorylation may reflect the fact that in cell extracts RsbS in found in a complex with RsbRA, RsbRB, and other proteins (9, 18), and some RsbS molecules in this complex could be inaccessible to the RsbT kinase. In contrast, RsbRA appears to be substantially phosphorylated in unstressed cells and slowly becomes more fully phosphorylated during the response. These trends are in accord with previous genetic data, from which it was inferred that phosphorylation of RsbS is sufficient to trigger the environmental stress response (16) and that phosphorylation of RsbRA is a prerequisite for this response (18), as we shall discuss.
While this overall picture of the in vivo phosphorylation state of RsbS and RsbRA is likely correct, we must also consider possible qualifications. Although our assay used a rapid harvesting procedure and yielded dependably reproducible results, it cannot be assured that the data shown in Fig. 3 precisely mirror the in vivo status of the tested regulators. This is particularly the case with RsbS-P, which achieves its highest measured level by the first time point, taken 1 min after the stress. Moreover, RsbS-P is labile to the action of the RsbX feedback phosphatase (9, 27, 30). RsbX expression is induced by stress in a σB-dependent manner (15), and RsbX activity may also increase in response to stress (24). Therefore, considering the rapid kinetics of both its phosphorylation and dephosphorylation, the amount of RsbS-P measured in Fig. 3 may be an underestimate. Nonetheless, our in vivo observations regarding the phosphorylation state of RsbS corroborate previous genetic analyses, from which it was deduced that the phosphorylated form of RsbS is closely associated with the transmission of environmental stress signals (16). In these genetic experiments the RsbS S59A variant, which cannot be phosphorylated, completely prevented environmental signaling. In contrast, the RsbS S59D variant, which is thought to mimic the phosphorylated state, promoted continuous environmental signaling.
There is also some uncertainty surrounding our results regarding RsbRA-P. Although we interpret the data shown in Fig. 3 to indicate that RsbRA is already 60 to 70% phosphorylated in unstressed cells, it remains possible that RsbRA is so readily phosphorylated by the RsbT kinase that even the rapid harvesting procedure we employed could elicit the results shown. Moreover, while the experiment in Fig. 4 indicates that the integrity of the T171 residue is important for the appearance of RsbRA-P, we cannot exclude the possibility that phosphorylation at T205 is also involved in environmental stress signaling. For example, a small amount of the T205-P isoform might be produced but go undetected in our assay, or T205-P might be extremely labile. However, the interpretation that RsbRA is primarily phosphorylated on T171 agrees with our genetic analyses reported elsewhere, from which we infer that this phosphorylation event is required to permit the efficient transmission of environmental stress signals (18).
This genetic analysis was conducted in strains from which the redundant RsbRA paralogs RsbRB, RsbRC, and RsbRD had been removed in order to clearly establish the effects of alterations at T171 and T205 of RsbRA (18). Notably, these studies found that the RsbRA T171A variant largely blocked environmental stress signaling, whereas the T171D variant had a normal stress response. These phenotypes are in accord with our interpretation of the data shown in Fig. 3 and 4—that in unstressed cells RsbRA is already substantially phosphorylated on T171, and the extent of this phosphorylation further increases after stress. If, as we surmise, the phenotype of the T171A alteration primarily reflects its effect on phosphorylation, these data imply that phosphorylation of T171 is normally a prerequisite for the environmental stress response, but it does not by itself trigger the response.
In contrast, the phenotypes caused by T205 alterations, together with our IEF studies, suggest that reversible phosphorylation of T205 is not part of the environmental stress response. In strains in which RsbRA was the only coantagonist present, the T205A alteration caused continuous environmental signaling, whereas the T205D alteration had a normal stress response (18). One explanation for these results is that phosphorylation of T205 is required for RsbRA to function as a coantagonist and that the inability to phosphorylate T205 leads to loss of coantagonist function and, consequently, to continuous environmental signaling. This explanation would require that T205 is normally phosphorylated in unstressed cells, which is contrary to our interpretation of the data shown in Fig. 2 to 4. Because the T205A alteration has no effect on the accumulation of RsbRA protein in vivo (18), we suggest that its signaling phenotype only shows that this residue is important for RsbRA function, perhaps reflecting the importance of a threonine (or aspartate) side chain to the RsbRA structure. In this view, the phosphorylation of T205 observed in vitro (12) does not normally occur in the in vivo environment.
Based on the sum of these results, we propose a new model of environmental signaling, shown in Fig. 5. The significant feature of this model is that a substantial fraction of the RsbRA coantagonist is phosphorylated in unstressed cells grown under the conditions used here. Following environmental stress, a small fraction of the RsbS antagonist is rapidly phosphorylated, leading to the release of RsbT and the activation of the general stress response. According to this model, the phosphorylation states of RsbS and RsbRA (and presumably the other members of the RsbR coantagonist family) are determined by the balance between the activities of the RsbT kinase and the RsbX feedback phosphatase. RsbS and RsbRA are known to be phosphorylated by the RsbT kinase in vitro (1, 9, 12, 30), and we have shown here that the appearance of the modified forms of RsbS and RsbRA is dependent on RsbT in vivo. Similarly, RsbS is known to be dephosphorylated by the RsbX phosphatase in vitro (30), and preliminary evidence points to an RsbX-dependent dephosphorylation of RsbS and RsbRA in vivo (T. J. Kim, T. A. Gaidenko, and C. W. Price, unpublished data). We therefore presume that the differential modification of RsbS and RsbRA noted in unstressed cells reflects a greater activity of the RsbT kinase toward RsbRA as a substrate, or perhaps a lesser activity of the RsbX phosphatase, compared to their activities toward RsbS.
FIG. 5.
New model for environmental stress signaling. The phosphorylation states of the RsbR coantagonist and RsbS antagonist proteins are controlled by the opposing activities of the RsbT kinase and the RsbX feedback phosphatase. In unstressed cells RsbRA is already partially phosphorylated on T171, and from the genetic analysis reported elsewhere (18) we propose that this phosphorylation is required for signaling. Following an environmental stress, RsbS becomes phosphorylated on S59, triggering the release of RsbT and the activation of the RsbU phosphatase. Phosphorylation of RsbRB, RsbRC, and RsbRD is not shown but is presumed to resemble that of RsbRA.
The present study supports the new model shown in Fig. 5, primarily by coupling the in vivo phosphorylation of the RsbS antagonist and RsbRA coantagonist to the active transmission of environmental stress signals. Adjacent genes encoding RsbS and RsbRA orthologs are common within the genomes of the Bacillales, including Listeria monocytogenes (13). And, as shown in Table 2, adjacent genes encoding RsbS and RsbRA orthologs, complete with conserved serine or threonine residues, are found in organisms representing evolutionarily distinct lineages, including the clostridia, actinomycetes, cyanobacteria, gliding bacteria, filamentous anoxygenic phototrophic bacteria, and the beta and gamma subdivisions of Proteobacteria. This widespread distribution suggests that phosphorylation events similar to those we have described here play an analogous role in diverse signaling pathways.
TABLE 2.
Adjacent RsbRA and RsbS ortholog genes in diverse prokaryotic genomes
| Organism | RsbRA orthologa | E valueb | RsbS orthologa | E valueb |
|---|---|---|---|---|
| Bacillus subtilis | RsbRA | 1e-149 | RsbS | 6e-52 |
| Listeria monocytogenes | NP_464415 | 7e-68 | NP_464416 | 1e-36 |
| Nostoc punctiforme | ZP_00107852 | 1e-32 | ZP_00107851 | 2e-20 |
| Burkholderia fungorum | ZP_00034596 | 2e-32 | ZP_00034597 | 2e-19 |
| Chloroflexus aurantiacus | ZP_00018003 | 4e-32 | Nonec | |
| Pseudomonas fluorescens | ZP_00084339 | 2e-31 | ZP_00084338 | 4e-16 |
| Cytophaga hutchinsonii | ZP_00117139 | 8e-30 | ZP_00117138 | 4e-20 |
| Xanthomonas campestris | NP_636545 | 2e-29 | NP_636546 | 1e-18 |
| Streptomyces coelicolor | NP_631378 | 1e-27 | NP_631377 | 2e-16 |
| Vibrio vulnificus | NP_762059 | 2e-26 | NP_762060 | 1e-09 |
| Thermoanaerobacter tengcongensis | NP_622693 | 1e-24 | NP_622694 | 5e-17 |
| Chromobacterium violaceum | NP_900548 | 2e-23 | NP_900549 | 1e-07 |
For organisms other than B. subtilis, each ortholog is indicated by its accession number in the NCBI Protein Database. Organisms are listed by similarity of their RsbRA orthologs to B. subtilis RsbRA.
E value for the alignment resulting from a BLASTP search (5) of proteins available in the NCBI Microbial Genomes Database, using the default parameters of the on-site program. Only the most similar ortholog for a given bacterium is shown.
Gene encoding the most similar C. aurantiacus RsbRA ortholog (ZP_00018003) is flanked by genes for additional RsbRA paralogs, from a total of 18 such genes in the organism. The gene for an RsbS ortholog (ZP_00018452; 5e-17) is not adjacent to any of these RsbRA-like genes.
Acknowledgments
We thank William Haldenwang for his generous gift of the anti-RsbRA and anti-RsbS antibodies and Kazuhiro Shiozaki for providing the rapid harvesting protocol.
This research was supported by Public Health Service grant GM42077 from the National Institute of General Medical Sciences.
REFERENCES
- 1.Akbar, S., T. A. Gaidenko, C. M. Kang, M. O'Reilly, K. Devine, and C. W. Price. 2001. New family of regulators in the environmental signaling pathway which activates the general stress factor σB of Bacillus subtilis. J. Bacteriol. 183:1329-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akbar, S., C. M. Kang, T. Gaidenko, and C. W. Price. 1997. Modulator protein RsbR regulates environmental signaling in the general stress pathway of Bacillus subtilis. Mol. Microbiol. 24:567-578. [DOI] [PubMed] [Google Scholar]
- 3.Alper, S., A. Dufour, D. A. Garsin, L. Duncan, and R. Losick. 1996. Role of adenosine nucleotides in the regulation of a stress response transcription factor in Bacillus subtilis. J. Mol. Biol. 260:165-177. [DOI] [PubMed] [Google Scholar]
- 4.Alper, S., L. Duncan, and R. Losick. 1994. An adenosine nucleotide switch controlling the activity of a cell type-specific transcription factor in B. subtilis. Cell 77:195-205. [DOI] [PubMed] [Google Scholar]
- 5.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benson, A. K., and W. G. Haldenwang. 1993. Bacillus subtilis σB is regulated by a binding protein (RsbW) that blocks its association with core RNA polymerase. Proc. Natl. Acad. Sci. USA 90:2330-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boylan, S. A., A. R. Redfield, M. S. Brody, and C. W. Price. 1993. Stress-induced activation of the σB transcription factor of Bacillus subtilis. J. Bacteriol. 175:7931-7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boylan, S. A., A. Rutherford, S. M. Thomas, and C. W. Price. 1992. Activation of Bacillus subtilis transcription factor σB by a regulatory pathway responsive to stationary-phase signals. J. Bacteriol. 174:3695-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, C.-C., R. J. Lewis, R. Harris, M. D. Yudkin, and O. Delumeau. 2003. A supramolecular signaling complex in the environmental signaling pathway of Bacillus subtilis. Mol. Microbiol. 49:1157-1669. [DOI] [PubMed] [Google Scholar]
- 10.Dufour, A., and W. G. Haldenwang. 1994. Interactions between a Bacillus subtilis anti-sigma factor (RsbW) and its antagonist (RsbV). J. Bacteriol. 176:1813-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dufour, A., U. Voelker, A. Voelker, and W. G. Haldenwang. 1996. Relative levels and fractionation properties of Bacillus subtilis σB and its regulators during balanced growth and stress. J. Bacteriol. 178:3701-3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaidenko, T. A., X. Yang, Y. M. Lee, and C. W. Price. 1999. Threonine phosphorylation of modulator protein RsbR governs its ability to regulate a serine kinase in the environmental stress signaling pathway in Bacillus subtilis. J. Mol. Biol. 284:569-578. [DOI] [PubMed] [Google Scholar]
- 13.Glaser, P., L. Frangeul, C. Buchreiser, C., Rusniok, A. Amend, et al. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 14.Hecker, M., and U. Völker. 2001. General stress response of Bacillus subtilis and other bacteria. Adv. Microb. Physiol. 44:35-91. [DOI] [PubMed] [Google Scholar]
- 15.Kalman, S., M. L. Duncan, S. M. Thomas, and C. W. Price. 1990. Similar organization of the sigB and spoIIA operons encoding alternate sigma factors of Bacillus subtilis RNA polymerase. J. Bacteriol. 172:5575-5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang, C. M., M. S. Brody, S. Akbar, X. Yang, and C. W. Price. 1996. Homologous pairs of regulatory proteins control activity of Bacillus subtilis transcription factor σB in response to environmental stress. J. Bacteriol. 178:3846-3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang, C. M., K. Vijay, and C. W. Price. 1998. Serine kinase activity of a Bacillus subtilis switch protein is required to transduce environmental stress signals but not to activate its target PP2C phosphatase. Mol. Microbiol. 30:189-196. [DOI] [PubMed] [Google Scholar]
- 18.Kim, T. J., T. A. Gaidenko, and C. W. Price. 2004. A multicomponent protein complex mediates environmental stress signaling in Bacillus subtilis. J. Mol. Biol. 341:135-150. [DOI] [PubMed] [Google Scholar]
- 19.Koonin, E. V., L. Aravind, and M. Y. Galperin. 2000. A comparative-genomic view of the microbial stress response, p. 417-444. In G. Storz and R. Hennge-Aronis (ed.), Bacterial stress responses. American Society for Microbiology, Washington, D.C.
- 20.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 21.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Mittenhuber, G. 2002. A phylogenomic study of the general stress response sigma factor σB of Bacillus subtilis and its regulatory proteins. J. Mol. Microbiol. Biotechnol. 4:427-452. [PubMed] [Google Scholar]
- 23.Price, C. W. 2002. General stress response, p. 369-384. In A. L. Sonenshein, R. Losick, and J. A. Hoch (ed.), Bacillus subtilis and its closest relatives: from genes to cells. American Society for Microbiology, Washington, D.C.
- 24.Scott, J. M., T. Mitchell, and W. G. Haldenwang. 2000. Stress triggers a process that limits activation of the Bacillus subtilis stress transcription factor σB. J. Bacteriol. 182:1452-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tatusov, R. L., D. A. Natale, I. V. Garkavtsev, T. A. Tatusova, U. T. Shankavaram, B. S. Rao, B. Kiryutin, M. Y. Galperin, N. D. Fedorova, and E. V. Koonin. 2001. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 29:22-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vijay, K., M. S. Brody, E. Fredlund, and C. W. Price. 2000. A PP2C phosphatase containing a PAS domain is required to convey signals of energy stress to the σB transcription factor of Bacillus subtilis. Mol. Microbiol. 35:180-188. [DOI] [PubMed] [Google Scholar]
- 27.Voelker, U., T. Luo, N. Smirnova, and W. G. Haldenwang. 1997. Stress activation of Bacillus subtilis σB can occur in the absence of the σB negative regulator RsbX. J. Bacteriol. 179:1980-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Voelker, U., A. Voelker, and W. G. Haldenwang. 1996. Reactivation of the Bacillus subtilis anti-σB antagonist, RsbV, by stress- or starvation-induced phosphatase activities. J. Bacteriol. 178:5456-5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wise, A., and C. W. Price. 1995. Four additional genes in the sigB operon of Bacillus subtilis that control activity of the general stress factor σB in response to environmental signals. J. Bacteriol. 177:123-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang, X., C. M. Kang, M. S. Brody, and C. W. Price. 1996. Opposing pairs of serine protein kinases and phosphatases transmit signals of environmental stress to activate a bacterial transcription factor. Genes Dev. 10:2265-2275. [DOI] [PubMed] [Google Scholar]
- 31.Zhuo, S., J. C. Clemens, D. J. Hakes, D. Barford, and J. E. Dixon. 1993. Expression, purification, and biochemical properties of a recombinant protein phosphatase. J. Biol. Chem. 268:17754-17761. [PubMed] [Google Scholar]