Abstract
The pathological accumulation of RNA-binding proteins (RBPs) within inclusion bodies is a hallmark of amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD). RBP aggregation results in both toxic gain and loss of normal function. Determining the protein binding partners and normal functions of disease-associated RBPs is necessary to fully understand molecular mechanisms of RBPs in disease. Herein, we characterized the protein-protein interactions (PPIs) of RBM45, a RBP that localizes to inclusions in ALS/FTLD. Using immunoprecipitation coupled to mass spectrometry (IP-MS), we identified 132 proteins that specifically interact with RBM45 within HEK293 cells. Select PPIs were validated by immunoblot and immunocytochemistry, demonstrating that RBM45 associates with a number of other RBPs primarily via RNA-dependent interactions in the nucleus. Analysis of the biological processes and pathways associated with RBM45-interacting proteins indicates enrichment for nuclear RNA processing/splicing via association with hnRNP proteins and cytoplasmic RNA translation via eiF2 and eiF4 pathways. Moreover, several other ALS-linked RBPs, including TDP-43, FUS, Matrin-3, and hnRNP-A1, interact with RBM45, consistent with prior observations of these proteins within intracellular inclusions in ALS/FTLD. Taken together, our results define a PPI network for RBM45, suggest novel functions for this protein, and provide new insights into the contributions of RBM45 to neurodegeneration in ALS/FTLD.
Keywords: RBM45, immunoprecipitation, mass spectrometry, protein-protein interaction, ALS
1. Introduction
The aggregation of RNA-binding proteins (RBPs) into inclusion bodies is one of the most prevalent and well-characterized pathological findings in amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD). The identification of cytoplasmic mis-localized TDP-43 (Neumann et al., 2006), and later FUS (Kwiatkowski et al., 2009; Vance et al., 2009), as primary components of ubiquitinated inclusions in motor neurons and glia in these disorders led to the “two-hit” hypothesis of RBP-mediated neurodegeneration. This model proposes that the pathological aggregation of RBPs confers toxicity by simultaneous gain of toxic function of the aggregates and the loss of normal functions served by these proteins in regulating gene expression. Ample experimental evidence now exists in support of this model, with studies consistently finding that under- or overexpression of numerous RBPs is sufficient to induce neuronal cell death in a variety of model systems (reviewed in (Ling et al., 2013).
This model of RBP-mediated neurodegeneration depends, in part, on the ability of RBPs to self-associate and interact with other RBPs within protein aggregates. Many ALS-linked RBPs, including TDP-43, FUS, hnRNP-A1, and TAF15 are aggregation prone as a result of prion-like domains contained within their protein sequence (Johnson et al., 2009; King et al., 2012). Mutations in the prion-like domain lead to familial forms of ALS/FTLD marked by the pathological aggregation of the mutant protein (reviewed in (Gitler and Shorter, 2011). In addition to self-aggregation, these proteins are capable of sequestering other proteins into aggregates/inclusions as a consequence of the normal functional associations between these proteins. For example, proteomic analysis of TDP-43 aggregates showed deposition of stress granule proteins G3BP and PABPC1 as well as paraspeckle proteins PSF and NONO (Dammer et al., 2012). Similar observations of paraspeckle proteins p54nrb and NONO in FUS-positive inclusions (Shelkovnikova et al., 2014) provide additional evidence in support of this concept. Thus, understanding the protein-protein interactions (PPIs) of ALS-linked RBPs is a necessary step towards defining the protein composition of inclusions in ALS/FTLD and new insight into mechanisms of disease.
Determining RBP PPIs is also essential for understanding the normal functions of RBPs, and how these functions may be compromised as a result of RBP aggregation in ALS/FTLD. Numerous RBP functions depend on the association of RBPs with protein/nucleic acid complexes. For example, FUS is a component of both nuclear gems, which participate in snRNP biogenesis, and paraspeckles, which are involved in cellular stress responses (Shelkovnikova et al., 2014; Yamazaki et al., 2012). The expression of mutant FUS reduces levels of these nuclear sub-structures, suggesting mechanisms by which loss of normal FUS function contributes to cell death in ALS/FTLD. In addition, many ALS/FTLD-linked RBPs also associate with cytoplasmic stress granules (Li et al., 2013), and disease-associated mutations tend to promote the excess formation of these structures (Kim et al., 2013). While stress granules normally aid in the response to cellular stress by protecting mRNAs and shifting gene expression towards a stress response, excessive stress granule formation promotes the formation of insoluble RBP aggregates that may be precursors to inclusion bodies (Kim et al., 2013; Li et al., 2013; Vance et al., 2013). This can lead to loss of other normal functions, such as impaired P-body formation that occurs in response to mutant FUS sequestration in stress granules (Takanashi and Yamaguchi, 2014). PPIs can be used to predict these and similar functional associations (Dammer et al., 2012). Defining RBP PPIs, therefore, helps uncover novel functions and candidate disease mechanisms related to these multifunctional proteins.
Given the diversity of RBP functions, which includes regulating transcription, RNA splicing/export, and miRNA biogenesis (Ling et al., 2013), a relatively high-throughput approach is preferable to identify candidate functions/binding partners for targeted validation. Immunoprecipitation coupled to mass spectrometry (IP-MS) offers tremendous promise towards identifying large sets of RBP protein-protein interactions (PPIs) and associated biological processes/pathways. The sensitivity of this approach can be further enhanced by the use of cross-linking methods, such as treatment with small cross-linking agents or formaldehyde, to detect low-affinity protein interactions (Li et al., 2015; Nittis et al., 2010). This approach has previously been used to identify proteins interacting with the ALS-linked Ewing Sarcoma (EWS) RBP (Pahlich et al., 2009), where interactions with hnRNPs and FUS are consistent with roles of EWS in mRNA splicing (Law et al., 2006) and inclusion formation in ALS/FTLD (Mackenzie and Neumann, 2012), respectively. Thus, IP-MS can identify multiple protein binding partners of a given target and this information can be used to predict novel functions and roles in disease.
Here, we applied this approach to RBM45, a recently characterized RNA-binding protein found in inclusions in ALS, FTLD, and Alzheimer’s disease (AD) (Collins et al., 2012). These inclusions are positive for TDP-43, and RBM45 physically interacts with TDP-43 and FUS in vitro (Li et al., 2015). RBM45 contains three RNA-recognition motifs (RRMs), a nuclear localization sequence (NLS), and a homo-oligomerization (HOA) domain that mediates self-association of the protein, and can localize to cytoplasmic stress granules (Bakkar et al., 2015; Li et al., 2015). The expression of RBM45 is developmentally regulated and the highest expression levels occur in the brain (Tamada et al., 2002). These properties make RBM45 a promising target for continued studies of ALS/FTLD, though at present little is known about the function of RBM45. To delineate protein binding partners of RBM45 and putative biological functions of the protein, we used an IP-MS approach to comprehensively characterize RBM45 protein-protein interactions (PPIs). We identified 132 RBM45 PPIs by IP-MS, including PPIs with many RBPs. Our results were used to associate RBM45 with biological processes and pathways. These were primarily related to nuclear mRNA processing and cytoplasmic RNA translation. Our IP-MS findings also indicate that RBM45 interacts with a number of ALS-linked proteins, including TDP-43, FUS, Matrin-3, hnRNP-A1, and hnRNP-A2/B1. Selected PPIs were externally validated via complementary techniques. Collectively, our results shed new light on RBM45 PPIs, biological functions, and contributions to neurodegeneration in ALS/FTLD.
2. Results
2.1. Identification of the RBM45 interacting proteins in HEK-293 cells
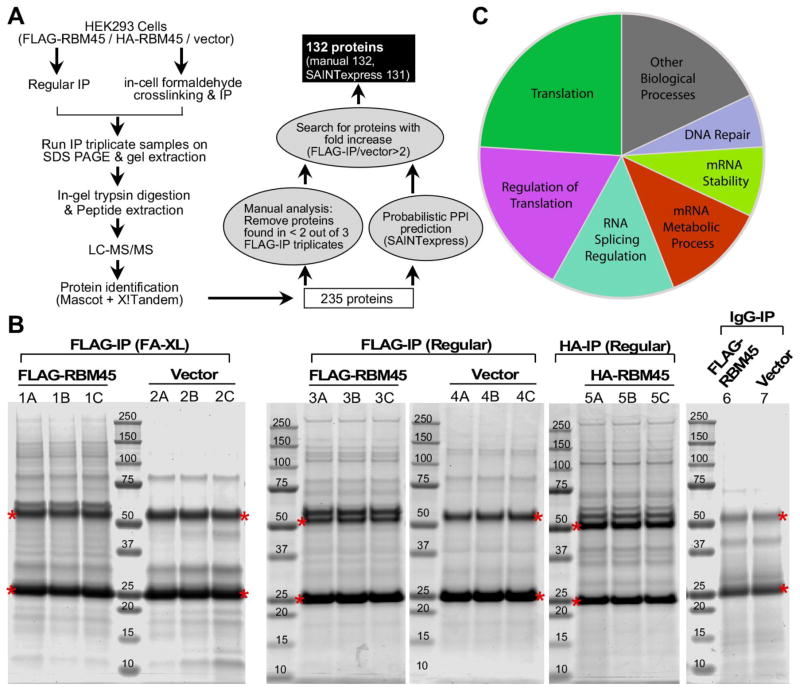
A schematic outline of the immunoprecipitation-mass spectrometry procedure used in this study is shown in Figure 1A. FLAG-RBM45 or empty vector was overexpressed in HEK293 cells and immunoprecipitated using whole cell lysates in triplicates. HA-tagged RBM45 was also included and used as a reference for the data analysis. HEK293 cells expressing empty vector alone served as negative controls. Regular IP and formaldehyde crosslinking IP were performed in parallel to identify strongly and weakly RBM45-associated proteins separately (Fig. 1A). Immunoblot analysis of the immunoprecipated fractions showed that tagged-RBM45 was enriched in the pulldown. In contrast, no RBM45 was detected in the pulldown in the vector control or IgG pulldown (Supplemental Fig. 1). These data demonstrate that tagged-RBM45 can be efficiently and specifically immunoprecipitated from cell extracts. Co-immunoprecipitated proteins were then separated using SDS-PAGE and stained (Fig. 1B). Coomassie staining of the gels loaded with RBM45-IP (sample 1, 3 and 5) identified several bands that were not present in vector (sample 2 and 4) or IgG controls (sample 6 and 7).
Figure 1. Identification of RBM45 interacting proteins.
(A) Diagram of the immunoprecipitation-mass spectrometry approach to identify the RBM45 interacting proteins.
(B) Triplicate immunoprecipitates from HEK293 cells stably expressing FLAG-RBM45, HA-RBM45 or empty vector were separated by SDS-PAGE and stained with Coomassie blue to visualize proteins. Immunoprecipitations with FLAG (sample 1, 2, 3, 4), HA (sample 5) antibody or IgG (sample 6, 7) were performed. Crosslinking IP (sample 1, 2) and regular IP (sample 3, 4, 5) were performed in parallel. For crosslinking IP, live cells were treated with 0.1% formaldehyde to cross-link proteins prior to cell lysis and immunoprecipitation. The crosslinking was reversed by heating in SDS-sample buffer prior to SDS-PAGE. The proteins along the entire length of the gel were extracted (excluding the IgG-heavy chain and IgG-light chain that are denoted by red *) and analyzed by LC/MS-MS.
(C) Pie-chart representation of functional classes of RBM45 interacting proteins.
Immunoprecipitated proteins were gel extracted, trypsin digested, and identified by liquid chromatography tandem mass spectroscopy (LC-MS/MS) (Fig. 1A, see Methods). We identified 235 unique proteins with a protein False Discovery Rate equal or lower than 1%. We then applied a manual thresholding approach and a probabilistic PPI prediction algorithm (SAINTexpress) to compute the most likely associations between each of these 235 proteins and RBM45, yielding 132 high-confidence candidates (Fig. 1A). These 132 candidate proteins were found in at least 2 out of the 3 FLAG-IP triplicates and were at least 2-fold more abundant compared to vector control, suggesting that they specifically associate with RBM45 (Table 1 and Supplemental Table 1, 2 and 3). Of these 132 proteins, 28 were found exclusively by regular-IP (Supplemental Table 4), 68 were found exclusively by crosslinking-IP (Supplemental Table 5), and 36 were found in both regular-IP and crosslinking-IP groups (Supplemental Table 6). Analysis of the average number of total spectrum counts by different immunoprecipitation group showed that in both regular IP and crosslinking IP, the proteins identified from empty vector groups were significantly lower than the proteins identified from FLAG-/HA-IP groups, providing further evidence of the specificity of the approach (Supplemental Fig. 2).
Table 1.
Merged list of identified RBM45 interacting proteins from manual analysis and SAINTexpress algorithma
| Proteins | Spectral Count Sum | Fold spectra increase | AvgP | Canonical Pathway | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Manual | SAINTexpress | SAINTexpress | EIF2 | eIF4/p70S6K | mTOR | Telomere Ext. | RAN | ||||||||
| Accession | Gene Name | Description | Regular | XL | Regular | XL | Regular | XL | Avg | ||||||
| Q8IUH3 | RBM45 | RNA-binding protein 45 | 969 | + + + | + | + + + | + | 1 | 1 | 1 | |||||
| P14866 | HNRNPL | Heterogeneous nuclear ribonucleoprotein L | 207 | + + + | + + + | + + | + + | 1 | 1 | 1 | |||||
| P22626 | HNRNPA2B1 | Heterogeneous nuclear ribonucleoproteins A2/B1 | 113 | + + + | + + + | + + | + + | 1 | 1 | 1 | x | ||||
| P43243 | MATR3 | Matrin-3 | 111 | + + + | + + + | + + | + + | 1 | 1 | 1 | |||||
| Q00839 | HNRNPU | Heterogeneous nuclear ribonucleoprotein U | 107 | + + + | + + + | + + | + + | 1 | 1 | 1 | |||||
| P61978 | HNRNPK | Heterogeneous nuclear ribonucleoprotein K | 105 | + + + | + + + | + + | + + | 1 | 1 | 1 | |||||
| P09651 | HNRNPA1 | Heterogeneous nuclear ribonucleoprotein A1 | 99 | + + + | + + + | + + | + + | 1 | 1 | 1 | x | ||||
| Q93008 | USP9X | Probable ubiquitin carboxyl-terminal hydrolase FAF-X | 97 | + + + | + + | 1 | 1 | ||||||||
| Q9NZI8 | IGF2BP1 | Insulin-like growth factor 2 mRNA-binding protein 1 | 63 | + | + | 1 | 1 | ||||||||
| Q8N163 | CCAR2 | Cell cycle and apoptosis regulator protein 2 | 62 | + + + | + + + | + + | + | 1 | 1 | 1 | |||||
| P52272 | HNRNPM | Heterogeneous nuclear ribonucleoprotein M | 60 | + + + | + + | 1 | 1 | ||||||||
| Q08211 | DHX9 | ATP-dependent RNA helicase A | 60 | + + + | + + + | + | + + | 1 | 1 | 1 | |||||
| P08107 | HSPA1A | Heat shock 70 kDa protein 1A/1B | 58 | + | + | 1 | 1 | ||||||||
| Q13263 | TRIM28 | Transcription intermediary factor 1-beta | 55 | + + + | + + | 1 | 1 | ||||||||
| P11940 | PABPC1 | Polyadenylate-binding protein 1 | 52 | + | + | 1 | 1 | x | x | ||||||
| P51991 | HNRNPA3 | Heterogeneous nuclear ribonucleoprotein A3 | 50 | + + + | + + + | + | + + | 1 | 1 | 1 | |||||
| P07910 | HNRNPC | Heterogeneous nuclear ribonucleoproteins C1/C2 | 48 | + + + | + + + | + | + + | 1 | 1 | 1 | |||||
| P38159 | RBMX | RNA-binding motif protein, X chromosome | 41 | + + + | + + + | + | + | 1 | 1 | 1 | |||||
| O43390 | HNRNPR | Heterogeneous nuclear ribonucleoprotein R | 32 | + + + | + + + | + | + | 1 | 1 | 1 | |||||
| Q13148 | TARDBP | TAR DNA-binding protein 43 | 27 | + + + | + | 1 | 1 | ||||||||
| Q15717 | ELAVL1 | ELAV-like protein 1 | 26 | + + + | + + + | + | + | 1 | 1 | 1 | |||||
| Q92841 | DDX17 | Probable ATP-dependent RNA helicase DDX17 | 25 | + + + | + + + | + | 1 | 1 | |||||||
| Q96PK6 | RBM14 | RNA-binding protein 14 | 25 | + ++ | + + + | + | + | 1 | 1 | 1 | |||||
| P23246 | SFPQ | Splicing factor, proline- and glutamine-rich | 23 | + + + | + | 1 | 1 | ||||||||
| Q13310 | PABPC4 | Polyadenylate-binding protein 4 | 23 | + + + | + | 1 | 1 | ||||||||
| Q14103 | HNRNPD | Heterogeneous nuclear ribonucleoprotein D0 | 23 | + + + | + + + | + | + | 1 | 1 | 1 | |||||
| Q9NZB2 | FAM120A | Constitutive coactivator of PPAR-gamma-like protein 1 | 21 | + + + | + | 1 | 1 | ||||||||
| P09874 | PARP1 | Poly [ADP-ribose] polymerase 1 | 20 | + + + | + | 1 | 1 | ||||||||
| P68104 | EEF1A1 | Elongation factor 1-alpha 1 | 20 | + + + | + | 1 | 1 | ||||||||
| P78347 | GTF2I | General transcription factor II-I | 20 | + + + | + | 1 | 1 | ||||||||
| Q96PU8 | QKI | Protein quaking | 20 | + + + | + | 1 | 1 | ||||||||
| O75643 | SNRNP200 | U5 small nuclear ribonucleoprotein 200 kDa helicase | 19 | + + + | + | 1 | 1 | ||||||||
| Q15366 | PCBP2 | Poly(rC)-binding protein 2 | 19 | + + + | + + + | + | + | 1 | 1 | 1 | |||||
| Q92945 | KHSRP | Far upstream element-binding protein 2 | 19 | + + + | + | 1 | 1 | ||||||||
| Q99729 | HNRNPAB | Heterogeneous nuclear ribonucleoprotein A/B | 19 | + + + | + + + | + | + | 1 | 1 | 1 | |||||
| O15042 | U2SURP | U2 snRNP-associated SURP motif-containing protein | 18 | + + + | + | 1 | 1 | ||||||||
| P17844 | DDX5 | Probable ATP-dependent RNA helicase DDX5 | 18 | + + + | + | 1 | 1 | ||||||||
| P13639 | EEF2 | Elongation factor 2 | 16 | + + + | + | 1 | 1 | ||||||||
| P60709 | ACTB | Actin, cytoplasmic 1 | 16 | + + + | + | 1 | 1 | ||||||||
| P62269 | HIST1H4A | Histone H4 | 15 | + + + | + | 1 | 1 | ||||||||
| O00425 | IGF2BP3 | Insulin-like growth factor 2 mRNA-binding protein 3 | 14 | + + + | + | 1 | 1 | ||||||||
| O43143 | DHX15 | Pre-mRNA-splicing factor ATP-dependent RNA helicase DHX15 | 14 | + + + | + | 1 | 1 | ||||||||
| Q9Y2L1 | DIS3 | Exosome complex exonuclease RRP44 | 14 | + + + | + | 1 | 1 | ||||||||
| P01614 | [ Ig kappa chain V-II region Cum ] | 13 | + + + | + | 1 | 1 | |||||||||
| P07437 | TUBB | Tubulin beta chain | 12 | + + + | + | 1 | 1 | ||||||||
| P35637 | FUS | RNA-binding protein FUS | 12 | + + + | + | 1 | 1 | ||||||||
| Q7L2E3 | DHX30 | Putative ATP-dependent RNA helicase DHX30 | 12 | + + + | + | 1 | 1 | ||||||||
| Q6PKG0 | LARP1 | La-related protein 1 | 11 | + + + | + | 1 | 1 | ||||||||
| Q9Y6M1 | IGF2BP 2 | Insulin-like growth factor 2 mRNA-binding protein 2 | 11 | + + + | + | 1 | 1 | ||||||||
| P62988 | UBB | Ubiquitin | 10 | + + + | + | 1 | 1 | ||||||||
| Q9UKM9 | RALY | RNA-binding protein Raly | 10 | + + + | + | 1 | 1 | ||||||||
| P25205 | MCM3 | DNA replication licensing factor MCM3 | 8 | + + + | + | 1 | 1 | ||||||||
| Q99623 | PHB2 | Prohibitin-2 | 8 | + + + | + | 1 | 1 | ||||||||
| O75688 | PPM1B | Protein phosphatase 1B | 64 | + + + | + | + | 0.97 | 7 | |||||||
| P62805 | HSPD1 | 60 kDa heat shock protein, mitochondrial | 15 | + | + | 0.88 | 0.88 | ||||||||
| P26599 | PTBP1 | Polypyrimidine tract-binding protein 1 | 20 | + + + | + + + | + | + | 0.67 | 1 | 0.835 | |||||
| Q13151 | HNRNP A0 | Heterogeneous nuclear ribonucleoprotein A0 | 20 | + + + | + + + | + | + | 1 | 0.67 | 0.835 | |||||
| P31943 | HNRNPH1 | Heterogeneous nuclear ribonucleoprotein H | 16 | + + + | + + + | + | + | 1 | 0.67 | 0.835 | |||||
| P10809 | RPS18 | 40S ribosomal protein S18 | 15 | + + + | + + + | + | + | 1 | 0.67 | 0.835 | x | x | x | ||
| Q14974 | KPNB1 | Importin subunit beta-1 | 14 | + + + | + + + | + | + | 1 | 0.67 | 0.835 | x | ||||
| P63241 | EIF5A | Eukaryotic translation initiation factor 5A-1 | 11 | + + + | + | 0.67 | 0.67 | ||||||||
| P12956 | XRCC6 | X-ray repair cross-complementing protein 6 | 9 | + + + | + | 0.67 | 0.67 | x | |||||||
| Q86U42 | PABPN1 | Polyadenylate-binding protein 2 | 9 | + + + | + | 0.67 | 0.67 | ||||||||
| O60814 | HIST1H2BK | Histone H2B type 1-K | 7 | + + + | + | 0.67 | 0.67 | ||||||||
| P62701 | RPS4X | 40S ribosomal protein S4, X isoform | 7 | + + + | + | 0.67 | 0.67 | x | x | x | |||||
| P04908 | HIST1H2AB | Histone H2A type 1-B/E | 6 | + + + | + | 0.67 | 0.67 | ||||||||
| P55060 | CSE1L | Exportin-2 | 6 | + + + | + | 0.67 | 0.67 | x | |||||||
| P62316 | SNRPD2 | Small nuclear ribonucleoprotein Sm D2 | 6 | + + + | + | 0.67 | 0.67 | ||||||||
| P84090 | ERH | Enhancer of rudimentary homolog | 6 | + + + | + | 0.67 | 0.67 | ||||||||
| P05388 | RPLP0 | 60S acidic ribosomal protein P0 | 5 | + + + | + | 0.67 | 0.67 | x | |||||||
| P08238 | HSP90AB1 | Heat shock protein HSP 90-beta | 5 | + + + | + | 0.67 | 0.67 | ||||||||
| P19338 | NCL | Nucleolin | 5 | + + + | + | 0.67 | 0.67 | ||||||||
| P49327 | FASN | Fatty acid synthase | 5 | + + + | + | 0.67 | 0.67 | ||||||||
| P52597 | HNRNPF | Heterogeneous nuclear ribonucleoprotein F | 5 | + + + | + | 0.67 | 0.67 | ||||||||
| P62314 | SNRPD1 | Small nuclear ribonucleoprotein Sm D1 | 5 | + + + | + | 0.67 | 0.67 | ||||||||
| P62318 | SNRPD3 | Small nuclear ribonucleoprotein Sm D3 | 5 | + + + | + | 0.67 | 0.67 | ||||||||
| Q15029 | EFTUD2 | 116 kDa U5 small nuclear ribonucleoprotein component | 5 | + + + | + | 0.67 | 0.67 | ||||||||
| Q8IY67 | RAVER1 | Ribonucleoprotein PTB-binding 1 | 5 | + + + | + | 0.67 | 0.67 | ||||||||
| Q9Y2W1 | THRAP3 | Thyroid hormone receptor-associated protein 3 | 5 | + + + | + | 0.67 | 0.67 | ||||||||
| P15927 | RPA2 | Replication protein A 32 kDa subunit | 4 | + + + | + | 0.67 | 0.67 | ||||||||
| Q12906 | ILF3 | Interleukin enhancer-binding factor 3 | 23 | + + + | + + + | + | + | 0.33 | 1 | 0.665 | |||||
| O60506 | SYNCRIP | Heterogeneous nuclear ribonucleoprotein Q | 21 | + + + | + + + | + | + | 1 | 0.33 | 0.665 | |||||
| P68363 | TUBA1B | Tubulin alpha-1B chain | 15 | + + + | + + + | + | + | 1 | 0.33 | 0.665 | |||||
| P52294 | KPNA1 | Importin subunit alpha-5 | 13 | + + + | + + + | + | + | 1 | 0.33 | 0.665 | |||||
| Q12905 | ILF2 | Interleukin enhancer-binding factor 2 | 11 | + + + | + + + | + | + | 0.66 | 0.67 | 0.665 | x | ||||
| P68032 | ACTC1 | Actin, alpha cardiac muscle 1 | 7 | + | + | 0.66 | 0.66 | ||||||||
| P01859 | IGHG2 | [ Ig gamma-2 chain C region ] | 10 | + | + + + | + | + | 0.97 | 0.33 | 0.65 | |||||
| P39019 | RPS19 | 40S ribosomal protein S19 | 6 | + | + | 0.65 | 0.65 | x | x | x | |||||
| O14979 | HNRNPDL | Heterogeneous nuclear ribonucleoprotein D-like | 14 | + + + | + + + | + | + | 0.67 | 0.33 | 0.5 | |||||
| P67809 | YBX1 | Nuclease-sensitive element-binding protein 1 | 12 | + + + | + + + | + | + | 1 | 0 | 0.5 | |||||
| P05141 | SLC25A5 | ADP/ATP translocase 2 | 9 | + + + | + + + | + | + | 0 | 1 | 0.5 | |||||
| P08779 | KRT16 | [ Keratin, type I cytoskeletal 16 ] | 13 | + | + | 0.33 | 0.33 | ||||||||
| P23396 | RPS3 | 40S ribosomal protein S3 | 7 | + + + | + + + | + | + | 0.66 | 0 | 0.33 | x | x | x | ||
| Q15233 | NONO | Non-POU domain-containing octamer-binding protein | 4 | + + + | + | 0.33 | 0.33 | ||||||||
| Q15365 | PCBP1 | Poly(rC)-binding protein 1 | 4 | + + + | + | 0.33 | 0.33 | ||||||||
| Q99459 | CDC5L | Cell division cycle 5-like protein | 4 | + + + | + | 0.33 | 0.33 | ||||||||
| Q99873 | PRMT1 | Protein arginine N-methyltransferase 1 | 4 | + + + | + | 0.33 | 0.33 | ||||||||
| Q9BXP5 | SRRT | Serrate RNA effector molecule homolog | 4 | + + + | + | 0.33 | 0.33 | ||||||||
| P09429 | HMGB1 | High mobility group protein B1 | 3 | + + + | + | 0.33 | 0.33 | ||||||||
| P42704 | LRPPRC | Leucine-rich PPR motif-containing protein, mitochondrial | 3 | + + + | + | 0.33 | 0.33 | ||||||||
| P43246 | MSH2 | DNA mismatch repair protein Msh2 | 3 | + + + | + | 0.33 | 0.33 | ||||||||
| P62851 | RPS25 | 40S ribosomal protein S25 | 3 | + + + | + | 0.33 | 0.33 | x | x | x | |||||
| Q04837 | SSBP1 | Single-stranded DNA-binding protein, mitochondrial | 3 | + + + | + | 0.33 | 0.33 | ||||||||
| Q13838 | DDX39B | Spliceosome RNA helicase DDX39B | 3 | + + + | + | 0.33 | 0.33 | ||||||||
| Q9BUJ2 | HNRNPUL1 | Heterogeneous nuclear ribonucleoprotein U-like protein 1 | 3 | + + + | + | 0.33 | 0.33 | ||||||||
| Q9UPT8 | ZC3H4 | Zinc finger CCCH domain-containing protein 4 | 3 | + + + | + | 0.33 | 0.33 | ||||||||
| P46782 | RPS5 | 40S ribosomal protein S5 | 5 | + | + | 0.32 | 0.32 | x | x | x | |||||
| P62913 | RPL11 | 60S ribosomal protein L11 | 9 | + | + + + | + | + | 0.59 | 0 | 0.295 | x | ||||
| P06748 | NPM1 | Nucleophosmin | 5 | + | + + + | + | + | 0 | 0 | 0 | |||||
| P26373 | RPL13 | 60S ribosomal protein L13 | 4 | + | + | 0 | 0 | x | |||||||
| P05387 | RPLP2 | 60S acidic ribosomal protein P2 | 3 | + | + | 0 | 0 | x | |||||||
| P14174 | MIF | Macrophage migration inhibitory factor | 3 | + + + | + | 0 | 0 | ||||||||
| P46783 | RPS10 | 40S ribosomal protein S10 | 3 | + + + | + | 0 | 0 | x | x | x | |||||
| P62263 | RPS14 | 40S ribosomal protein S14 | 3 | + + + | + | 0 | 0 | x | x | x | |||||
| P62917 | RPL8 | 60S ribosomal protein L8 | 3 | + + + | + | 0 | 0 | x | |||||||
| P82673 | MRPS35 | 28S ribosomal protein S35, mitochondrial | 3 | + + + | + | 0 | 0 | ||||||||
| P84103 | SRSF3 | Serine/arginine-rich splicing factor 3 | 3 | + + + | + | 0 | 0 | ||||||||
| Q13283 | G3BP1 | Ras GTPase-activating protein-binding protein 1 | 3 | + + + | + | 0 | 0 | ||||||||
| Q16576 | RBBP7 | Histone-binding protein RBBP7 | 3 | + + + | + | 0 | 0 | ||||||||
| Q7Z5L9 | IRF2BP 2 | Interferon regulatory factor 2-binding protein 2 | 3 | + + + | + | 0 | 0 | ||||||||
| Q9HAV4 | XPO5 | Exportin-5 | 3 | + + + | + | 0 | 0 | ||||||||
| P05386 | RPLP1 | 60S acidic ribosomal protein P1 | 2 | + + + | + | 0 | 0 | x | |||||||
| P06313 | [ Ig kappa chain V-IV region JI ] | 2 | + + + | + | 0 | 0 | |||||||||
| P31942 | HNRNPH3 | Heterogeneous nuclear ribonucleoprotein H3 | 2 | + + + | + | 0 | 0 | ||||||||
| P38919 | EIF4A3 | Eukaryotic initiation factor 4A-III | 2 | + + + | + | 0 | 0 | x | x | x | |||||
| P60842 | EIF4A1 | Eukaryotic initiation factor 4A-I | 2 | + + + | + | 0 | 0 | x | x | x | |||||
| P60866 | RPS20 | 40S ribosomal protein S20 | 2 | + + + | + | 0 | 0 | x | x | x | |||||
| P62081 | RPS7 | 40S ribosomal protein S7 | 2 | + + + | + | 0 | 0 | x | x | x | |||||
| P62249 | RPS16 | 40S ribosomal protein S16 | 2 | + + + | + | 0 | 0 | x | x | x | |||||
| P62277 | RPS13 | 40S ribosomal protein S13 | 2 | + + + | + | 0 | 0 | x | x | x | |||||
| Q14498 | RBM39 | RNA-binding protein 39 | 2 | + + + | + | 0 | 0 | ||||||||
| Q1KMD3 | HNRNPUL2 | Heterogeneous nuclear ribonucleoprotein U-like protein 2 | 2 | + + + | + | 0 | 0 | ||||||||
| Q93009 | USP7 | Ubiquitin carboxyl-terminal hydrolase 7 | 2 | + + + | + | 0 | 0 | ||||||||
| Q9BQ04 | RBM4B | RNA-binding protein 4B | 2 | + + + | + | 0 | 0 | ||||||||
| Q9H074 | PAIP1 | Polyadenylate-binding protein-interacting protein 1 | 2 | + + + | + | 0 | 0 | x | x | ||||||
| P35908 | * KRT2 | [ Keratin, type II cytoskeletal 2 epidermal ] | 41 | + + + | + + + | ||||||||||
| O15523 | * DDX3Y | Cluster of ATP-dependent RNA helicase DDX3Y | 4 | + + + | |||||||||||
| P13647 | * KRT5 | [ Keratin, type II cytoskeletal 5 ] | 2 | + + + | |||||||||||
Proteins listed were detected in either regular IP or crosslinking IP. The proteins were detected by either manual analysis or SAINTexpress algorithm. Candidate RBM45 protein interactors are identified using thresholding analysis and probabilistic scoring of associations (SAINTexpress). Proteins are sorted by decreasing AvgP, higher values predict high likelihood of interaction. Fold-changes are computed from positive purifications against negative control purifications (empty vector) are categorized as follows: + 2.5 – 100, + + 101 – 1000, + + + >1000. Fold-change values are provided in the Supplemental Material section. Associations with top five canonical pathways are highlighted (EIF2 Signaling (EIF2), Regulation of eIF4 and p7056K Signaling (eIF4/p7056K), mTOR Signaling (mTOR), Telomere Extension by Telomerase (Telomere Ext.), and RAN Signaling (RAN)). Starred proteins were only identified in the thresholding analysis. Bracketed proteins were identified as putative contaminants in the CRAPome database (v1.1). Bolded proteins highlight interactions verified by immunoblot in the current or in the previous study (Li et al., 2015).
Gene = manual analysis only (all others were found in both methods)
[ Description ] = putative contaminant
Bold = Externally Validated
“+” = fold change 2.5 – 100
“+ +” = fold change 101 – 1000
“+ + +” = fold change > 1000
2.2. Validation of select RBM45 PPIs
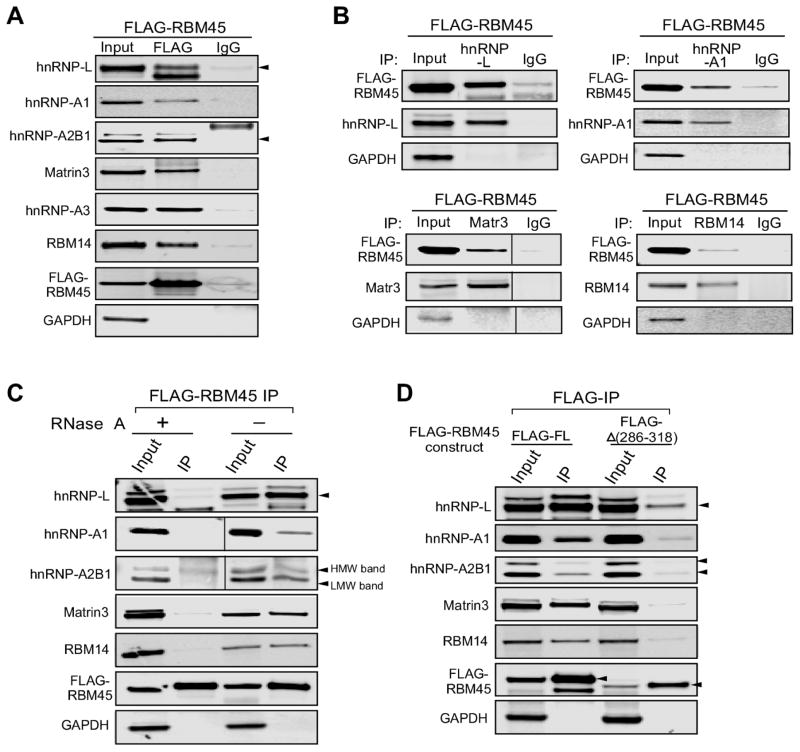
We have previously demonstrated that FLAG-RBM45 associates with ALS-linked proteins TDP-43 and FUS in HEK293 cells by immunoblot (Li et al., 2015). As expected, both TDP-43 and FUS were detected in the current IP-MS study (Table 1 and Supplemental Table 1). We next confirmed specific interactions of several identified candidate proteins with RBM45 by co-immunoprecipitation (co-IP) followed by immunoblot (Fig. 2A): hnRNP-L, hnRNP-A1, hnRNP-A2B1, Matrin-3, hnRNP-A3, and RBM14. All of these candidate proteins were in the top 20% highest interaction probability and abundance (spectral counts), and identified in both the regular and crosslinking IP experiments (Table 1). We stably expressed FLAG-RBM45 in HEK293 cells and performed co-IP from whole cell lysates. To detect transient or weak interactions, the cells were treated with formaldehyde to cross-link associated proteins prior to co-IP analysis. Anti-FLAG co-IP experiments demonstrated that all the proteins tested co-purified with FLAG-RBM45 but not with IgG (Fig. 2A). GAPDH, which was not identified by mass spectrometry, was used as negative control to further demonstrate the specificity of the observed interactions. As expected, we failed to detect association of GAPDH and FLAG-RBM45 (Fig. 2A, bottom). Moreover, we performed reciprocal co-IP assays using whole cell lysate from HEK293 cells expressing FLAG-RBM45 and antibodies against the selected candidate proteins. The reciprocal co-IP analysis demonstrated that FLAG-RBM45 co-purified with endogenous hnRNP-L, hnRNP-A1, Matrin-3 and RBM14 proteins (Fig. 2B). Taken together, these data provide evidence of the validity of the IP-MS approach and confirm specific interactions of selected candidate proteins with RBM45.
Figure 2.
Verification of RBM45 interacting proteins. (A) Pull-down of selected proteins with FLAG-RBM45 expressed in HEK293 cells. Crosslinking immunoprecipitations with FLAG antibody or IgG were performed. The IP fractions were immunoblotted with hnRNP-L, hnRNP-A1, hnRNP-A2B1, Matrin-3, hnRNP-A3 and RBM14 antibodies. The same IP fractions were also immunobloted with FLAG (FLAG-RBM45) and GAPDH (negative IP control) antibodies. (B) Immunoprecipitations of endogenous candidate proteins in FLAG-RBM45 expressing cells shows that FLAG-RBM45 co-purified with the tested endogenous proteins. The endogenous candidate proteins were immunoprecipitated with hnRNP-L, hnRNP-A1, Matrin-3 or RBM14 antibody, while IgG pull-down was used for IP control. The immunoblots were detected with FLAG antibody (FLAG-RBM45), tested endogenous protein specific antibodies, and GAPDH antibody (negative IP control). Similar validation studies for TDP-43 and FUS were previously reported (Li et al., 2015). (C) In-cell RNase treatment and crosslinking-IP were performed on cells expressing FLAG-RBM45. The amount of hnRNP-L, hnRNP-A1, the lower-molecular-weight band of hnRNP-A2B1, Matrin-3 and RBM14 that co-purified with FLAG-RBM45 was reduced upon the RNase treatment. (D) The RBM45-(Δ286-318) construct, i.e. the homo-oligomerization assembly (HOA) domain deficient construct (Li et al., 2015), exhibits significantly reduced binding to the tested candidate proteins when compared to full-length-RBM45. Full-length FLAG-RBM45 or FLAG-Δ(286-318) construct were expressed in HEK293 cells. FLAG-IP was performed as described previously. Immunoblot analysis shows that all the tested candidate proteins displayed reduced co-IP% with FLAG-Δ(286-318) as compared with the full-length FLAG-RBM45.
Since RBM45 contains three RRM domains, it may associate with its interacting proteins through RNA-protein interactions. To determine if associations between RBM45 and the previously tested proteins are RNA-dependent, we used in-cell RNase treatment prior to the cross-linking and anti-FLAG IP (Li et al., 2015). The in-cell RNase treatment significantly reduced the amounts of the hnRNP-L, hnRNP-A1, Matrin-3 and RBM14 proteins that co-purified with FLAG-RBM45 (Fig. 2C). Interestingly, the amount of the lower-molecular-weight species (arrows, Fig. 2C) of the hnRNP-A2B1 protein that was co-purified with FLAG-RBM45 reduced upon RNase treatment. However, the co-purified amount of the higher-molecular-weight species of hnRNP-A2B1 was not affected by RNase treatment (Fig. 2C). This result suggests that many RBM45 PPIs are RNA-dependent.
2.3. RBM45 homo-oligomerization mediates association with a large number of proteins
We previously reported that the RBM45 homo-oligomer assembly (HOA) domain mediates association with TDP-43 and FUS (Li et al., 2015). We hypothesize that the HOA domain serves as a general protein-protein interaction mediator. To test this hypothesis, we expressed FLAG-tagged constructs of either the full-length RBM45 or the Δ(286-318) construct with the majority of the HOA domain deleted and incapable of homo-oligomerization (Li et al., 2015). Anti-FLAG co-IP analysis showed that the tested candidate proteins co-purified efficiently with the FLAG-full-length RBM45. However, the FLAG-Δ(286-318) construct exhibited significantly reduced binding to all the tested candidate proteins (Fig. 2D). These results suggest that the HOA domain is an important mediator of RBM45 PPIs and that homo-oligomerization of RBM45 is required for many RBM45 PPIs.
2.4. Gene ontology and pathway analysis
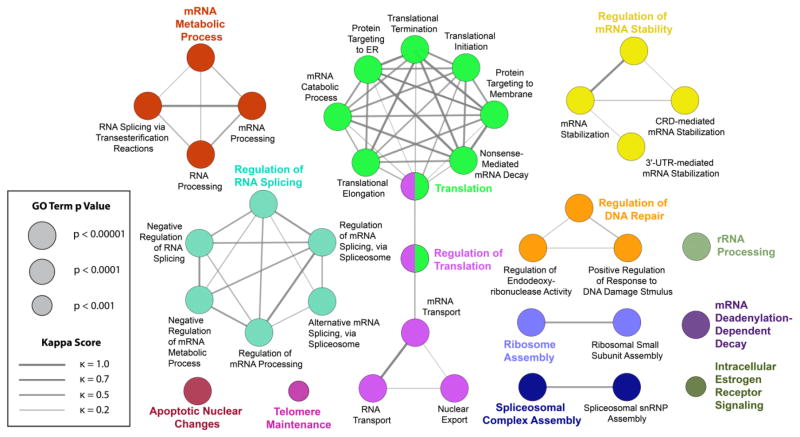
To identify putative biological processes associated with RBM45-interacting proteins, we performed enrichment analysis in the Gene Ontology (GO) domain “Biological Process” (Fig. 3). The results of this analysis identified two predominant themes: (1) nuclear RNA processing and (2) cytoplasmic RNA translation. RNA processing terms were chiefly related to splicing (e.g., “regulation of RNA splicing”, “alternative mRNA splicing”). Other nuclear RNA-associated terms included “mRNA transport”, “regulation of mRNA stability”, and “nuclear export”. Cytoplasmic translational themes were more diverse and included events directly to mRNA translation (“translation initiation”, “translation termination”), as well as downstream processing events (“protein targeting to ER”, “nonsense mediated mRNA decay”). Finally, terms unrelated to these phenomena and unconnected to any nodes included “apoptotic nuclear changes” and “telomere maintenance” (Fig. 3).
Figure 3.
Enriched GO Biological Process Terms. RBM45-interacting proteins were tested for GO Biological Process enrichment using the right-sided hypergeometric test with Benjamini-Hochberg post-hoc p value correction. Terms with a p value of 0.001 or less were visualized in a network layout, where node size corresponds to term p value. The proportion of shared proteins between terms was evaluated using the kappa statistic and nodes with a kappa score (κ) of at least 0.4 were connected with edges on the graph, with edge width proportional to kappa score. Leading terms, those terms with the highest number of proteins, are colored for emphasis.
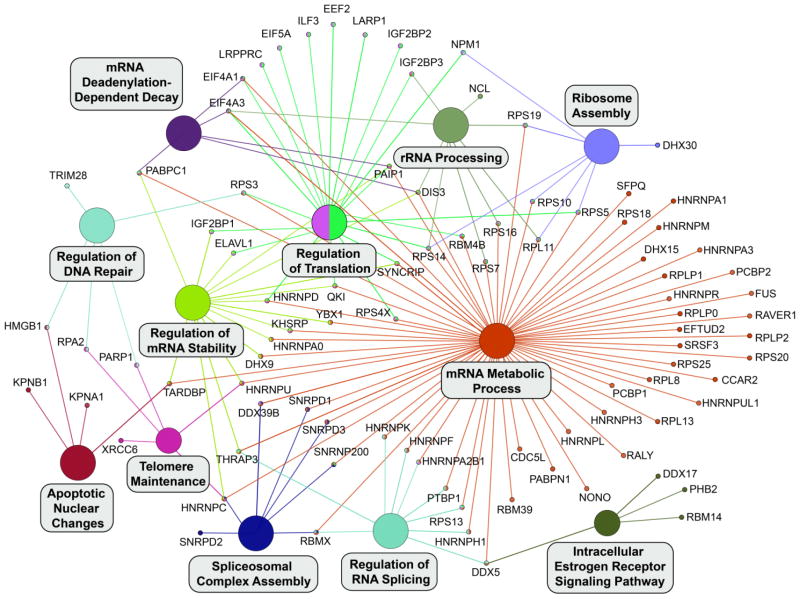
To provide further insights into the biological processes identified by this approach, we took leading terms, those terms with the highest number of associated proteins, from our results and visualized these terms with their associated proteins in a network layout where edges connect proteins to an associated biological process (Fig. 4). The results show the individual proteins that result in the identification of an enriched biological process. For example, the identification of the “mRNA metabolic process” and “regulation of RNA splicing” terms results in large part from the many hnRNP proteins in our list of RBM45-interacting proteins. Conversely, the enrichment for “regulation of translation” results from the presence of initiation and elongation factors (e.g., eIF proteins) in our list of RBM45-interacting proteins (Fig. 4).
Figure 4.
Leading Terms with Associated Proteins. Leading terms from Figure 3 were placed into a separate network and all associated proteins from the list of RBM45-interacting proteins were visualized as nodes and connected to the appropriate term. Where a protein is associated with multiple terms, multiple edges emanate from that protein and edges are color-matched to their associated terms.
Major canonical pathways associated with RBM45-interacting proteins were identified using Ingenuity Pathway Analysis (IPA®, QIAGEN Redwood City). Out of a total of 103 pathways, 28 were significantly enriched (p-value lower than 0.05). The top 5 pathways, ranked by significance and percent overlap are “EIF2 Signaling”, “Regulation of eIF4 and p70S6K Signaling”, “mTOR Signaling”, “Telomere Extension by Telomerase”, and “RAN Signaling” (Table 1 and Supplemental Table 7). These results were consistent with associations found in the gene ontology analysis. Collectively, this view emphasizes the diverse array of biological functions served by RBM45-interacting proteins.
2.5. Co-localization analysis
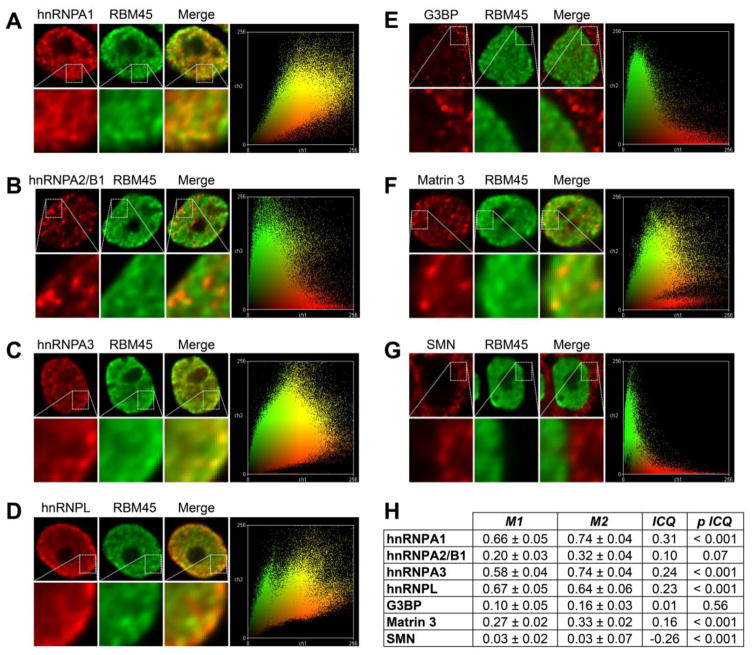
To assess the association of RBM45 and selected interacting proteins in cells, we used immunocytochemistry of our FLAG-RBM45 stable HEK293 cells together with digital deconvolution and co-localization analysis. The results of this analysis are shown in Figure 5 and Supplemental Figure 3. Because the staining we observe for the majority of the proteins analyzed is predominantly nuclear (Supplemental Fig. 3), multiple methods were used to provide a quantitative measure of the extent of co-localization. We thus analyzed the co-localization of FLAG-RBM45 and selected proteins using Manders coefficients (Bolte and Cordelieres, 2006) and the intensity correlation quotient (ICQ) (Li et al., 2004), together with pixel intensity scatter plots. The ICQ evaluates the co-variation of pixel intensities for each protein and provides a correlation-based metric (the ICQ, range −0.5 to 0.5) that reflects the degree to which protein staining intensities vary in synchrony and associated statistical significance. If staining intensities vary in synchrony (co-localization), the ICQ is large, positive, and statistically significant. For proteins with subcellular segregation, the ICQ is large, negative, and statistically significant, while for random variations in intensity, the ICQ = ~0, p > 0.05.
Figure 5.
Co-localization analysis. (A–G) The co-localization of RBM45 and the indicated proteins were evaluated using immunocytochemistry together with image deconvolution and co-localization analysis. Representative images and pixel intensity scatter plots are shown with cutouts at higher magnification to highlight detail. (H) Statistical analysis of protein co-localization. M1 = RBM45 overlap with indicated protein. M2 = indicated protein overlap with RBM45. ICQ = intensity correlation quotient. p ICQ = p value of ICQ. Manders coefficients (M1 and M2) measure the proportion of co-localizing proteins in each channel of a two-channel image and are shown as mean ± SEM. The intensity correlation quotient (ICQ) has a range of −0.5 (perfect segregation) to 0.5 (perfect co-localization), with random intensity variation resulting in a value ~0. The statistical significance of each ICQ value is shown at far right. SMN staining was used as a negative control.
The results of this analysis are shown in Figure 5B. We used SMN as a negative control, as the staining for this protein is predominantly cytoplasmic. As shown in Figure 5B, by either measure of co-localization, the association between RBM45 and SMN is low, reflecting subcellular segregation, as anticipated. We then evaluated the extent of co-localization between RBM45 and several RBM45-interacting proteins. FLAG-RBM45 staining was exclusively nuclear and we evaluated the co-localization of RBM45 with several nuclear hnRNP proteins. By both methods, the highest degree of co-localization was observed between RBM45 and hnRNP-A1 (Fig. 5H). RBM45 also exhibited statistically significant co-localization with hnRNP-A3, hnRNP-L, and Matrin 3, in descending order of extent of co-localization (Fig. 5B). By contrast, RBM45 co-localization with hnRNP-A2/B1 by either approach was lesser and did not reach statistical significance, despite a nuclear localization for both proteins (Fig. 5A). This finding highlights the utility of digital deconvolution and quantitative co-localization measures for assessing the true extent of association between proteins by immunocytochemistry. We also observed a lack of co-localization between RBM45 and G3BP, the latter of which was predominantly cytoplasmic. The absence of statistically significant co-localization between RBM45 and hnRNP-A2/B1 and G3BP may reflect the absence of required stimuli/signaling events necessary for the interaction of these proteins. The association of RBM45 and G3BP, for example, most likely occurs in cytoplasmic stress granules that not observed under basal conditions (Li et al., 2015).
3. Discussion
We used IP-MS to identify RBM45 PPIs and gain insight into the biological functions of this ALS/FTLD-associated RNA-binding protein (RBP) in HEK293 cells. By employing two complementary IP methods, regular IP and formaldehyde crosslinking-IP, we detected 132 RBM45 PPIs with high confidence. Our ability to identify numerous RBM45 PPIs with high confidence was a result of our stringent IP-MS approach. We identified 132 “true” interactors along with another 6 proteins matched to putative contaminants in the CRAPome database (Mellacheruvu et al., 2013). Triplicate IPs were analyzed by mass spectrometry. Identified proteins were subjected to a manual thresholding approach (resulting in 132 hits) and a probabilistic approach (resulting in 131 hits) to remove non-specifically bound proteins and predict putative PPIs. The resulting candidate proteins overlapped at 98.9%, highlighting the robustness of analytical method. RBPs were the most prominent protein family identified by our analytical approach, both in overall number of proteins and individual protein spectral counts. Taking the list of RBM45 PPIs, we next used enrichment and pathway analysis to link RBM45 PPIs to putative biological functions and pathways. The results showed enrichment for nuclear RNA processing via hnRNPs and cytoplasmic translation functions via eiF2 and eiF4 pathways. Taken together, these results provide new insights into the PPIs, biological functions, and roles in ALS/FTLD of RBM45.
These insights are necessary to further understand the role of RBM45 (and RBPs more generally) in ALS/FTLD. RBM45 is a component of ubiquitinated inclusions in neurons and glial cells in ALS, FTLD, and AD patients (Collins et al., 2012). The mechanisms mediating the protein’s incorporation into inclusions are poorly understood, however. RBM45 is distinct from other inclusion forming RBPs, such as TDP-43, FUS, TAF15, and hnRNP-A1, in that it does not possess a prion-like domain (King et al., 2012). RBM45 does, however, contain a homo-oligomerization (HOA) domain that mediates RBM45 self-association and association with other RBPs, including TDP-43 and FUS, suggesting a role for this domain in RBM45 inclusion formation (Li et al., 2015). Consistent with this notion, we identify numerous inclusion-forming RBPs that bind to RBM45 via our IP-MS approach, including hnRNP-A1, hnRNP-A2/B1, TDP-43 and FUS (reviewed in (Peters et al., 2015). For several of these proteins, the HOA domain is requisite for interaction (Fig. 2D). Thus, while the HOA domain is likely necessary for normal RBM45 functions, its role in mediating RBM45 oligomerization and association with other RBPs suggests this domain also contributes to the pathological aggregation of RBM45 and other RBPs in ALS/FTLD. The presence of prion-like domains in many RBM45 interacting proteins and the lack of a prion domain in RBM45 also suggests that RBM45 aggregation may be driven by its association with other aggregation-prone RBPs, as has been observed for RBPs such as PSF and NONO found in TDP-43/FUS positive aggregates (Dammer et al., 2012; Shelkovnikova et al., 2014). Matrin 3 is a nuclear matrix protein implicated in binding and stabilizing mRNA and matrin 3 mutations have been linked to ALS (Johnson et al., 2014). While matrin 3 has not been associated with cytoplasmic inclusions in ALS, interactions between RBM45 and matrin 3 within the nucleus may contribute to the regulation of mRNA stability and transport within the nucleus. The identification of numerous ALS-associated proteins within our RBM45 PPI list suggests that RBM45 can directly contribute to disease by virtue of its association with these proteins.
The aggregation of RBPs in ALS/FTLD confers toxicity both by aggregation-induced toxic gain of function as well as aggregation-induced loss of normal RBP function. Thus, understanding the normal functions of RBPs is critical to identifying molecular mechanisms of disease and potential therapeutic targets. RBPs are typically multifunctional and act in both the nucleus and cytoplasm, influencing transcription, RNA splicing, RNA export, translation, and transport of mRNAs (Dreyfuss et al., 2002). Interestingly, RBM45 associates with many of the validated binding proteins via RNA-mediated interactions (Fig. 2), suggesting that RBM45 and its binding proteins share the regulation of specific RNA targets. We used our list of RBM45-interacting proteins to generate a list of putative RBM45 biological functions and associated pathways using Gene Ontology and pathway analysis. Two major themes emerged: nuclear RNA processing/splicing via hnRNPs and cytoplasmic translation via the eiF2 and eiF4 pathways (Figs. 3 and 4; Supplemental Table 7). The many splicing-associated proteins in our list (Fig. 4) suggest a role for RBM45 in the regulation of splicing events. Dysregulation of RNA splicing is a well-characterized phenomenon in ALS/FTLD and can result from RBP cytoplasmic mis-localization, aggregation, or both (Walsh et al., 2015). Loss of individual RBP function due to these phenomena can have profound effects on transcriptional regulation. For example, TDP-43 and FUS bind to more than 50% of the human transcriptome and the loss of these proteins results in substantial global alterations in transcription and splicing (Lagier-Tourenne et al., 2012; Polymenidou et al., 2011; Tollervey et al., 2011). We anticipate that future studies directly examining the role of RBM45 in the regulation of transcription and RNA splicing will likewise reveal widespread RBM45 binding across the transcriptome and substantial influence on mRNA splicing decisions.
In further support of this notion, the identification of RBM45 PPIs with 19 members of the hnRNP family suggests considerable functional overlap between RBM45 and this diverse class of proteins. Spectral count values for many of these proteins were among the highest observed in our study (Table 1) and we accordingly predict considerable functional overlap between RBM45 and the hnRNP family. hnRNPs participate in a variety of mRNA processing/maturation processes, including mRNA maturation, splicing, nuclear export, and 3′-end processing (Kim and Dreyfuss, 2001). Abnormalities in the expression/function of hnRNPs are associated with a number of human diseases, including ALS by virtue of the recent demonstration that mutations in the prion domains of hnRNP-A2/B1 and hnRNP-A1 cause familial forms of ALS (Kim et al., 2013). Our analysis of the co-localization of RBM45 and these proteins demonstrates that RBM45 co-localizes most highly with hnRNP-A1, followed by hnRNP-A3 and hnRNP-L, with low, non-significant co-localization observed with hnRNP-A2/B1 (Fig. 5).
The association of RBM45 with hnRNP-A1, together with the aggregation-prone prion-like domain of hnRNP-A1, may thus mediate both the function and aggregation of RBM45. We observe a high degree of nuclear co-localization between these proteins (Fig. 5) and confirmed their physical, RNA-dependent interaction via co-immunoprecipitation (Fig. 2C). hnRNP-A1 serves many purposes in the nucleus, including regulating the transcription of numerous genes (Jean-Philippe et al., 2013). Transcriptional regulation by hnRNP-A1 is, in part, conferred by its ability to bind and relax G-quadruplex nucleic acid structures, including the fALS-linked c9ORF72 GGGGCC hexanucleotide repeat expansion (Cooper-Knock et al., 2014; Fukuda et al., 2002). RBM45 may thus be sequestered to c9ORF72 repeat expansion G-quadruplex structures in c9-linked fALS cases, causing a loss of normal RBM45 functions. Indeed, we identify numerous c9ORF72 repeat expansion binding proteins, including FUS, ELAVL1, hnRNP-K, hnRNP-L, hnRNP-Q, and hnRNP-U, as RBM45 PPIs (Table 1) (Cooper-Knock et al., 2014; Mori et al., 2013). Despite its high affinity for poly(G)/(C) RNA (Tamada et al., 2002), RBM45 binding to c9ORF72 has not been shown, although discrepancies between experimental approaches and results suggest that additional c9-binding RBPs remain as yet unidentified (Cooper-Knock et al., 2014; Mori et al., 2013).
We also found significant co-localization of RBM45 with hnRNP-L and hnRNP-A3 in the nucleus (Fig. 5). hnRNP-L is a multifunctional protein that regulates transcript splicing (Hui et al., 2003b), stability (Hui et al., 2003a), and translation (Majumder et al., 2009). The protein affects splice site decisions for a large number of transcripts and is capable of inhibiting spliceosome assembly via coordinated action with hnRNP-A1 (Chiou et al., 2013; Hung et al., 2008). These results, together with their RNA-dependent physical interaction (Fig. 2C) and co-localization of RBM45 with these proteins (Fig. 5), provides further evidence of a role for RBM45 in mRNA splicing decisions. hnRNP-A3 is involved in the nucleocytoplasmic trafficking of mRNA (Ma et al., 2002) and is involved in telomere maintenance and protection by virtue of its direct binding to telomeres (Huang et al., 2010; Tanaka et al., 2007). The protein is also a component of p62 positive/TDP-43 negative inclusions in c9ORF72-linked fALS motor neurons (Mori et al., 2013). hnRNP-A3 is a component mRNP complexes that act to stabilize mRNA (Papadopoulou et al., 2012). The co-localization of RBM45 and hnRNP-A3 within distinct nuclear foci (Fig. 5, Supplemental Fig. 3) suggests a possible role for RBM45 in this process as well.
A variety of cytoplasmic RBP functions also contribute to cellular function and studies have repeatedly shown that loss of these functions negatively impact cellular viability. TDP-43, for example, associates with cytoplasmic stress granules (Colombrita et al., 2009), regulates local mRNA translation (Wang et al., 2008), and participates in RNA transport (Narayanan et al., 2013). Our results likewise suggest important cytoplasmic functions for RBM45 in both normal cellular homeostasis and disease. We identified RNA transport as a biological process putatively regulated by RBM45 (Fig. 3). The interaction of RBM45 with ELAVL1, a known RNA transport protein (Kraushar et al., 2014), is consistent with a role for RBM45 in the transport of mRNA and local translation (Fig. 4). We also identified enrichment in numerous biological processes directly and indirectly related to cytoplasmic translation. A direct role for RBM45 in translation is predicted from the identification of numerous elongation and initiation factors (e.g., eiF4a, eiF5A, EEF2, … [Fig. 4]) as RBM45 interactors. Twelve percent of the eiF2 signaling pathway responsible for charged tRNA delivery to the ribosome and start site recognition is mapped by PPIs with RBM45, highlighting a possible role of RBM45 in early translational events (Supplemental Table 7). Indirect contributions to translation included the GO biological process “Protein Targeting to ER” (Fig. 3). ER stress occurs in ALS (Lautenschlaeger et al., 2012) and RNA-binding proteins may directly associate with ER to modulate its functions in certain cell/tissue types (Gautrey et al., 2005). Despite these findings, immunocytochemical analysis shows an exclusively nuclear staining pattern for RBM45 in HEK293 cells (Supplemental Fig. 3). However we hypothesize that RBM45 can mediate nuclear mRNA export via its association with nucleocytoplasmic shuttling hnRNP’s, such as hnRNP-A1, hnRNP-L, and hnRNP-K (Kim et al., 2000).
One limitation of the current approach is that our analyses were performed exclusively in HEK293 cells. HEK293 cells have a unique gene expression profile, rapidly divide, and have an unstable karyotype. Each of these properties could influence the list of RBM45 PPIs detected in the present work. Future studies are necessary to determine cell-type and phenotype-specific RBM45 PPIs and how these contribute to cellular physiology. One area of particular interest is the role of RBM45 in cell division and cell type specification. The initial characterization of RBM45 demonstrated developmental regulation and neuronal enrichment of RBM45 expression, suggesting that RBM45 and, by extension, RBM45 PPIs contribute to cell division and organismal development. Delineating which RBM45 PPIs occur in differentiated cell populations, such as neurons, may likewise yield insight into RBM45 PPIs and cellular functions that lead to its incorporation into inclusions in ALS/FTLD. While stress is known to induce cytoplasmic stress granules to modulate translation, chronic stress has been proposed to induce the generation of cytoplasmic inclusions from stress granules (Wolozin and Apicco, 2015). Further studies examining the RBM45 protein complexes under stress conditions may identify biological pathways relevant to the induction of RBM45 aggregation and inclusion formation.
Finally, we used multiple immunoprecipitation methods coupled with mass spectrometry to increase the confidence of our results. Two different tagged RBM45 constructs as well as the presence or absence of a formaldehyde crosslinking method were used for immunoprecipitation. We used a combination of cross-linking and regular IP to distinguish weak and strong interactions, respectively. While commonly used to identify PPIs, regular IP may also yield non-physiological protein associations resulting from artefactual, non-specific binding after cell lysis (Mili and Steitz, 2004). Formaldehyde is a mild, cell-permeable and reversible crosslinker with very short spacer length (2.3–2.7 Å) and cross-links only closely associated proteins (Klockenbusch and Kast, 2010). In vivo formaldehyde crosslinking-IP can help reduce interaction artifacts introduced after cell lysis and help preserve transient and/or weak protein-protein interactions and has been used for discovering novel protein-protein interactions in many proteomics studies (Corgiat et al., 2014; Klockenbusch and Kast, 2010; Miernyk and Thelen, 2008; Sutherland et al., 2008; Zhang et al., 2009). Crosslinking-IP also facilitates stringent immunoprecipitations via increased detergent concentration, sonication and extensive washes. Identification of a protein only in crosslinking-IP experiments suggests that the interaction with RBM45 is weak. However, one cannot predict the biologic significance of the interaction with RBM45 based solely in whether the interaction is strong or weak. Of the identified 132 proteins, 68 proteins were found solely in crosslinking-IP, while only 28 proteins were found exclusively in regular IP. It is possible that the protein-binding sites in these 28 proteins were masked by the crosslinking reaction and thus not detected by crosslinking-IP.
Collectively, our results demonstrate that RBM45 associates with a large and functionally diverse set of protein binding partners. Functions served by these proteins, particularly the hnRNPs, suggest plausible and previously unknown biological functions for RBM45. The identification of these functions and the association of RBM45 with numerous ALS-associated RBPs points to RBM45-mediated mechanisms of disease in ALS/FTLD and provides further insight into the pathological aggregation of RBM45 occurring in neurodegenerative disease. The association of RBM45 with the set of proteins identified herein provides new directions for future studies of RBM45’s role in neuronal development, the regulation of gene expression, and neurodegeneration.
4. Experimental Procedure
4.1. Cell culture and plasmid construction
HEK293 (FreeStyle™ 293-F Cells, Invitrogen) cells were cultured in DMEM medium with 10% FBS and 1% Pen-Strep at 37°C with 5% CO2. Transfection was performed using the Lipofectamine 2000 (Life technologies) and stable cell lines were selected in the presence of 500 μg/ml G418 (Life Technologies) 48 hours post-transfection. The RBM45 cDNA clone plasmid, cGST-hRBM45 (HsCD00356971), was obtained from the DNASU Plasmid Repository at Arizona State University, Tempe. The cDNA was amplified by PCR using Phusion High-Fidelity DNA Polymerase (NEB) and sub-cloned into the pcDNA3 vector (Invitrogen). The 3xFLAG tag (DYKDHDGDYKDHDIDYKDDDDK) or 2xHA tag (DYPYDVPDYAGGAAYPYDVPDYA) was appended to the N-terminus of specific proteins to generate the 3xFLAG- or 2xHA-tagged construct.
4.2. LC-MS/MS protein identification
4.2.1. Immunoprecipitation
Each immunoprecipitation (IP) was carried out in triplicate. Stable cell lines expressing FLAG-RBM45, HA-RBM45, or pcDNA3 vector were grown on 10cm plates till 90% confluent and harvested. For regular IP, cells from one 10cm plate were lysed with 500 μl of 0.5% NP40 lysis buffer (50 mM HEPES pH 7.6, 150 mM KCl, 2 mM EDTA, 0.5% NP40, 0.5 mM DTT and protease (Sigma P8340)/phosphatase (Calbiochem 524629)/RNase inhibitors (Ambion AM2694)) at 4°C for 15 min. For formaldehyde crosslinking-IP, formaldehyde in-cell crosslinking was performed prior to IP as previously reported (Li et al., 2015). Cells from one 10cm plate were suspended in 1 ml PBS containing 0.1% formaldehyde and incubated at room temperature for 7 min with gentle agitation. The suspension was spun for 3 min at 1,800 g at room temperature and the supernatant was discarded. The pellet was washed with 1 ml 1.25 M glycine in cold PBS twice to quench the crosslinking reaction. The pellet was further washed in PBS, lysed with 500 μl of 1% NP40 lysis buffer (50 mM HEPES pH 7.6, 150 mM KCl, 2 mM EDTA, 1% NP-40, 0.5 mM DTT and protease/phosphatase inhibitors) at 4°C and sonicated in a water bath sonicator (Misonix Sonicator 3000) at level 2 for 4 cycles (15 sec on/30 sec off).
The lysates were first cleared by spinning at 16,000 g at 4°C for 15 min to remove cell debris, pre-cleared using IgG-Agarose (Sigma A0919) for 1 hour and further centrifugated. 3 μg of total protein was used for immunoprecipitation with 50 μg of either pre-crosslinked antibody or IgG. FLAG-IP was performed using anti-FLAG M2 Affinity Gel (Sigma A2220), HA-IP was performed using anti-HA Agarose (Sigma A2095), and IgG-IP control was performed using Mouse IgG-Agarose (Sigma A0919). IPs were performed at 4°C for 2 hr and the beads were washed six times in IP buffer. The proteins were eluted with SDS sample buffer and heated at 95°C for 5 min for regular IP samples and heated for 20 min for formaldehyde crosslinking IP samples. The samples were then run on the Bolt 4–12% Bis-Tris Plus Gel (Life Technologies), and stained using Bio-Safe Coomassie Stain (BioRad).
4.2.2. Protein digestion
Gel lanes in the molecular weight range between 10 kDa and greater than 250 kDa were excised into individual fractions, excluding the stained IgG-H (52kDa) and IgG-L (25kDa) bands. Bands fractions were then further reduced into cubes of 1–2mm3, destained, washed, dried and further processed using an established method (Shevchenko et al., 2006). Briefly, each fraction was reduced using 10mM DTT (6°C for 30 min) and alkylated using 55mM iodoacetamide (room temperature for 30 min, in the dark), using multiple hydration and dehydration cycles of the acrylamide gel. Fractions were then digested using 20 ng/mL of Trypsin Gold (Promega) (37°C, overnight). Finally, peptides were extracted, concentrated to dryness under vacuum and stored at −20°C until LC-MS analysis.
4.2.3. LC-MS analysis
Each fraction was reconstituted in 0.1% formic acid and analyzed using online liquid chromatography on a nanoAcquity-UPLC coupled to a Thermo LTQ Orbitrap Velos mass-spectrometry. Samples were loaded onto a 100-μm diameter column (length 100 mm) packed with 3 μm Reprosil Pur C18 AQ resin. Solvent A and B were 0.1% formic acid in water and acetonitrile, respectively. The gradient was 3% B to 40% B in 17 min followed by 40% B to 90% B in 0.5 min, then 90% B for 2 min and final re-equilibration for 10.5 min. The flow rate was set to 500 nL/min The mass spectrometer was operated in positive ion mode using a spray voltage of 1.8 kV, and a capillary temperature of 200°C. Data were acquired in top-15, data-dependent acquisition mode using a collision voltage of 30 V.
4.3. Protein identification
Mass spectra were extracted, deconvolved and deisotoped using Proteome Discoverer 1.4.1.14 (Thermo Fisher Scientific, Waltham, MA) and searched against a concatenated database (Homo sapiens, Mus musculus, UniprotKB/Swissprot) using Mascot (Matrix Science, London, UK; version 1.4.1.14). Oxidation (Met), carbamidomethylation (Cys) were specified as variable modifications. Peptides were allowed maximum two trypsin missed cleavages with a mass tolerance of ±10 ppm, and a fragment ion mass tolerance of ±0.8 Da. Search results were imported into Scaffold (Proteome Software Inc., Portland, OR), and identifications were confirmed by X!Tandem (The GPM, v2010.12.01.1). Only proteins with probabilities equal or higher than 99.0% were retained for analysis (one or more peptide per protein contributing to a positive match). Computation of putative PPIs (manual and SAINTexpress) were based on exclusive spectrum counts, as determined by Scaffold.
4.4. Bioinformatics, pathway analysis and gene ontology analysis
A combination of an unsupervised probabilistic approach (SAINTexpress, (Choi et al., 2012)) and a manual approach was used to identify proteins potentially interacting with RBM45. For each protein-protein interaction, SAINTexpress predicted an individual probability based on spectral counts and reported average probabilities across all replicates (AvgP), average fold-change, average spectral counts and a Bayesian False Discovery Rate (BFDR) (Teo et al., 2014). Empty vector IPs were used as experimental controls to provide a background list of proteins binding non-specifically to the construct. The interactions provided by SAINTexpress were filtered for protein fold change equal or greater than 2, for proteins observed in at least 2 out of 3 replicates and with an AvgP equal or greater than 0.7, as recommended (Choi et al., 2012).
For manual elucidation of candidate PPIs, only proteins observed in at least 2 out of 3 replicates were retained in RBM45 IPs. Fold-change was calculated as the sum of exclusive spectral counts across RBM45 replicates divided by the sum of the exclusive spectral counts of that protein in the vector control replicates. Any protein with a fold-change smaller than 2 was filtered out.
PPIs with the highest degree of confidence, e.g. valid across the unsupervised and manual approaches were then analyzed using Ingenuity Pathway Analysis software (IPA®, QIAGEN Redwood City). The default IPA parameters were utilized along with Uniprot identifiers for mapping proteins within IPA. The reference set for analysis was the Ingenuity Knowledge Base. Direct and indirect relationships were included but only from proteins that were experimentally observed. IPA mapped 127 out of 131 proteins to known pathways. P-value and percent overlap were used to rank potentially significant pathways.
To identify biological processes associated with the list of RBM45 interacting proteins, we performed enrichment analysis in the Gene Ontology (GO) Biological Process domain using Cytoscape (Shannon et al., 2003) together with the ClueGo plugin (Bindea et al., 2009). We performed enrichment analysis using the right-sided hypergeometric test with Benjamini-Hochberg post-hoc correction. GO terms were considered significant at the p < 0.001 level and the resultant significant terms were visualized in a network layout where GO Biological Process terms were visualized as color-coded circular nodes, with node size corresponding to enrichment p value. The overlap of proteins associated with any two Biological Process terms was evaluated using the kappa statistic and nodes were connected where the κ value was ≥ 0.4 using edges, with edge thickness corresponding to kappa score. We then took leading terms, those GO Biological Process terms with the highest number of associated proteins, and visualized these in a network layout where Biological Process terms were connected by edges to their associated proteins. All final figures were assembled using Adobe Illustrator CS5 (Adobe Systems; San Jose, CA, USA).
4.5. Reciprocal immunoprecipitation
Cells were cultured and processed as described previously. In-cell RNase treatment was performed as described in (Li et al., 2015). 500 μg total protein and 2 μg antibody was first incubated at 4 °C for 1 hour, and the entire mixture was added to 15 μl Protein A/G Agarose (Pierce) and rotated at 4 °C for 3 hr. The immunoprecipitates were washed 4 times and analyzed for immunoblot. The antibodies used for immunoprecipitations are as follows: mouse monoclonal hnRNP-L antibody (Novus Biological NB120-6106), rabbit monoclonal hnRNP-A1 antibody (Cell Signaling 8443S), rabbit polyclonal Matrin-3 antibody (Abcam ab70336), rabbit polyclonal RBM14 antibody (Proteintech 10196-1-AP). IgG-IP control was performed using rabbit IgG (Sigma I5006) and mouse IgG (Sigma I5381).
4.6. Immunoblot
Protein samples were mixed with 4x SDS loading buffer and denatured by heating (95°C for 5 min for regular IP samples and 95°C for 20 min for crosslinking IP samples), resolved on the Bolt 4–12% Bis-Tris Plus Gel (Life Technologies), and transferred to Immobilon-FL PVDF membrane (Millipore). The membranes were blocked with Odyssey Blocking Buffer (LiCOR) for 1 hr. The antibodies were diluted in Odyssey Blocking Buffer with 0.1% Tween-20. Primary antibody incubation was performed at room temperature for 1 hr or 4°C overnight. The IRDye-conjugated secondary antibody (LiCOR) incubation was performed at room temperature for 1 hr. The membranes were scanned using the Odyssey CLx Infrared Imaging System (LiCOR). The primary antibodies used for immunoblot are as follows: mouse monoclonal FLAG M2 antibody (Sigma F3165, 1:5000), rabbit monoclonal RBM45 C-terminal antibody (custom-made, 1:3000), rabbit monoclonal hnRNP-A1 antibody (Cell Signaling 8443S, 1:3000), mouse monoclonal hnRNP-L antibody (Novus Biological NB120-6106, 1:10000), mouse monoclonal hnRNP-A2B1 antibody (Santa Cruz sc-32316, 1:3000), rabbit monoclonal Matrin-3 antibody (Abcam ab151714, 1:10000), rabbit polyclonal RBM14 antibody (Proteintech 10196-1-AP, 1:5000), rabbit polyclonal TDP-43 antibody (Proteintech 10782-2-AP, 1:3000), rabbit monoclonal GAPDH antibody (Cell Signaling 2118S, 1:5000). The appropriate secondary antibodies conjugated with LiCOR IRDye 800CW or IRDye 680RD antibodies made in goat (1:15000) were used for immunoblot experiments.
4.7. Immunocytochemistry
For immunocytochemistry, HEK293 cells were grown on number 1.5 glass coverslips. Cells were washed with 1X PBS and fixed in 4% paraformaldehyde for 10 min. After fixation and further washing, cells were permeabilized by immersion in 1X PBS containing 0.1% Triton X-100 for 15 min. After further washing, cells were blocked by incubation in SuperBlock (Scytek) for 1 hr. Subsequently, primary antibody solutions were applied and allowed to incubate for 2 hr. Following primary antibody incubations, coverslips were washed four times in a 1:10 mixture of SuperBlock:1X PBS. Secondary antibodies were applied following these washes, allowed to incubate for 1 hr, and washed four times as above. Cell nuclei were visualized by staining with a 300 nM DAPI solution for 10 min followed by washing with 1X PBS. Coverslips were mounted on glass slides using 2,2′-thiodiethanol (TDE) according to the method of (Staudt et al., 2007). In brief, coverslips were immersed in a series of increasing concentrations of TDE (10%, 25%, 50%, 97%). The final TDE solution has a refractive index of 1.518 to match that of the immersion oil used in imaging the slides.
The primary antibodies used for immunofluorescence were as follows: rabbit monoclonal RBM45 C-terminal antibody (custom-made, 1:250), rabbit monoclonal hnRNP-A1 antibody (Cell Signaling 8443S, 1:800), mouse monoclonal hnRNP-A2B1 antibody (Santa Cruz sc-32316, 1:250), rabbit polyclonal hnRNP-A3 antibody (Sigma AV41195, 1:200), mouse monoclonal hnRNP-L antibody (Novus Biological NB120-6106, 1:1000), rabbit monoclonal Matrin-3 antibody (Abcam ab151714, 1:500), mouse monoclonal G3BP antibody (BD Transduction Laboratories, 1:250), mouse monoclonal SMN antibody (Sigma S2944, 1:400), and mouse monoclonal FLAG M2 antibody (Sigma F3165, 1:1000). The secondary antibodies used for immunofluorescence were goat-anti-Cy2 (rabbit) and goat-anti-Cy5 (Mouse) (Millipore, 1:1000 for both).
4.8. Microscopy, digital deconvolution, and co-localization analysis
An Observer Z1 microscope (Zeiss) was used for all image acquisitions using a 63x (1.4 NA) objective and LED light source. Images were acquired as three-dimensional stacks with a Z sampling interval of 0.240 μm. Images were shading corrected and background subtracted. Following acquisition, images were deconvolved using Huygens Essential deconvolution software (SVI). Deconvolution and chromatic shift correction were performed using a measured PSF obtained by volume imaging of 200 μm fluorescent beads (Life Technologies) together with the Huygens Essential PSF Distiller application. Deconvolution was performed using the software’s classic maximum likelihood estimation algorithm. Deconvolved images were used to analyze the co-localization of RBM45 and selected RBM45 interacting proteins identified by IP-MS. Co-localization analysis was performed using ImageJ (Schneider et al., 2012) in conjunction with the JaCoP plugin (Bolte and Cordelieres, 2006). Images were automatically thresholded for analysis using the method of (Costes et al., 2004) and the M1 and M2 overlap coefficients (Bolte and Cordelieres, 2006) and intensity correlation quotient (ICQ) (Li et al., 2004) were calculated. Statistical significance of the ICQ was evaluated using the normal approximation of the sign test as in (Li et al., 2004).
Supplementary Material
Highlights.
The protein-protein interaction (PPI) network of RBM45 is characterized by IP-MS.
Many RBM45 PPIs are RNA-dependent and mediated by the homo-oligomerization domain.
Select PPIs were validated and interaction networks constructed.
RBM45 PPIs are enriched for nuclear RNA splicing and cytoplasmic translation pathways.
Several ALS-linked RNA-binding proteins physically interact with RBM45.
Acknowledgments
This work was supported by National Institutes of Health grants R01NS061867, R56NS061867, and R01NS068179 to RB, NIH grant F31NS080614 to MC, an award from the Achievement Rewards for College Scientists Foundation, Inc. Pittsburgh Chapter to MC, and the ALS Association Milton Safenowitz Post-Doctoral Fellowship for ALS Research to YL.
Appendix A
Supplemental material: Supplemental data associated with this article can be found in the online version.
Footnotes
Author contributions
YL and RB designed the project. YL and PP designed the IP-MS experiments. YL performed the validation IP-WB experiments. KG, PP and YL performed the mass spectrometry data analysis. MC performed the microscopy experiments and constructed the GO networks. JA, KT and RG helped with the IP-MS experiments. TT performed the LC-MS analysis. YL, MC, PP and RB wrote the manuscript.
Competing financial interests
RB is founder of Iron Horse Diagnostics, Inc., a biotechnology company focused on developing diagnostic and prognostic tests for neurologic diseases.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bakkar N, et al. RBM45 Modulates the Antioxidant Response in Amyotrophic Lateral Sclerosis through Interactions with KEAP1. Mol Cell Biol. 2015;35:2385–99. doi: 10.1128/MCB.00087-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindea G, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–3. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224:213–32. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
- Chiou NT, Shankarling G, Lynch KW. hnRNP L and hnRNP A1 induce extended U1 snRNA interactions with an exon to repress spliceosome assembly. Mol Cell. 2013;49:972–82. doi: 10.1016/j.molcel.2012.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H, et al. SAINT-MS1: protein-protein interaction scoring using label-free intensity data in affinity purification-mass spectrometry experiments. J Proteome Res. 2012;11:2619–24. doi: 10.1021/pr201185r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins M, et al. The RNA-binding motif 45 (RBM45) protein accumulates in inclusion bodies in amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration with TDP-43 inclusions (FTLD-TDP) patients. Acta Neuropathol. 2012;124:717–32. doi: 10.1007/s00401-012-1045-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombrita C, et al. TDP-43 is recruited to stress granules in conditions of oxidative insult. J Neurochem. 2009;111:1051–61. doi: 10.1111/j.1471-4159.2009.06383.x. [DOI] [PubMed] [Google Scholar]
- Cooper-Knock J, et al. Sequestration of multiple RNA recognition motif-containing proteins by C9orf72 repeat expansions. Brain. 2014;137:2040–51. doi: 10.1093/brain/awu120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corgiat BA, Nordman JC, Kabbani N. Chemical crosslinkers enhance detection of receptor interactomes. Front Pharmacol. 2014;4:171. doi: 10.3389/fphar.2013.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costes SV, et al. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys J. 2004;86:3993–4003. doi: 10.1529/biophysj.103.038422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammer EB, et al. Coaggregation of RNA-binding proteins in a model of TDP-43 proteinopathy with selective RGG motif methylation and a role for RRM1 ubiquitination. PLoS One. 2012;7:e38658. doi: 10.1371/journal.pone.0038658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- Fukuda H, et al. Unfolding of quadruplex structure in the G-rich strand of the minisatellite repeat by the binding protein UP1. Proc Natl Acad Sci U S A. 2002;99:12685–90. doi: 10.1073/pnas.152456899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautrey H, et al. Polarised distribution of the RNA-binding protein Staufen in differentiated intestinal epithelial cells. FEBS Lett. 2005;579:2226–30. doi: 10.1016/j.febslet.2005.02.074. [DOI] [PubMed] [Google Scholar]
- Gitler AD, Shorter J. RNA-binding proteins with prion-like domains in ALS and FTLD-U. Prion. 2011;5:179–87. doi: 10.4161/pri.5.3.17230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang PR, Hung SC, Wang TC. Telomeric DNA-binding activities of heterogeneous nuclear ribonucleoprotein A3 in vitro and in vivo. Biochim Biophys Acta. 2010;1803:1164–74. doi: 10.1016/j.bbamcr.2010.06.003. [DOI] [PubMed] [Google Scholar]
- Hui J, Reither G, Bindereif A. Novel functional role of CA repeats and hnRNP L in RNA stability. RNA. 2003a;9:931–6. doi: 10.1261/rna.5660803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui J, et al. HnRNP L stimulates splicing of the eNOS gene by binding to variable-length CA repeats. Nat Struct Biol. 2003b;10:33–7. doi: 10.1038/nsb875. [DOI] [PubMed] [Google Scholar]
- Hung LH, et al. Diverse roles of hnRNP L in mammalian mRNA processing: a combined microarray and RNAi analysis. RNA. 2008;14:284–96. doi: 10.1261/rna.725208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean-Philippe J, Paz S, Caputi M. hnRNP A1: the Swiss army knife of gene expression. Int J Mol Sci. 2013;14:18999–9024. doi: 10.3390/ijms140918999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BS, et al. TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J Biol Chem. 2009;284:20329–39. doi: 10.1074/jbc.M109.010264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JO, et al. Mutations in the Matrin 3 gene cause familial amyotrophic lateral sclerosis. Nat Neurosci. 2014;17:664–666. doi: 10.1038/nn.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495:467–73. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, et al. Protein-protein interaction among hnRNPs shuttling between nucleus and cytoplasm. J Mol Biol. 2000;298:395–405. doi: 10.1006/jmbi.2000.3687. [DOI] [PubMed] [Google Scholar]
- Kim VN, Dreyfuss G. Nuclear mRNA binding proteins couple pre-mRNA splicing and post-splicing events. Mol Cells. 2001;12:1–10. [PubMed] [Google Scholar]
- King OD, Gitler AD, Shorter J. The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease. Brain research. 2012;1462:61–80. doi: 10.1016/j.brainres.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klockenbusch C, Kast J. Optimization of formaldehyde cross-linking for protein interaction analysis of non-tagged integrin beta1. J Biomed Biotechnol. 2010;2010:927585. doi: 10.1155/2010/927585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraushar ML, et al. Temporally defined neocortical translation and polysome assembly are determined by the RNA-binding protein Hu antigen R. Proc Natl Acad Sci U S A. 2014;111:E3815–24. doi: 10.1073/pnas.1408305111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski TJ, Jr, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–8. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- Lagier-Tourenne C, et al. Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nature neuroscience. 2012;15:1488–97. doi: 10.1038/nn.3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lautenschlaeger J, Prell T, Grosskreutz J. Endoplasmic reticulum stress and the ER mitochondrial calcium cycle in amyotrophic lateral sclerosis. Amyotroph Lateral Scler. 2012;13:166–77. doi: 10.3109/17482968.2011.641569. [DOI] [PubMed] [Google Scholar]
- Law WJ, Cann KL, Hicks GG. TLS, EWS and TAF15: a model for transcriptional integration of gene expression. Brief Funct Genomic Proteomic. 2006;5:8–14. doi: 10.1093/bfgp/ell015. [DOI] [PubMed] [Google Scholar]
- Li Q, et al. A syntaxin 1, Galpha(o), and N-type calcium channel complex at a presynaptic nerve terminal: analysis by quantitative immunocolocalization. J Neurosci. 2004;24:4070–81. doi: 10.1523/JNEUROSCI.0346-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, et al. RBM45 homo-oligomerization mediates association with ALS-linked proteins and stress granules. Sci Rep. 2015;5:14262. doi: 10.1038/srep14262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YR, et al. Stress granules as crucibles of ALS pathogenesis. J Cell Biol. 2013;201:361–72. doi: 10.1083/jcb.201302044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling SC, Polymenidou M, Cleveland DW. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron. 2013;79:416–38. doi: 10.1016/j.neuron.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma AS, et al. Heterogeneous nuclear ribonucleoprotein A3, a novel RNA trafficking response element-binding protein. J Biol Chem. 2002;277:18010–20. doi: 10.1074/jbc.M200050200. [DOI] [PubMed] [Google Scholar]
- Mackenzie IR, Neumann M. FET proteins in frontotemporal dementia and amyotrophic lateral sclerosis. Brain Res. 2012;1462:40–3. doi: 10.1016/j.brainres.2011.12.010. [DOI] [PubMed] [Google Scholar]
- Majumder M, et al. The hnRNA-binding proteins hnRNP L and PTB are required for efficient translation of the Cat-1 arginine/lysine transporter mRNA during amino acid starvation. Mol Cell Biol. 2009;29:2899–912. doi: 10.1128/MCB.01774-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellacheruvu D, et al. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat Methods. 2013;10:730–6. doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miernyk JA, Thelen JJ. Biochemical approaches for discovering protein-protein interactions. Plant J. 2008;53:597–609. doi: 10.1111/j.1365-313X.2007.03316.x. [DOI] [PubMed] [Google Scholar]
- Mili S, Steitz JA. Evidence for reassociation of RNA-binding proteins after cell lysis: implications for the interpretation of immunoprecipitation analyses. RNA. 2004;10:1692–4. doi: 10.1261/rna.7151404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, et al. hnRNP A3 binds to GGGGCC repeats and is a constituent of p62-positive/TDP43-negative inclusions in the hippocampus of patients with C9orf72 mutations. Acta Neuropathol. 2013;125:413–23. doi: 10.1007/s00401-013-1088-7. [DOI] [PubMed] [Google Scholar]
- Narayanan RK, et al. Identification of RNA bound to the TDP-43 ribonucleoprotein complex in the adult mouse brain. Amyotroph Lateral Scler Frontotemporal Degener. 2013;14:252–60. doi: 10.3109/21678421.2012.734520. [DOI] [PubMed] [Google Scholar]
- Neumann M, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–3. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- Nittis T, et al. Revealing novel telomere proteins using in vivo cross-linking, tandem affinity purification, and label-free quantitative LC-FTICR-MS. Mol Cell Proteomics. 2010;9:1144–56. doi: 10.1074/mcp.M900490-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pahlich S, et al. Analysis of Ewing sarcoma (EWS)-binding proteins: interaction with hnRNP M, U, and RNA-helicases p68/72 within protein-RNA complexes. J Proteome Res. 2009;8:4455–65. doi: 10.1021/pr900235t. [DOI] [PubMed] [Google Scholar]
- Papadopoulou C, et al. Expression profile and interactions of hnRNP A3 within hnRNP/mRNP complexes in mammals. Arch Biochem Biophys. 2012;523:151–60. doi: 10.1016/j.abb.2012.04.012. [DOI] [PubMed] [Google Scholar]
- Peters OM, Ghasemi M, Brown RH., Jr Emerging mechanisms of molecular pathology in ALS. J Clin Invest. 2015;125:2548. doi: 10.1172/JCI82693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymenidou M, et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci. 2011;14:459–68. doi: 10.1038/nn.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–5. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelkovnikova TA, et al. Compromised paraspeckle formation as a pathogenic factor in FUSopathies. Hum Mol Genet. 2014;23:2298–312. doi: 10.1093/hmg/ddt622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, et al. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1:2856–60. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- Staudt T, et al. 2,2′-thiodiethanol: a new water soluble mounting medium for high resolution optical microscopy. Microsc Res Tech. 2007;70:1–9. doi: 10.1002/jemt.20396. [DOI] [PubMed] [Google Scholar]
- Sutherland BW, Toews J, Kast J. Utility of formaldehyde cross-linking and mass spectrometry in the study of protein-protein interactions. J Mass Spectrom. 2008;43:699–715. doi: 10.1002/jms.1415. [DOI] [PubMed] [Google Scholar]
- Takanashi K, Yamaguchi A. Aggregation of ALS-linked FUS mutant sequesters RNA binding proteins and impairs RNA granules formation. Biochem Biophys Res Commun. 2014;452:600–7. doi: 10.1016/j.bbrc.2014.08.115. [DOI] [PubMed] [Google Scholar]
- Tamada H, et al. cDNA cloning and characterization of Drb1, a new member of RRM-type neural RNA-binding protein. Biochemical and biophysical research communications. 2002;297:96–104. doi: 10.1016/s0006-291x(02)02132-0. [DOI] [PubMed] [Google Scholar]
- Tanaka E, et al. HnRNP A3 binds to and protects mammalian telomeric repeats in vitro. Biochem Biophys Res Commun. 2007;358:608–14. doi: 10.1016/j.bbrc.2007.04.177. [DOI] [PubMed] [Google Scholar]
- Teo G, et al. SAINTexpress: improvements and additional features in Significance Analysis of INTeractome software. J Proteomics. 2014;100:37–43. doi: 10.1016/j.jprot.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey JR, et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci. 2011;14:452–8. doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance C, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–11. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance C, et al. ALS mutant FUS disrupts nuclear localisation and sequesters wild-type FUS within cytoplasmic stress granules. Human molecular genetics. 2013 doi: 10.1093/hmg/ddt117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh MJ, et al. Invited review: decoding the pathophysiological mechanisms that underlie RNA dysregulation in neurodegenerative disorders: a review of the current state of the art. Neuropathol Appl Neurobiol. 2015;41:109–34. doi: 10.1111/nan.12187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang IF, et al. TDP-43, the signature protein of FTLD-U, is a neuronal activity-responsive factor. J Neurochem. 2008;105:797–806. doi: 10.1111/j.1471-4159.2007.05190.x. [DOI] [PubMed] [Google Scholar]
- Wolozin B, Apicco D. RNA binding proteins and the genesis of neurodegenerative diseases. Adv Exp Med Biol. 2015;822:11–5. doi: 10.1007/978-3-319-08927-0_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki T, et al. FUS-SMN protein interactions link the motor neuron diseases ALS and SMA. Cell Rep. 2012;2:799–806. doi: 10.1016/j.celrep.2012.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, et al. Identification of protein-protein interactions and topologies in living cells with chemical cross-linking and mass spectrometry. Mol Cell Proteomics. 2009;8:409–20. doi: 10.1074/mcp.M800232-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.