Abstract
The availability of a large number of complete genome sequences raises the question of how many genes are essential for cellular life. Trying to reconstruct the core of the protein-coding gene set for a hypothetical minimal bacterial cell, we have performed a computational comparative analysis of eight bacterial genomes. Six of the analyzed genomes are very small due to a dramatic genome size reduction process, while the other two, corresponding to free-living relatives, are larger. The available data from several systematic experimental approaches to define all the essential genes in some completely sequenced bacterial genomes were also considered, and a reconstruction of a minimal metabolic machinery necessary to sustain life was carried out. The proposed minimal genome contains 206 protein-coding genes with all the genetic information necessary for self-maintenance and reproduction in the presence of a full complement of essential nutrients and in the absence of environmental stress. The main features of such a minimal gene set, as well as the metabolic functions that must be present in the hypothetical minimal cell, are discussed.
INTRODUCTION
Complete genome sequences are becoming available for a large number of diverse bacterial species. Comparative genomics shows that most bacterial proteins are highly conserved in evolution, allowing predictions to be made about the functions of most products of uncharacterized genomes based on model organisms, such as Escherichia coli and Bacillus subtilis (gram-positive and gram-negative bacteria, respectively), for which abundant high-quality genetic and biochemical information has been obtained in the past.
One important question raised by the availability of complete genomic sequences is how many genes are essential for cellular life. Although bacterial genomes differ vastly in their sizes and gene repertoires, no matter how small, they must contain all the information to allow the cell to perform many essential (housekeeping) functions that give the cell the ability to maintain metabolic homeostasis, reproduce, and evolve, the three main properties of living cells (53). Cells usually can import metabolites but not functional proteins; therefore, they have to rely on their own gene products to perform such essential functions.
The determination of the minimal set of protein-coding genes necessary to maintain a living cell is becoming an increasingly appealing issue, considering that such minimal gene set should include “the smallest possible group of genes that would be sufficient to sustain a functioning cellular life form under the most favorable conditions imaginable, that is, in the presence of a full complement of essential nutrients and in the absence of environmental stress” (48). Reconstruction of the minimal gene set can take advantage of the increasing knowledge of completely characterized genomes. In recent years, several research groups have tried to define the essential set of survival protein-encoding genes in bacteria by different experimental and computational methods (reviewed in references 22, 24, 38, and 79). Three different experimental approaches have been used to identify genes that are essential under particular growth conditions: massive transposon mutagenesis strategies (the most widely used approach), the use of antisense RNA to inhibit gene expression (22, 37), and the systematic inactivation of each individual gene present in a genome (31, 45, 64; http://www.genome.wisc.edu/functional/tnmutagenesis.htm). However, all these approaches have limitations. Transposon mutagenesis might overestimate the set by misclassification of nonessential genes that slow growth without arresting it but can also miss essential genes that tolerate transposon insertions. The use of antisense RNA is limited to the genes for which an adequate expression of the inhibitory RNA can be obtained in the organism under study. Finally, inactivation of single genes does not detect essential functions encoded by redundant genes, and the essential gene set is not the same as the minimal genome, since it is clear that genes that are individually dispensable may not be simultaneously dispensable.
Computational analysis has also been extensively used to try to get closer to the minimal gene set (28, 48, 65, 81). For this purpose, the smallest bacterial genomes fully sequenced, from bacterial parasites or endosymbionts, have been very useful, since they must retain all genes involved in housekeeping functions and a minimum amount of metabolic transactions for cellular survival and replication in their given niche. The smallest complete bacterial genome reported thus far corresponds to the human pathogen Mycoplasma genitalium, with only 480 protein-coding genes in a 540-kb genome (23), which can be considered an upper-limit estimate for the minimal bacterial gene set. However, smaller genomes have been described for other bacteria, such as some strains of Buchnera aphidicola, the endosymbiotic bacteria of aphids, whose genomes have an estimated size of around 450 kb, which can contain about 400 protein-coding genes (27).
The two first bacterial genomes completely sequenced were those from the parasitic bacteria Haemophilus influenzae (21) and M. genitalium (23), gram-negative and gram-positive bacteria, respectively, with reduced genomes (1.83 Mb and 0.58 Mb respectively). Soon after that, Mushegian and Koonin performed a computational comparison between them, assuming that genes conserved across the large phylogenetic distances that exist between these two species are likely to be essential (65). The authors suggested a list of 256 conserved genes as a close estimate of the minimal gene set. However, some of the genes considered essential by this approach can be disrupted by transposon insertion (34), and it is reasonable to consider that enlargening the number of genomes used in the comparison will significatively reduce the number of genes considered to be essential.
In the last few years, complete genomes of five bacterial endosymbionts of insects have been described (2, 28, 77, 80, 83). These bacteria have a permanent host-associated life-style, and therefore they have relaxed selection on the maintenance of genes that are not required for survival in their protected environments. Based on the above-mentioned assumption that the genes shared by multiple genomes are likely to be essential and to be good candidates for inclusion in the minimal gene set, we performed a comparative analysis of all five endosymbiont genomes, which revealed that they share only 277 orthologous protein-coding genes (28). The obtained minimal endosymbiont gene set was then compared with the genome of M. genitalium; this resulted in the identification of 180 housekeeping genes that were shared by all six genomes. The number of shared genes among endosymbiotic and parasitic bacteria was further reduced to 156 when the parasites Rickettsia prowazekii and Chlamydia trachomatis were added to the comparison (44). However, computational analysis has also limitations, since it is likely to underestimate the minimal set because it takes into account only the genes that have remained similar enough during the course of evolution to be recognized as true orthologues. Therefore, it will not include genes with a high rate of evolution, which may not show their relationship in comparisons of distant taxons. In the above-mentioned study, the displacement of unrelated but functionally analogous protein-coding genes was not considered, thus presenting only a core of the possible minimum gene set necessary to sustain life.
In an attempt to get closer to the minimal bacterial genome, in this paper we present an enhanced review of all previously used strategies for addressing this issue. Taking as a start our previous computational comparison of gene orthologues for the five completely sequenced endosymbionts, we added to the comparison functional equivalent genes that do not present sequence similarity. The obtained gene set was compared with the B. subtilis essential genes determined by systematic inactivation (45) and with the proposed essential genes for E. coli based on the genome-scale genetic footprinting performed by Gerdes et al. (25), the information found at the PEC (for “Profiling E. coli Chromosome”) website (http://www.shigen.nig.ac.jp/ecoli/pec), and the results obtained by F.R. Blattner and coworkers (http://www.genome.wisc.edu/functional/tnmutagenesis.htm). It should be noticed that the PEC and Blattner's E. coli sequencing-project databases are incomplete, and in some cases there are discrepancies about the essentiality of E. coli genes depending on the source. Some differences might be due to the different growth conditions used in the experiments, while others might be reflecting the inability of a mutant with a severe decrease in fitness to maintain an enough vigorous growth to be isolated in the footprinting experiments (25). We also took into account the computationally derived minimal gene set proposed for M. genitalium (65) and the results of the global transposon mutagenesis for mycoplasmas (34) in order to detect genes that appear to be dispensable. Genes that are present in all five endosymbionts, and for which no transposon insertion could be detected in mycoplasmas, were considered essential in reduced genomes, even if they appear to be nonessential in bacteria with larger genomes. We have also included in our minimal gene set some of the genes that were disrupted by transposon insertion in mycoplasmas but were proven to be essential in other bacteria and are present in all five endosymbionts under study. We also compared our set with the list of essential genes identified in Staphylococcus aureus by antisense RNA experiments (22, 37) and the recently sequenced genome of Phytoplasma asteris (70). Finally, the resulting gene list was reanalyzed to detect gaps in metabolic pathways that could be considered essential to maintain a reasonable metabolic homeostasis for any living cell.
The following gene and protein servers and databases were used to identify the corresponding orthologous genes and protein functions, as well as to reconstruct the metabolic pathways: BRENDA (for “Braunschweig Enzyme Database”) (http://www.brenda.uni-koeln.de/index.php4), COG (for “Clusters of Orthologous Groups of Proteins”) (http://www.ncbi.nlm.nih.gov/COG/), ExPASy (for “Expert Protein Analysis System”) molecular biology server (http://us.expasy.org), KEGG (for “Kyoto Encyclopedia of Genes and Genomes”) (http://www.genome.ad.jp/kegg2.html), MBGD (for “Microbial Genome Databasefor Comparative Analysis”) (http://mbgd.genome.ad.jp), andPfam (for “Protein Families Database of Alignments and HMMs”) (http://www.sanger.ac.uk/Software/Pfam).
The results of this analysis are summarized in Table 1 and, with more detail, in Table S2 presented in the supplemental material. The functional classification of the genes is based on the categories used in the sequencing work on Aquifex aeolicus (19). The number of genes included in each category is indicated parenthetically in the tables. The gene name column generally indicates the most common synonymous for the corresponding gene in gram-negative bacteria (except when no gene with such function has been identified in E. coli), although in some cases it does not correspond with the given name for the gram-positive bacteria orthologous gene. Table S2 in the supplemental material also includes the homologous genes present in the six completely sequenced reduced genomes (M. genitalium, B. aphidicola strains BAp, BSg and BBp, Candidatus Blochmannia floridanus, and Wigglesworthia glossinidia) and the two model bacteria used in this analysis (E. coli and B. subtilis). The B. subtilis gene numbers are those given by the Japanese and European Consortium involved in its sequencing(http://bacillus.genome.ad.jp/ and http://genolist.pasteur.fr/SubtiList/) (50) and can also be found in the KEGG database. In the cases of nonorthologous or nonhomologous genes encoding proteins with the same function, we have included in Table S2 (in the supplemental material) the gene that was present in more analyzed organisms.
TABLE 1.
Protein-coding genes in the hypothetical minimal cell
| Category | Subcategory | Gene | E.C. no. | Protein function |
|---|---|---|---|---|
| DNA metabolism (16 genes) | Basic replication machinery (13 genes) | dnaB | 3.6.1.- | Replicative DNA helicase |
| dnaE | 2.7.7.7 | DNA polymerase III, α subunit | ||
| dnaG | 2.7.7.- | DNA primase | ||
| dnaN | 2.7.7.7 | DNA polymerase III, β subunit | ||
| dnaQ | DNA polymerase III, ɛ subunit | |||
| dnaX | 2.7.7.7 | DNA polymerase III, γ and τ subunits | ||
| gyrA | 5.99.1.3 | DNA gyrase, A subunit | ||
| gyrB | 5.99.1.3 | DNA gyrase, B subunit | ||
| holA | 2.7.7.7 | DNA polymerase III, δ subunit | ||
| holB | 2.7.7.7 | DNA polymerase III, δ′ subunit | ||
| hupA | DNA binding protein | |||
| lig | 6.5.1.2 | DNA ligase (NAD dependent) | ||
| ssb | SSB | |||
| DNA repair, restriction, and modification (3 genes) | nth | 4.2.99.18 | Endonuclease III | |
| polA | 3.1.11.- | 5′-3′ exonuclease domain of DNA polymerase I | ||
| ung | 3.2.2.- | Uracil-DNA glycosylase | ||
| RNA metabolism (106 genes) | Basic transcription machinery (8 genes) | deaD | ATP-dependent RNA helicase | |
| greA | Transcription elongation factor | |||
| nusA | Transcription-translation coupling | |||
| nusG | Transcription antitermination protein | |||
| rpoA | 2.7.7.6 | RNA polymerase, α subunit | ||
| rpoB | 2.7.7.6 | RNA polymerase, β subunit | ||
| rpoC | 2.7.7.6 | RNA polymerase, β′ subunit | ||
| rpoD | RNA polymerase major σ factor | |||
| Translation: aminoacyl-tRNA synthesis (21 genes) | alaS | 6.1.1.7 | Alanyl-tRNA synthase | |
| argS | 6.1.1.19 | Arginyl-tRNA synthase | ||
| asnS | 6.1.1.22 | Asparaginyl-tRNA synthase | ||
| aspS | 6.1.1.12 | Aspartyl-tRNA synthase | ||
| cysS | 6.1.1.16 | Cysteinyl-tRNA synthase | ||
| glnS | 6.1.1.18 | Glutaminyl-tRNA synthase | ||
| gltX | 6.1.1.17 | Glutamyl-tRNA synthase | ||
| glyS | 6.1.1.14 | Glycyl-tRNA synthase, b subunit | ||
| hisS | 6.1.1.21 | Histidyl-tRNA synthase | ||
| ileS | 6.1.1.5 | Isoleucyl-tRNA synthase | ||
| leuS | 6.1.1.4 | Leucyl-tRNA synthase | ||
| lysS | 6.1.1.6 | Lysyl-tRNA synthase | ||
| metS | 6.1.1.10 | Methionyl-tRNA synthase | ||
| pheS | 6.1.1.20 | Phenylalanyl-tRNA synthase, a subunit | ||
| pheT | 6.1.1.20 | Phenylalanyl-tRNA synthase, b subunit | ||
| proS | 6.1.1.15 | Prolyl-tRNA synthase | ||
| serS | 6.1.1.11 | Seryl-tRNA synthase | ||
| thrS | 6.1.1.3 | Threonyl-tRNA synthase | ||
| trpS | 6.1.1.2 | Tryptophanyl-tRNA synthase | ||
| tyrS | 6.1.1.1 | Tyrosyl-tRNA synthase | ||
| valS | 6.1.1.9 | Valyl-tRNA synthase | ||
| Translation: tRNA maturation and modification (6 genes) | iscS | 4.4.1- | Cysteine desulfurase-NifS homolog | |
| mnmAa | 2.1.1.61 | tRNA (5-methylaminomethyl-2-thiouridylate) methyltransferase | ||
| mnmEb | GTP binding protein involved in biosynthesis of 5-methilaminomethyl-2-thiouridine | |||
| mnmGc | Glucose-inhibited division protein A, involved in biosynthesis of 5-methylaminomethyl-2-thiouridine | |||
| pth | 3.1.1.29 | Peptidyl-tRNA hydrolase | ||
| rnpA | 3.1.26.5 | Protein component of RNa P | ||
| Translation: ribosomal proteins (50 genes) | rplA | 50S ribosomal protein L1 | ||
| rplB | 50S ribosomal protein L2 | |||
| rplC | 50S ribosomal protein L3 | |||
| rplD | 50S ribosomal protein L4 | |||
| rplE | 50S ribosomal protein L5 | |||
| rplF | 50S ribosomal protein L6 | |||
| rplI | 50S ribosomal protein L9 | |||
| rplJ | 50S ribosomal protein L10 | |||
| rplK | 50S ribosomal protein L11 | |||
| rplL | 50S ribosomal protein L12 | |||
| rplM | 50S ribosomal protein L13 | |||
| rplN | 50S ribosomal protein L14 | |||
| rplO | 50S ribosomal protein L15 | |||
| rplP | 50S ribosomal protein L16 | |||
| rplQ | 50S ribosomal protein L17 | |||
| rplR | 50S ribosomal protein L18 | |||
| rplS | 50S ribosomal protein L19 | |||
| rplT | 50S ribosomal protein L20 | |||
| rplU | 50S ribosomal protein L21 | |||
| rplV | 50S ribosomal protein L22 | |||
| rplW | 50S ribosomal protein L23 | |||
| rplX | 50S ribosomal protein L24 | |||
| rpmA | 50S ribosomal protein L27 | |||
| rpmB | 50S ribosomal protein L28 | |||
| rpmC | 50S ribosomal protein L29 | |||
| rpmE | 50S ribosomal protein L31 | |||
| rpmF | 50S ribosomal protein L32 | |||
| rpmG | 50S ribosomal protein L33 | |||
| rpmH | 50S ribosomal protein L34 | |||
| rpmI | 50S ribosomal protein L35 | |||
| rpmJ | 50S ribosomal protein L36 | |||
| rpsB | 30S ribosomal protein S2 | |||
| rpsC | 30S ribosomal protein S3 | |||
| rpsD | 30S ribosomal protein S4 | |||
| rpsE | 30S ribosomal protein S5 | |||
| rpsF | 30S ribosomal protein S6 | |||
| rpsG | 30S ribosomal protein S7 | |||
| rpsH | 30S ribosomal protein S8 | |||
| rpsI | 30S ribosomal protein S9 | |||
| rpsJ | 30S ribosomal protein S10 | |||
| rpsK | 30S ribosomal protein S11 | |||
| rpsL | 30S ribosomal protein S12 | |||
| rpsM | 30S ribosomal protein S13 | |||
| rpsN | 30S ribosomal protein S14 | |||
| rpsO | 30S ribosomal protein S15 | |||
| rpsP | 30S ribosomal protein S16 | |||
| rpsQ | 30S ribosomal protein S17 | |||
| rpsR | 30S ribosomal protein S18 | |||
| rpsS | 30S ribosomal protein S19 | |||
| rpsT | 30S ribosomal protein S20 | |||
| Translation: ribosome function, maturation and modification (7 genes) | cspR | 2.1.1.- | Ribosomal methytransferase | |
| engA | GTP binding protein | |||
| era | GTP binding protein | |||
| ksgA | 2.1.1.- | Dimethyladenosine transferase | ||
| obg | GTP binding protein | |||
| rbfA | Ribosome binding factor A | |||
| ychF | GTP binding protein | |||
| Translation factors (12 genes) | efp | Elongation factor P | ||
| fusA | 3.6.1.48 | Elongation factor G | ||
| frr | Ribosome-recycling factor | |||
| hemK | 2.1.1.- | N5-glutamine methyltransferase, modulation of release factor activity | ||
| infA | Initiation factor IF-1 | |||
| infB | Initiation factor IF-2 | |||
| infC | Initiation factor IF-3 | |||
| lepA | GTP binding elongation factor | |||
| prfA | Peptide chain release factor 1 (RF1) | |||
| smpB | tmRNA binding protein | |||
| tsf | Elongation factor Ts | |||
| tufA | 3.6.5.3 | Elongation factor Tu | ||
| RNA degradation (2 genes) | pnp | 2.7.7.8 | Polyribonucleotide nucleotidyltransferase | |
| rnc | 3.1.26.3 | Ribonuclease III | ||
| Protein processing, folding, and secretion (15 genes) | Protein posttranslational modification (2 genes) | map | 3.4.11.18 | Methionine aminopeptidase |
| pepA | 3.4.11.1 | Aminopeptidase A/I | ||
| Protein folding (5 genes) | dnaJ | Hsp70 cochaperone | ||
| dnaK | Chaperone Hsp70 | |||
| groEL | Class I heat shock protein | |||
| groES | Class 1 heat shock protein | |||
| grpE | Hsp70 cochaperone | |||
| Protein translocation and secretion (5 genes) | ffh | Protein component of signal recognition particle | ||
| ftsY | Signal recognition particle receptor | |||
| secA | Preprotein translocase subunit (ATPase) | |||
| secE | Membrane-embedded preprotein translocase subunit | |||
| secY | Membrane-embedded preprotein translocase subunit | |||
| Protein turnover (3 genes) | gcp | 3.4.24.57 | Probable O-sialoglycoprotein endopeptidase | |
| hflB | 3.4.24.- | ATP-dependent protease | ||
| Ion | 3.4.21.53 | ATP-dependent protease La | ||
| Cellular processes (5 genes) | Cell division (1 gene) | ftsZ | Cytoskeletal cell division protein | |
| Transport (4 genes) | pitA | Low-affinity inorganic phosphate transporter | ||
| ptsG | 2.7.1.69 | PTS glucose-specific enzyme II | ||
| ptsH | Histidine-containing phosphocarrier protein of PTS | |||
| ptsI | PTS enzyme I | |||
| Energetic and intermediary metabolism (56 genes) | Glycolysis (10 genes) | eno | 4.2.1.11 | Enolase |
| fbaA | 4.1.2.13 | Fructose-1,6-bisphosphate aldolase | ||
| gapA | 1.2.1.12 | Glyceraldehyde-3-phosphate dehydrogenase | ||
| gpmA | 5.4.2.1 | Phosphoglycerate mutase | ||
| ldh | 1.1.1.27 | l-Lactate dehydrogenase | ||
| pfkA | 2.7.1.11 | 6-Phosphofructokinase | ||
| pgi | 5.3.1.9 | Glucose-6-phosphate isomerase | ||
| pgk | 2.7.2.3 | Phosphoglycerate kinase | ||
| pykA | 2.7.1.40 | Pyruvate kinase | ||
| tpiA | 5.3.1.1 | Triose-phosphate isomerase | ||
| Proton motive force generation (9 genes) | atpA | 3.6.3.14 | ATP synthase α chain | |
| atpB | 3.6.3.15 | ATP synthase A chain | ||
| atpC | 3.6.3.16 | ATP synthase ɛ chain | ||
| atpD | 3.6.3.17 | ATP synthase β chain | ||
| atpE | 3.6.3.18 | ATP synthase C chain | ||
| atpF | 3.6.3.19 | ATP synthase B chain | ||
| atpG | 3.6.3.20 | ATP synthase γ chain | ||
| atpH | 3.6.3.21 | ATP synthase δ chain | ||
| yidC | Essential for proper integration of ATPase into the membrane | |||
| Penthose phosphate pathway (3 genes) | rpe | 5.1.3.1 | Ribulose-phosphate 3-epimerase | |
| rpiA | 5.3.1.6 | Ribose 5-phosphate isomerase | ||
| tkt | 2.2.1.1 | Transketolase | ||
| Lipid metabolism (7 genes) | cdsA | 2.7.7.41 | Phosphatidate cytidylyltransferase | |
| fadD | 6.2.1.3 | Acyl-CoA synthase | ||
| gpsA | 1.1.1.94 | sn-Glycerol-3-phosphate dehydrogenase | ||
| plsB | 2.3.1.15 | sn-Glycerol-3-phosphate acyltransferase | ||
| plsC | 2.3.1.51 | 1-Acyl-sn-glycerol-3-phosphate acyltransferase | ||
| psd | 4.1.1.65 | Phosphatidylserine decarboxylase | ||
| pssA | 2.7.8.8 | Phosphatidylserine synthase | ||
| Biosynthesis of nucleotides (15 genes) | adk | 2.7.4.3 | Adenylate kinase | |
| dcd | 3.5.4.13 | dCTP deaminase | ||
| gmk | 2.7.4.8 | Guanylate kinase | ||
| hpt | Hypoxanthine phosphoribosyltransferase | |||
| ndk | 2.7.4.6 | Nucleoside diphosphate kinase | ||
| nrdE | 1.17.4.1 | Ribonucleoside diphosphate reductase (major subunit) | ||
| nrdF | 1.17.4.1 | Ribonucleoside diphosphate reductase (minor subunit) | ||
| ppa | 3.6.1.1 | Inorganic pyrophosphatase | ||
| prsA | 2.7.6.1 | Phosphoribosylpyrophosphate synthase | ||
| pyrG | 6.3.4.2 | CTP synthase | ||
| thyA | 2.1.1.45 | Thymidylate synthase | ||
| tmk | 2.7.4.9 | Thymidylate kinase | ||
| trxA | Thioredoxin | |||
| trxB | 1.8.1.9 | Thioredoxin reductase | ||
| upp | 2.4.2.9 | Uracil phosphoribosyltransferase | ||
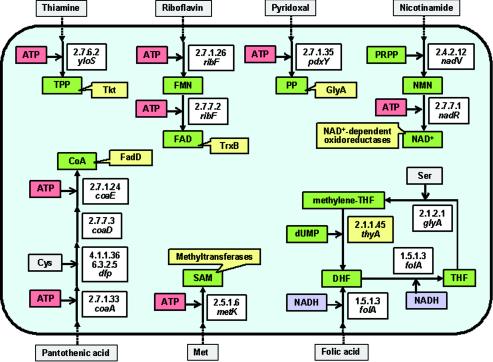
| Biosynthesis of cofactors (12 genes) | coaA | 2.7.1.33 | Pantothenate kinase | |
| coaD | 2.7.7.3 | 4′-Phosphopantetheine adenylyltransferase | ||
| coaE | 2.7.1.24 | Dephospho-CoA kinase | ||
| dfp | 6.3.2.5 | Phosphopantothenate cysteine ligase | ||
| 4.1.1.36 | 4′-Phospho-pantothenyl-l-cysteine decarboxylase | |||
| folA | 1.5.1.3 | Dihydrofolate reductase | ||
| glyA | 2.1.2.1 | Glycine hydroxymethyltransferase | ||
| metK | 2.5.1.6 | Methionine adenosyltransferase | ||
| nadR | 2.7.7.1 | Adenylyltransferase | ||
| nadV | Nicotinamide phosphoribosyltransferase | |||
| pdxY | 2.7.1.35 | Pyridoxal kinase | ||
| ribF | 2.7.1.26 | Riboflavin kinase | ||
| 2.7.7.2 | Flavin mononucleotide adenylyltransferase | |||
| yloS | 2.7.6.2 | Thiamine pyrophosphokinase | ||
| Poorly characterized (8 genes) | mesJ | Conserved hypothetical protein | ||
| mraW | 2.1.1.- | Methyltrasferase | ||
| ybeY | Conserved hypothetical protein | |||
| ycfF | HIT family | |||
| ycfH | 3.1.21.- | Putative deoxyribonuclease, tatD family | ||
| yoaE | Conserved hypothetical protein | |||
| yqgF | Conserved hypothetical protein | |||
| yraL | Conserved hypothetical protein |
Old gene name: trmU.
Old gene name: thdF.
Old gene name: gidA.
The notion of a minimal cell cannot be sharply defined, since different essential functions can be defined depending on the environmental conditions, and numerous versions of minimal gene sets can be conceived to fulfill such functions even for the same set of conditions (49). Nevertheless, it is possible to try to define which functions should be performed in any living cell and to list the genes that would be necessary to maintain such functions. The number of these genes, but not the number of functions, could show some minor variation associated with putative gene fusions. In this work, we present the hypothetical core of one of the possible minimal protein-coding gene sets able to sustain a functional bacterial cell under ideal conditions, and we examine the main features of the proposed minimal gene set. The rationale followed to decide which genes should be included in it is described in detail.
MAIN FEATURES OF THE MINIMAL SET
Information Storage and Processing
DNA metabolism. (i) Basic replication machinery.
One of the most important housekeeping functions in all living cells is DNA replication, a multistage reaction with four basic steps. First, a replication origin sequence within the genome is recognized by protein components. Second, initiation of replication occurs through the recruitment of replisomal proteins at the origin. Third, the general replication reaction duplicates both strands of DNA. Fourth, replication terminates and the two daughter chromosomes are separated.
The specific mechanism for replication initiation depends both on the structure of the replication origin and on the nature of the initiation protein (47). Several mechanisms have been described in bacteria (46), but the analysis of the genome of B. floridanus, the endosymbiotic bacteria of carpenter ants, revealed that it does not encode any of the proposed initiation proteins (DnaA, RecA, and PriA). Another recruitment protein (DnaC in E. coli, DnaI in B. subtilis) is also required to form a complex with the helicase DnaB (EC 3.6.1.-) to prevent the indiscriminate binding of the helicase to single-stranded DNA, transferring it only in the correct DNA location which is not coated by Single-Stranded DNA Binding protein. (SSB). However, DnaC is absent in B. floridanus and W. glossinidia (endosymbiont of tsetse flies), indicating that it is also dispensable in these bacteria. Hence, it can be assumed that DNA replication can take place without the need for these initiation and recruiting proteins under some conditions, which might be the case for small genomes, or that there are other, as yet unidentified proteins that perform such functions in these reduced genomes. The recruitment and loading of helicase at the replication origin generally requires the presence of a histone-like protein for the destabilization of the duplex DNA near the origin of replication. At least one histone-like protein is also encoded by all five analyzed endosymbiont genomes, although none of these proteins is conserved in all of them. In M. genitalium, MG353 appears to encode a nucleoid DNA binding protein similar to hupA. Since this gene is also present in all endosymbionts analyzed except B. floridanus, we decided to include this histone-like protein-coding gene in our minimal set.
To begin DNA replication, the helicase DnaB (EC 3.6.1.-) attracts the primase DnaG (EC 2.7.7.-) to the replication fork, both of them present in all bacteria with reduced genomes analyzed in this study. Once the DNA is melted and primed, the general DNA replication begins, with the action of DNA polymerase III (EC 2.7.7.7). Genes encoding the α, ɛ, γ and τ, δ, δ', and β subunits of DNA polymerase III, all subunits that are present both in gram-negative and gram-positive bacteria, are also conserved. It should be noticed that in gram-positive bacteria there are two genes encoding the α subunits of this holoenzyme, both of them essential in B. subtilis, but one of them (dnaE) has been proven to be dispensable in M. genitalium, while polC encodes both the α and ɛ subunits of DNA polymerase III. Two more proteins required at this stage, gyrase (EC 5.99.1.2, encoded by gyrA and gyrB) and ligase (EC 6.5.1.2, encoded by lig), are also conserved, thus allowing the completion of the third stage of DNA replication. Although lig was previously annotated as a pseudogene in B. aphidicola BSg due to the absence of the C-terminal protein end (80), it has been reannotated as a functional gene encoding the catalytic domain of the protein but with the absence of the BRCT domain (28). On the other hand, the gene encoding the protein necessary to stop replication by inhibiting helicase translocation (tus in E. coli, rtp in B. subtilis) cannot be found in the reduced genomes under study. Several topoisomerases that are present in M. genitalium and essential in B. subtilis and E. coli to allow independent segregation of the two daughter chromosomes (EC 5.99.1.2, encoded by topA, and EC 5.99.1.-, encoded by parC and parE) must also be considered dispensable, since parC and parE have been lost in all endosymbionts analyzed, while topA is only present in two strains of B. aphidicola. It is possible that the above-mentioned GyrA/B, which is present in all these bacteria, performs all the topoisomerase functions required for DNA replication and segregation, since it has been suggested that type IA (TopA) and type II (GyrA/B and ParD/E) topoisomerases may use comparable mechanisms to bind and cleave DNA, based on similarities in several structural elements (6). Other proteins involved in chromosome condensation that are essential in B. subtilis (SMC, ScpA, and ScpB) are not conserved either, and they have not been included in the minimal set. SMC proteins are conserved in most (but not all) prokaryotes, and E. coli mutants lacking the encoding gene are viable (85).
(ii) DNA repair, restriction, and modification.
A common feature of reduced genomes is the loss of most genes involved in DNA recombination and repair, which has been extensively studied in endosymbiotic bacteria. It has been proposed that the loss of DNA repair mechanisms at the beginning of the symbiotic relationship started a process of continuous degeneration of these genomes (62) while the loss of genes involved in homologous recombination has probably contributed to the chromosome stability of reduced genomes (78, 80). However, a rudimentary system of DNA repair is still maintained in every endosymbiont genome and in M. genitalium. As in B. aphidicola, M. genitalium has lost the DNA polymerase domain of polA (which encodes DNA polymerase I), and only the portion with the 5′-3′ exonuclease activity of the protein (EC 3.1.11.-) has been conserved, indicating that the truncated protein must be involved only in DNA repair mechanisms. Furthermore, endonuclease III (EC 4.2.99.18, encoded by nth), which repairs DNA at apurinic or apyrimidinic sites, is present in all endosymbionts, while M. genitalium retains the nfo gene, which encodes an endonuclease IV (EC 3.1.21.2) with similar activity. The exonuclease V (EC 3.1.11.5), encoded by the recBCD system in gram-negative bacteria, is present in all endosymbionts under study, but its gram-positive counterpart, encoded by the addAB system, is not present in M. genitalium. Instead, M. genitalium retains the UvrABC system, although uvrA has proven to be dispensable (34). None of them seems to be essential for B. subtilis and E. coli, probably due to the existence of several overlapping DNA repair mechanisms in large genomes or because they are dispensable in the absence of selective pressure. The ung gene, encoding the DNA repair enzyme uracil-DNA glycosylase (EC 3.2.2.-), is also present in all reduced genomes that have been analyzed, although it is not considered essential in B. subtilis and E. coli. Taking everything into account, we propose that a minimal gene set should include at least the genes that encode one endonuclease (nth or nfo), one exonuclease (encoded by a truncated version of polA), and ung. The presence of these genes would allow the cell to maintain the rate of mutation at tolerable levels.
RNA metabolism.
RNA metabolism refers to all processes that involve RNA, including transcription, processing, and modification of transcripts; translation; and RNA degradation and its regulation. It is the central and most evolutionarily conserved part of cell physiology. The genes involved in these pathways represent more than 50% of the total number of genes included in our proposed minimal set (107 of 206 genes).
(i) Basic transcription machinery.
Five genes that encode components of the basic transcription machinery (rpoA, rpoB, rpoC, and rpoD, related to RNA polymerase function [EC 2.7.7.6], and nusA, involved in the coupling between translation and termination of RNA synthesis) are essential in B. subtilis and are conserved in all genomes analyzed in this study. They have all been included in the minimal gene set, although, surprisingly, rpoD (encoding the major σ factor of RNA polymerase; which promotes the transcription of a wide variety of genes) can be disrupted in M. pneumoniae. However, the authors recognize that there might exist some nonidentified problems with the method of mutagenesis used, which result in leakage for some of the mutants (34). We have also included in the minimal gene set four more genes involved in transcription which are present in all five endosymbionts and M. genitalium: deaD (ATP-dependent RNA helicase) and greA and nusG, encoding two transcription factors. Although none of them are essential in B. subtilis and some experiments indicate that they might not be essential in E. coli either, all of them seem to be essential in M. genitalium, since none of them have been disrupted in the massive transposon knockout experiment performed by Hutchison et al. (34). Furthermore, deaD is the only gene encoding an RNA helicase that has been preserved in the reduced genomes analyzed.
Two more genes that are essential in B. subtilis, members of a two-component system involved in regulation of RNA synthesis (yycF and yycG, EC 2.7.3.-), are nonessential in E. coli and are absent in the reduced genomes under study. We have not included in the minimal gene set any transcription regulator, since it does not seem to be an essential function in bacteria with reduced genomes.
(ii) Translation.
The largest category of preserved genes corresponds to those involved in protein synthesis (78 genes), represented mainly by aminoacyl-tRNA synthases and ribosomal proteins.
Aminoacyl-tRNA synthases for all amino acid present in proteins are represented in all genomes with the exception of only glutaminyl-tRNA synthase (glnS, EC 6.1.1.18), which is absent in B. subtilis and in M. genitalium. These two gram-positive bacteria contain instead the genes that encode the different subunits of the glutamyl-tRNA amidotransferase (EC 6.3.5.-), gatA, gatB, and gatC, the last of which is missing in M. genitalium. It has been proposed that only the A and B subunits of the heterotrimeric protein are necessary for its activity, since only homologues of the gatA and gatB genes can be found in all the archaea, gram-positive bacteria, and organelles analyzed (18). In all genomes analyzed in this work, the glycyl-tRNA synthase is an heterotetramer composed of two α and two β subunits, except in M. genitalium, where it is composed only of two β subunits. Thus, it can be considered that the smallest set of genes necessary to encode all 20 aminoacyl-tRNA synthases (EC 6.1.1.-) will contain the genes that are present in all five endosymbionts, but considering that a functional glycyl-tRNA synthase can be encoded only by the glyS gene, as in M. genitalium. The fmt gene, required for the formylation of methionyl tRNA (EC 2.1.2.9), is also preserved in all the genomes under study, is essential in B. subtilis, and E. coli strains lacking this gene have severely impaired growth rates (30). However, it was not identified in the recently sequenced genome of Phytoplasma asteris (70), and formylation has proven not to be essential for all bacteria, since it is dispensable in Pseudomonas aeruginosa (66), while several E. coli, S. aureus, and S. pneumoniae mutants simultaneously lacking fmt and def (encoding a peptide deformylase) have been obtained (13, 55, 56, 59). Therefore, it has not been included in the minimal gene set.
Among the genes involved in tRNA modification that have been described as essential in B. subtilis, only rnpA (encoding the protein component of RNase P, essential for tRNA processing; EC 3.1.26.5), pth (which encodes a peptidyl-tRNA hydrolase, EC 3.1.1.29), and trmU (encoding a tRNA methyltransferase that participates in the hypermodification of tRNA, EC 2.1.1.61) seem to be essential in small genomes. rnpA also appears to be essential in E. coli (39), but published reports disagree about whether pth and trmU (renamed mnmA [52]) are essential in E. coli (16, 25, 29, 40). mnmA can be disrupted in M. genitalium (34), but it is also essential in S. aureus (22). With respect to the other B. subtilis essential genes, cca (which encodes a tRNA nucleotidyl transferase involved in tRNA repair, EC 2.7.7.25), which is also essential in E. coli, is absent in M. genitalium. The loss of trmD (encoding a tRNA methylase, EC 2.1.1.31) reduces the growth rate in E. coli (72), but trmD appears as a pseudogene in W. glossinidia and is absent in M. genitalium. Three more genes in this category that are not considered essential in B. subtilis and E. coli have been conserved in all five analyzed endosymbionts and M. genitalium: gidA (glucose-inhibited division protein A), thdF (GTPase protein involved in hypermodification of tRNA, renamed mnmE [52]), and truA (site-specific pseudouridine synthase, EC 4.2.1.70), although the last of these can be disrupted in M. genitalium (five different mutants were found in the global transposon mutagenesis experiment [34]) and therefore has not been included in our minimal set. The gidA gene was previously described as a gene involved in cell division, but it appears to be also involved in the hypermodification of tRNAs and has been renamed mnmG (7). MnmA, MnmE, and MnmG are involved in the first steps of the biosynthesis of the hypermodified nucleoside 5-methylaminomethyl-2-thiouridine, which is found in the wobble position of some tRNAs. Mutations in mnmE result in excessive frameshifting during protein synthesis. However, in E. coli some of the mutations can be tolerated in some genetic backgrounds, which proves that it can be nonessential depending on features of other elements involved in translation (9). iscS, the gene that encodes a cysteine desulfurase (EC 4.4.1.-), which has also been involved is in vitro 2-thiouridine biosynthesis in E. coli as a sulfur transferase (40), is also present in all analyzed reduced genomes. Since these four genes have been preserved in reduced genomes and since the 2-thiouridine modification of tRNAs stabilizes anticodon structure, confers ribosome binding ability to tRNA, and improves open reading frame maintenance, we decided to include them in the minimal set.
The largest number of conserved genes encode ribosomal proteins. Four of the ribosomal proteins present in all five endosymbionts (L25, L30, S1, and S21) are not encoded by the genome of M. genitalium, and disruptions in the gene that encodes L28 are viable in this microorganism, although the authors consider that this finding does not prove that L28 is not essential (34). Therefore, we assume that the minimal number of ribosomal proteins required for proper functioning of the ribosome corresponds to the gene set present in M. genitalium, which includes 31 proteins for the large ribosomal subunit and 19 proteins for the small one.
Among the genes involved in ribosome maturation and modification, three genes encoding the GTP-binding proteins EngA, Era, and Obg, whose function is not yet well understood, have been described as being essential in B. subtilis and E. coli. The GTPase superfamily of cellular regulators is well represented in bacteria, and a small number of GTPases are universally conserved over the entire range of bacterial species, suggesting that they play important roles in bacterial cellular systems (11). Although the function of most of the universally conserved bacterial GTPases is poorly understood (with the exception of the factors necessary for protein synthesis and secretion, which are described in the corresponding section), recent studies support the idea that GTPases of the Obg and Era groups regulate and coordinate ribosome function, cell cycle activity, and DNA partitioning and segregation. Two Obg-like GTPases, Obg and YchF, are present in all analyzed bacteria with reduced genomes. Furthermore, obg appears to be essential in E. coli, B. subtilis, and S. aureus (22, 37), and ychF is also essential in E. coli and S. aureus (22). Very little is known about YchF, although it is also widely distributed. It has been found that its coding gene is cotranscribed with pth in E. coli (15), which links this bacterial GTPase to regulation of protein synthesis and ribosome function. The orthologous gene can be disrupted in M. pneumoniae, which probably indicates that another GTPase can perform its function in this microorganism. Nevertheless, since no direct M. genitalium mutant was obtained in the transposon insertion experiment (34), we decide to include this gene in our minimal set. Among the GTPases of the Era group, only ThdF (already described in this section) and EngA are present in all reduced genomes analyzed. EngA is a unique GTPase that contains two GTP-binding domains arranged in tandem and that has been involved in ribosome assembly or stability. It is essential in E. coli and S. aureus (22). The era gene is essential in E. coli and B. subtilis and was included in the minimal gene set proposed by Mushegian and Koonin (65). The orthologous gene can be found in all three analyzed strains of B. aphidicola but not in B. floridanus and W. glossinidia, where another conserved GTPase might perform its function. Therefore, we have included both era and engA in our minimal gene set.
The rRNA methyltransferase CspR (EC 2.1.1.-) is also essential in B. subtilis. This protein is also present in M. genitalium and E. coli but not in the five analyzed endosymbionts. Instead, ftsJ (renamed rrmJ [10]), encoding a 23S rRNA methyltransferase, has no orthologous gene identifiable in the B. subtilis and M. genitalium genomes, but some studies indicate that it is essential in E. coli (69), and it is present in all five endosymbionts analyzed. It has been proposed that methylation could modulate rRNA maturation, affect rRNA stability, or alter translation rates, although the real function is poorly understood. At least one 23S rRNA methylase has been identified in all reduced genomes analyzed, and therefore we decided to include cspR in our minimal set, since this gene can be found in both gram-positive and gram-negative bacteria. Four more genes in this category that are nonessential in B. subtilis are conserved in all five endosymbionts, and three of these genes also have an orthologue in M. genitalium (ksgA [EC 2.1.1.-], rbfA, and rluD [EC 4.2.1.70]). KsgA and RbfA are required for efficient processing of the 16S rRNA. However, rluD, whose product is involved in 23S rRNA maturation, appears to be dispensable in M. genitalium (34) and has not been included in the minimal gene set.
All the essential translation factors in B. subtilis are present in all five endosymbionts analyzed. infA, infB, and infC are required for translation initiation and are also essential in E. coli (17, 25) and S. aureus (22). The status of the infC gene in B. aphidicola BSg, initially annotated as a pseudogene due to the absence of a translation start codon (80), was recently changed (28) because, as in other enteric bacteria, it possesses an unusual AUU initiator codon. The genes tufA (EC 3.6.5.3), tsf, and fusA (EC 3.6.1.48), which encode elongation factors, are also present in all analyzed reduced genomes and are essential in S. aureus (22, 37). tufA has been proven to be dispensable in E. coli (88) and Salmonella enterica serovar Typhimurium (33), because these microorganisms, like many other gram-negative bacteria, have a duplication (tufA and tufB). prfA and prfB encode the two codon-specific bacterial release factors RF1 and RF2. prfB has been lost in M. genitalium, since it recognizes the UGA codon that, in this microorganism, has been reassigned to the tryptophan codon during evolution. Since both the prfA and prfB genes are paralogous, since there is only one release factor in eukaryotes, and since it has been proven that a single mutation in RF2 can allow the new protein encoded to recognize all three stop codons (36), it can be assumed that a single ancestral RF existed for the direct reading of the three stop codons, and therefore only one prf gene has been included in the minimal set. The last conserved B. subtilis essential gene, frr, is required for ribosome recycling. Two more elongation factors, efp and lepA, and a modulator of the release factors activity, hemK, which are nonessential in B. subtilis, are present in all reduced genomes under study, which probably reveals that they are essential in bacteria with small genomes. All of them have orthologous genes in B. subtilis and E. coli. Furthermore, efp and hemK are essential in E. coli, and lepA is essential in S. aureus (22, 37). The smpB gene is also present in all resident genomes analyzed. Although it can be disrupted in M. pneumoniae and is nonessential in B. subtilis and E. coli, it has been proven to be essential in S. aureus (22) and H. influenzae (1). smpB encodes the small protein B, essential for the activity of tmRNA in releasing stalled ribosomes from damaged mRNAs and targeting incompletely synthesized protein fragments for degradation. Since tmRNA has also been identified in all prokaryotic genomes sequenced (41), we have included smpB in the minimal gene set.
(iii) RNA degradation.
The catabolism of RNA molecules is known to encompass a wide variety of reactions and to require a large number of distinct RNases, although many of them appear to overlap functionally.
Exoribonucleases belonging to three different superfamilies (87) have been identified in the bacteria analyzed in this study, although there is no single conserved exoribonuclease-coding gene. The RNR family is composed of the RNase II (EC 3.1.13.1) and RNase R (EC 3.1.-.-) types. Although both types can normally be found in γ-proteobacteria, B. floridanus and W. glossinidia lack both of them. M. genitalium contains only one RNase R (encoded by MG104, homologous to vacB). RNase T (EC 3.1.13.-) and oligoribonuclease (EC 3.1.-.-), members of the DEDD family, are exoribonucleases that are found in only some bacteria (including γ-proteobacteria) and are represented in all five endosymbionts analyzed. The PDX family includes polynucleotide phosphorylase (EC 2.7.7.8, encoded by the pnp gene), a highly conserved protein that has been found in every sequenced bacterial genome except those of mycoplasmas.
Some endoribonucleases are also present in small genomes. However, only rnc, an essential gene in B. subtilis that encodes RNase III, is present in all analyzed reduced genomes. RNase III (EC 3.1.26.3) is a multifunctional endoribonuclease involved in the processing of rRNA precursors and some mRNAs and in the maturation and decay of RNAs, and it also cleaves double-stranded RNA. M. genitalium also contains RNase HII (EC 3.1.26.4), which acts on RNA-DNA hybrids but is absent in most endosymbionts analyzed. On the other hand, RNase E, the major endoribonuclease participating in mRNA turnover, is present in all five endosymbionts but not in M. genitalium.
RNA degradation must be one of the essential functions in any living cell. However, only the endonuclease encoded by rnc is conserved in all bacteria with reduced genomes analyzed in this study. Nevertheless, at least one exoribonuclease must be also included in the minimal gene set. In fact, some species seem to need only one exoribonuclease (87). Therefore, we have also included in our minimal set the widely conserved exoribonuclease encoded by pnp.
Protein Processing, Folding, and Secretion
Two genes are essential for posttranslational modification, both of which are preserved in all analyzed reduced genomes: map, which encodes a methionine aminopeptidase (EC 3.4.11.18), and def, which encodes a polypeptide deformylase (EC 3.5.1.88). Two alternative gene products can carry out deformylation in B. subtilis, and so it is not essential for this microorganism. Furthermore, def is not present in P. asteris and can be disrupted in M. genitalium. As mentioned above, cells simultaneously lacking def and fmt are still viable (13, 55, 56, 59), and therefore none of these genes have been included in our minimal set. The gene coding for the aminopeptidase PepA (EC 3.4.11.1), which is present in all resident genomes analyzed, has been included it in the minimal genome, although it is nonessential in E. coli and B. subtilis.
Several genes encoding molecular chaperones are also present in all genomes analyzed. In fact, molecular chaperones are quite abundant in reduced genomes, probably being essential in a genomic environment that lacks most repair mechanisms, to avoid the effect of slightly deleterious mutations (20). The best-conserved chaperones, GroEL and GroES, have also been described as essential in E. coli and B. subtilis. Surprisingly, it was possible to disrupt groEL in M. pneumoniae, although the survival conditions of mutants lacking this protein might be diminished (48). The DnaK-DnaJ-GrpE chaperone system, involved in many processes such as protein folding, translocation of proteins through biological membranes, oligomeric assembly of proteins, and their degradation, is nonessential in the free-living bacteria used in this comparison (3, 71) but is conserved in the five endosymbionts and M. genitalium. Therefore, we have chosen to include these genes in the minimal set.
Preproteins depend on chaperones to maintain their translocation-competent state. SRP (for “bacterial Signal Recognition Particle”) is a ribosome-bound chaperone that binds to emerging nascent preproteins and interacts with its receptor FtsY, activating the translocase machinery. Ffh (the protein component of the signal recognition particle) and FtsY are essential in E. coli and B. subtilis and are present in all analyzed bacteria with reduced genomes. The preprotein translocase system in bacteria is a large and complex system. The essential subunits of the translocase are the dissociable peripheral ATPase SecA and the integral membrane proteins SecY and SecE, which form the preprotein-conducting channel (60). All three genes are present in the reduced genomes analyzed and are essential in E. coli (25, 75) and B. subtilis.
Two signal peptidase are present in all five endosymbionts and essential in E. coli; one of these, LspA (EC 3.4.23.36), is also present in M. genitalium. However, LspA encodes a lipoprotein signal peptidase. Since no lipoproteins are encoded in our proposed minimal set, it has not been included either.
Protein turnover is another essential function that needs to be considered in a minimal cell. However, only one gene involved in protein turnover, gcp (EC 3.4.24.57), was found to be essential in B. subtilis. This gene was eliminated from the minimal genome computationally derived from a comparison of the H. influenzae and M. genitalium genomes because it was considered parasite specific. Nevertheless, it also appears to be essential in E. coli (4) and S. aureus (22), and it is present in all analyzed resident genomes; therefore, we have included it in the minimal gene set. Five genes in this category that are nonessential in B. subtilis have been conserved in all five endosymbionts, but only two have orthologues in M. genitalium: hflB and lon, which encode two ATP-dependent proteases. HflB (EC 3.4.24.-) has been involved in degradation of some integral membrane proteins, and it is essential in S. aureus (22, 37), while the protease La (EC 3.4.21.53), encoded by lon, degrades short-lived regulatory and abnormal proteins. Both of them have also been included in our minimal set.
Cell Structure and Cellular Processes
Cell wall.
In our comparison, we are taking into account gram-positive and gram-negative bacteria simultaneously. Thus, none of the genes that are essential in B. subtilis for the synthesis of teichoic acids has an orthologue in the five gram-negative endosymbionts. Furthermore, the endosymbionts have obvious defects in the peptidoglycan structure, since many of the genes involved in its synthesis are absent or have become pseudogenes in some of them. The presence of a well-structured cell wall is probably not necessary for microorganisms living inside other cells. In addition, M. genitalium has no cell wall. Therefore, it can be assumed that, in a protected environment, the cell wall might not be necessary for cellular structure, and thus we have not included any gene of this category in our proposed minimal set.
Cell shape and division.
Eleven essential genes involved in cell shape and division have been identified in B. subtilis; six of them are also essential in E. coli. Among them, only ftsZ is present in all resident genomes analyzed, and it has been included in the minimal gene set. FtsZ, a cytoskeletal protein, is by far the most highly conserved known cell division protein (54), and although it is not present in the obligate chlamidia Ureaplasma urealyticum or in P. asteris (70), it appears that another protein might be performing the same function, at least in the chlamidiae (8). The other genes involved in septum formation are absent in M. genitalium due to its lack of cell wall, suggesting that FtsZ is sufficient for cell division and that the other proteins serve to coordinate cell wall and septum synthesis. It has been proposed that FTsZ itself can provide the constrictive force necessary to split the cells. The five endosymbionts also contain the minCDE system, one of the main determinants of division site placement. MinC and MinD act together to regulate the assembly of the FtsZ ring, while MinE negatively regulates the action of MinCD. The DivIVA protein acts as the MinE equivalent in B. subtilis. Nevertheless, MinE is present in only a few characterized species, and MinC and MinD are absent in some species, including M. genitalium, H. influenzae, and many cocci (54). Furthermore, it has been proven that mutant E. coli strains lacking the Min system are still viable (86). The GTP binding protein EngB, essential in B. subtilis and E. coli, where it is necessary for normal cell division and for the maintenance of normal septation, can be disrupted in M. genitalium and is absent in B. floridanus. The other essential B. subtilis genes necessary for determination of cell shape are not present in all endosymbionts either.
Substrate transport.
Bacteria with small genomes are unable to synthesize many of their essential metabolites, and they must rely on the environment to obtain them. Thus, it could be expected that many transport systems must be present in order to obtain the necessary molecules, as it has been proven for P. asteris (70). In fact, the presence of many specific transporters in this bacterium can help to explain why many metabolic pathways for the synthesis of small molecules are missing in this microorganism but have been preserved in its close relative, M. genitalium, with a smaller genome. However, B. aphidicola has a reduced number of genes devoted to such function, a number that is slightly increased in the other endosymbiotic bacteria analyzed and in M. genitalium. This fact might be related to the presence of a less highly structured cell envelope, which might be more permeable to small metabolites. Furthermore, the analyzed bacteria with small genomes differ in the transport systems that have been preserved from the reductive process suffered by their genomes, and they probably compensate for the reduced transporter spectrum by encoding transporters with broadened specificity. Several porins and low-affinity active transporters, as well as a few ATP binding cassette (ABC) transporters have been identified in the five endosymbionts under study, while M. genitalium has a larger number of ABC transporters. ABC transporters are heterotrimeric transport systems made up of a specificity (ligand binding) subunit, a permease, and an ATP binding protein. Many ATP binding subunits appear to be “orphan” proteins with unknown partners, which are apparently overrepresented compared to the other two subunits in all genomes sequenced thus far (32), including the reduced genomes under study. However, none of them has been preserved in all analyzed reduced genomes simultaneously.
Phosphate transport is thought to be an essential function. However, only the low-affinity inorganic phosphate transporter encoded by pitA is present in all endosymbionts under study, although it is not present in the smallest described B. aphidicola genome, which is being sequenced in our laboratory (unpublished results). No orthologous gene has been found in M. genitalium, which instead contains an ABC phosphate transporter, although two of its three subunits (MG410 and MG411) can be disrupted. It appears that another yet unidentified phosphate transport system exists in mycoplasmas and in B. aphidicola BCce.
Several more or less specific transporters for mono- and divalent cations have also been identified in every microorganism analyzed in this study, but none of them is shared in all cases.
All the components of a phosphoenolpyruvate-dependent sugar phosphotransferase system (PTS), a major carbohydrate active-transport system that catalyzes sugar phosphorylation and transport across the cytoplasmic membrane, are present in all bacteria with reduced genomes analyzed except W. glossinidia, although the specific permease differs among them. The B. floridanus genome encodes the mannose permease, which can transport a wide variety of sugar monomers, while B. aphidicola encodes the genes for intake of mannitol and glucose and M. genitalium contains the corresponding glucose and fructose permeases. It is not known if the presence of different permeases is due to different metabolic requirements and/or environmental conditions or if such permeases are less specific in these microorganisms and can thus be used to transport other sugars.
Based on the present analysis, it is clear that no single transport system is shared by all analyzed bacteria. B. aphidicola appears to be close to the minimal “free diffusing cell” described by Luisi et al. (53), a cell in which the low-molecular-mass compounds, including nucleotides and amino acids, can be provided by the environment and are able to permeate the cell membrane. Nevertheless, it appears that at least one or several ABC transporters, a PTS for glucose transport, and some more or less specific mono- and divalent cation transporters must be part of a minimal cell genome. However, with the available data, it is not possible to define which ones should be such transporters, and different alternative transporters can be included in a minimal cell depending on the nutrients available in the environment or the cell membrane characteristics. Without detailed information about such possible alternatives, we propose that our hypothetical minimal cell must be able to internalize small molecules that are not highly charged, and we have included in our proposed minimal set only a PTS for glucose (since glucose can enter the cell by using different PTS in all analyzed bacteria with reduced genomes, and the PTS delivers phosphorylated glucose to the cell interior, making glucose available as a metabolic substrate) and pitA (to provide the phosphate that is necessary in metabolic reactions).
Energetic and Intermediary Metabolism
The definition of the essential metabolic functions that must be present in a minimal cell is a difficult part of the analysis of a minimal genome, since it is highly dependent on the repertoire of metabolites present in the environment. We are assuming by definition that a minimal cell lives in a nutrient-rich medium and that therefore the major metabolites (amino acids, nitrogenous bases, fatty acids, precursors of coenzymes, and glucose) are available without limitation. This situation makes unnecessary the function of entire anabolic and assimilatory pathways of the main biomolecules, i.e., de novo biosyntheses of amino acids, fatty acids, or cofactors. In that case, even the synthesis of a central metabolite such as acetyl coenzyme A (acetyl-CoA) turns out to be dispensable. As discussed below, the bioenergetics of our minimal cell is basically fermentative, and the only metabolic role for acetyl-CoA would be the synthesis of fatty acids; however, we postulate that these essential biomolecules are present and available in the medium. Furthermore, as discussed in the previous section, our proposed minimal cell does not contain specific transporters for charged molecules (such as mono- or dinucleotides), and such molecules must be synthesized by the cell from intermediary metabolites provided by the environment and its own metabolic machinery. Therefore, some of the metabolic pathways that we include as part of our hypothetical minimal cell might not be fully present in bacteria with small genomes that contain the corresponding specific transporter or a cell membrane that allows the internalization of such biomolecules.
To define which metabolic pathways should be considered essential in a minimal cell, we first considered those that have been preserved in the different bacteria with reduced genomes analyzed. Some metabolic pathways (such as the biosynthesis of purine nucleotides or redox coenzymes) have been preserved in all analyzed microorganisms, but they use different alternative routes, revealing degenerative processes that are randomly affecting different genes in each genome. In those cases, the loss of some genes conditioned the essentiality of other genes that became necessary to conserve a specific metabolic function. To approach the minimal genome, we choose to include in our minimal set the costless pathway that will allow the cell to preserve such metabolic function. Other metabolic pathways are incomplete. Although some of the genes involved in these pathways may still be present in all genomes under study, they have not been included in the minimal set.
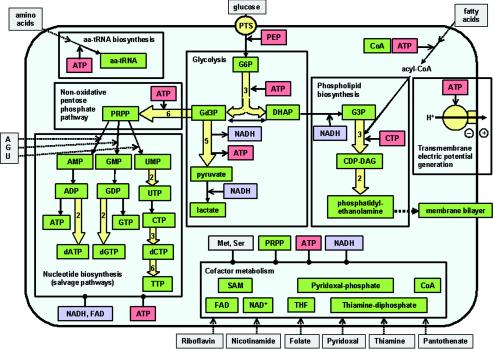
A general outline of the metabolic abilities of the proposed minimal cell is presented in Fig. 1.
FIG. 1.
A minimal metabolism. The minimal cell can obtain its more basic components from the environment: glucose, fatty acids, amino acids, adenine, guanine, uracil, and coenzyme precursors (nicotinamide, riboflavin, folate, pantothenate, and pyridoxal). Each box includes the metabolic transformations classified in major groups of pathways: glycolysis, phospholipid biosynthesis, nonoxidative pentose-phosphate pathway, nucleotide biosynthesis, synthesis of enzymatic cofactors, and synthesis of protein precursors, i.e., aminoacyl-tRNAs (aa-tRNA). Arrows with discontinuous lines represent incorporation from the environment. Single continuous arrows represent single enzymatic steps, whereas wide arrows represent several enzymatic steps (the number within the arrow indicates the number of steps). Lines with a final black point indicate the necessity of metabolites for some of the transformations inside the corresponding box. Metabolic intermediates and final pathway products are in green boxes. Metabolites acting as a source of chemical energy are in red boxes. Reducing-power cofactors are in light blue boxes. Abbreviations (besides the accepted symbols and those defined in the text): PEP, phosphoenolpyruvate; G6P, glucose-6-phosphate; Gd3P, glyceraldehyde-3-phosphate; DHAP, dihydroxyacetonephosphate; G3P, sn-glycerol-3-phosphate; CDP-DAG, CDP-diacylglycerol; SAM, S-adenosylmethionine; THF, tetrahydrofolate. Metabolic precursors of external origin are in gray boxes.
Glycolysis, gluconeogenesis, pyruvate metabolism, and the TCA cycle.
All genes involved in glycolysis, except the one responsible for the phosphorylation of glucose, are present in all endosymbionts and in M. genitalium. The exception is pfkA (EC 2.7.1.11), which has not been identified in W. glossinidia. In this case, it is remarkable that the absence of phosphofructokinase is accompanied by the presence of two idiosyncratic activities of the gluconeogenic pathway: fructosebisphosphatase (fbp, EC 3.1.3.11) and phosphoenolpyruvate carboxykinase (pck, EC 4.1.1.49). This is a strong indication that W. glossinidia, unlike the other described endosymbionts, synthesizes hexoses from C3 compounds derived mainly from amino acids. The phosphorylation of glucose in the other endosymbionts and M. genitalium can be achieved if we consider the action of the PTS transporters. Since no other pathway for the glucose degradation has been identified in the small genome organisms under study and since glycolysis also provides precursor metabolites for some anabolic pathways, we have included all genes involved in this pathway in the minimal set.
Pyruvate, the final product of glycolysis, is converted into acetyl-CoA by the action of pyruvate dehydrogenase, a multienzymatic complex that is composed of three subunits encoded by aceE (EC 1.2.4.1, E1 subunit), aceF (EC 2.3.1.12, E2 subunit), and lpdA (EC 1.8.1.4, E3 subunit) in gram-negative bacteria. All three genes are present in the five endosymbionts analyzed. Gram-positive pyruvate dehydrogenase subunit E1 is a heterodimer of α and β subunits encoded by pdhA and pdhB, respectively. Both the α and β subunits are present in M. genitalium, but only the α subunit appears to be essential in B. subtilis. The third activity, dihydrolipoamide dehydrogenase, is also required by 2-oxoglutarate dehydrogenase, an enzyme involved in the tricarboxylic acid (TCA) cycle that is maintained in all endosymbionts but not in M. genitalium and whose E1 (EC 1.2.4.2) and E2 (EC 2.3.1.61) subunits are encoded by sucA and sucB, respectively. Only the orthologous genes of sucB (odhB) and aceE (pdhA) are essential in B. subtilis, although adding their respective metabolites to the culture medium can restore the growth of the mutant strains (45). These observations, together with the phylogenetic analysis of the bacterial genes encoding the E1 and E2 subunits, led to the conclusion that these two enzymes and the branched-chain 2-oxoacid dehydrogenases (EC 1.2.4.4) form a family of paralogous genes. It should be noticed that 2-oxoglutarate dehydrogenase is the only enzyme of the TCA cycle that is present in all analyzed endosymbionts. Thus, only the components of the pyruvate dehydrogenase appear to be good candidates for inclusion in the minimal set. Acetyl-CoA, the product of this enzyme, is an essential substrate for the synthesis of amino acids and fatty acids. Since our proposed minimal cell can obtain these biomolecules from the environment, acetyl-CoA is not necessary. Therefore, the genes that encode the pyruvate dehydrogenase have not been included in the minimal set. Instead, we propose that, in our hypothetical minimal cell, the pyruvate generated by the glycolysis would be reduced to lactate by the l-lactate dehydrogenase (EC 1.1.1.27), thus regenerating NAD+. The gene encoding this enzyme (ldh) is present in M. genitalium but not in the five endosymbionts analyzed, since these endosymbionts maintain different segments of the electron transport chain as a means of oxidizing NADH (as discussed in the next section).
Electron transport chain and proton motive force generation.
Electron transport chains and oxidative phosphorylation also appear to be essential processes as a source of energy in the five analyzed endosymbionts, but not in M. genitalium. NADH:quinone oxidoreductase (EC 1.6:5.3) is present in the three genomes of B. aphidicola and in B. floridanus, whereas the succinate:quinone oxidoreductase complex (EC 1.3.99.1) is present in both B. floridanus and W. glossinidia. The cytochrome o:quinol oxidoreductase complex (EC 1.10.3.-), structurally related to the aa3-type family of cytochrome c oxidases (12), and the complete set of genes for an F0F1-type ATP synthase (EC 3.6.3.14) are present in all endosymbionts. The NADH dehydrogenase function is performed by a complex of 13 proteins (EC 1.6.5.3) in B. aphidicola and B. floridanus, while W. glossinidia has only the ndh gene, which encodes an NADH dehydrogenase independent of ATP (EC 1.6.99.3). Thus, we can consider that all five endosymbionts can perform electron transport to energize the membrane from NADH to molecular oxygen (B. aphidicola), from NADH and succinate to molecular oxygen (B. floridanus), or from succinate to molecular oxygen (W. glossinidia), in all three instances via the reduced quinone pool. In all the cases, both NADH:quinone oxidoreductase and cytochrome o:quinol oxidoreductase conserve the redox energy as a proton motive force. Many of the genes in this category that are essential in B. subtilis are required for menaquinone biosynthesis, but most of them have been lost in the five endosymbionts analyzed or appear as pseudogenes in B. aphidicola BBp, which indicates that all the endosymbionts are dependent on the host quinones. On the other hand, in M. genitalium, the major route of ATP synthesis might be through substrate-level phosphorylation, since no NADH dehydrogenase, succinate:menaquinone oxidoreductase, or cytochrome oxidase are present. Therefore, as suggested by Mushegian and Koonin (65), the minimal cell we propose should be able to obtain its energy through substrate-level phosphorylation, and no genes involved in oxidative phosphorylation have been included in the minimal set. Nevertheless, all the components of the ATP synthase have also been preserved in M. genitalium, where it most probably functions as a proton pump consuming ATP. Although these genes are absent in the recently sequenced genome of P. asteris (70), we decided to include them in the proposed minimal gene set, since a proton motive force could be necessary to generate and maintain a negative transmembrane potential, which is necessary for physiologically normal function of the cell membrane. The ydiC gene, which seems to be essential for proper integration of the ATP synthase into the membrane (82), is present in all reduced genomes analyzed and has also been included in the minimal set.
Pentose phosphate pathway.
B. aphidicola and B. floridanus contain two of three enzymes of the oxidative branch of the pentose phosphate cycle (a pathway that produces reducing power, i.e., NAD[P]H), and pentoses from hexoses and/or trioses), encoded by zwf (glucose-6-phosphate dehydrogenase, EC 1.1.1.49) and gnd (6-phosphogluconate dehydrogenase, EC 1.1.1.44). The third activity in the standard pathway, 6-phosphogluconolactonase (EC 3.1.1.31), is missing in the above-mentioned endosymbionts. This absence in itself should not represent an interruption of the metabolic flux, since it is well known that 6-phosphogluconolactone undergoes a rapid spontaneous hydrolysis (61). On the other hand, the five endosymbionts analyzed present all the enzymes involved in the nonoxidative branch of the pentose phosphate pathway, encoded by rpe (EC 5.1.3.1, ribulose-phosphate 3-epimerase), rpiA (EC 5.3.1.6, ribose 5-phosphate isomerase A), tktA or tktB (EC 2.2.1.1 transketolase), and talA (EC 2.2.1.2, transaldolase A). Only the last gene is missing in M. genitalium. The pathway allows the synthesis of carbohydrates with different number of carbons. Moreover, tkt is also essential in B. subtilis and S. aureus (37). The absence of transaldolase in M. genitalium is not a problem, since the action of all the above enzymes except this one can also allow the interconversion of sugars of different lengths, particularly hexoses (or trioses) and pentoses, as is performed by the nonreductive phase of the Calvin cycle (67). The pathway is completely missing in P. asteris (70), which indicates that this plant pathogen is using the host pentoses. We propose that a minimal cell can perform the synthesis of pentoses from trioses or hexoses only with the activities of the nonoxidative branch of the pentose phosphate pathway encoded by rpe, rpiA, and tkt.
Biosynthesis of amino acids
Several genes necessary for the synthesis of amino acids of the aspartate family are included among the essential genes of B. subtilis, probably reflecting the lack of such amino acids in the culture media or the absence of the proper transporters to import them into the cell. B. aphidicola and B. floridanus provide their host insect with essential amino acids, and thus the genes involved in the biosynthetic pathways for those amino acids are present in their genomes. However, W. glossinidia has lost most of these genes, and the only gene in this category found in M. genitalium is glyA. This gene is also involved in the biosynthesis of folate derivatives and has been included in the section an biosynthesis of cofactors (below). Thus, it can be assumed that amino acids can be provided by the environment in the small genome microorganisms analyzed, and no genes of this category need to be included in the minimal set.
Biosynthesis of lipids
Fatty acid biosynthesis, the first stage in membrane lipid biogenesis, is catalyzed in most bacteria by the type II fatty acid synthase system, composed of a series of small, soluble proteins that are each encoded by a discrete gene (74). Although many of the genes involved in this pathway are essential in E. coli and B. subtilis, all the genes involved in the synthesis of malonyl-CoA have been lost in B. aphidicola and M. genitalium. In fact, the fatty acid biosynthesis pathway is incomplete in most reduced genomes analyzed, since only acpP and acpS (EC 2.7.8.7) are present in all of them. These findings probably imply that fatty acids can be provided by the environment in bacteria with small genomes, and that ACP (the acyl carrier protein, encoded by acpP) is involved in fatty acids uptake and activation (i.e., synthesis of acyl-CoA). However, since the only metabolic destination of the fatty acids would be the biosynthesis of phospholipids, we propose that the activation should take place through a single step catalyzed by acyl-CoA synthase (EC 6.2.1.3) encoded by fadD (76).
One of the most surprising findings when the first B. aphidicola genome was sequenced was that this bacterium lacks all genes responsible for phospholipid biosynthesis (except cls, the gene that encodes cardiolipin synthase, EC 2.7.8.-). The sequencing of two more B. aphidicola genomes corroborated this finding. However, a set of 10 genes coding for a complete biosynthetic pathway of the major bacterial phospholipids (i.e., phosphatidylethanolamine, phosphatidylglycerol, and cardiolipin) from triosephosphate and acyl-CoA is present in W. glossinidia, all except one (plsB, EC 2.3.1.15) are present in B. floridanus, and some of them are essential in B. subtilis. A hypothetical phospholipid biosynthetic pathway from glycerol and exogenous fatty acids has been proposed by Mushegian and Koonin (65) for M. genitalium, albeit with an unreasonably small number of steps. These facts probably indicate that the cell surface in B. aphidicola is fragile, maybe due to its prolonged intracellular life inside vacuole-like host-derived organelles. In contrast, pathogenic bacteria, such as M. genitalium, and endosymbionts that live free in the cytosol of the host cell (which is the case for B. floridanus and W. glossinidia) need a more structured and flexible surface to protect themselves from the host cell. Since phospholipids are an indispensable component of the formation of the membrane lipid bilayer, it seems reasonable to assume that B. aphidicola imports them from the host cell. However, we consider phospholipid biosynthesis one of the essential functions to be performed by a minimal cell, and therefore we have included in our minimal gene set all the genes necessary for the biosynthesis of phosphatidylethanolamine from dihydroxyacetone phosphate and activated fatty acids. We assume for our minimal cell a theoretical lipid bilayer made only of phosphatidylethanolamine, the main phospholipid present in bacterial membranes (it represents 70 to 80% of all zwitterionic phospholipids in E. coli). The observation that E. coli cells lacking the quantitatively major acidic phospholipids (phosphatidylglycerol and cardiolipin) are viable (42) also supports this choice. The nonessentiality of some metabolic steps or the absence of some genes can be explained by metabolic redundancy, nonorthologous substitutions, or even the presence of other kinds of phospholipids acting as functional surrogates (14).
lgt, which encodes a prolipoprotein diacylglycerol transferase (EC 2.4.99.-) involved in the first step of the synthesis of lipoproteins, is conserved in all analyzed reduced genomes but is absent in P. asteris (70). Although it was annotated as a pseudogene in B. aphidicola BSg (80), the corresponding open reading frame contains a frameshift 29 nucleotides after the start codon of the orthologous gene, but there is an alternative start a few nucleotides downstream, rendering a protein of 246 amino acids that has lost only the first transmembrane domain. The presence of this gene in the small genomes analyzed can be explained because all such genomes contain genes that encode at least one lipoprotein. However, since none of them has been included in the minimal set, lgt has not been included either.
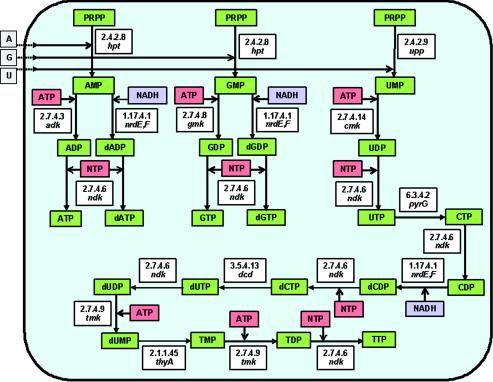
Biosynthesis of nucleotides.
Among the genes related to nucleic acids metabolism, several genes involved in the synthesis of purines and pyrimidines are essential in B. subtilis and are present in all analyzed genomes; these include nrdE and nrdF (or the homologous genes nrdA and nrdB, encoding the two subunits of the ribonucleoside diphosphate reductase, EC 1.17.4.1), together with trxA and trxB (thioredoxin and thioredoxin reductase, EC 1.8.1.9), adk (adenylate kinase, EC 2.7.4.3), gmk (guanylate kinase, EC 2.7.4.8), tmk (thymidylate kinase, EC 2.7.4.9), and prs (phosphoribosylpyrophosphate synthase, EC 2.7.6.1). The nrdI gene, encoding a putative ribonucleoside diphosphate reductase that is essential in B. subtilis, has no orthologue in the five endosymbionts analyzed. However, its presence in the M. genitalium genome may explain why nrdE seems to be nonessential in mycoplasmas (34).
All bacteria with small genomes analyzed in this study can synthesize purine nucleotides, but with different metabolic solutions. The enzyme ribose-phosphate diphosphokinase (EC 2.7.6.8) synthesizes 5-phosphoribosyl-alpha-1-diphosphate (PRPP) from ribose 5-phosphate and ATP, the starting point for nucleotide biosynthesis. This activity is encoded by the prsA gene, orthologous to the essential prs gene in B. subtilis, that is present in all the reduced genomes analyzed. The ppa gene, encoding an inorganic pyrophosphatase (EC 3.6.1.1), is also present in all analyzed genomes and is essential in B. subtilis and E. coli. The gene product drives anabolic fluxes by pyrophosphate hydrolysis in various biochemical reactions, including all reactions that involve PRPP. Purine monophosphate nucleotides can then be synthesized by using two alternative pathways. In all five endosymbionts, 1-(5′-phosphoribosyl)-5-amino-4-imidazolecarboxamide (AICAR) can be synthesized de novo and used for the synthesis of the purine nucleotides. In W. glossinidia, AICAR biosynthesis is performed by a standard pathway starting with PRPP and glutamine, involving the products of the purF (EC 2.4.2.14), purD (EC 6.3.4.13), purL (EC 6.3.5.3), purM (EC 6.3.3.1), purE and purK (EC 4.1.1.21), purC (EC 6.3.2.6), and purB (EC 4.3.2.2) genes, although the step usually catalyzed by the protein encoded by purN (EC 2.1.1.2) has been replaced by the enzyme 5′-phosphoribosylglycinamide transformylase 2 (EC 2.1.2.-) (68) encoded by purT, a homologue of purN (58). On the other hand, both B. aphidicola and B. floridanus synthesize AICAR through the four first steps of the histidine biosynthetic pathway starting with PRPP and ATP, using the set of activities encoded by hisG (EC 2.4.2.17), hisI (EC 3.6.1.31 and 3.5.4.19), and his A (EC 5.3.1.16). AMP and GMP are then synthesized from AICAR through a pathway involving the products of the purA (EC 6.3.4.4), purB (EC 4.3.2.2), and purH (EC 3.5.4.10) genes and the guaC (EC 1.7.1.7) (in B. aphidicola) or guaA (EC 1.1.1.205) and guaB (EC 6.3.5.2) (in B. floridanus and W. glossinidia) genes. M. genitalium has a different and costless metabolic solution, since it can use the enzyme hypoxanthine phosphoribosyltransferase (EC 2.4.2.8, encoded by the gene hpt) for the synthesis of GMP and AMP from PRPP and guanine or adenine, respectively, as may also be the case for B. aphidicola BAp and BSg. The kinases encoded by adk and gmk are conserved in all reduced genomes analyzed, allowing the synthesis of nucleoside diphosphates from the nucleoside monophosphates. Purine nucleotide diphosphate can be phosphorylated by the kinase encoded by ndk or by the glycolytic enzyme pyruvate kinase (EC 2.7.1.40), encoded by pykA, in B. aphidicola and M. genitalium, which lack ndk. The ribonucleoside diphosphate reductase function can be performed by the products of the nrdA and nrdB genes or the nrdE and nrdF genes (depending on the analyzed organism), with the help of the products of trxA and trxB, to obtain the corresponding deoxyribonucleoside diphosphate (Fig. 2).
FIG. 2.
A minimal nucleotide metabolism based on salvage pathways. Activated ribonucleotides and deoxyribonucleotides are obtained from free bases (A, G, and U), PRPP, ATP-dependent phosphorylating reactions, and NADH-dependent reduction. White boxes indicate the individual enzymatic activity (EC number and coding gene). Other colors are used as in Fig. 1.
Pyrimidine metabolism is not so highly conserved in the analyzed bacteria with small genomes. Only a few genes involved in the synthesis of nucleoside monophosphates have been maintained in all of them simultaneously: tmk, pyrG (CTP synthase, EC 6.3.4.2), and dut (EC 3.6.1.23) (or its corresponding suggested homologue in M. genitalium, MG268) (65). The upp gene (uracil phosphoribosyl transferase, EC 2.4.2.9), necessary for the incorporation of free uracil for the synthesis of activated ribonucleotides, is present only in B. floridanus and M. genitalium. It appears that B. aphidicola and W. glossinidia can obtain the pyrimidine nucleotides from the cytoplasm of the host cell, probably as nucleoside monophosphates. However, since we have not included specific transporters for such molecules in our minimal cell, we consider that in a hypothetical minimal cell, free uracil can be provided by the environment and used to synthesize all the necessary pyrimidine nucleotides, using the costless pathway as described in Fig. 2. The conversion of pyrimidine monophosphate to pyrimidine diphosphate needs the action of one kinase, encoded by cmk (EC 2.7.4.14) or tmk (EC 2.7.4.9). At least one of these genes appears in all genomes sequenced to date; i.e., chlamydiae and mycobacteria contain only cmk, while archaea, B. aphidicola, and some δ- and ɛ-proteobacteria have only tmk. Therefore, only tmk has been included in the minimal gene set. Pyrimidine nucleoside diphosphates cannot be phosphorylated by the kinase encoded by pykA, and therefore in our metabolic map the activity encoded by ndk plays an important role in catalyzing the transfer of the gamma-phosphate from the nucleoside triphosphate to a variety of nucleoside diphosphates. This broad specificity has been demonstrated in several experimental systems including prokaryotic cells (see the primary literature in the BRENDA database).
Biosynthesis of cofactors.
Most of the genes involved in the biosynthesis of cofactors have been lost in the reduced genomes analyzed, and only a few genes included in this category that are essential in B. subtilis or E. coli are present in all of them. However, the presence of biosynthetic pathways in bacteria with reduced genomes or their essentiality in laboratory-cultured bacteria reflects the chemical complexity of the environment. If a microorganism has free availability of coenzyme precursors (i.e., vitamins), it can use short and simple salvage pathways rather than long and complicated de novo biosyntheses. As a general criterion, we propose that our minimal metabolic chart starts from elaborate molecules, such as vitamins, and it contains only the minimum number of steps to incorporate them as enzymatic cofactors (Fig. 3). Only the cofactors that are necessary for the correct catalytic function of the enzymes present in our proposed minimal cell have been included.
FIG. 3.
Biosynthesis of cofactors. A metabolism of essential cofactors used by the minimal cell starting with precursors (i.e., vitamins): nicotinamide, riboflavin, thiamine, pyridoxal, patothenic acid, methionine, and folic acid. Yellow boxes indicate the enzymes that need each cofactor for their correct function. Other colors and symbols are as in Fig. 1 and 2. PP, pyridoxal-phosphate.
The folA (EC 1.5.1.3) and glyA (EC 2.1.2.1) genes encode enzymes involved in the monocarbon folate pool interconversion, and they are present in all analyzed reduced genomes. The detection of one glyA mutant in M. genitalium by Hutchison et al. (34) may be explained by the presence of an alternative pathway through the use of ThyA (EC. 2.1.1.45). Although both genes are absent in U. urealyticum (26), the pathway may be closed in this species by the presence of the gene fthS (EC. 6.3.4.3), which encodes the enzyme responsible for the transformation of tetrahydrofolate to 10-formyltetrahydrofolate. In our proposed minimal metabolism, the reaction catalyzed by GlyA is essential for charging tetrahydrofolate with monocarbon units (i.e., the synthesis of 5,10-methylenetetrahydrofolate with serine as the monocarbon unit donor). folC is also present in all five endosymbionts. Although it is not present in M. genitalium, Musheguian and Koonin suggested that several proteins with unknown function that have an ortholog in H. influenzae may be candidates to perform such function (65). However, the gene is not essential in B. subtilis and E. coli (43). The dihydrofolate synthase (EC 6.3.2.17 and 6.3.2.12) is necessary only for the last step in the de novo synthesis of folate, but considering that this coenzyme precursor can also be provided by the host cell, we have not included it in our minimal set.
Another gene of this category that has been conserved is ppnK, involved in the phosphorylation of NAD+ to produce NADP+ (EC 2.7.1.23). The biosynthesis of the essential redox coenzymes NAD(P)+ can occur by both de novo and salvage pathways (5). The de novo biosynthesis from aspartate is present in E. coli, B. subtilis, and W. glossinidia. Three different salvage pathways that are operative in different bacterial groups have been described so for (51). Two strains of B. aphidicola (BAp and BSg) can perform salvage pathway I (synthesis of nicotinate mononucleotide [NMN] from nicotinate and PRPP). In B. floridanus, only the above-mentioned ppnK gene can be detected, indicating either that this organism synthesizes or recycles NAD+ by an unidentified mechanism or that all the steps except one have suffered nonorthologous substitutions. Salvage pathway III, present in Enterobacteriaceae, Pasteurellaceae, and other gram-negative bacteria, requires extracellular enzymes and membrane transporters to recycle exogenous nicotinamide nucleotides. For our minimal gene set, we propose the shortest pathway to synthesize NAD+ from PRPP and nicotinamide, i.e., salvage pathway II. First, NMN is synthesized from free nicotinamide and PRPP by the recently described nicotinamide phosphoribosyltransferase (EC 2.4.2.12) encoded by nadV (57). Then adenylyl transferase (EC 2.7.7.1), encoded by nadR (51), catalyzes the synthesis of NAD+ from NMN. Finally, the synthesis of NADP+ from NAD+ can be catalyzed by kinases with a wide bacterial distribution, such as the ppnK product. However, we have not included this last gene in our final set, since we assume that all the reductive steps in the minimal metabolism can use NADH. This is the case, at least in in vitro assays, for the reductases involved in the synthesis of ribonucleotides and the monocarbon metabolism (see the primary references in the BRENDA database).
Thiamine diphosphate (cofactor of transketolase) can be synthesized from thiamine (vitamin B1) by the action of thiamine pyrophosphokinase (EC 2.7.6.2). The product of the B. subtilis gene yloS, whose function has not been characterized, might be performing this function in this microorganism. The synthesis of flavin adenine dinucleotide (cofactor of thiredoxin reductase) from riboflavin (vitamin B2) requires the action of riboflavin kinase (EC 2.7.1.26) and flavin mononucleotide adenylyltransferase (EC 2.7.7.2). Both activities are encoded by the gene ribF, which is present in B. aphidicola (BAp and BSg strains), W. glossinidia, and B. floridanus. The synthesis of pyridoxal-5-phosphate (cofactor of glycine hydroxymethyltransferase) from pyridoxal (vitamin B6) is catalyzed by pyridoxal kinase (EC 2.7.1.35). The synthesis of S-adenosylmethionine, the methyl donor in methyltransferases, from methionine is catalyzed by methionine adenosyltransferase (metK, EC 2.5.1.6). We suppose that our minimal cell cannot recycle the S-adenosyl-l-homocysteine produced by methyltransferases. Finally, the synthesis of CoA from pantothenic acid (vitamin B5) requires the action of five activities encoded by four genes (coaA, dfp, coaD, and coaE), all of which are present in W. glossinidia.
Poorly Characterized Genes
There are many poorly characterized genes that were identified as essential in B. subtilis. Among them, only mesJ, a gene that encodes a hypothetical conserved protein of unknown function, is also present in all analyzed genomes. A few more genes that encode proteins of unknown function but are not essential in B. subtilis are also present in all analyzed reduced genomes, probably indicating that they perform an essential function in small genomes. Therefore, we have decided to include them in our minimal set, except for ycfF and ycfH, which appear to be nonessential in M. genitalium, since they can be disrupted (34). Some other genes in this category that are present in all five endosymbionts analyzed do not have an orthologue in gram-positive bacteria. We have decided not to include them in a minimal gene set, although another gene of unknown function that appears to be essential in B. subtilis and conserved in M. genitalium could be performing the same function. Further studies need to be done in order to characterize their gene products.
CONCLUSIONS
Based on the conjoint analysis of several computational and experimental strategies designed to define the minimal set of protein-coding genes that are necessary to maintain a functional bacterial cell, we propose a minimal gene set composed of 206 genes. Such a gene set will be able to sustain the main vital functions of a hypothetical simplest bacterial cell with the following features.
(i) A virtually complete DNA replication machinery, composed of one nucleoid DNA binding protein, SSB, DNA helicase, primase, gyrase, polymerase III, and ligase. No initiation and recruiting proteins seem to be essential, and the DNA gyrase is the only topoisomerase included, which should perform both replication and chromosome segregation functions.
(ii) A very rudimentary system for DNA repair, including only one endonuclease, one exonuclease, and a uracyl-DNA glycosylase.
(iii) A virtually complete transcriptional machinery, including the three subunits of the RNA polymerase, a σ factor, an RNA helicase, and four transcriptional factors (with elongation, antitermination, and transcription-translation coupling functions). Regulation of transcription does not appear to be essential in bacteria with reduced genomes, and therefore the minimal gene set does not contain any transcriptional regulators.
(iv) A nearly complete translational system. It contains the 20 aminoacyl-tRNA synthases, a methionyl-tRNA formyltransferase, five enzymes involved in tRNA maturation and modification, 50 ribosomal proteins (31 proteins for the large ribosomal subunit and 19 proteins for the small one), six proteins necessary for ribosome function and maturation (four of which are GTP binding proteins whose specific function is not well known), 12 translation factors, and 2 RNases involved in RNA degradation.
(v) Protein-processing, -folding, secretion, and degradation functions are performed by at least three proteins for posttranslational modification, two molecular chaperone systems (GroEL/S and DnaK/DnaJ/GrpE), six components of the translocase machinery (including the signal recognition particle, its receptor, the three essential components of the translocase channel, and a signal peptidase), one endopeptidase, and two proteases.
(vi) Cell division can be driven by FtsZ only, considering that, in a protected environment, the cell wall might not be necessary for cellular structure.
(vii) A basic substrate transport machinery cannot be clearly defined, based on our current knowledge. Although it appears that several cation and ABC transporters are always present in all analyzed bacteria, we have included in the minimal set only a PTS for glucose transport and a phosphate transporter. Further analysis should be performed to define a more complete set of transporters.
(viii) The energetic metabolism is based on ATP synthesis by glycolytic substrate-level phosphorylation.
(ix) The nonoxidative branch of the pentose pathway contains three enzymes (ribulose-phosphate epimerase, ribose-phosphate isomerase, and transketolase), allowing the synthesis of pentoses (PRPP) from trioses or hexoses.
(x) No biosynthetic pathways for amino acids, since we suppose that they can be provided by the environment.
(xi) Lipid biosynthesis is reduced to the biosynthesis of phosphatidylethanolamine from the glycolytic intermediate dihydroxyacetone phosphate and activated fatty acids provided by the environment.
(xii) Nucleotide biosynthesis proceeds through the salvage pathways, from PRPP and the free bases adenine, guanine, and uracil, which are obtained from the environment.
(xiii) Most cofactor precursors (i.e., vitamins) are provided by the environment. Our proposed minimal cell performs only the steps for the syntheses of the strictly necessary coenzymes tetrahydrofolate, NAD+, flavin aderine dinucleotide, thiamine diphosphate, pyridoxal phosphate, and CoA.
Several attempts have been made to try to define the characteristics of a minimal cell. A computer model of a such minimal cell was performed a few years ago and was called the E-CELL Project (81). The proposed virtual self-surviving cell (SSC) contained only 105 protein-coding genes that allowed the cell to maintain protein and membrane structure. The only functions considered in this virtual cell were glycolysis (using exogenous glucose to obtain ATP), phospholipid biosynthesis (from exogenous fatty acids and glycerol), transcription, and translation. Therefore, this virtual minimal cell is able to maintain metabolic homeostasis but not to reproduce and evolve. Among the 105 genes proposed for SSC, 97 are included in our proposed minimal set. The only differences involve the fmt gene (methionyl-tRNA formyltransferase) and the genes proposed in SSC for phospholipid biosynthesis, since we have included a different pathway for this purpose. Another computational approach obtained by comparative analysis of 21 complete genomes of bacteria, archaea, and eukaryotes (48) suggested that a bare-bones set of about 150 genes would be sufficient to maintain basal systems for transcription, translation, and replication, a reduced repair machinery, a small set of molecular chaperones, an intermediate metabolism reduced to glycolysis, a primitive transport system, and no cell wall. Our proposed minimal cell has a similar number of genes devoted to such functions. However, we also included in our minimal set the genes involved in a nonoxidative pentose phosphate pathway, the maintenance of the membrane proton motive force, and nucleotide and coenzyme syntheses, although some of these biosynthetic pathways are not present in some bacteria with reduced genomes. The lack of such pathways reflects that these bacteria are very dependent on their hosts due to an irreversible degenerative process of their metabolic abilities. Therefore, we think they should be present in a hypothetical minimal cell able to perform all the necessary reactions to maintain a minimal and coherent metabolic functionality, although it remains questionable whether such a minimal cell could survive under any realistic conditions.
At any rate, we should accept that there is no conceptual or experimental support for the existence of one particular form of minimal cell, at least from a metabolic point of view. In this sense, our conclusions must be regarded as provisional. Different approaches, ours among others, should converge in several solutions (35, 49). Finally, one must keep in mind that this kind of research has little relevance for the study of the origin of life, since it is impossible to identify any of the above-mentioned diverse solutions with the one adopted by the more primitive cells (63). This is especially true in the cases where a bacterium-centered approach is followed, as described in this paper. Any attempt to universalize the conclusions would necessarily include the comparison with archaeal genomes, more specifically the smallest ones (84). We are sure that future studies will highlight a diversity of minimal ecologically dependent metabolic charts supporting a universal genetic machinery. In a more technological vein, the development of more sophisticated techniques for genomic engineering, together with the continued efforts in defining the minimal gene set, could help to achieve the exciting goal of experimentally constructing a minimal living cell (73).
Supplementary Material
Acknowledgments
We thank Antonio Lazcano and Amparo Latorre for their suggestions and critical reading of the manuscript.
This work was supported by Ministerio de Ciencia y Tecnología, Spain (project BFM2003-00305), and Generalitat Valenciana, Spain (project grupos 03/204). R.G. is a recipient of a contract in the Ramon y Cajal Program from the Ministerio de Ciencia y Tecnología, Spain.
Footnotes
Supplemental material for this article may be found at http://mmbr.asm.org.
REFERENCES
- 1.Akerley, B. J., E. J. Rubin, V. L. Novick, K. Amaya, N. Judson, and J. J. Mekalanos. 2002. A genome-scale analysis for identification of genes required for growth or survival of Haemophilus influenzae. Proc. Natl. Acad. Sci. USA 99:966-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akman, L., A. Yamashita, H. Watanabe, K. Oshima, T. Shiba, M. Hattori, and S. Aksoy. 2002. Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat. Genet. 32:402-407. [DOI] [PubMed] [Google Scholar]
- 3.Ang, D. and C. Georgopoulos. 1989. The heat-shock-regulated grpE gene of Escherichia coli is required for bacterial growth at all temperatures but is dispensable in certain mutant backgrounds. J. Bacteriol. 171:2748-2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arigoni, F., F. Talabot, M. Peitsch, M. D. Edgerton, E. Meldrum, E. Allet, R. Fish, T. Jamotte, M. L. Curchod, and H. Loferer. 1998. A genome-based approach for the identification of essential bacterial genes. Nat. Biotechnol. 16:851-856. [DOI] [PubMed] [Google Scholar]
- 5.Begley, T. P., C. Kinsland, R. A. Mehl, A. Osterman, and P. Dorrestein. 2001. The biosynthesis of nicotinamide adenine dinucleotides in bacteria. Vitam. Horm. 61:103-109. [DOI] [PubMed] [Google Scholar]
- 6.Berger, J. M., D. Fass, J. C. Wang, and S. C. Harrison. 1998. Structural similarities between topoisomerases that cleave one or both strands. Proc. Natl. Acad. Sci. USA 95:7876-7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brégeon, D., V. Colot, M. Radman, and F. Taddei. 2001. Translational misreading: a tRNA modification counteracts a +2 ribosomal frameshift. Genes Dev. 15:2295-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown, W. J. and D. D. Rockey. 2000. Identification of an antigen localized to an apparent septum within dividing chlamidiae. Infect. Immun. 68:708-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cabedo, H., F. Macián, M. Villarroya, J. C. Escudero, M. Martínez-Vicente, E. Knecht, and M. E. Armengod. 1999. The Escherichia coli trmE (mnmE) gene, involved in tRNA modification, codes for an evolutionarily conserved GTPase with unusual biochemical properties. EMBO J. 18:7063-7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caldas, T., E. Binet, P. Bouloc, A. Costa, J. Desgres, and G. Richarme. 2000. The FtsJ/RrmJ heat shock protein of Escherichia coli is a 23 S ribosomal RNA methyltransferase. J. Biol. Chem. 22:16414-16419. [DOI] [PubMed] [Google Scholar]
- 11.Caldon, C. E., and P. E. March. 2003. Function of the universally conserved bacterial GTPases. Curr. Opin. Microbiol. 6:135-139. [DOI] [PubMed] [Google Scholar]
- 12.Chepuri, V., L. Lemieux, D. C. Au, and R. B. Gennis. 1990. The sequence of the cyo operon indicates substantial structural similarities between cytochrome o ubiquinol oxidase of Escherichia coli and the aa3-type family of cytochrome c oxidases. J. Biol. Chem. 265:11185-11192. [PubMed] [Google Scholar]
- 13.Clements, J. M., R. P. Beckett, A. Brown, G. Catlin, M. Lobell, S. Palan, W. Thomas, M. Whittaker, S. Wood, S. Salama, P. J. Baker, H. F. Rodgers, V. Barynin, D. W. Rice, and M. G. Hunter. 2001. Antibiotic activity and characterization of BB-3497, a novel peptide deformylase inhibitor. Antimicrob. Agents Chemother. 45:563-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cronan, J. E. 2003. Bacterial membrane lipids: where do we stand? Annu. Rev. Microbiol. 57:203-224. [DOI] [PubMed] [Google Scholar]
- 15.Cruz-Vera, L. R., J. M. Galindo, and G. Guarneros. 2002. Transcriptional analysis of the gene encoding peptidyl-tRNA hydrolase in E. coli. Microbiology 148:3457-3466. [DOI] [PubMed] [Google Scholar]
- 16.Cruz-Vera, L. R., I. Toledo, J. Hernandez-Sanchez, and G. Guarneros. 2000. Molecular basis for the temperature sensitivity of Escherichia coli pth(Ts). J. Bacteriol. 182:1523-1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cummings, H. S., and J. W. Hershey. 1994. Translation initiation factor IF1 is essential for cell viability in Escherichia coli. J. Bacteriol. 176:198-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Curnow, A. W., K. W. Hong, R. Yuan, S. I. Kim, O. Martins, W. Winkler, T. M. Henkin, and D. Söll. 1997. Glu-tRNAGln amidotransferase: a novel heterotrimeric enzyme required for correct decoding of glutamine codons during translation. Proc. Natl. Acad. Sci. USA 94:11819-11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deckert, G., P. V. Warren, T. Gaasterland, W. G. Young, A. L. Lenox, D. E. Graham, R. Overbeek, M. A. Snead, M. Keller, M. Aujay, R. Huber, R. A. Feldman, J. M. Short, G. J. Olsen, and R. V. Swanson. 1998. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature 392:353-358. [DOI] [PubMed] [Google Scholar]
- 20.Fares, M. A., M. X. Ruiz-González, A. Moya, S. F. Elena, and E. Barrio. 2002. GroEL as an enabler for bacterial endosymbiotic lifestyle. Nature 417:398. [DOI] [PubMed] [Google Scholar]
- 21.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. F. Tomb, B. A. Dougherty, J. M. Merrick, K. McKenney, G. Sutton, W. FitzHugh, C. Fields, J. D. Gocayne, J. Scott, R. Shirley, L.-I. Liu, A. Glodek, J. M. Kelley, J. F. Weidman, C. A. Phillips, T. Spriggs, E. Hedblom, M. D. Cotton, T. R. Utterback, M. C. Hanna, D. T. Nguyen, D. M. Saudek, R. C. Brandon, L. D. Fine, J. L. Fritchman, J. L. Fuhrmann, N. S. M. Geoghagen, C. L. Gnehm, L. A. McDonald, K. V. Small, C. M. Fraser, H. O. Smith, and J. C. Venter. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 22.Forsyth, R. A., R. J. Haselbeck, K. L. Ohlsen, R. T. Yamamoto, H. Xu, J. D. Trawick, D. Wall, L. Wang, V. Brown-Driver, J. M. Froelich, G. C. Kedar, P. King, M. McCarthy, C. Malone, B. Miniser, D. Robbins, Z. Tan, Z.-Y. Zhu, G. Carr, D. A. Mosca, C. Zamudio, J. G. Foulkes, and J. W. Zyskind. 2002. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol. Microbiol. 43:1387-1400. [DOI] [PubMed] [Google Scholar]
- 23.Fraser, C. M., J. D. Gocayne, O. White, M. D. Adams, R. A. Clayton, R. D. Fleischmann, C. J. Bult, A. R. Kerlavage, G. Sutton, J. M. Kelley, J. L. Fritchman, J. F. Weidman, K. V. Small, M. Sandusky, J. Fuhrmann, D. Nguyen, T. R. Utterback, D. M. Saudek, C. A. Phillips, J. M. Merrick, J. F. Tomb, B. A. Dougherty, K. F. Bott, P. C. Hu, T. S. Lucier, S. N. Peterson, H. O. Smith, C. A. Hutchison III, and J. C. Venter. 1995. The minimal gene complement of Mycoplasma genitalium. Science 270:397-403. [DOI] [PubMed] [Google Scholar]
- 24.Gerdes, S. Y., M. D. Schole, M. D'Souza, A. Bernal, M. V. Baev, M. Farrell, O. V. Kurnasov, M. D. Daugherty, F. Mseeh, B. M. Polanuyer, J. W. Campbell, S. Anantha, K. Y. Shatalin, S. A. K. Chowdhury, M. Y. Fonstein, and A. L. Osterman. 2002. From genetic footprinting to antimicrobial drug targets: examples in cofactor biosynthetic pathways. J. Bacteriol. 184:4555-4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerdes, S. Y., M. D. Scholle, J. W. Campbell, G. Balazsi, E. Ravasz, M. D. Daugherty, A. L. Somera, N. C. Kyrpides, I. Anderson, M. S. Gelfand, A. Bhattacharya, V. Kapatral, M. D'Souza, M. V. Baev, Y. Grechkin, F. Mseeh, M. Y. Fonstein, R. Overbeek, A. L. Barabasi, Z. N. Oltvai, and A. L. Osterman. 2003. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J. Bacteriol. 185:5673-5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glass, J. I., E. J. Lefkowitz, J. S. Glass, C. R. Heiner, E. Y. Chen, and G. H. Cassell. 2000. The complete sequence of the mucosal pathogen Ureaplasma urealyticum. Nature 407:757-762. [DOI] [PubMed] [Google Scholar]
- 27.Gil, R., B. Sabater-Muñoz, A. Latorre, F. J. Silva, and A. Moya. 2002. Extreme genome reduction in Buchnera spp.: towards the minimal genome needed for symbiotic life. Proc. Natl. Acad. Sci. USA 99:4454-4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gil, R., F. J. Silva, E. Zientz, F. Delmotte, F. González-Candelas, A. Latorre, C. Rausell, J. Kamerbeek, J. Gadau, B. Hölldobler, R. C. H. J. van Ham, R. Gross, and A. Moya. 2003. The genome sequence of Blochmannia floridanus: comparative analysis of reduced genomes. Proc. Natl. Acad. Sci. USA 100:9388-9393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Green, S. M., T. Malik, I. G. Giles, and W. T. Drabble. 1996. The purB gene of Escherichia coli K-12 is located in an operon. Microbiology 142:3219-3230. [DOI] [PubMed] [Google Scholar]
- 30.Guillon, J. M., Y. Mechulam, J. M. Schmitter, S. Blanquet, and G. Fayat. 1992. Disruption of the gene for Met-tRNA(fMet) formyltransferase severely impairs growth of Escherichia coli. J. Bacteriol. 174:4294-4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herring, C. D., J. D. Glasner, and F. R. Blattner. 2003. Gene replacement without selection: regulated suppression of amber mutations in Escherichia coli. Gene 331:153-163. [DOI] [PubMed] [Google Scholar]
- 32.Higgins, C. F. 2001. ABC transporters: physiology, structure and mechanism—an overview. Res. Microbiol. 152:205-210. [DOI] [PubMed] [Google Scholar]
- 33.Hughes, D. 1990. Both genes for EF-Tu in Salmonella typhimurium are individually dispensable for growth. J. Mol. Biol. 215:41-51. [DOI] [PubMed] [Google Scholar]
- 34.Hutchison, C. A., III, S. N. Peterson, S. R. Gill, R. T. Cline, O. White, C. M. Fraser, H. O. Smith, and J. C. Venter. 1999. Global transposon mutagenesis and a minimal mycoplasma genome. Science 286:2165-2169. [DOI] [PubMed] [Google Scholar]
- 35.Huynen, M. 2000. Constructing a minimal genome. Trends Genet. 16:116. [Google Scholar]
- 36.Ito, K., M. Uno, and Y. Nakamura. 1998. Single amino acid substitution in prokaryote polypeptide release factor 2 permits it to terminate translation at all three stop codons. Proc. Natl. Acad. Sci. USA 95:8165-8169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ji, Y., B. Zhang, S. F. van Horn, P. Warren, G. Woodnutt, M. K. R. Burnham, and M. Rosemberg. 2001. Identification of critical staphylococcal genes using conditional phenotypes generated by antisense RNA. Science 293:2266-2269. [DOI] [PubMed] [Google Scholar]
- 38.Judson, N., and J. J. Mekalanos. 2000. Transposon-based approaches to identify essential bacterial genes. Trends Microbiol. 8:521-526. [DOI] [PubMed] [Google Scholar]
- 39.Jung, Y. H., I. Park, and Y. Lee. 1992. Alteration of RNA I metabolism in a temperature-sensitive Escherichia coli rnpA mutant strain. Biochem. Biophys. Res. Commun. 186:1463-1470. [DOI] [PubMed] [Google Scholar]
- 40.Kambampati, R., and C. T. Lauhon. 2003. MnmA and IscS are required for in vitro 2-thiouridine biosynthesis in Escherichia coli. Biochemistry 42:1109-1117. [DOI] [PubMed] [Google Scholar]
- 41.Keiler, K. C., L. Shapiro, and K. P. Williams. 2000. tmRNAs that encode proteolysis-inducing tags are found in all known bacterial genomes: a two-piece tmRNA functions in Caulobacter. Proc. Natl. Acad. Sci. USA 97:7778-7783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kikuchi, S., I. Shibuya, and K. Matsumoto. 2000. Viability of an Escherichia coli pgsA null mutant lacking detectable phosphatidylglycerol and cardiolipin. J. Bacteriol. 182:371-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kimlova, L. J., C. Pyne, K. Keshavjee, J. Huy, G. Beebakhee, and A. L. Bognar. 1991. Mutagenesis of the folC gene encoding folylpolyglutamate synthetase-dihydrofolate synthetase in Escherichia coli. Arch. Biochem. Biophys. 284:9-16. [DOI] [PubMed] [Google Scholar]
- 44.Klasson, L., and S. G. Andersson. 2004. Evolution of minimal-gene-sets in host-dependent bacteria. Trends Microbiol. 12:37-43. [DOI] [PubMed] [Google Scholar]
- 45.Kobayashi, K., S. D. Ehrlich, A. Albertini, G. Amati, K. K. Andersen, M. Arnaud, K. Asai, S. Ashikaga, S. Aymerich, P. Bessieres, F. Boland, S. C. Brignell, S. Bron, K. Bunai, J. Chapuis, L. C. Christiansen, A. Danchin, M. Débarbouillé, E. Dervyn, E. Deuerling, K. Devine, S. K. Devine, O. Dreesen, J. Errington, S. Fillinger, S. J. Foster, Y. Fujita, A. Galizzi, R. Gardan, C. Eschevins, T. Fukushima, K. Haga, C. R. Harwood, M. Hecker, D. Hosoya, M. F. Hullo, H. Kakeshita, D. Karamata, Y. Kasahara, F. Kawamura, K. Koga, P. Koski, R. Kuwana, D. Imamura, M. Ishimaru, S. Ishikawa, I. Ishio, D. Le Coq, A. Masson, C. Mauël, R. Meima, R. P. Mellado, A. Moir, S. Moriya, E. Nagakawa, H. Nanamiya, S. Nakai, P. Nygaard, M. Ogura, T. Ohanan, M. O'Reilly, M. O'Rourke, Z. Pragai, H. M. Pooley, G. Rapoport, J. P. Rawlins, L. A. Rivas, C. Rivolta, A. Sadaie, Y. Sadaie, M. Sarvas, T. Sato, H. H. Saxild, E. Scanlan, W. Schumann, J. F. M. L. Seegers, J. Sekiguchi, A. Sekowska, S. J. Séror, M. Simon, P. Stragier, R. Studer, H. Takamatsu, T. Tanaka, M. Takeuchi, H. B. Thomaides, V. Vagner, J. M. van Dijl, K. Watabe, A. Wipat, H. Yamamoto, M. Yamamoto, Y. Yamamoto, K. Yamane, K. Yata, K. Yoshida, H. Yoshikawa, U. Zuber, and N. Ogasawara. 2003. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. USA 100:4678-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kogoma, T. 1997. Stable DNA replication: interplay between DNA replication, homologous recombination, and transcription. Microbiol. Mol. Biol. Rev. 61:212-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Konieczny, I. 2003. Strategies for helicase recruitment and loading in bacteria. EMBO Rep. 4:37-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koonin, E. V. 2000. How many genes can make a cell: the minimal-gene-set concept. Annu. Rev. Genomics Hum. Genet. 1:99-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koonin, E. V. 2003. Comparative genomics, minimal gene-sets and the last universal common ancestor. Nat. Rev. Microbiol. 1:127-136. [DOI] [PubMed] [Google Scholar]
- 50.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessières, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S.-K. Choi, J.-J. Codani, I. F. Connerton, N. J. Cummings, R. A. Daniel, F. Denizot, K. M. Devine, A. Düsterhöft, S. D. Ehrlich, P. T. Emmerson, K. D. Entian, J. Errington, C. Fabret, E. Ferrari, D. Foulger, C. Fritz, M. Fujita, Y. Fujita, S. Fuma, A. Galizzi, N. Galleron, S.-Y. Ghim, P. Glaser, A. Goffeau, E. J. Golightly, G. Grandi, G. Guiseppi, B. J. Guy, K. Haga, J. Haiech, C. R. Harwood, A. Hénaut, H. Hilbert, S. Holsappel, S. Hosono, M.-F. Hullo, M. Itaya, L. Jones, B. Joris, D. Karamata, Y. Kasahara, M. Klaerr-Blanchard, C. Klein, Y. Kobayashi, P. Koetter, G. Koningstein, S. Krogh, M. Kumano, K. Kurita, A. Lapidus, S. Lardinois, J. Lauber, V. Lazarevic, S.-M. Lee, A. Levine, H. Liu, S. Masuda, C. Mauël, C. Médigue, N. Medina, R. P. Mellado, M. Mizuno, D. Moestl, S. Nakai, M. Noback, D. Noone, M. O'reilly, K. Ogawa, A. Ogiwara, B. Oudega, S.-H. Park, V. Parro, T. M. Pohl, D. Portetelle, S. Porwollik, A. M. Prescott, E. Presecan, P. Pujic, B. Purnelle, G. Rapoport, M. Rey, S. Reynolds, M. Rieger, C. Rivolta, E. Rocha, B. Roche, M. Rose, Y. Sadaie, T. Sato, E. Scanlan, S. Schleich, R. Schroeter, F. Scoffone, J. Sekiguchi, A. Sekowska, S. J. Seror, P. Serror, B.-S. Shin, B. Soldo, A. Sorokin, E. Tacconi, T. Takagi, H. Takahashi, K. Takemaru, M. Takeuchi, A. Tamakoshi, T. Tanaka, P. Terpstra, A. Tognoni, V. Tosato, S. Uchiyama, M. Vandenbol, F. Vannier, A. Vassarotti, A. Viari, R. Wambutt, E. Wedler, H. Wedler, T. Weitzenegger, P. Winters, A. Wipat, H. Yamamoto, K. Yamane, K. Yasumoto, K. Yata, K. Yoshida, H.-F. Yoshikawa, E. Zumstein, H. Yoshikawa, and A. Danchin. 1997. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 51.Kursanov, O. V., B. M. Polanuyer, S. Ananta, R. Sloutsky, A. Tam, S. Y. Gerdes, and A. L. Ostedrman. 2002. Ribosylnicotinamide kinase domain of NadR protein: identification and implications in NAD biosynthesis. J. Bacteriol. 184:6906-6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leung, H. C. E., T. G. Hagervall, G. R. Björk, and M. E. Winkler. 1998. Genetic locations and database accession numbers of RNA-modifying and -editing enzymes, p. 561-567. In H. Grosjean and R. Benne (ed.), Modification and editing of RNA. American Society for Microbiology, Washington, D.C.
- 53.Luisi, P. L., T. Oberholzer, and A. Lazcano. 2002. The notion of a DNA minimal cell: a general discourse and some guidelines for an experimental approach. Helv. Chim. Acta 85:1759-1777. [Google Scholar]
- 54.Margolin, W. 2000. Themes and variations in prokaryotic cell división. FEMS Microbiol. Rev. 24:531-548. [DOI] [PubMed] [Google Scholar]
- 55.Margolis, P., C. Hackbarth, S. Lopez, M. Maniar, W. Wang, Z. Yuan, R. White, and J. Trias. 2001. Resistance of Streptococcus pneumoniae to deformylase inhibitors is due to mutations in defB. Antimicrob. Agents Chemother. 45:2432-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Margolis, P. S., C. J. Hackbarth, D. C. Young, W. Wang, D. Chen, Z. Yuan, R. White, and J. Trias. 2000. Peptide deformylase in Staphylococcus aureus: resistance to inhibition is mediated by mutations in the formyltransferase gene. Antimicrob. Agents Chemother. 44:1825-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martin, P. R., R. J. Shea, and M. H. Mulks. 2001. Identification of a plasmid- encoded gene from Haemophilus ducreyi which confers NAD independence. J. Bacteriol. 183:1168-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Masslewski, A., J. M. Smith, and S. J. Benkovic. 1994. Cloning and characterization of a new purine biosynthetic enzyme: a non-folate glycinamide ribonucleotide transformylase from Escherichia coli. Biochemistry 33:2531-2537. [DOI] [PubMed] [Google Scholar]
- 59.Mazel, D., S. Pochet, and P. Marliere. 1994. Genetic characterization of polypeptide deformylase, a distinctive enzyme of eubacterial translation. EMBO J. 13:914-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Meyer, T. H., J. F. Ménétret, R. Breitling, K. R. Miller, C. W. Akey, and T. A. Rapoport. 1999. The bacterial SecY/E translocation complex forms channel-like structures similar to those of the eukaryotic Sec61p complex. J. Mol. Biol. 285:1789-1800. [DOI] [PubMed] [Google Scholar]
- 61.Miclet, E., V. Stoven, P. A. M. Michels, F. R. Opperdoes, J. Y. Lallemand, and F. Duffieux. 2001. NMR spectroscopic analysis of the first two steps of the pentose-phosphate pathway elucidates the role of 6-phosphogluconolactonase. J. Biol. Chem. 276:34840-34846. [DOI] [PubMed] [Google Scholar]
- 62.Moran, N. A., and J. J. Wernegreen. 2000. Lifestyle evolution in symbiotic bacteria: insights from genomics. Trends Ecol. Evol. 15:321-326. [DOI] [PubMed] [Google Scholar]
- 63.Morange, M. 2003. La vie expliquée? 50 ans après la double helix. Odile Jacob, Paris, France.
- 64.Mori, H., K. Isono, T. Horiuchi, and T. Miki. 2000. Functional genomics of Escherichia coli in Japan. Res. Microbiol. 151:121-128. [DOI] [PubMed] [Google Scholar]
- 65.Mushegian, A. R., and E. V. Koonin. 1996. A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc. Natl. Acad. Sci. USA 93:10268-10273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Newton, D. T., C. Creuzenet, and D. Mangroo. 1999. Formylation is not essential for initiation of protein synthesis in all eubacteria. J. Biol. Chem. 274:22143-22146. [DOI] [PubMed] [Google Scholar]
- 67.Nuño, J. C., I. Sánchez-Valdenebro, C. Pérez-Iratxeta, E. Meléndez-Hevia, and F. Montero. 1997. Network organization of cell metabolism: monosaccharide interconversion. Biochem. J. 325:103-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nygaard, P., and J. M. Smith. 1993. Evidence for a novel glycinamide ribonucleotide transformylase in Escherichia coli. J. Bacteriol. 175:3591-3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ogura, T., T. Tomoyasu, T. Yuki, S. Morimura, K. J. Begg, W. D. Donachie, H. Mori, H. Niki, and S. Hiraga. 1991. Structure and function of the ftsH gene in Escherichia coli. Res. Microbiol. 142:279-282. [DOI] [PubMed] [Google Scholar]
- 70.Oshima, K., S. Kakizawa, H. Nishigawa, H.-Y. Jung, W. Wei, S. Suzuki, R. Arashida, D. Nakata, S. Miyata, M. Ugaki, and S. Namba. 2004. Reductive evolution suggested from the complete genome sequence of a plant-pathogenic phytoplasma. Nat. Genet. 36:27-29. [DOI] [PubMed] [Google Scholar]
- 71.Paciorek, J., K. Kardys, B. Lobacz, and K. I. Wolska. 1997. Escherichia coli defects caused by null mutations in dnaK and dnaJ genes. Acta Microbiol. Pol. 46:7-17. [PubMed] [Google Scholar]
- 72.Persson, B. C., G. O. Bylund, D. E. Berg, and P. M. Wikstrom. 1995. Functional analysis of the ffh-trmD region of the Escherichia coli chromosome by using reverse genetics. J. Bacteriol. 177:5554-5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pohorille, A., and D. Deamer. 2002. Artificial cells: prospects for biotechnology. Trends Biotechnol. 20:123-128. [DOI] [PubMed] [Google Scholar]
- 74.Rock, C. O., and S. Jackowski. 2002. Forty years of bacterial fatty acid synthesis. Biochem. Biophys. Res. Commun. 292:1155-1166. [DOI] [PubMed] [Google Scholar]
- 75.Schatz, P. J., K. L. Bieker, K. M. Ottemann, T. J. Silhavy, and J. Beckwith. 1991. One of three transmembrane stretches is sufficient for the functioning of the SecE protein, a membrane component of the E. coli secretion machinery. EMBO J. 10:1749-1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schmelter, T., B. L. Trigatti, G. E. Gerber, and D. Mangroo. 2004. Biochemical demonstration of the involvement of fatty acyl-CoA synthetase in fatty acid translocation across the plasma membrane. J. Biol. Chem. 279:24163-24170. [DOI] [PubMed] [Google Scholar]
- 77.Shigenobu, S., H. Watanabe, M. Hattori, Y. Sakaki, and H. Ishikawa. 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407:81-86. [DOI] [PubMed] [Google Scholar]
- 78.Silva, F. J., A. Latorre, and A. Moya. 2003. Why are the genomes of endosymbiotic bacteria so stable? Trends Genet. 19:176-180. [DOI] [PubMed] [Google Scholar]
- 79.Smalley, D. J., M. Whiteley, and T. Conway. 2003. In search of the minimal Escherichia coli genome. Trends Microbiol. 11:6-8. [DOI] [PubMed] [Google Scholar]
- 80.Tamas, I., L. Klasson, B. Canbäck, A. K. Näslund, A. S. Eriksson, J. J. Wernegreen, J. P. Sandström, N. A. Moran, and S. G. E. Andersson. 2002. 50 million years of genomic stasis in endosymbiotic bacteria. Science 296:2376-2379. [DOI] [PubMed] [Google Scholar]
- 81.Tomita, M., K. Hashimoto, K. Takahashi, T. S. Shimizu, Y. Matsuzaki, F. Miyoshi, K. Saito, S. Tanida, K. Yugi, J. C. Venter, and C. A. Hutchison III. 1999. E-CELL: software environment for whole-cell simulation. Bioinformatics 15:72-84. [DOI] [PubMed] [Google Scholar]
- 82.van der Laan, M., M. L. Urbanus, C. M. ten Hagen-Jongman, N. Nouwen, B. Oudega, N. Harms, A. J. M. Driessen, and J. Luirink. 2003. A conserved function of YidC in the biogenesis of respiratory chain complexes. Proc. Natl. Acad. Sci. USA 100:5801-5806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.van Ham, R. C. H. J., J. Kamerbeek, C. Palacios, C. Rausell, F. Abascal, U. Bastolla, J. M. Fernández, L. Jiménez, M. Postigo, F. J. Silva, J. Tamames, E. Viguera, A. Latorre, A. Valencia, F. Morán, and A. Moya. 2003. Reductive genome evolution in Buchnera aphidicola. Proc. Natl. Acad. Sci. USA 100:581-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Waters, E., M. J. Hohn, I. Ahel, D. E. Graham, M. D. Adams, M. Barnstead, K. Y. Beeson, L. Bibbs, R. Bolanos, M. Keller, K. Kretz, X. Lin, E. Mathur, J. Ni, M. Podar, T. Richardson, G. G. Sutton, M. Simon, D. Soll, K. O. Stetter, J. M. Short, and M. Noordewier. 2003. The genome of Nanoarchaeum equitans: insights into early archaeal evolution and derived parasitism. Proc. Natl. Acad. Sci. USA 100:12984-12988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Weitao, T., K. Nordström, and S. Dasgupta. 1999. Mutual suppression of mukB and seqA phenotypes might arise from their opposing influences on the Escherichia coli nucleoid structure. Mol. Microbiol. 34:157-168. [DOI] [PubMed] [Google Scholar]
- 86.Yu, X.-C., and W. Margolin. 1999. FtsZ ring clusters in min and partition mutants: role of both the Min system and the nucleoid in regulating FtsZ ring localization. Mol. Microbiol. 32:315-326. [DOI] [PubMed] [Google Scholar]
- 87.Zuo, Y., and M. P. Deutscher. 2001. Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic Acids Res. 29:1017-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zuurmond, A. M., A. K. Rundlof, and B. Kraal. 1999. Either of the chromosomal tuf genes of E. coli K-12 can be deleted without loss of cell viability. Mol. Gen. Genet. 260:603-607. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.